Abstract
We have constructed the first genetic linkage map for the North American arboviral vector Culex tarsalis. 120 F2 offspring from a cross between two colonies were genotyped using 25 microsatellites and six inter-simple sequence repeat (ISSR) markers. We resolved four linkage groups which likely correspond to two full-length chromosomes and two arms of the final chromosome. The longest linkage group contains the sex locus and corresponds to chromosome 3. Recombination rates around the sex locus were dramatically higher in females compared to males. The majority of microsatellite loci share sequence identity with regions of the Culex quinquefasciatus genome, whose assembly should aid in anchoring linkage groups to physical chromosomes. This map will aid in identification of loci involved with variable phenotypes in C. tarsalis including WNV susceptibility.
Keywords: Culex tarsalis, linkage map, West Nile virus, QTL
Introduction
The mosquito Culex tarsalis is a major vector of West Nile Virus (WNV), Western Equine Encephalitis Virus (WEEV) and St. Louis Encephalitis Virus (SLEV) in North America. The species distribution of C. tarsalis spans from the west coast to the Mississippi River and extends into portions of Canada and Mexico (Darsie & Ward, 1981). It is one of the most efficient vectors of WNV known (Goddard et al., 2002, 2003), reaches very large population sizes (Reisen & Reeves, 1987; Reisen & Lothrop, 1995) and serves as both an amplifying vector in bird reservoirs (Reisen et al., 2004) and a bridge vector to human hosts in rural areas (California DHS, 2008).
Decades of extensive field and laboratory research on C. tarsalis have revealed a great deal of variation in its physiology, vector competence and vectorial capacity. Autogeny (the ability to generate eggs without a bloodmeal) is commonly observed in both laboratory and field-caught females, and is known to vary both temporally (Spadoni et al., 1974) and spatially (Hardy & Reeves, 1973; Reisen, 1995). The ability to diapause varies within populations (Reisen & Reeves, 1987) while body size and early season survival has been shown to vary among populations when reared under controlled conditions (Reisen, 1995). Perhaps most importantly, C. tarsalis exhibits high intra-species variation in its ability to transmit arboviruses. Early studies on transmission of WEE indicated that populations from different parts of California varied greatly in ID50 (viral dose at which 50% of mosquitoes were infected 14 days postinoculation) and infection rate by oral challenge (Hardy et al., 1976; Hardy & Reeves, 1987). More recent work on WNV has shown that susceptibility, oral and vertical transmission of West Nile virus varies significantly among populations (Goddard et al., 2003).
While phenotypic variation in this mosquito has been well-described, the dearth of molecular tools for C. tarsalis has made it difficult to investigate the relationship between phenotypic and genotypic variation in laboratory colonies or wild populations. Until recently, genetic studies in C. tarsalis were limited to crude linkage associations of a limited number of morphological mutations (Asman et al., 1987) and a single population study using allozymes in California and Nevada (Gimnig et al., 1999). The characterization of genetic variation in this mosquito has recently been facilitated by the development and validation of a large panel of microsatellite markers (Rasgon et al., 2006; Venkatesan et al., 2007a). A preliminary analysis of microsatellite variation across populations in five states showed significant genetic differentiation in southern California, New Mexico and Nebraska (Venkatesan et al., 2007b). These results are consistent with the possibility that genetic variation may condition phenotypic variation among individuals and/or populations. The presence of genetic structure along with phenotypic differences in WNV susceptibility and transmission in C. tarsalis suggest that genetic polymorphism at particular loci may condition WNV infection phenotypes, and that genetically disparate populations of C. tarsalis may also be phenotypically distinct with respect to vector competence for WNV.
Quantitative trail loci (QTL) associated with pathogen susceptibility and transmission have been identified in other mosquitoes and arboviral systems. Linkage mapping has been used extensively to identify dengue virus QTL in Aedes aegypti involved in barriers to both midgut infection and dissemination (Bosio et al., 2000; Gomez-Machorro et al., 2004; Bennett et al., 2005). QTL have also been identified for transovarial (Graham et al., 2003) and oral (Anderson et al., 2005) transmission of La Crosse virus in Ochlerotatus triseriatus. However, virus-related QTL have not yet been identified for vectors of WNV or for any members of the genus Culex, which includes several significant, globally distributed arboviral vector species.
In order to lay the foundation for QTL mapping of phenotypic traits such as virus susceptibility, autogeny and diapause in C. tarsalis, we have generated the first molecular genetic linkage map for this species. Linkage groups were derived from an F2 intercross between two laboratory colonies using microsatellite markers (Table 1) and intersimple sequence repeats (ISSRs; Table 2). We anticipate that this map will facilitate genetic characterization of traits of medical and biological interest in this important arboviral vector. Additionally, we discuss the possibility of physically anchoring multiple loci mapped in our cross based on orthologous regions in the recently sequenced Culex quinquefasciatus genome.
Table 1.
