Summary
Copper (Cu) is an essential metal that is toxic at high concentrations. Thus, pathogens often rely on host Cu for growth, but host cells can hyper-accumulate Cu to exert anti-microbial effects. The human fungal pathogen Cryptococcus neoformans encodes various Cu-responsive genes but their role in infection is unclear. We determine that pulmonary C. neoformans infection results in Cu-specific induction of genes encoding the Cu-detoxifying metallothionein (Cmt) proteins. Mutant strains lacking CMTs or expressing Cmt variants defective in Cu-coordination exhibit severely attenuated virulence and reduced pulmonary colonization. Consistent with the up-regulation of Cmt proteins, C. neoformans pulmonary infection results in increased serum Cu concentrations and respectively increases and decreases alveolar macrophage expression of the Cu importer, Ctr1, and ATP7A, a transporter implicated in phagosomal Cu compartmentalization. These studies indicate that the host mobilizes Cu as an innate anti-fungal defense but that C. neoformans senses and neutralizes toxic Cu to promote infection.
Introduction
Copper (Cu) has a long history as an anti-microbial agent, employed to sterilize wounds by the ancient Egyptians, to ward off cholera in the 19th century, and as an anti-fungal agent in Bordeaux mixture in vineyards (Cassat and Skaar, 2012; Hodgkinson and Petris, 2012; Hood and Skaar, 2012; Samanovic et al., 2012). More recently, Cu surfaces are utilized in healthcare settings to reduce nosocomial infections (Schmidt et al., 2012). While the precise mechanisms by which Cu exerts anti-microbial activity are not well understood, the redox properties of this metal foster the generation of toxic hydroxyl radicals (•OH) and hydroxyl anions (OH−), which can cause DNA and protein damage (Halliwell and Gutteridge, 1985). Furthermore, Cu hyper-accumulation has been shown to interfere with Fe-S clusters that are critical to enzymes involved in a plethora of essential biochemical processes (Chillappagari et al., 2010; Liochev, 1996; Macomber and Imlay, 2009; Macomber et al., 2007).
The phagosomal compartment of innate immune cells presents a hostile environment to invading microbial pathogens via the generation of reactive oxygen and nitrogen species, the elaboration of proteases and other degradative enzymes, acidification of the phagosomal lumen and by nutritional limitation of metals such as Fe, Zn and Mn that are essential for microbial growth (Hood and Skaar, 2012; Nathan and Shiloh, 2000). While phagocytic cells sequester these metals from invading pathogens, macrophages infected with Mycobacterium species hyper-accumulate Cu within the phagosome (Wagner et al., 2005). Moreover, macrophage cell lines that have been activated with IFN-γ elevate expression of both the plasma membrane Cu+ importer, Ctr1, and the ATP7A vesicular Cu pump (White et al., 2009). As ATP7A is thought to traffic to the phagosomal membrane in these cells, and ATP7A depletion enhances E. coli survival to macrophage killing, these observations suggest that elevated luminal Cu is microbiocidal (White et al., 2009).
Cryptococcus species such as C. neoformans are pathogenic fungi that cause cryptococcosis in both immunodeficient and immunocompetent individuals. C. neoformans is acquired from the environment through inhalation, disseminates through the bloodstream to the brain and causes ~600,000 deaths annually from lethal meningitis (Heitman, 2011; Kronstad et al., 2012; Kronstad et al., 2011). Previous studies demonstrated that the metals Fe and Cu play important roles in C. neoformans virulence, because they are directly involved in many key biochemical processes (Jung et al., 2009; Jung et al., 2008; Jung et al., 2006; Salas et al., 1996; Walton et al., 2005; Williamson, 1994). In particular, Fe is critical for heme biosynthesis, oxidative phosphorylation and serves as a critical cofactor for dozens of enzymatic reactions. Cu functions in melanin formation, Fe uptake, reactive oxygen detoxification and respiration (Ding et al., 2011; Jung et al., 2009; Jung et al., 2008; Jung et al., 2006; Kronstad et al., 2012; Samanovic et al., 2012; Williamson, 1994). Melanin, a protective pigment and virulence factor, is synthesized by C. neoformans via the secreted Cu-dependent oxidase laccase, using host brain catecholamines as substrate (Williamson, 1994). Accordingly, deletion of the genes encoding laccase, or the secretory compartment Cu importer Ccc2, severely compromised C. neoformans virulence (Salas et al., 1996; Walton et al., 2005). The C. neoformans Cu metalloregulatory transcription factor Cuf1 has also been demonstrated to be important for virulence (Waterman et al., 2007). Since Cuf1 plays a critical role in activating expression of the CTR4 gene, encoding a high affinity plasma membrane Cu+ importer, Cu acquisition was proposed to underlie the requirement for Cuf1 for virulence (Waterman et al., 2007). However, additional studies demonstrated that cuf1Δ mutants exhibit both Cu sensitivity phenotypes and growth defects under Cu deficient conditions (Ding et al., 2011; Lin et al., 2006). Accordingly, we demonstrated that Cuf1 activates the transcription of genes encoding the Cu acquisition machinery (CTR1 and CTR4), or genes encoding the Cu detoxification machinery (CMT1 and CMT2), under Cu limitation or Cu excess, respectively (Ding et al., 2011). Given the role of Cuf1 target genes in both Cu acquisition and detoxification, it is important to clarify the specific functions of the Cuf1 regulon in virulence.
Here we report that live animal imaging studies using specific Cu-activated reporters responsive to either high Cu, or Cu deficiency, demonstrate that C. neoformans high Cu-induced reporter is dramatically induced during initial respiratory colonization. We demonstrate that the C. neoformans metallothioneins, which are induced in a Cu-specific manner and have a high capacity for Cu binding, play a critical role in virulence. Analysis of host Cu homeostasis proteins in bronchoalveolar lavage (BAL) cells from infected animals showed a dramatic increase in the high affinity mammalian Cu importer, Ctr1, and decreased abundance of the ATP7A Cu transporter that has been implicated in phagosomal Cu compartmentalization.
Results
C. neoformans metallothionein gene expression is activated in lung infection
We previously demonstrated that C. neoformans genes encoding metallothioneins or Cu transporters are strongly induced under high or low Cu conditions, respectively, in a Cu concentration dependent manner (Ding et al., 2011). Here, using qRT-PCR as a sensitive and quantitative assay, Cmt1 mRNA levels were induced ~800 fold in response to Cu and Ctr4 mRNA levels were induced ~600 fold in response to the Cu+-specific chelator BCS (Figure 1A). To ascertain whether these genes are directly regulated by Cuf1 in response to Cu levels, a FLAG-epitope tagged Cuf1 allele was generated and expressed in cuf1Δ cells for use in Chromatin immunoprecipitation (ChIP) experiments. Cuf1 was tagged with 2 copies of the FLAG sequence at the carboxyl-terminus, and cuf1Δ strains transformed with this expression plasmid are fully complemented with respect to the Cu and BCS sensitive phenotype of cuf1Δ cells, demonstrating that this is a functional Cuf1-Flag protein (Figure S1A). ChIP assays followed by qPCR analysis of promoter sequences from the CMT1/2 and CTR1/4 genes showed strong Cu-regulated Cuf1 binding to the CTR1 and CTR4 promoters under Cu deficiency as compared to high Cu conditions. In contrast, Cuf1 binding to the CMT1 promoter was induced under high Cu conditions, with binding to CMT2 observed under both conditions (Figure 1B). These results demonstrate that Cuf1 plays a direct role in the activation of Cu detoxification genes and Cu acquisition genes, when cells encounter distinct Cu environments.