Microsatellite markers used in this cross
| Locus | Accession no. | Forward Primer | Reverse Primer | Allele size range (bp) |
Sequence identity to Cx. pip genome |
|
|---|---|---|---|---|---|---|
| Linkage group 1 (Chromosome 3) | ||||||
| CUTC105 | DQ296487 | 5′-GCCGGTTGTTGTTGTTGTAC-′3 | 5′-TCCTCGTCAATTTCATCGAC-′3 | 209 | 230 | |
| CUTB210* | DQ682690 | 5′-ACCCACTGTTTGCGTATGAA-′3 | 5′-ACACTCACACCACCTTGTGC-′3 | 262 | 280 | X |
| CUTB218* | DQ682694 | 5′-TGCTGAGGCCGTTTTACC-′3 | 5′-CCCTGGAAAAGCATCAAACT-′3 | 150 | 214 | X |
| CUTB228* | DQ682697 | 5′-CATCACCATCAATCGTTTCC-′3 | 5′-GAAAACTTCCGGCACACAC-′3 | 145 | 195 | X |
| CUTB224* | – | 5′-CGAAGAGCAACAACATTCCA-′3 | 5′-CTCTGAAATCGATACACCAAGC-′3 | 204 | 232 | |
| CUTB101* | DQ682680 | 5′-GGGGTTCTTCGTGAGTTC-′3 | 5′-AGCAAGCGATTTCCCTAC-′3 | 208 | 254 | |
| CUTA220* | DQ682674 | 5′-TGAGCACGGGTGAGTTACAC-′3 | 5′-CCAATCGACGGGAAATTACA-′3 | 148 | 168 | X |
| CUTC203* | DQ682700 | 5′-AGGCCATGCAACATCCTTAC-′3 | 5′-CGACTTTATCTAGGCGCTCTC-′3 | 192 | 225 | X |
| CUTB112 | DQ682681 | 5′-AACCCCAGATTCTTAATGGC-′3 | 5′-GGAATTGGCTCAAACAACC-′3 | 154 | 184 | |
| Linkage group 2 | ||||||
| CUTB203* | DQ682687 | 5′-ACGAACGCGAAAGAAGAGAG-′3 | 5′-CACACCCGATTGTAGAGTGC-′3 | 230 | 256 | X |
| CUTA6 | DQ682664 | 5’-ACTCACACCCGATTGTAGAG-′3 | 5’-AGCCAGTCAGTCAGTCAGTG-′3 | 259 | 307 | X |
| CUTC12 | DQ296486 | 5′-GTGGAGAACCCGTATTCAAC-′3 | 5′-TACAATCACGACTCGCACATA-′3 | 184 | 208 | X |
| CUTB223* | DQ682696 | 5′-CGATATTTTGCTCCCACTTTG-′3 | 5′-AACTCCTTCGGGCTACACTG-′3 | 145 | 177 | X |
| CUTC102* | DQ682698 | 5′-GGAACCACAATCATCATAACC-′3 | 5′-GCAACAAACGAATCTTAGAAAC-′3 | 255 | 282 | X |
| CUTD211* | DQ682706 | 5′-TTCTGTTGTTGGGATTGCTG-′3 | 5′-GTCCGCACCCTGAATTGTA-′3 | 251 | 278 | X |
| CUTB212* | DQ682691 | 5′-TGTCGAGGTGAAACAACCAG-′3 | 5′-CCGAACGAAAAGCAAAAGTC-′3 | 148 | 176 | X |
| CUTD114 | DQ296492 | 5’AGGAAGAGTGGTTCGTTTTC′3 | 5’GGGTAAGTTTCAGGGCTATC-′3 | 180 | 198 | X |
| CUTD102* | DQ682703 | 5′-CAGTTCCAGCAGCAGTCA-′3 | 5′-CAGGTGATGGGGGTGTAG-′3 | 117 | 142 | X |
| CUTD113 | DQ296491 | 5′-ATCATACCACTGCCCATAGTC-′3 | 5′-AACCAGCAGGGACAAGTC-′3 | 159 | 185 | X |
| CUTA109.7* | DQ682667 | 5′-CCATCACATTGAACATCACTT-′3 | 5′-CGAGTTGCCGATAGAAGAT-′3 | 236 | 288 | X |
| Linkage group 3 | ||||||
| CUTB1 | DQ296484 | 5′-GAAAAAAAGGCGCAACAT-T-′3 | 5′-GAAGGTGCCAGCCTACTTG-′3 | 104 | 138 | X |
| CUTB214* | DQ682693 | 5′-GCAGTAGCTGGAACGTGCT-′3 | 5′-GCGCATAAAATACACAGCAAA-′3 | 156 | 190 | |
| CUTA105.7* | DQ682666 | 5′-TCGCCTTACTTCCCACAT-′3 | 5′-AGGACCCAACAACAGCAC-′3 | 244 | 282 | |
| Linkage group 4 | ||||||
| CUTD120 | DQ296493 | 5′-TACCCTCGCAAACAAAACAA-′3 | 5′-GTCGGCTTCCATTCCACTAC-′3 | 159 | 183 | X |
| CUTD203* | DQ682708 | 5′-TATCCGGCAGCAGAACTTG-′3 | 5′-ACAAGCACCACAGCAAACTG-′3 | 214 | 241 | X |
Allele size range includes 26 bp 5′ M13 tag.
Table 2.
ISSR primers and loci used in this cross
| Primer | Scorable loci | Segregating loci | Mapped loci | Label | Location |
|---|---|---|---|---|---|
| (AG)8CG | 4 | 2 | 1 | (AG)8CG_B | Linkage group 1 (Chromosome 3) |
| (AC)8TGA | 9 | 2 | 1 | (AC)8TGA_B | Linkage group 1 (Chromosome 3) |
| (AG)8TGA | 8 | 6 | 2 | (AG)8TGA_A (AG)8TGA_E |
Linkage group 4 Linkage group 3 |
| (AC)8TCT | 3 | 2 | 2 | (AC)8TCT_A (AC)8TCT_B |
Linkage group 4 Linkage group 2 |
Results
F2 cross
High mortality in 4th instar F2 larvae led us to select multiple F2 families for the mapping population. A total of 120 F2 offspring (41 females, 79 males) from the five largest families were successfully reared to adulthood and used for linkage analysis. Family 3 contained 22 individuals (five females, 17 males), Family 5 contained 12 (five females, seven males), Family 6 contained 18 (eight females, 10 males), Family 12 contained 23 (12 females, 11 males) and Family 22 contained 45 (11 females, 34 males).
Microsatellites
Parental F0 individuals were successfully genotyped at 53 of the 57 available microsatellite loci. 28 markers were identical in the two F0 individuals and were thus uninformative for linkage analysis. Of the remaining 25 microsatellites, 13 loci segregated fully between the two F0 parentals and 12 segregated partially, with one allele varying between F0’s. Twenty of the 25 markers were informative for all five families with a total of 22 microsatellites in Family 5, 25 in Families 5 and 6, and 24 in both Families 12 and 24.
In general, microsatellite loci conformed to expected Mendelian inheritance ratios (Table 3). After correcting for multiple tests, seven in 123 comparisons demonstrated statistically significant deviations. No family-specific deviation pattern was observed and none of the loci exhibited significant deviations in more than a single family. The newly described locus CUTB224 appears to segregate in accordance with expected Mendelian inheritance ratios (Table 3) but has not yet been tested for deviation from Hardy-Weinberg allele frequencies in natural populations.
Table 3.
Chi-squared values of Mendelian inheritance ratios for microsatellite and ISSR markers
| Chi-squared values |
||||||
|---|---|---|---|---|---|---|
| Family 3 | Family 5 | Family 6 | Family 12 | Family 22 | ||
| Locus | No. individuals | 22 | 12 | 18 | 23 | 45 |
| CUTC105 | Linkage group 1 (Chromosome 3) | 3.45 | 2.00 | 10.89 | 0.80 | 5.95 |
| CUTB210 | 6.55 | 0.82 | 0.00 | 0.47 | 11.52 | |
| CUTB218 | 35.10* | 1.00 | 3.00 | 5.68 | 3.42 | |
| CUTB228 | 7.36 | 6.00 | 2.00 | 3.38 | 2.33 | |
| CUTB224 | 0.55 | 4.00 | 6.00 | 2.00 | 6.91 | |
| CUTB101 | 8.52 | 0.20 | 3.00 | 0.50 | 10.22 | |
| CUTA220 | 0.18 | 5.18 | 1.89 | 1.00 | 14.58 | |
| (AG)8CG_B | 10.24 | 8.33 | 10.89 | 0.53 | 23.27 | |
| CUTC203 | 2.91 | 5.33 | 2.00 | 1.31 | 26.27 | |
| (AC) 8TGA_B | 0.06 | 4.00 | 2.00 | 0.00 | 12.30 | |
| CUTB112 | 3.57 | 3.18 | 24.18 | 0.16 | 7.10 | |
| (AC)8TCT_B | Linkage group 2 | 16.40 | 0.09 | 0.06 | 4.00 | 3.76 |
| CUTB203 | – | 4.45 | 0.06 | 4.00 | 0.21 | |
| CUTA6R.7 | – | 5.33 | 0.06 | – | 0.03 | |
| CUTC12 | 5.18 | 5.33 | 8.22 | 2.41 | 2.12 | |
| CUTB223 | 3.60 | 4.67 | 14.44 | 0.23 | 3.82 | |
| CUTC102 | 5.18 | 8.00 | 9.11 | 1.00 | 2.76 | |
| CUTD211 | – | 7.25 | 0.89 | 8.07 | 0.68 | |
| CUTB212 | 8.91 | 7.36 | 0.89 | 9.00 | 1.14 | |
| CUTD114 | 1.64 | 4.45 | 6.00 | 27.60 | 2.00 | |
| CUTD102 | 0.59 | 3.00 | 0.89 | 8.00 | 1.52 | |
| CUTD113 | 0.00 | 0.33 | 0.89 | 11.84 | 1.19 | |
| CUTA109.7 | 4.55 | 3.60 | 5.44 | 0.40 | 1.19 | |
| CUTB1 | Linkage group 3 | – | 0.50 | 3.67 | 0.30 | 0.24 |
| CUTB214 | 19.48 | 5.67 | 5.35 | 4.14 | 2.60 | |
| (AG)8TGA_E | 0.18 | 0.33 | 21.41 | 2.58 | 0.56 | |
| CUTA105.7 | 2.91 | 0.00 | 0.22 | 0.47 | – | |
| (AG)8TGA_A | Linkage group 4 | 14.73 | 1.78 | 0.07 | 6.37 | 11.76 |
| CUTD120 | 12.55 | 12.82 | 7.71 | 17.00 | 2.00 | |
| CUTD203 | 3.45 | 1.00 | 1.80 | 15.13 | 21.21 | |
| (AC)8TCT_A | 0.05 | 0.81 | 0.02 | 30.42 | 0.20 | |
Chi-squared values that deviate significantly from expected Mendelian inheritance ratios are shown in bold.
ISSR markers
Each of the 12 ISSR primers produced between three and nine scorable bands. Two to six bands per primer segregated in the F0 parentals (Table 2), producing 12 informative loci. Six of the 12 loci were mapped (Fig. 1) while the other six were excluded from the map because they did not exhibit linkage to any other markers or linkage groups.
Figure 1.
ISSR loci used in this cross. A total of 6 bands from 4 primers were mapped. Mapped band positions are indicated by arrows. Genotypes of the F0 male and F0 female are shown along with 4 F2 males and 4 F2 females (1–8). m = male, f = female.
Mendelian inheritance ratio patterns were similar to those observed in microsatellites. All six ISSR loci deviated from expected ratios in at least one of the five families (Table 3; Table S1), but no family-specific patterns were observed. Eight of 30 total comparisons exhibited deviation from expected Mendelian inheritance ratios after a correction for multiple tests.
Linkage Map
Genotypic information from 32 loci (25 microsatellites + 6 ISSRs + sex locus) in 120 F2 progeny from five families was used to construct a genetic map. Family-specific linkages along with a composite multi-family linkage analysis integrating recombination information from each family were constructed. The composite map resolved four linkage groups (Fig 2 and Fig 3). The total map length was 510 cM, with an average recombination distance between markers of 18.22 ± 2.12 cM (s.e.). Family-specific maps showed similar marker order to each other and to the composite map, although there was significant variation in calculated recombination rates and map lengths between families, likely due to sample size effects (Fig. 2).
Figure 2.
Family-specific and composite linkage maps of Culex tarsalis based on recombination frequencies observed in an F2 cross. Maps were derived from recombination estimates using the Kosambi mapping function. Map distances are listed in cM. Those few markers that were uninformative in family-specific maps (Table 3) were placed by linear interpolation for comparative purposes.
Figure 3.
Composite F2 linkage map of Culex tarsalis showing loci mapped by homology to the sequenced C. quinquefasciatus genome (indicated by asterisks). Map distances are listed in cM. Map was constructed using the Kosambi function.
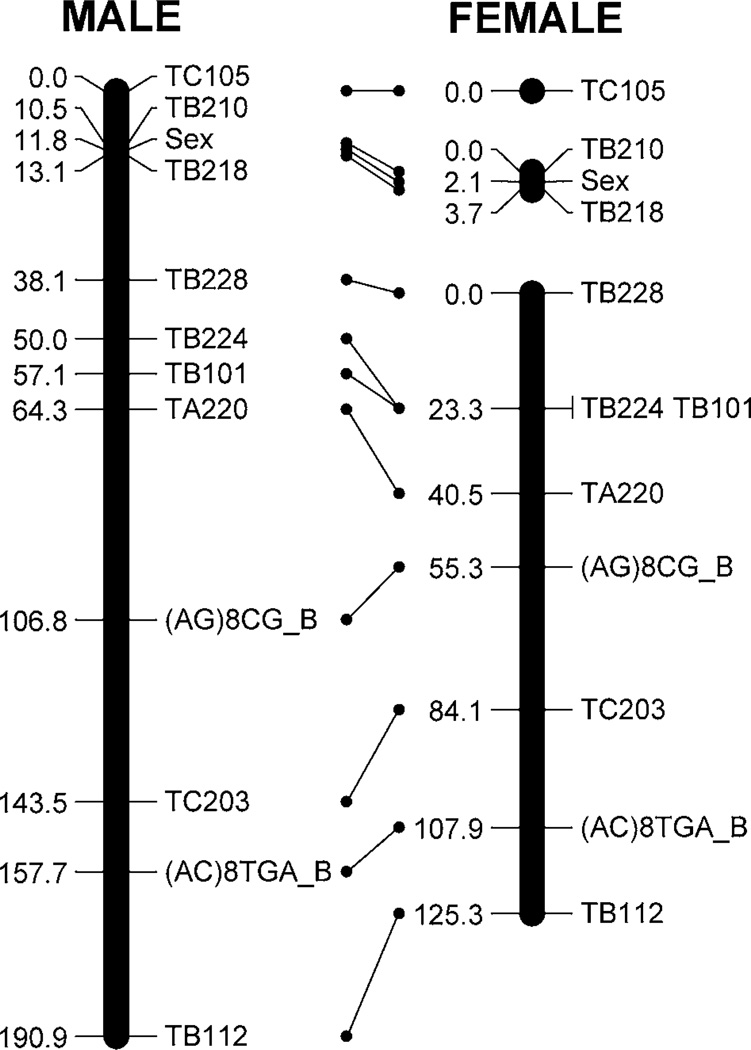
Across most of the genome, there was no significant difference in recombination rates between sexes, and male and female-specific maps resolved identical linkage groups and marker order, except in the area directly flanking the sex locus. In this region only, the recombination rate was dramatically higher in females compared to males, resulting in the region containing the sex locus being unlinked from the rest of the linkage group in females (Fig. 4).