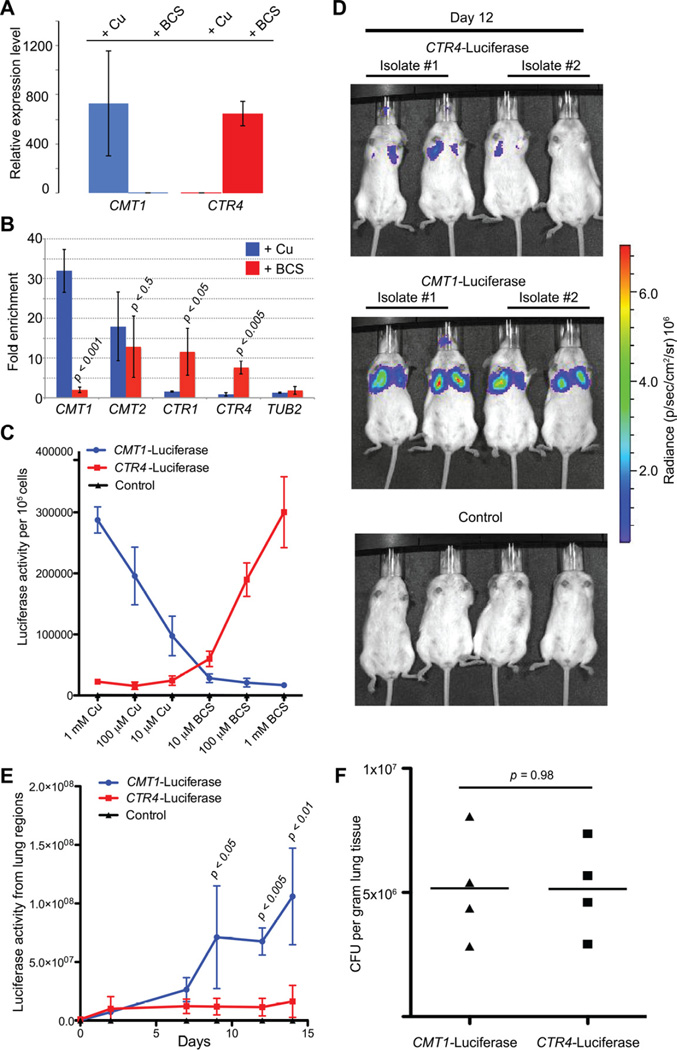
Figure 1. Cu sensing reporter systems in C. neoformans (See also Figure S1 and Table S1).
(A) Expression of CMT1 and CTR4 was quantitated using qRT-PCR. Cell cultures were subcultured in SC medium supplemented with 1 mM Cu or BCS, and incubated at 37°C for 3 hrs. Expression levels were normalized to ACT1. Error bars indicate standard deviation (SD).
(B) ChIP was performed in cuf1Δ/CUF1-2xFLAG strains after growth in the presence of 1 mM Cu or BCS. Quantitative PCR was performed to measure enrichment of promoter sequences from CMT1, CMT2, CTR1, CTR4 and TUB2 (negative control). Statistical analysis was performed using the student t test. Error bars indicate SD.
(C) Luciferase activities from fungal cells harboring reporter genes for CMT1-Luciferase, CTR4-Luciferase or wildtype (negative control) were quantified. Cells were grown in SC medium supplemented with Cu or BCS, 37°C for 9 hrs. Luciferase activities were measured using the Luciferase Reporter Assay (Qiagen). Error bars indicate SD.
(D) Luciferase activities from four mice each infected with CMT1-Luciferase, CTR4-Luciferase or wild type were measured using live animal imaging. Two independent isolates carrying CMT1-Luc or CTR4-Luc, or control wild type cells were used for intranasal mouse infections and luciferase activity scans performed at days 0, 2, 7, 9, 12 and 14, and day 12-post infection is shown.
(E) Luciferase activity from the lungs of each mouse (in D) was measured and analyzed using Living Image 4.2 (Caliper, PerkinElmer). Statistical analysis was performed using the student t test. Error bars indicate SD.
(F) Fungal cell burden assessed by colony forming units (CFU) from mouse lung homogenates derived from animals in Figure 1D.
To assess the potential Cu environment in host tissue sensed in the initial stages of C. neoformans infection through its natural respiratory route of infection, two Cu-responsive reporter plasmids were constructed in which luciferase expression is driven by the C. neoformans CTR4 promoter (CTR4-Luc) in response to Cu limiting conditions or the CMT1 promoter (CMT1-Luc) in response to elevated Cu. CMT1- and CTR4- driven luciferase protein expression and activities from each reporter was confirmed by luciferase enzyme assays and immunoblotting (Figure 1C and S1B). In C. neoformans cells luciferase activity from the CMT1 promoter is induced 17-fold in response to increasing Cu, while activity from the CTR4 promoter is induced 14-fold in response to BCS treatment; no activity was detected from cells lacking a luciferase reporter (Figure 1C). Intranasal infection of mice was carried out with independent isolates of C. neoformans carrying an integrated copy of the CTR4-Luc or CMT1-Luc reporter, followed by live animal imaging and luciferase activity quantitation (Figure 1D and E). After 2 days weak activity was detected in animals infected with cells harboring the CMT1 or CTR4 reporters, but not with control cells. While luciferase activity for the CTR4-luc infection remained low and unchanged throughout the subsequent 14 day infection period, there was a time-dependent increase in CMT1-driven luciferase activity in lung tissue. To ascertain whether the difference in luciferase activity between the two reporter strains is due to impaired lung colonization by cells harboring the CTR4-Luciferase reporter, we performed fungal burden assays and detected no difference in lung fungal cell burden between the two reporter strains (Figure 1F). These results suggest that the Cmt1 gene is induced, when C. neoformans is acquired by the respiratory route, the natural route of infection in humans.
C. neoformans MTs are critical factors for lung colonization and virulence
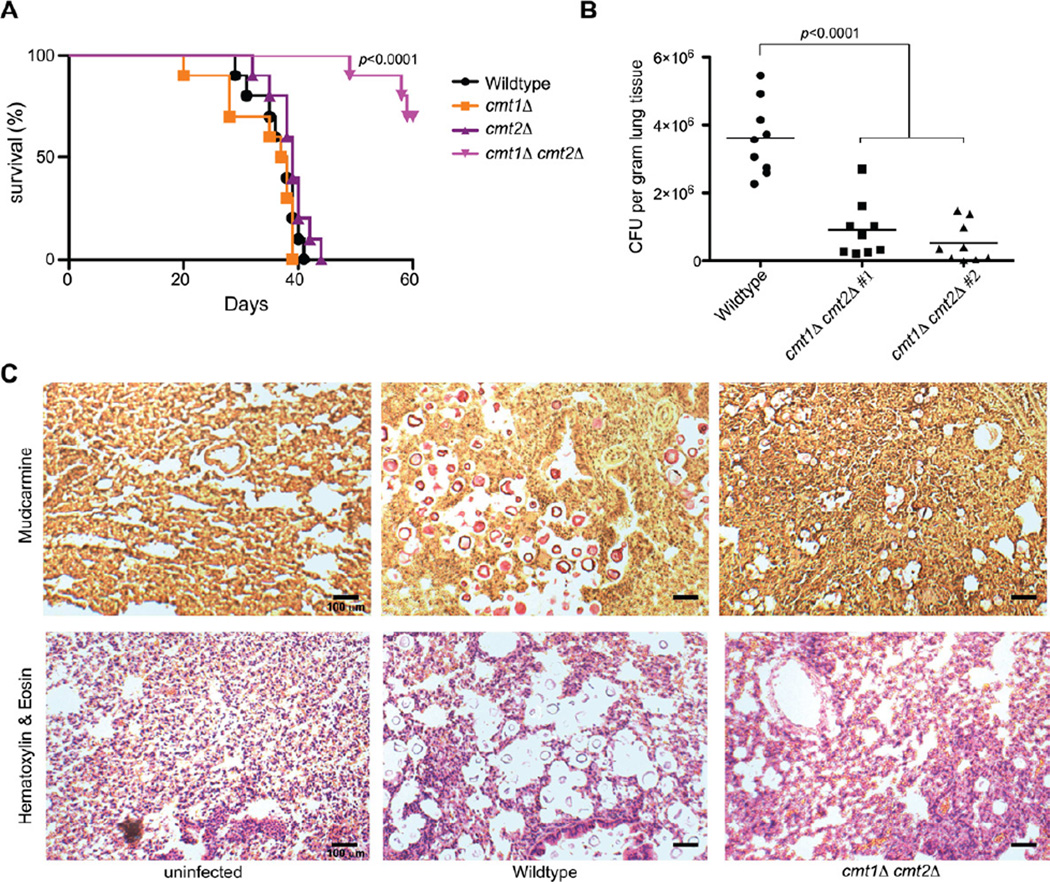
Expression from the C. neoformans CMT1 promoter is activated in pulmonary infection, implying that fungal cells sense elevated Cu in the lung, and Cuf1 directly activates transcription of the CMT1 and CMT2 genes, whose encoded proteins are required for Cu detoxification in C. neoformans (Figure 1). Consequently, the potential role of CMT1 and CMT2 in C. neoformans virulence was investigated by infecting mice with wild type or isogenic cmt1Δ, cmt2Δ or cmt1Δ cmt2Δ mutants (Figure 2A). While CMT1 and CMT2 are functionally redundant for Cu detoxification (Ding et al., 2011), an cmt1Δ cmt2Δ mutant is over 30-fold more Cu-sensitive than WT cells in vitro (IC50 for WT of 2.3 mM vs. 73 µM for cmt1Δ cmt2Δ). Deletion of either CMT1 or CMT2 did not alter mouse survival compared to the parental strain. However, the cmt1Δ cmt2Δ strain was strongly attenuated in virulence (Figure 2A). Two independently generated cmt1Δ cmt2Δ strains were evaluated for lung tissue burden 14 days post infection, with both cmt1Δ cmt2Δ strains showing a dramatic decrease in lung tissue fungal burden in comparison to the wild type strain (Figure 2B). This observation was validated by staining tissue sections for C. neoformans with the capsule-specific stain mucicarmine, which showed a reduction in cmt1Δ cmt2Δ cells as compared to wild type cells, with no defect in melanin production, capsule formation or phagocytosis observed between wild type and cmt1Δ cmt2Δ cells (Figure S2). Furthermore, a corresponding decrease in host lung tissue damage was evident as determined by H&E staining of lung tissue sections (Figure 2C). Taken together, these results are correlate with the strong expression of CMT1 in lung observed in live animal imaging studies, and demonstrate that C. neoformans Cmts are required for full fungal virulence when acquired via the respiratory route, the natural route of infection.