Figure 4.
Comparison of sex-specific composite maps of linkage group 1/chromosome 3, highlighting the dramatically increased recombination rate around the sex locus in females. Recombination rates across the remainder of the genome did not vary significantly between sexes.
As it contains the sex locus, the first linkage group likely corresponds to chromosome 3 (McDonald et al., 1978). Nine microsatellites and two ISSRs mapped to linkage group 1 at an average genetic distance of 20.38 cM ± 4.03 cM and a total length of 224 cM. The sex locus mapped to 1.4 cM from CUTB218, representing the shortest distance between markers on linkage group 1. The greatest distance occurred between microsatellite loci CUTB218 and CUTB228.
The second largest linkage group, spanning 131.6 cM, group consisted of 11 microsatellites and one ISSR with an average distance of 11.96 cM ± 2.44 cM between markers. Initially, an additional microsatellite CUTA6R.7, was mapped to this linkage group in the exact same position as CUTA11. Further examination revealed that the primer sets for CUTA6 (Accession no. DQ682664) and CUTA11 (Accession no. DQ296483) amplify the same microsatellite locus. CUTA11 was subsequently removed from the analysis and a note was made to GenBank to correct this error.
The third and fourth linkage groups were the smallest, spanning 82.5 cM and 71.8 cM, respectively. Linkage group 3 consisted of three microsatellites and one ISSR with an average distance of 27.5 cM ± 1.77 cM). Linkage group 4 encompassed two microsatellites and two ISSRs spaced with an average distance of 24 cM ± 1.11 cM.
Synteny with C. quinquefasciatus
Microsatellite-containing sequences from C. tarsalis, obtained during initial marker development described in Rasgon et al. (2006), were compared to the C. quinquefasciatus genome (www.vectorbase.org) using BlastN. Flanking sequences of 19 of the 25 mapped microsatellites exhibited high similarity to regions of the C. quinquefasciatus genome (Fig. 3; Table 4). Aligned regions showed 70%–96% sequence similarity in ∼100–600 bp segments. The actual microsatellite repeat motif was present in 17 of the C. quinquefasciatus sequences. CUTA109.7 showed similar sequence identity to portions of two separate supercontigs (Table 4).
Table 4.
Mapped microsatellite loci in Culex tarsalis with orthologs to flanking regions in the Culex quinquefasciatus genome
| Corresponding regions in C. quinquefasciatus genome |
||||||
|---|---|---|---|---|---|---|
| Locus | Location | Alignment length (no. of bp) |
Supercontig | From (bp) | To (bp) | Sequence identity |
| CUTB210 | Linkage group 1 (Chromosome 3) | 257 | 3.22 | 1047680 | 1047937 | 0.91 |
| CUTB218* | 92 | 3.10 | 749087 | 749179 | 0.89 | |
| CUTB228 | 481 | 3.355 | 192600 | 193081 | 0.72 | |
| CUTA220 | 435 | 3.16 | 1042459 | 1042894 | 0.86 | |
| CUTC203 | 214 | 3.198 | 92748 | 92962 | 0.71 | |
| CUTB203 | Linkage group 2 | 459 | 3.185 | 332456 | 332915 | 0.81 |
| CUTA6R.7 | 476 | 3.185 | 332456 | 332932 | 0.81 | |
| CUTC12 | 375 | 3.369 | 252315 | 252690 | 0.80 | |
| CUTB223 | 315 | 3.561 | 223790 | 224105 | 0.74 | |
| CUTC102** | 469 | 3.384 | 128435 | 128904 | 0.78 | |
| CUTD211 | 240 | 3.50 | 892111 | 892351 | 0.84 | |
| CUTB212 | 400 | 3.297 | 90515 | 90915 | 0.70 | |
| CUTD114 | 299 | 3.805 | 61858 | 62157 | 0.88 | |
| CUTD102 | 628 | 3.258 | 59592 | 60220 | 0.91 | |
| CUTD113 | 251 | 3.1 | 2851920 | 2852171 | 0.96 | |
| CUTA109.7*** | 268 | 3.1233 | 65213 | 65481 | 0.69 | |
| 355 | 3.1424 | 55752 | 56107 | 0.70 | ||
| CUTB1** | Linkage group 3 | 151 | 3.32 | 76478 | 76629 | 0.78 |
| CUTD120 | Linkage group 4 | 317 | 3.49 | 490661 | 490978 | 0.79 |
| CUTD203 | 407 | 3.1235 | 59619 | 60026 | 0.71 | |
All sequence positions in C. quinquefasciatus refer to genome version CpipJ1, released in March 2007
Corresponding supercontig in C. quinquefasciatus begins at position 371 of C. tarsalis microsatellite-containing sequence.
Microsatellite repeat region absent in corresponding C. quinquefasciatus sequence.
CUTA109 shares similarity to two regions in the C. quinquefasciatus genome.
Discussion
We present in this study the first genetic linkage map of C. tarsalis using modern molecular markers. We resolved four linkage groups using a total of 32 loci. Based on estimates of the genome sizes of C. quinquefasciatus at 580 megabases (Mb) (CpipJ1.2 genome assembly; http://cpipiens.vectorbase.org/SequenceData/Genome/), mapped loci in the C. tarsalis genome occur at an approximate frequency of one marker per every 18 Mb assuming that the genome sizes of C. quinquefasciatus and C. tarsalis are similar.
Our map at 510 cM is larger than those constructed for other mosquitoes. To confirm that this result was not due to problems with the mapping algorithms implemented by R/ qtl, we validated the maps by separate analyses with the CRI-MAP software package, which is widely used in human genetics (Green et al., 1990). Analysis with CRI-MAP required the omission of a small portion of the genotypes at dominant markers, as the software was constructed for use with co-dominant markers only. Nevertheless, the maps constructed with CRI-MAP exactly matched those constructed with R/qtl to within 0.1 cM except for a single 30 cM interval on linkage group 3, for which the distance estimates differed by 2 cM (data not shown).
Variation among family-specific maps due to sample size effects could in part account for the relatively large composite map distances observed in this study. However, at approximately 500 cM, the C. tarsalis map is not unreasonably larger than those generated for some other mosquitoes. For instance, both the Anopheles funestus map and a map generated in a hybrid cross between Ochlerotatus trisariatus/ hendersoni were almost 400 cM (Anderson et al., 2005; Wondji et al., 2005). Future mapping experiments will serve to clarify the issue of map size for C. tarsalis.
An early study of sex linkage in C. tarsalis showed that the sex locus is located on the longest chromosome (McDonald et al., 1978), corresponding to chromosome 3. Convention holds that mosquito chromosomes are labeled as 1, 2, and 3 in order by increasing size, chromosome 1 being the shortest and chromosome 3 being the longest (Rai, 1963). Here we term our longest linkage group, which also contains the sex locus, as linkage group 1/chromosome 3. In contrast, the sex locus in C. pipiens, Ae. aegypti and most culicines is located on chromosome 1, the shortest chromosome (McDonald & Rai, 1970; Jost & Laven, 1971). Interestingly, in some populations of Culex tritaeniorhynchus, the sex locus is found on chromosome 1 (Baker et al., 1971; Selinger, 1972) while in others it is found on chromosome 3 (Baker & Sakai, 1976; Baker et al., 1977; Mori et al., 2001).
Variation among linkage groups in marker number and length suggests that apart from linkage group 1/chromosome 3, another chromosome likely corresponding to linkage group 2 has been well-resolved. We suspect that the smaller linkage groups 3 and 4 correspond to two arms of the remaining chromosome. This phenomenon is not uncommon in lower-resolution linkage analyses of other mosquitoes where chromosomal arms appear to assort independently or are separated by large genetic distances (An. funestus; Wondji et al., 2005; Ochlerotatus spp.; Anderson et al., 2006), particularly if centromere-spanning markers are lacking. Full resolution of the remaining chromosome may require higher marker density and/or a larger mapping population. Alternatively, since microsatellite homologues from each of the two smallest linkage groups exist in the C. quinquefasciatus genome, we may gain insight into the identity of these linkage groups in C. tarsalis when the C. quinquefasciatus genome is fully assembled.
Sex-specific recombination rates are known to vary in insects including Bombyx mori (Rasmussen, 1977) and Drosophila species (Clements, 1992). Many studies have found that C. tritaeniorhynchus females do not undergo recombination (e.g. Baker & Rabbani, 1970; Baker & Sakai, 1973a,b; Mori et al., 2001), though recombination occurs in both sexes in Culex pipiens (D. Severson, pers. comm.; Rasgon & Scott, 2004). We did not observe evidence of sex-specific differences in recombination for the majority of markers in C. tarsalis. However, recombination does appear to vary significantly between males and females in the region directly adjacent to the sex locus, where females exhibited dramatically higher rates of recombination. This phenomenon warrants further investigation.
ISSR markers are commonly used in plant linkage mapping (Irzykowska et al., 2002; Hashuzime et al., 2003; Irzykowska & Wolko, 2004) and have been tested for population variation in invertebrates (Abbot, 2001). In our hands, 12 ISSR bands segregated clearly and reliably in F0’s, F1’s and F2’s, suggesting that ISSR markers are variable and informative for mapping purposes. However, only six of the 12 mapped to the four defined linkage groups. The reasons for such poor mapping performance are unclear, but this phenomenon does not necessarily appear to be specific to the cross presented here or to insect mapping in general. Mapping studies of various plant species, where ISSRs have been used extensively, show a high percentage (up to 80%) of non-mapping ISSR markers (Kojima et al. 1998; Duran et al. 2004; Yu et al. 2006; Gupta et al. 2007). We recommend that ISSRs be used in future insect mapping studies only if a large number of segregating bands can be identified, since a significant proportion may not map to any linkage group.
Of the 53 reliably amplifying microsatellite primers, nearly half were informative in the cross. The mapping population arose from a cross between two California colonies which were established from sites 240 miles apart and reared independently for at least four years. Recent evidence suggests that there is moderately differentiation between populations in the two sites (Venkatesan et al., 2007b); yet the majority of microsatellite loci did not segregate between the F0 individuals. If possible, future studies should attempt to cross colonies established from populations at an even greater genetic distance from each other to increase the number of informative loci from the available panel of markers.
The total number of mapped loci, at 32, lies well within the range of marker number for QTL analysis in other mosquito species such as An. funestus, at 49 or 56 loci (Wondji et al., 2005, 2007), Ochlerotatus sp., at 25 loci (Anderson et al., 2005, 2006) C. tritaeniorhynchus, at 14 loci (Mori et al., 2001) and C. pipiens, at 9–13 loci (Mori et al., 2007). While the availability and density of mapping markers may increase in future efforts, these studies suggest that the existing set of 60 + available loci and linkage map of 32 loci should be sufficient to identify QTL in C. tarsalis.
Several of the flanking sequences of mapped microsatellites in C. tarsalis exhibit high similarity to regions of the recently released C. quinquefasciatus genome (Fig. 3; Table 4). Seventeen of the 19 repeat motifs are present in C. quinquefasciatus, suggesting that microsatellite loci are well-conserved between the two species. Additionally, others have used microsatellite loci developed for C. tarsalis to investigate population genetic questions in the C. pipiens species complex (Kent et al., 2007). Future assignment of C. quinquefasciatus supercontigs to chromosomes will assist in validating marker placement in our linkage map and contribute to an improved physical positioning of microsatellites in Cx. tarsalis. Additionally, genome assembly should allow us to make estimates of physical distance among markers for comparison with linkage distances, explore synteny between the two species and aid in cloning and characterization of genes of interest.