Figure 2. C. neoformans metallothioneins are virulence factors (See also Figure S2 and Table S1).
(A) Ten A/J female mice were infected intranasally with wildtype, cmt1Δ, mt2Δ, or cmt1Δ cmt2Δ cells and animals monitored for viability over 60 days. Shown is a Kaplan-Meier survival plot.
(B) Mice were infected with wildtype, or two independent cmt1Δ cmt2Δ mutants for 14 days and lung tissues were isolated, homogenized and CFU were quantitated and normalized with respect to tissue weight. Statistical analysis was performed using ANOVA.
(C) Lung tissue from uninfected, wildtype, or cmt1Δ cmt2Δ infected mice were isolated, fixed, and stained with mucicarmine or hematoxylin and eosin.
C. neoformans MTs are atypical metallothioneins specifically activated by Cu
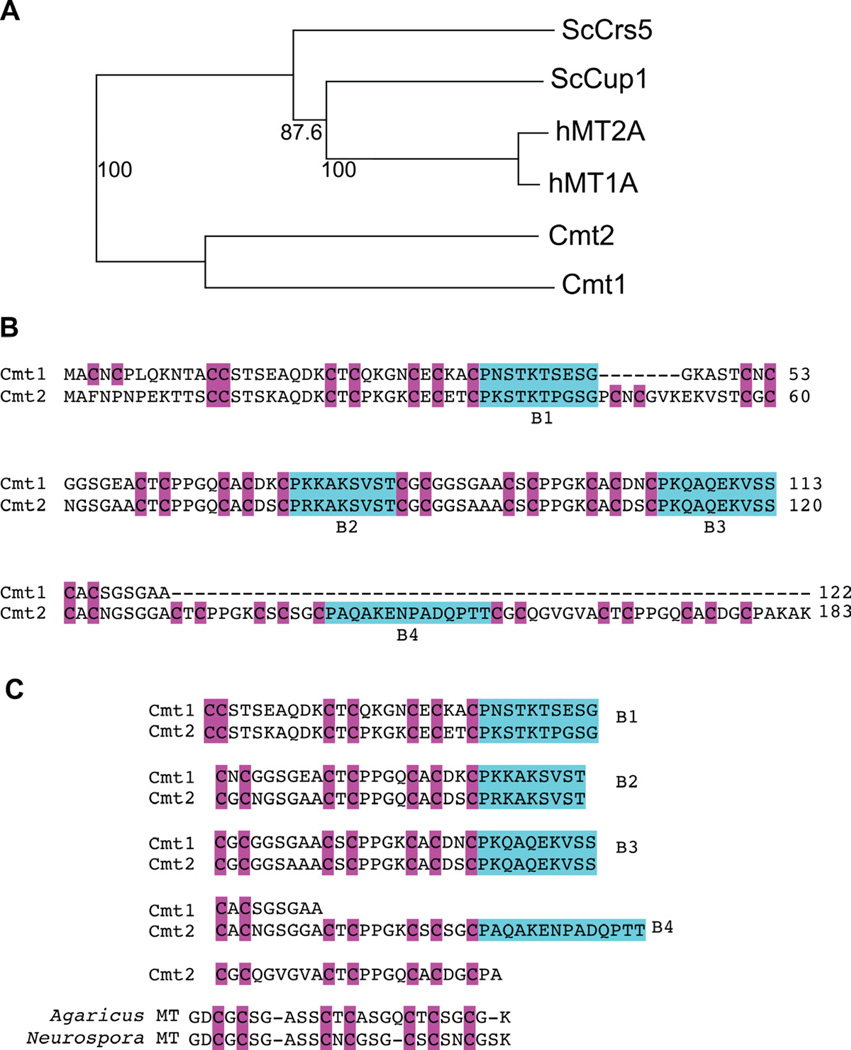
MTs are expressed in organisms from prokaryotes to humans, which bind metals through Cys thiolate bonds (Butt et al., 1984; Kagi and Hunziker, 1989; Szczypka and Thiele, 1989; Winge and Nielson, 1984; Winge et al., 1985). We previously reported that Cmts from C. neoformans possess Cu binding motifs typical of MTs (CxC), yet they are atypical MTs compared with those from other species (Ding et al., 2011). Both Cmt1 and Cmt2 are much larger proteins, where Cmt1 is 122 amino acids and Cmt2 is 183 amino acids long as compared to human MT1A of 61 amino acids. Multiple sequence alignments demonstrated that metallothioneins from Cryptococcus share homology with the MTs from S. cereviaise and humans (Figure S3). Phylogenetic analysis suggests that Cmt1 and Cmt2 share the same evolutionary origin, but are distantly related to the Crs5 and Cup1 MTs from Saccharomyces cerevisiae and human MT1A and MT2A (Figure 3A). Comparison of protein sequences between C. neoformans MT1 and MT2 reveals that both proteins are divided into multiple Cys-rich sequence segments by spacer sequences termed B1 to B4 (Figure 3B), with Cmt1 divided into three segments by three spacer regions and Cmt2 harboring three Cys segments separated by four spacer regions. The spacer regions between Cmt1 and Cmt2 share high similarity for B1 and are identical for B2 and B3. Interestingly, the Cys-rich motif resembles that found in MTs from other fungi such as Agaricus and Neurospora and may imply evolutionary divergence from a common ancestor among these species (Figure 3C).
Figure 3. Atypical C. neoformans metallothioneins (See also Figure S3).
(A). A phylogenetic MT tree was generated as described previously (Ding et al., 2011), with percentage of confidence (bootstrap) shown in numbers. Both C. neoformans MTs are distantly related to those from S. cerevisiae (Sc) and human (h).
(B). Protein sequences from CMT1 and CMT2 were aligned. The homologous cysteine residues are shaded in purple, and spacer boxes are shaded in green. Both Cmts contain spacer regions (B1 to B3 for Cmt1 and B1 to B4 for Cmt2). The spacer sequences share high similarity between Cmt1 and Cmt2 and divide each Cmt into multiple cysteine-rich segments, resulting in a peculiar architecture of 3 Cys-rich segments for Cmt1 and 5 for Cmt2.
(C). Cmt1 and Cmt2 share a common motif Cys-X-Cys-X6-Cys-X-Cys-X4-Cys-X-Cys-X2-Cys motif in their Cys-rich segments. These motifs are separated by 3 spacer regions in Cmt1 and 4 in Cmt2, and this motif is similar to that found in MTs in other fungi such as Agaricus and Neurospora.
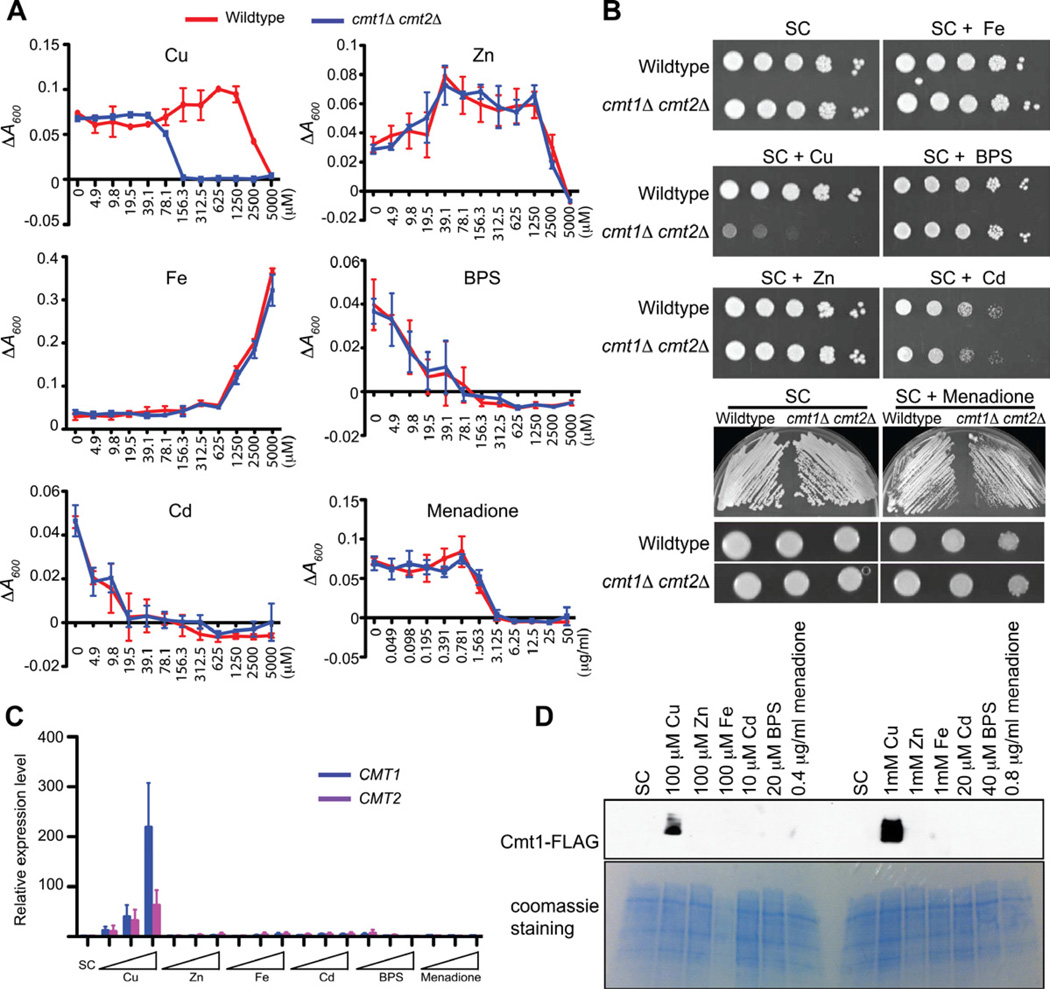
Mammalian MT genes are transcriptionally induced by metals that include Zn, Cd, Cu and Ag and protect cells against these and other metals (Durnam and Palmiter, 1984, 1987; Kagi and Hunziker, 1989). To decipher the specificity of metal detoxification with respect to Cryptococcus MTs, we measured cell growth in liquid medium supplemented with a range of metal concentrations including Cu, Zn, and Cd. The effect of high and low Fe on cell growth was also tested because Fe acquisition via the Fe permease is directly dependent on a multi-Cu oxidase in C. neoformans (Jung and Kronstad, 2008; Kronstad et al., 2012). A potential role for the C. neoformans MTs for growth in the presence of reactive oxygen species was also tested using the superoxide generator, menadione. We observed a striking growth defect of cmt1Δ cmt2Δ cells in the presence of Cu, with no significant difference in the presence of Cd, Zn, Fe, the Fe chelator BPS or the superoxide generator menadione (Figure 4A). Agar spotting assays were also performed to confirm the liquid growth experiments, and similar cell growth phenotypes were observed (Figure 4B). Consequently, we determined whether RNA and protein expression of C. neoformans MTs are elevated in response to these conditions by qRT-PCR and immunoblot assays. As shown in Figures 4C and 4D, expression of both CMT1 and CMT2 mRNA and FLAG-epitope-tagged protein is strongly elevated in response to Cu exposure in a dose-dependent manner, but not in response to any concentration of Zn, Fe, Cd, BPS or Menadione tested. Taken together, these results demonstrate that, of all conditions tested, the C. neoformans MT genes are Cu responsive and function specifically in Cu detoxification.
Figure 4. C. neoformans MTs are Cu responsive Cu detoxification proteins (See also Table S1).
(A). C. neoformans cell growth assays were performed in SC medium in 96 well plates. Cell cultures of wildtype and cmt1Δ cmt2Δ were diluted to an A600 of 0.002, supplemented with metals. A600 was measured after overnight growth. ΔA600 was calculated by subtracting from blank (medium without cells). Graphs show average of three biological replicates. Error bars indicate SD.
(B). C. neoformans metal sensitivity assays on agar medium. Cell cultures of wild type and cmt1Δ cmt2Δ cells were diluted in water to an A600 of 1.0. 10-fold serial dilutions cells were spotted onto SC agar or agar supplemented with 400 µM Cu, Zn, Fe, 100 µM Cd, 40 µM BPS, or 10 µg/ml menadione. Plates were incubated for 2 days (6 days for menadione spotting assay) and photographed.
(C). Expression of CMT1 and CMT2 was quantitated by qRT-PCR. Cell cultures were diluted to an A600 of 0.2 in SC medium at 37°C for 1hr, supplemented with the indicated concentrations (selected according to the results from Figure 4A. 10, 100, 1000 µM For Cu, Zn, Fe; 5, 10, 20 µM for Cd; 10, 20, 40 µM for BPS; 0.2, 0.4, 0.8 µg/ml for menadione). Error bars indicate SD.
(D). Protein expression of Cmt1-FLAG was confirmed by immunoblotting. Cells were grown as described as Figure 4C. Protein extracts were treated with TCEP, resolved by SDS-PAGE and anti-FLAG mouse antibody was used for immunoblotting. Coomassie staining was used as a loading control.
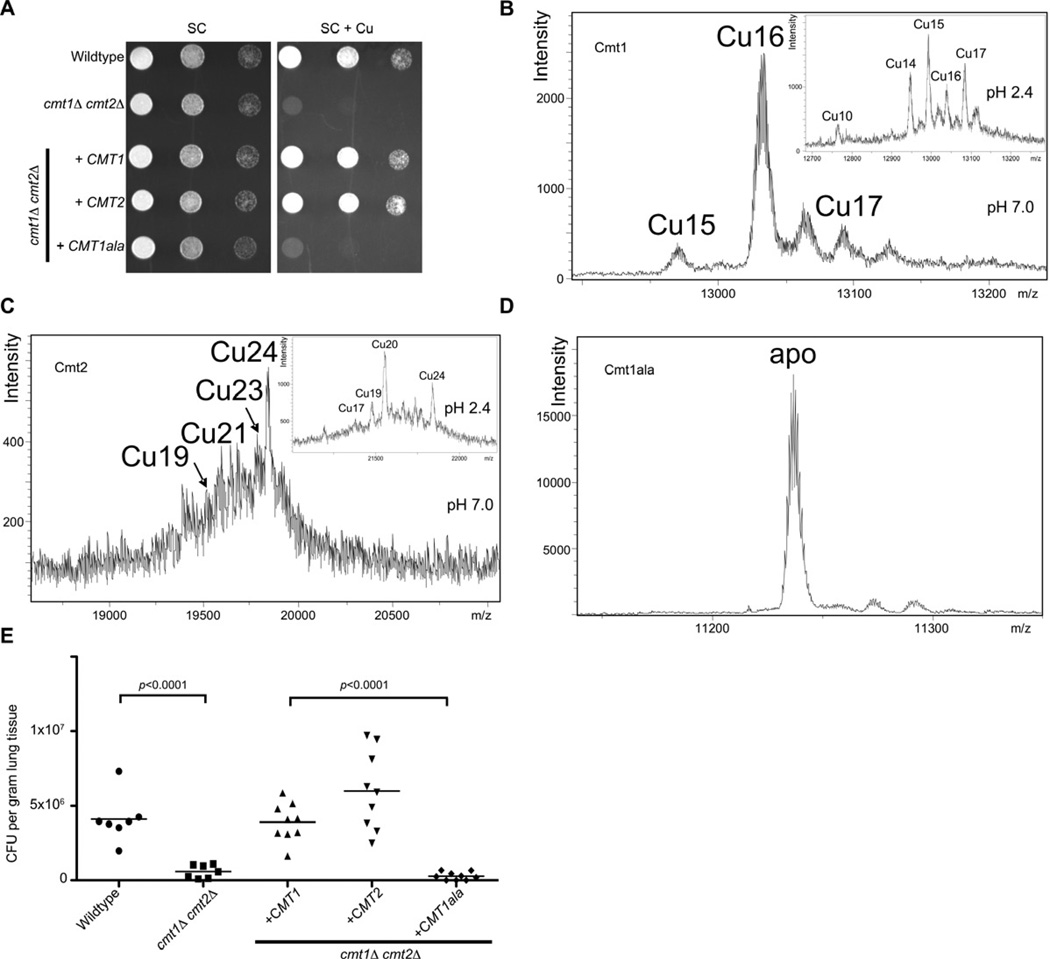
To evaluate the importance of Cu binding by Cmt1 and Cmt2 to Cu detoxification, the importance of the Cmt1 Cys residues in protecting cells from Cu toxicity was tested. A DNA sequence encoding a CMT1 allele in which all Cys residues were converted to Ala was synthesized, cloned under control of the CMT1 promoter and introduced into cmt1Δ cmt2Δ cells to generate the Cmt1ala strain. Using qRT-PCR, the expression of CMT1ala was confirmed to be robust and Cu responsive, as the fold induction of expression between Cu and BCS treatment is comparable to that observed for wild type CMT1 (Figure S4A). Cell growth assays demonstrated that the Cmt1 Cu-coordinating Cys residues are essential for Cu resistance (Figure 5A).