The linkage map presented in this report is an important tool for genetic studies of C. tarsalis and comparative analysis of related Culex species. We anticipate that it will be useful as a basis for QTL studies of variable phenotypes such as autogeny, insecticide resistance and susceptibility to arboviruses in this important vector mosquito.
Experimental procedures
Mosquito strains and rearing conditions
The two Culex tarsalis strains used in the cross were KNWR, colonized from the Kern National Wildlife Refuge in Kern, CA and CTC from Coachella, CA in 2003 by W. Reisen (UC Davis). Colonies were acquired by our group at the Johns Hopkins School of Public Health in 2005 (KNWR) and 2007 (CTC) and maintained independently. Colonies were reared at 27 °C and 90% relative humidity on a 16:8 light-dark cycle. Larvae were fed a 1:2:2 mixture of ground rabbit pellets, liver powder and fish flakes at 7.5 mg/individual. Adults were provided with 10% sucrose solution ad libitum. KNWR females were maintained autogenously while CTC and F1 cross females were allowed to bloodfeed on anesthetized mice according to JHU Animal Welfare Assurance protocol A3272-01.
crosses
Harem crosses were conducted in pint cages with six to eight virgin F0 CTC females (2–3 days old) and a single F0 KNWR male per cage. Due to an insectary lighting malfunction, mating in the attempted reciprocal cross was unsuccessful. Females were bloodfed after five days and placed in individual cages to oviposit. Each F1 egg raft was reared independently and offspring were siblingharem-mated as described above. Mated F1 females were bloodfed up to three times to produce the F2 generation. F2 families were reared to adulthood and preserved for subsequent DNA extraction along with the original F0 parents and productive F1 mosquitoes.
DNA preparation and microsatellite genotyping
The five largest F2 families were selected for genotyping. DNA was extracted by salt extraction/ethanol precipitation as previously described (Black & DuTeau, 1997) and suspended in nucleasefree water at a concentration of 30 ng/uL. We successfully genotyped F0 mosquitoes at 53 of 57 available microsatellite loci (Rasgon et al ., 2006; Venkatesan et al ., 2007a) using previously described methods (Boutin-Ganache et al., 2001; Venkatesan et al. 2007a). PCR products were resolved on an ABI Prism Genetic Analyzer 3100 Avant (Applied Biosystems, Foster City, CA). Allele sizes were automatically determined with an internal ROX-500 size standard (Applied Biosystems) using GeneScan v. 3.1 and Genotyper software (Applied Biosystems). Loci informative in the F0’s were genotyped in F1’s and 120 F2’s. Microsatellite markers used in the cross are shown in Table 1 and include a previously unpublished microsatellite, the (AG) repeat CUTB224. 5′ M13-labelled amplification of CUTB224 was carried out as described in Venkatesan et al., 2007a. This locus appears to segregate in accordance with expected Mendelian inheritance ratios (Table 3) but has not yet been tested for deviation from Hardy-Weinberg allele frequencies in natural populations.
ISSR genotyping and scoring
The segregating population was genotyped at 12 inter-simple sequence repeat (ISSR) loci derived from four 3′ anchored repeat primers shown in Table 2. The 25 uL PCR mixture contained 2.5 μl of 10X reaction buffer (New England Biolabs, Ipswich, MA, USA), 30–40 ng of DNA template, 0.8 μM of a single ISSR primer, 2.0 mM MgCl2, 0.2 mM of each dNTP and 1 μl (5 units) of Taq DNA Polymerase. PCR products were amplified using a DNA Engine thermal cycler (Biorad, Hercules, CA, USA), under the following conditions adapted from Abbot (2001): an initial denaturation step at 94 ° for 2 min, 13 cycles of 94 ° for 30 s, 68 ° for 30 s with a 0.7 ° reduction per cycle and 72 ° for 1 min, followed by 36 cycles of 94 ° for 30 s, 55 ° for 30 s and 72 ° for 1 min, finishing with a 10 min final extension at 72 °. 20 μl of each PCR product was loaded into 2% agarose gels buffered with 1X TBE. Gels were run at 60 V for 300 minutes. Marker sizes were estimated using a 100-bp DNA ladder. All PCRs and gels were run twice to ensure reproducibility of the markers. Reproducible bands were scored as present or absent in each individual (Fig. 1). Parental (F0) PCR products for each primer were run in each set of gels for band size comparison. Amplicons of the same size were assumed to be the same locus regardless of band intensity.
Statistical analysis and map construction
Deviation from expected Mendelian inheritance ratios for all markers within each family was determined using Chi-square analysis. Mapping calculations were performed with R/qtl (Broman et al., 2003), an add-on package to the R statistical software (Ihaka and Gentleman, 1996). The Kosambi mapping function was used for all maps. We focused on microsatellite markers for initial map construction. For each pair of markers, the recombination fraction between them, r, was estimated and a LOD score was calculated for the test of r = 0.5. Initial linkage groups were formed on the basis of the pairwise marker linkage information, where two markers were placed in the same linkage groups if LOD > 3. For each linkage group, a rough initial marker order was established by a greedy algorithm: markers were added, one at a time, in the position giving the maximum likelihood, until all markers had been placed. An improved marker order was identified by considering all possible shuffles of a sliding window of eight markers, and choosing the order with the minimal number of obligate crossovers. The final marker order was chosen by maximum multipoint likelihood, considering all possible shuffles of a sliding window of four markers. Multipoint calculations were performed assuming a genotyping error rate of 1%. Once the genetic map for the microsatellite markers was constructed, the placement of the dominant ISSR markers was considered. All possible F1 genotypes, and all possible positions for each marker, were considered. The dominant markers were placed, one at a time, in the position giving the highest likelihood, provided that there was good evidence for linkage between the marker and a linkage group. Linkage maps were graphically depicted using MapChart v. 2.2 (Voorrips, 2002).
Supplementary Material
Acknowledgments
We thank Dr W. Reisen and H Lothrop for providing the mosquito strains used in this study. We thank Dr. William Black and an anonymous reviewer for suggestions that significantly improved the manuscript. This work was funded by NIH/NIAID grant R01AI067371 and funds from the Bloomberg Family Foundation to JLR, NIH/NIGMS grant R01GM074244 to KWB and NIH/NIEHS Training Grant T32ES07141 to MV.