Figure 5. C. neoformans MT Cu binding capacity is critical for virulence (See also Figure S4 and Table S1).
(A) Cu-resistance growth assays in SC medium supplemented with 1 mM Cu with the cmt1Δ cmt2Δ mutant expressing CMT1, CMT2 or the CMT1ala mutant. Growth assays were performed as described in Figure 4B.
(B) ESI-MS spectra at pH 7.0 and pH 2.4 (insets) of purified Cmt1, (C) Cmt2 and the Cmt1ala mutant (D).
(E) Lung tissue fungal burden (colony forming units, CFU) from cmt1Δ cmt2Δ cells transformed with plasmids expressing CMT1, CMT2 or CMT1ala. Experiments and statistical analysis were performed as described in Figure 2B.
To quantify the Cu binding capacity of Cmt1 and Cmt2, the Cmts were synthesized in and purified from E. coli, after which spectroscopic analysis of Cmt1, Cmt2 and Cmt1ala was performed. These experiments showed a high, preferential Cu+ binding capacity yielding major homonuclear Cu16-Cmt1 and Cu24-Cmt2 complexes (Figure 5B and C). In vitro Zn/Cu replacement experiments using recombinantly synthesized Zn-Cmt1 and Zn-Cmt2 complexes fully corroborated these stoichiometries (Figure S4B and C) and pointed to the progressive and cooperative formation of several Cu5 ion clusters (3 for Cmt1 and 5 for Cmt2), in accordance with the peculiar protein architecture in the same number of Cys-rich regions (Figure 3). Furthermore, the CD spectra of the complexes and the products of recombinant synthesis were nearly identical (Figure S4D and E), indicating equivalent folding. Consistent with the inability of the Cmt1ala mutant to support Cu-resistance in cmt1Δ cmt2Δ cells (Figure 5A), the Cmt1ala protein was defective in Cu+ binding and was isolated exclusively in the apo form (Figure 5D). These results establish Cu+ binding to the C. neoformans MTs with high stoichiometry that is dependent on Cys thiolate bonds. To test whether the cysteine residues of CMT1 are required for virulence, mice were infected with wild type C. neoformans cells, isogenic cmt1Δ cmt2Δ cells, or the same mutant strain expressing either CMT1, CMT2 or CMT1ala and fungal burdens were evaluated in host lung tissue. Consistent with the Cu binding results, C. neoformans expressing a Cmt1 protein that is incompetent for Cu binding (Cmt1ala) exhibited poor survival as evidenced by decreased fungal burden in lung tissue from infected mice (Figure 5E). These results demonstrate the essential role of the Cmt1 Cys residues, required for Cu+ coordination, for virulence in mouse lung infection.
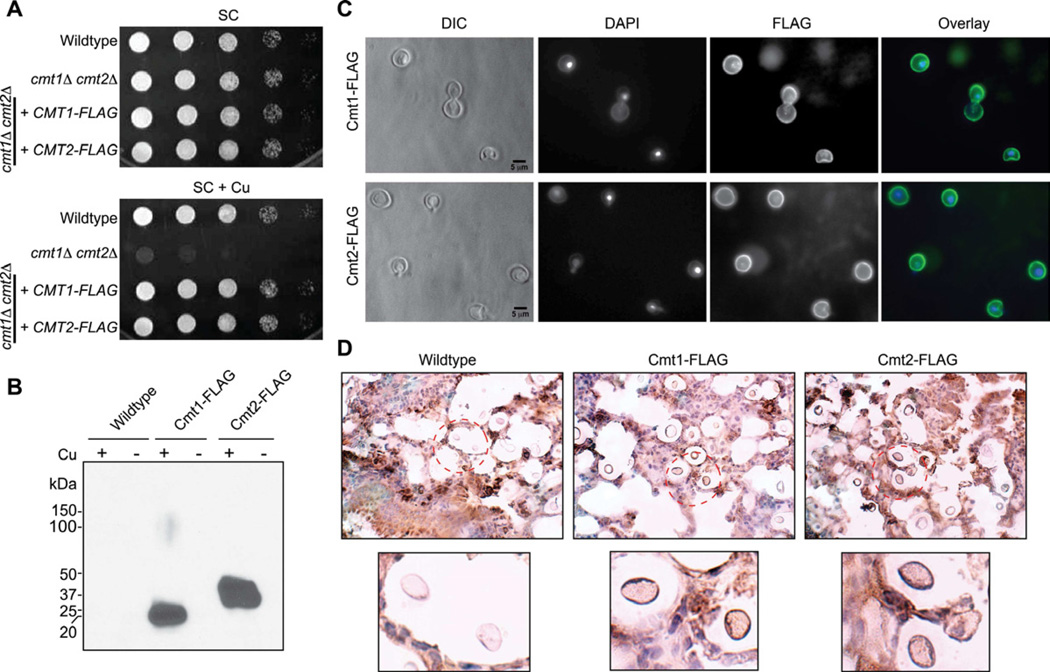
Although MTs have been previously localized to the cytosol of fungal and mammalian cells (Banerjee et al., 1982; Hamer, 1986; Winge and Nielson, 1984), the subcellular distribution of Cmt1 and Cmt2 was determined in cells cultured in vitro. The expression of functional FLAG-epitope tagged Cmt1 and Cmt2 (Figure 6A) was confirmed by immunoblotting experiments in which an ~20 KDa polypeptide was detected for Cmt1-FLAG and ~37 KDa species for Cmt2-FLAG (Figure 6B. Subcellular localization experiments by indirect immunofluorescence microscopy of Cu-treated C. neoformans cell cultures demonstrated that Cmt1-FLAG and Cmt2-FLAG concentrate at the cell periphery (Figure 6C). This observation was recapitulated by immunohistochemistry analysis of lung tissue infected with C. neoformans cells expressing either Cmt1-FLAG or Cmt2-FLAG (Figure 6D). Taken together, these experiments demonstrate that a Cu-binding Cmts is critical for both C. neoformans Cu resistance in vitro and virulence in mouse infections. Furthermore, distinct from the pancellular localization of Cmts observed in other eukaryotes (Hamer, 1986), the C. neoformans atypical MT proteins concentrate at the cellular periphery.
Figure 6. C. neoformans MTs concentrate at the cell periphery (See also Table S1).
(A) cmt1Δ cmt2Δ CMT1-FLAG and cmt1Δ cmt2Δ CMT2-FLAG cells were generated and Cu sensitive phenotype assays performed by spotting 10-fold serial dilutions on SC agar or SC agar supplemented with 1 mM Cu.
(B) Immunoblotting confirmed expression of Cmt1-FLAG and Cmt2-FLAG. C. neoformans cells (wildtype, cmt1Δ cmt2Δ CMT1-FLAG and cmt1Δ cmt2Δ CMT2-FLAG) were incubated in the presence of 200 µM Cu (+) or BCS (−) in SC medium for 3 hrs and immunoblotting was performed as described in Figure 4D. Ponceau S staining confirmed equal protein loading.
(C) Cmt1-Flag and Cmt2-Flag proteins localized by indirect immunefluorescence microscopy with anti-Flag antibody and the DNA stains DAPI for localizing nuclei.
(D) Lung tissue from mice infected (14 day-post infection) with wildtype, cmt1Δ cmt2Δ CMT1-FLAG or cmt1Δ cmt2Δ CMT2-FLAG was analyzed by H&E staining and immunohistochemistry using anti-Flag antibody.
C. neoformans infection alters host Cu mobilization and Cu transporter expression
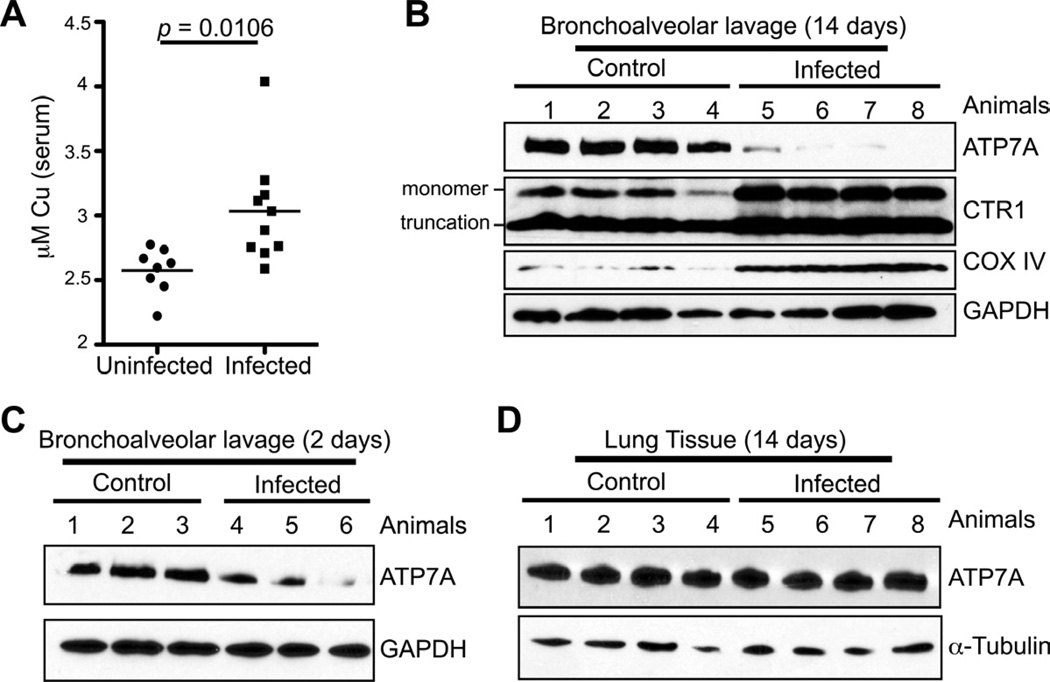
Bronchial alveolar macrophages are phagocytic cells that provide the first line of defense against C. neoformans infection in the lung (Brummer, 1998; Kronstad et al., 2011). Previous in vitro studies demonstrated that macrophage-like cell lines infected with Mycobacterium spp. accumulate Cu in the phagosomal compartment and activation of macrophage cell lines with LPS induces expression of the ATP7A Cu+-transporting P-type ATPase and the Ctr1 high affinity Cu+ importer (Wagner et al., 2005; White et al., 2009). Macrophage cell lines with reduced expression of ATP7A are deficient in killing E. coli, consistent with a potential role for ATP7A in phagosomal microbiocidal Cu+ loading (White et al., 2009). Because activated macrophages have elevated ATP7A and Ctr1 levels, and we show here that C. neoformans senses high Cu and activates the MT1 promoter during lung infection, we ascertained whether there are changes in host circulating Cu levels and in expression of the host Cu homeostatic machinery in response to C. neoformans infection. Serum Cu levels were significantly increased, suggesting a mobilization of host Cu in response to C. neoformans infection (Figure 7A). Moreover, cells from mouse bronchoalveolar lavage (BAL) 14 days after infection, of which the dominant cell type has been shown to be alveolar macrophages (Giles et al., 2007), displayed a strong decrease in the steady-state levels of ATP7A (Figure 7B) that was observed to a lesser extent 2 days post-infection (Figure 7C). In contrast, infected mice exhibited no apparent change in lung tissue ATP7A levels compared to uninfected controls (Figure 7D). After 14 days of infection the levels of the Ctr1 high affinity Cu+ importer, and the Cox IV subunit of mitochondrial cytochrome oxidase whose levels correlate with intracellular Cu availability, were strongly elevated (Figure 7B). Taken together, these observations suggest that in response to C. neoformans infection via the respiratory route, host mobilize Cu into the circulation and lung alveolar cells may re-orient Cu away from vesicular compartments and toward the mitochondria or other pools.
Figure 7. C. neoformans infection alters host Cu transport machinery.
(A) Mice were infected with wild type C. neoformans cells and serum was isolated at day 14-post infection. Cu were measured using ICPMS and shown for uninfected and C. neoformans infected mice.
(B) Bronchoalveolar lavage cells (BAL) were isolated from uninfected mice and mice infected with wildtype cells 14 post-infection. Protein was extracted from the BAL and ATP7A, Ctr1, Cox IV and GAPDH levels analyzed by SDS-PAGE and immunoblotting.
(C) BAL protein extract was analyzed 2 days after infection by immunoblotting for ATP7A and GAPDH.
(D) Lung tissue from mice 14-days after infection was analyzed for ATP7A and tubulin levels by immunoblotting.
Discussion
Prokaryotic Cu detoxification mechanisms involving Cu-responsive transcription factors and Cu efflux pumps are emerging as critical virulence factors for organisms such as M. tuberculosis, E. coli, S. enterica and others (Achard et al., 2010; Osman and Cavet, 2011; Samanovic et al., 2012; Schwan et al., 2005; Wagner et al., 2005; White et al., 2009; Wolschendorf et al., 2011). In line with these observations are studies that demonstrate the compartmentalization of Cu within the macrophage phagosome in response to infection (Wagner et al., 2005; White et al., 2009), in a manner that correlates with elevated expression of the mammalian Ctr1 plasma membrane Cu+ importer and the ATP7A Cu+ transporting ATPase (White et al., 2009). As depletion of ATP7A renders macrophages more permissive for E. coli survival (White et al., 2009), and mice receiving dietary Cu supplements more effectively clear M. tuberculosis (Wolschendorf et al., 2011), these and other experimental results point to phagosomal Cu compartmentalization, that may involve ATP7A, as a potent anti-microbial weapon against infectious disease (Hodgkinson and Petris, 2012; Rowland and Niederweis, 2012; Samanovic et al., 2012; Wolschendorf et al., 2011).
Pathogenic fungi such as C. neoformans and C. albicans are quite resistant to Cu levels in vitro (Ding et al., 2011; Weissman et al., 2000), with C. neoformans H99 resistant to ~2 mM Cu in liquid medium and clinical isolates of C. albicans able to tolerate ~20 mM Cu (Weissman et al., 2000). An important question is why C. neoformans, or other pathogenic fungi, are tolerant to such high Cu concentrations and is this relevant to the concentrations of Cu they encounter during infection? In contrast to Cu detoxification in prokaryotes, Cu acquisition has been implicated in virulence by C. neoformans in mouse infection models (Waterman et al., 2007). Cuf1 was previously implicated in virulence by mouse tail-vein administration studies, and its known activation of CTR4 implied a requirement for Cu for virulence (Waterman et al., 2007). More recent studies using URA5 to disrupt CTR4 resulted in C. neoformans cells with a pleiotropic nutritional deficiency that was not corrected by external Cu and resulted in a reduction in virulence in mice (Waterman et al., 2012). However, given that such a growth phenotype has not been observed in Cu transporter knockouts of Ctr1 or Ctr4 in C. neoformans by us (Ding et al., 2011), or in response to inactivation of other Cu importer genes in S. cerevisiae, S. pombe or C. albicans in other studies (Beaudoin et al., 2006; Dancis et al., 1994; Marvin et al., 2003; Pena et al., 2000; Zhou and Thiele, 2001), it is not clear why this pleiotropic phenotype was observed. One possibility is that use of the URA5 marker for gene disruption in C. neoformans, and the URA3 marker in C. albicans and C. parapsilosis, has been shown to cause defects in adhesion, colony morphology and virulence that are unrelated to the target genes of interest (Bain et al., 2001; Ding and Butler, 2007; Kirsch and Whitney, 1991; Kwon-Chung et al., 1992; Lay et al., 1998; Staab and Sundstrom, 2003). While it is possible that the Cu acquisition machinery may contribute to the colonization of lung and brain due to a requirement to activate Cu, Zn SOD (Bermingham-McDonogh et al., 1988; Furukawa et al., 2004), the Cu dependent oxidase involved in Fe uptake (Dancis et al., 1994; Jung and Kronstad, 2008), and for the synthesis of melanin from laccase (Walton et al., 2005; Williamson, 1994) using host catecholamine as substrate, the reasons behind these discrepant studies merits further investigation.