Footnotes
Research conducted at The W. Harry Feinstone Department of Molecular Microbiology and Immunology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland, 21205
Supporting Information Additional Supporting Information may be found in the online version of this article:
Table S1. Observed and expected Mendelian inheritance ratios for microsatellites and ISSRs
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abbot P. Individual and population variation in invertebrates revealed by Inter-simple Sequence Repeats (ISSRs) J Insect Sci. 2001;1:8. [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Schneider JR, Grimstad PR, Severson DW. Quantitative genetics of vector competence for La Crosse virus and body size in Ochlerotatus hendersoni and Ochlerotatus triseriatus interspecific hybrids. Genetics. 2005;169:1529–1539. doi: 10.1534/genetics.104.033639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Schneider JR, Grimstad PR, Severson DW. Identification of quantitative trait loci for larval morphological traits in interspecific hybrids of Ochlerotatus triseriatus and Ochlerotatus hendersoni (Diptera: Culicidae) Genetica. 2006;127:163–175. doi: 10.1007/s10709-005-4227-9. [DOI] [PubMed] [Google Scholar]
- Asman SM, Milby MM, Reeves WC. Genetics of Culex tarsalis . In: Reeves WC, editor. Epidemiology and control of mosquito-borne arboviruses in California, 1943–1987. Sacramento, CA: California mosquito and vector control association; 1987. pp. 330–356. [Google Scholar]
- Baker RH, Rabbani MG. Complete linkage in females of Culex tritaeniorhynchus mosquitoes. J Hered. 1970;61:59–61. doi: 10.1093/oxfordjournals.jhered.a108039. [DOI] [PubMed] [Google Scholar]
- Baker RH, Sakai RK. The genetics of rose, an allele of the white locus in a mosquito. J Hered. 1973a;64:19–23. doi: 10.1093/oxfordjournals.jhered.a108328. [DOI] [PubMed] [Google Scholar]
- Baker RH, Sakai RK. Genetic studies on two new mutants in linkage group III of the mosquito, Culex tritaeniorhynchus . Ann Trop Med Parasitol. 1973b;67:467–473. doi: 10.1080/00034983.1973.11686915. [DOI] [PubMed] [Google Scholar]
- Baker RH, Sakai RK. Male determining factor on chromosome 3 in the mosquito, Culex tritaeniorhynchus . J Hered. 1976;67:289–294. doi: 10.1093/oxfordjournals.jhered.a108733. [DOI] [PubMed] [Google Scholar]
- Baker RH, Sakai RK, Mian A. Linkage group chromosome correlation in Culex tritaeniorhynchus . Science. 1971;171:585–587. doi: 10.1126/science.171.3971.585. [DOI] [PubMed] [Google Scholar]
- Baker RH, Saifuddin UT, Sakai RK. Variations in the linkage of the sex allele in laboratory colonies of the mosquito Culex tritaeniorhynchus . Jpn J Genet. 1977;52:425–430. [Google Scholar]
- Bennett KE, Flick D, Fleming KH, Jochim R, Beaty BJ, Black WC4th. Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti . Genetics. 2005;170:185–94. doi: 10.1534/genetics.104.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WC, IV, DuTeau NM. RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors. Cambridge: Cambridge University Press; 1997. pp. 361–373. [Google Scholar]
- Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WC. Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti . Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques. 2001;31:25–28. [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL crossing in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- California State mosquito-borne virus surveillance and response plan. [Online] California Department of Health Services. [accessed 15 July 2008]; Available at: http://westnile.ca.gov/downloads.php? download_id=820&filename=2008_CA_Mosq_Surv.pdf.
- Clements AN. The Biology of Mosquitoes. Vol. 1. London: Chapman & Hall; 1992. pp. 29–32. [Google Scholar]
- Darsie RF, Ward RA. Identification and geographical distribution of the mosquitoes of North America, north of Mexico. Mosq Syst Suppl. 1981;1:1–313. [Google Scholar]
- Duran Y, Fratini R, Garcia P, Perez de la Vega M. An intersubspecific genetic map of Lens . Theor Appl Genet. 2004;108:1265–1273. doi: 10.1007/s00122-003-1542-3. [DOI] [PubMed] [Google Scholar]
- Gimnig JE, Reisen WK, Eldridge BF, Nixon KC, Schutz SJ. Temporal and spatial genetic variation within and among populations of the mosquito Culex tarsalis (Diptera: Culicidae) from California. J Med Entomol. 1999;36:23–29. doi: 10.1093/jmedent/36.1.23. [DOI] [PubMed] [Google Scholar]
- Goddard LB, Roth AE, Reisen WK, Scott TW. Vector competence of California mosquitoes for West Nile virus. Emerg. Infect Dis. 2002;8:1385–1391. doi: 10.3201/eid0812.020536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard LB, Roth AE, Reisen WK, Scott TW. Vertical transmission of West Nile virus by three California Culex (Diptera: Culicidae) species. J Med Entomol. 2003;40:743–766. doi: 10.1603/0022-2585-40.6.743. [DOI] [PubMed] [Google Scholar]
- Gomez-Machorro C, Bennett KE, de Lourdes Munoz M, Black WC. Quantitative trait loci affecting dengue midgut infection barriers in an advanced intercross line of Aedes aegypti . Insect Mol Biol. 2004;13:637–648. doi: 10.1111/j.0962-1075.2004.00522.x. [DOI] [PubMed] [Google Scholar]
- Graham DH, Holmes JL, Beaty BJ, Black WC. Quantitative trait loci conditioning vertical transmission of La Crosse virus in the eastern treehole mosquito, Ochlerotatus triseriatus . Insect Mol Biol. 2003;12:307–318. doi: 10.1046/j.1365-2583.2003.00412.x. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation for CRI-MAP, version 2.4. 1990 [Google Scholar]
- Gupta S, Pandey-Rai S, Srivastava S, Chandra Naithani S, Prasad M, Kumar S. Construction of genetic linkage map of the medicinal and ornamental plant Catharanthus roseus . J Gen. 2007;86:259–268. doi: 10.1007/s12041-007-0033-8. [DOI] [PubMed] [Google Scholar]
- Hardy JL, Reeves WC. Emerging concepts of factors that limit the competence of Culex tarsalis to vector encephalitis viruses. Proc Calif Mosq Contr Assoc. 1973;41:7–10. [Google Scholar]
- Hardy JL, Reeves WC. Experimental studies on infection in vectors. In: Reeves WC, editor. Epidemiology and Control of Mosquito-Borne Arboviruses in California, 1943–1987. Sacramento, CA: California mosquito and vector control association; 1987. pp. 145–253. [Google Scholar]