A recent study using a CTR4-Cherry reporter suggested that CTR4 is strongly expressed in macrophages in vitro and in lung and brain tissue (Waterman et al., 2012). However, CTR4-driven expression of mCherry in this study was compared to that of C. neoformans cells harboring an empty vector without the mCherry gene. In this work we used C. neoformans cells harboring high and low Cu-responsive reporter plasmids, as well as negative control cells, to quantitatively ascertain, over the course of a 14-day intranasal infection, whether C. neoformans is exposed to a high or low Cu environment. While both CTR4-luc and CMT1-luc are expressed in lung during the initial phase of the infection, the CMT1-luc reporter was activated in a time-dependent manner while the CTR4-luc remained low and constant. The strong induction of the CMT1-luc reporter in lung implies that C. neoformans senses elevated Cu levels in the lung. Consistent with this observation, we demonstrated that the CMT1 and CMT2, and a Cu-binding competent Cmt1 protein, are required for virulence at the natural site of acquisition, the lungs. Indeed, in contrast to mammalian MTs, the C. neoformans MTs, and other fungal MTs, are transcriptionally activated in response to Cu, rather than to any other metal tested, suggesting a role that is specific under conditions of high Cu. Moreover, we show that the C. neoformans MTs are longer and have an exceptionally high Cu binding capacity compared to other MT proteins, perhaps due to evolutionary pressure to evolve by tandem amplification of a basic Cu binding unit similar to that found in well characterized fungal metallothioneins. The concentration of Cmt1 and Cmt2 to the cell periphery, via currently uncharacterized targeting mechanisms, could provide a means to efficiently capture Cu+ immediately after it enters cells, prior to engaging in redox chemistry, interfering with Fe-S clusters or targeting other mechanisms for toxicity (Chillappagari et al., 2010; Liochev, 1996; Macomber and Imlay, 2009; Macomber et al., 2007).
The results presented here showing a requirement for CMT1 and CMT2 for virulence are consistent with macrophages in the lung and other tissues using Cu as an antimicrobial condition within the lumen of the phagosome. As the expression of ATP7A and Ctr1 was shown to be elevated in activated macrophage cell lines, and a fraction of ATP7A was found in the phagosomal membrane, this Cu+ pump is implicated as a potential driver of phagosomal compartmentalization of anti-microbial Cu (White et al., 2009). Complementary to the elevation of ATP7A in INF-γ activated macrophages in vitro, we found that C. neoformans infection caused a time-dependent down regulation of ATP7A in lung lavage cells, an environment reported to be composed predominantly of phagocytic cells (Giles et al., 2007). In addition to the Cuf1-dependent Cu detoxification genes, this could provide a survival advantage to C. neoformans within the phagosomal compartment that ultimately allows this organism to escape into the cytoplasm by vomocytosis (Nicola et al., 2011). While the mechanism for ATP7A dampening, but maintenance of high Ctr1 levels, is currently unknown, C. neoformans infection is known to cause a reduction in host pro-inflammatory cytokines and an increase in NF-κB activity, via glucuronoxylomannan in the outer capsule (Ben-Abdallah et al., 2012; Piccioni et al., 2013), which may reduce ATP7A expression. We speculate that as the Ctr1 promoter, but not that of ATP7A, contains a putative NF-κB binding site (http://genome.ucsc.edu/ENCODE) (Dunham et al., 2012), this could maintain Ctr1 expression while ATP7A levels are tuned down. It is likely that the regulation of Ctr1 and ATP7A expression is due to a complex interplay between C. neoformans and the host immune system and this should be explored in more detail. The increased levels of circulating Cu and the high levels of Ctr1 and Cox IV in the host could suggest that C. neoformans infection results in the down-regulation of the host Cu compartmentalization machinery, potentially reorienting available Cu to other intracellular targets such as the mitochondria. The use of C. neoformans mutants, in concert with mouse models with altered Cu homeostasis, could help elucidate the detailed mechanisms by which Cu functions at the host-pathogen axis.
Experimental Procedures
Strains and media
Cryptococcus neoformans H99 strains (Table S1) were routinely grown as previously described (Ding et al., 2011). YPD agar supplemented with 100 mg/L G418 or 200 U/ml hygromycin B was used for colony selection. Mutants were generated as described in Supplemental procedures.
Chromatin Immunoprecipitation
Cells expressing Cuf1-FLAG were treated with 1 mM Cu or BCS for 3 hrs. Cell fixation and ChIP were performed as described previously (Pondugula et al., 2009), except buffer (50 mM HEPES, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease inhibitors) was used to lyse cells and M2 beads (Sigma) were used for immunoprecipitation. Promoter sequences from CMT1, CMT2, CTR1, CTR4 and TUB2 was analyzed using qPCR. Primer sequences are described in the Supplemental procedures.
Luciferase assays and live animal imaging
Strains transformed with luciferase reporter genes were diluted to an A600 of 0.2 in SC medium supplemented with Cu or BCS and incubated at 37°C for 9 hrs. Cell cultures were washed and resuspended in PBS.10 µl of cell suspension was mixed with 100 µl with luciferase reporter reagent (Promega) and activity was measured using a bioilluminator Victor (PerkinElmer). The samples were then measured at A600 for cell number.
A/J mice were infected with wildtype, CTR4-Luc, or CMT1-Luc strains intranasally. Mice were anesthetized using 2.5% of isoflurane. Luciferin was introduced intranasally into each animal (no signal was observed when luciferin was administered intraperitoneally). Animals were placed in a Caliper IVIS Spectrum (PerkinElmer) chamber at 37°C. The scan was performed exactly 5 min after introducing luciferin. Scanning was performed at day 0, 2, 7, 9, 12 and 14. Animals were sacrificed at day 14 for CFU analysis. All images were analyzed using Living Image 4.2 (Caliper, PerkinElmer). The lung region from each animal was cropped and total luciferase signal intensity in the cropped area was extracted using Living Image 4.2. Statistical analysis was performed using the student t-test.
Animal infection, fungal burden assay and histopathology
Animal infections were performed as described previously (Crabtree et al., 2012). Histology of uninfected or infected lung tissue was processed and mucicarmine or hematoxylin/eosin staining was performed.
Spectroscopic analyses and Electrospray Ionization Mass Spectrometry (ESI-MS)
Cmt proteins were expressed in E. coli BL21 strain and purified using GST fusion as described in the Supplemental procedures. The S, Zn and Cu content of the Zn- and Cu-Cmt preparations was analysed by Inductively Coupled Plasma Atomic Emission Spectroscopy (ICP-AES) as described previously (Bongers et al., 1988; Capdevila et al., 2005).
Molecular weight determinations were performed by electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS). The calibration was attained with 0.2 g NaI dissolved in 100 ml of a 1: H2O: isopropanol mixture. Samples containing Zn, Cu-Cmt complexes with divalent metal ions were analysed under the following conditions: 20 µL of protein solution injected through a PEEK (polyether heteroketone) column at 40 µl/min; capillary counter-electrode voltage 5 kV for Zn and 3.5 kV for Cu; desolvation temperature 80–110°C; dry gas 6 L/min; spectra collection range 80 0–2500 m/z. The carrier buffer was a 5:95 mixture of acetonitrile:ammonium acetate (15 mM, pH 7.0) for Zn and a 10:90 mixture for Cu. Analysis of apo-Cmt and Cu-Cmt at low pH was performed using a 5:95 mixture of acetonitrile:formic acid at pH 2.4. Under all the conditions assayed, the error associated with the mass measurements was always lower than 0.1%.
Antibodies
ATP7A antibody was a gift from Dr. Michael Petris, University of Missouri. Cox IV and luciferase antibodies were purchased from Abcam. GAPDH antibody was purchased from Santa Cruz. FLAG antibody was purchased from Sigma. Immunofluorescence microscopy and immunohistochemistry were performed as previously described (Ding et al., 2011). All microscopy images were taken using a Zeiss Axio Imager wide field fluorescence microscope (ZEISS).
Bronchoalveolar lavage (BAL) isolation from animals
BAL isolation was performed as described previously (Okagaki et al., 2010), except the fluid was centrifuged and resuspended in ACK lysis buffer (NH4Cl, KHCO3 and EDTA) to lyse red blood cells, and then washed 3 times with PBS.