- Hardy JL, Reeves WC, Sjogren RD. Variation in the susceptibility of field and laboratory populations of Culex tarsalis to experimental infection with western equine encephalomyelitis virus. Am J Epidemiol. 1976;103:498–505. doi: 10.1093/oxfordjournals.aje.a112251. [DOI] [PubMed] [Google Scholar]
- Hashuzime T, Shimamoto I, Hirai M. Construction of a linkage map and QTL analysis of horticultural traits for watermelon (Citrullus lanatus) using RAPD, RFLP and ISSR markers. Theoretic Appl Gen. 2003;106:779–785. doi: 10.1007/s00122-002-1030-1. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comp Graphl Stat. 1996;5:299–314. [Google Scholar]
- Irzykowska L, Wolko B. Interval mapping of QTLs controlling yield-related traits and seed protein content in Pisum sativum . J Applied Gen. 2004;45:297–306. [PubMed] [Google Scholar]
- Irzykowska L, Wolko B, Swiecicki WK. Interval mapping of QTLs controlling some morphological traits in the pea. Cel Mol Biol Let. 2002;7:417–422. [PubMed] [Google Scholar]
- Jost E, Laven H. Meiosis in translocation heterozygotes in the mosquito Culex pipiens (Diptera: Culicidae) Chromosoma. 1971;35:184–205. doi: 10.1007/BF00285736. [DOI] [PubMed] [Google Scholar]
- Kent RJ, Harrington LC, Norris DE. Genetic differences between Culex pipiens f. molestus and Culex pipiens pipiens (Diptera: Culicidae) in New York. J Med Entomol. 2007;44:50–59. doi: 10.1603/0022-2585(2007)44[50:gdbcpf]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Nagaoka T, Noda K, Ogihara Y. Genetic linkage map of ISSR and RAPD markers in einkorn wheat in relation to that of RFLP markers. Theor Appl Genet. 1998;96:37–45. [Google Scholar]
- McDonald PT, Rai KS. Correlation of linkage groups with chromosomes in the mosquito Aedes aegypti . Genetics. 1970;66:473–83. doi: 10.1093/genetics/66.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PT, Asman SM, Terwedow HA., Jr. Sex-linked translocations in Culex tarsalis: chromosomal linkage-group correlation and segregation patterns. J Heredity. 1978;69:304–310. [Google Scholar]
- Mori A, Tomita T, Hidoh H, Kono Y, Severson DW. Comparative linkage map development and identification of an autosomal locus for insensitive acetylcholinesterase-mediated insecticide resistance in Culex tritaeniorhynchus . Insect Mol Biol. 2001;10:197–203. doi: 10.1046/j.1365-2583.2001.00255.x. [DOI] [PubMed] [Google Scholar]
- Mori A, Romero-Severson J, Severson DW. Genetic basis for reproductive diapause is correlated withlife history traits within the Culex pipiens complex. Insect Mol Bioly. 2007;16:515–524. doi: 10.1111/j.1365-2583.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- Rai KS. A comparative study of mosquito karyotypes. Ann Entomol Soc Am. 1963;56:160–170. [Google Scholar]
- Rasgon JL, Scott TW. Crimson: a novel sex-linked eye color mutant of Culex pipiens L. (Diptera: Culicidae) J Med Entomol. 2004;41:385–391. doi: 10.1603/0022-2585-41.3.385. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Venkatesan M, Westbrook CJ, Hauer CM. Polymorphic microsatellite loci from the West Nile virus vector Culex tarsalis . Mol Ecol Notes. 2006;6:680–682. [Google Scholar]
- Rasmussen SW. Meiosis in Bombyx mori females. Philos Trans R Soc Lond B Biol Sci. 1977;277:343–350. doi: 10.1098/rstb.1977.0022. [DOI] [PubMed] [Google Scholar]
- Reisen WK. Effect of temperature on Culex tarsalis (Diptera: Culicidae) from the Coachella and San Joaquin Valleys of California. J Med Entomol. 1995;32:636–645. doi: 10.1093/jmedent/32.5.636. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Lothrop H. Population ecology and dispersal of Culex tarsalis (Diptera: Culicidae) in the Coachella Valley of California. J Med Entomol. 1995;32:490–502. doi: 10.1093/jmedent/32.4.490. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Reeves WC. Bionomics and ecology of Culex tarsalis and other potential mosquito vector species. In: Reeves WC, editor. Epidemiology and Control of Mosquito-Borne Arboviruses in California, 1943–1987. Sacramento, CA: California mosquito and vector control association; 1987. pp. 254–329. [Google Scholar]
- Reisen WK, Lothrop H, Chiles R, Madon M, Cossen C, Woods L, et al. West Nile virus in California. Emerg Infect Dis. 2004;10:1369–1378. doi: 10.3201/eid1008.040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger R. Inherited semisterility for control of harmful insects. V. Translocations in Culex tritaeniorhynchus . Experientia. 1972;28:481–482. doi: 10.1007/BF02008355. [DOI] [PubMed] [Google Scholar]
- Spadoni RD, Nelson RL, Reeves WC. Seasonal occurrence,egg production, and blood-feeding activity of autogenous Culex tarsalis . Ann Entomol Soc Am. 1974;67:895–902. [Google Scholar]
- Venkatesan M, Hauer MC, Rasgon JL. Using fluorescently labelled M13-tailed primers to isolate 45 novel microsatellite loci from the arboviral vector Culex tarsalis . Med Vet Ent. 2007a;21:204–208. doi: 10.1111/j.1365-2915.2007.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M, Westbrook CJ, Hauer MC, Rasgon JL. Evidence for a population expansion in the West Nile Virus vector Culex tarsalis . Mol Biol Evol. 2007b;24:1208–1218. doi: 10.1093/molbev/msm040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wondji CS, Hunt RH, Pignatelli P, Steen K, Coetzee M, Besansky N, et al. An integrated genetic and physical map for the malaria vector Anopheles funestus . Genetics. 2005;171:1779–1787. doi: 10.1534/genetics.105.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondji CS, Morgan J, Coetzee M, Hunt RH, Steen K, Black WC, 4th, et al. Mapping a quantitative trait locus (QTL) conferring pyrethroid resistance in the African malaria vector Anopheles funestus . BMC Genomics. 2007;8:34. doi: 10.1186/1471-2164-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-K, Kantety RV, Graznak E. A genetic linkage map for tef [Eragrostis tef (Zucc.) Trotter.] Theor Appl Genet. 2006;113:1093–1102. doi: 10.1007/s00122-006-0369-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.