Supplementary Material
C. neoformans senses a high Cu environment during pulmonary colonization
Cryptococcus Cu detoxification machinery is important for fungal virulence
Cryptococcus metallothioneins are Cu specific detoxification proteins
Acknowledgments
We thank G. Sempowski and K. Riebe for animal imaging assistance, M.J. Petris for ATP7A antibody, and the Thiele lab for critical comments. C.D. and R.A.F. acknowledge the Duke Scholars in Infectious Disease Program. We acknowledge support from the NIH (GM48140-24 to DJT, 2P30 AI064518-06 to YLC and AI50438 to JH), “Ministerio de Ciencia e Innovación” grants BIO2009-12513-C02-01 to S.A. and BIO2009-12513-C02-02 to M.C. and Serveis Científico-Tècnics and the Servei d’Anàlisi Química and European Union support via FEDER. S.A. and M.C. acknowledge 2009SGR-1457 “Grup de Recerca de la Generalitat de Catalunya.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard ME, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG. The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect Immun. 2010;78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Stubberfield C, Gow NA. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett. 2001;204:323–328. doi: 10.1111/j.1574-6968.2001.tb10905.x. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Onosaka S, Cherian MG. Immunohistochemical localization of metallothionein in cell nucleus and cytoplasm of rat liver and kidney. Toxicology. 1982;24:95–105. doi: 10.1016/0300-483x(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Beaudoin J, Laliberte J, Labbe S. Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology. 2006;152:209–222. doi: 10.1099/mic.0.28392-0. [DOI] [PubMed] [Google Scholar]
- Ben-Abdallah M, Sturny-Leclere A, Ave P, Louise A, Moyrand F, Weih F, Janbon G, Memet S. Fungal-induced cell cycle impairment, chromosome instability and apoptosis via differential activation of NF-kappaB. PLoS Pathog. 2012;8:e1002555. doi: 10.1371/journal.ppat.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Gralla EB, Valentine JS. The copper, zincsuperoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc Natl Acad Sci U S A. 1988;85:4789–4793. doi: 10.1073/pnas.85.13.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers J, Walton CD, Richardson DE, Bell JU. Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur. Anal Chem. 1988;60:2683–2686. doi: 10.1021/ac00175a008. [DOI] [PubMed] [Google Scholar]
- Brummer E. Human defenses against Cryptococcus neoformans: an update. Mycopathologia. 1998;143:121–125. doi: 10.1023/a:1006905331276. [DOI] [PubMed] [Google Scholar]
- Butt TR, Sternberg EJ, Gorman JA, Clark P, Hamer D, Rosenberg M, Crooke ST. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci U S A. 1984;81:3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila M, Domenech J, Pagani A, Tio L, Villarreal L, Atrian S. Zn- and Cd-metallothionein recombinant species from the most diverse phyla may contain sulfide (S2-) ligands. Angew Chem Int Ed Engl. 2005;44:4618–4622. doi: 10.1002/anie.200501183. [DOI] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin Immunopathol. 2012;34:215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol. 2010;192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K. Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun. 2012;80:3776–3785. doi: 10.1128/IAI.00507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A, Yuan DS, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner RD. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- Ding C, Butler G. Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryot Cell. 2007;6:1310–1319. doi: 10.1128/EC.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Yin J, Tovar EM, Fitzpatrick DA, Higgins DG, Thiele DJ. The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol Microbiol. 2011;81:1560–1576. doi: 10.1111/j.1365-2958.2011.07794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnam DM, Palmiter RD. Induction of metallothionein-I mRNA in cultured cells by heavy metals and iodoacetate: evidence for gratuitous inducers. Mol Cell Biol. 1984;4:484–491. doi: 10.1128/mcb.4.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnam DM, Palmiter RD. Analysis of the detoxification of heavy metal ions by mouse metallothionein. Experientia Suppl. 1987;52:457–463. doi: 10.1007/978-3-0348-6784-9_45. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Torres AS, O'Halloran TV. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. Embo J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Zaas AK, Reidy MF, Perfect JR, Wright JR. Cryptococcus neoformans is resistant to surfactant protein A mediated host defense mechanisms. PLoS One. 2007;2:e1370. doi: 10.1371/journal.pone.0001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. The importance of free radicals and catalytic metal ions in human diseases. Mol Aspects Med. 1985;8:89–193. doi: 10.1016/0098-2997(85)90001-9. [DOI] [PubMed] [Google Scholar]
- Hamer DH. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Heitman J. Cryptococcus: from human pathogen to model yeast. Washington DC: ASM press; 2011. [Google Scholar]
- Hodgkinson V, Petris MJ. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012;287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W, Kronstad JW. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot Cell. 2009;8:1511–1520. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Kronstad JW. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol. 2008;10:277–284. doi: 10.1111/j.1462-5822.2007.01077.x. [DOI] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagi JH, Hunziker P. Mammalian metallothionein. Biol Trace Elem Res. 1989;21:111–118. doi: 10.1007/BF02917243. [DOI] [PubMed] [Google Scholar]
- Kirsch DR, Whitney RR. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, Geddes JM, Griffiths EJ, Choi J, Cadieux B, et al. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell. 2012;11:109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Attarian R, Cadieux B, Choi J, D'Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A, Edman JC, Bennett JE. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J Med Vet Mycol. 1992;30:61–69. [PubMed] [Google Scholar]
- Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE, Becker JM. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun. 1998;66:5301–5306. doi: 10.1128/iai.66.11.5301-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SL. The role of iron-sulfur clusters in in vivo hydroxyl radical production. Free Radic Res. 1996;25:369–384. doi: 10.3109/10715769609149059. [DOI] [PubMed] [Google Scholar]
- Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin ME, Williams PH, Cashmore AM. The Candida albicans CTR1 gene encodes a functional copper transporter. Microbiology. 2003;149:1461–1474. doi: 10.1099/mic.0.26172-0. [DOI] [PubMed] [Google Scholar]
- Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AM, Robertson EJ, Albuquerque P, Derengowski Lda S, Casadevall A. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. MBio. 2011;2 doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D, Cavet JS. Metal sensing in Salmonella: implications for pathogenesis. Adv Microb Physiol. 2011;58:175–232. doi: 10.1016/B978-0-12-381043-4.00005-2. [DOI] [PubMed] [Google Scholar]
- Pena MM, Puig S, Thiele DJ. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J Biol Chem. 2000;275:33244–33251. doi: 10.1074/jbc.M005392200. [DOI] [PubMed] [Google Scholar]
- Piccioni M, Monari C, Kenno S, Pericolini E, Gabrielli E, Pietrella D, Perito S, Bistoni F, Kozel TR, Vecchiarelli A. A Purified Capsular Polysaccharide Markedly Inhibits Inflammatory Response during Endotoxic Shock. Infect Immun. 2013;81:90–98. doi: 10.1128/IAI.00553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondugula S, Neef DW, Voth WP, Darst RP, Dhasarathy A, Reynolds MM, Takahata S, Stillman DJ, Kladde MP. Coupling phosphate homeostasis to cell cycle-specific transcription: mitotic activation of Saccharomyces cerevisiae PHO5 by Mcm1 and Forkhead proteins. Mol Cell Biol. 2009;29:4891–4905. doi: 10.1128/MCB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland JL, Niederweis M. Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis (Edinb) 2012;92:202–210. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MG, Attaway HH, Sharpe PA, John J, Jr, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, et al. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J Clin Microbiol. 2012;50:2217–2223. doi: 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan WR, Warrener P, Keunz E, Stover CK, Folger KR. Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. Int J Med Microbiol. 2005;295:237–242. doi: 10.1016/j.ijmm.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 2003;11:69–73. doi: 10.1016/s0966-842x(02)00029-x. [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Thiele DJ. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol Cell Biol. 1989;9:421–429. doi: 10.1128/mcb.9.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE, 3rd, Honer Zu Bentrup K, Russell DG, Bermudez LE. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Hacham M, Hu G, Zhu X, Park YD, Shin S, Panepinto J, Valyi-Nagy T, Beam C, Husain S, et al. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest. 2007;117:794–802. doi: 10.1172/JCI30006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Park YD, Raja M, Qiu J, Hammoud DA, O'Halloran TV, Williamson PR. Role of CTR4 in the Virulence of Cryptococcus neoformans. MBio. 2012;3 doi: 10.1128/mBio.00285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman Z, Berdicevsky I, Cavari BZ, Kornitzer D. The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc Natl Acad Sci U S A. 2000;97:3520–3525. doi: 10.1073/pnas.97.7.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge DR, Nielson KB. Formation of the metal-thiolate clusters of rat liver metallothionein. Environ Health Perspect. 1984;54:129–133. doi: 10.1289/ehp.8454129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge DR, Nielson KB, Gray WR, Hamer DH. Yeast metallothionein. Sequence and metal-binding properties. J Biol Chem. 1985;260:14464–14470. [PubMed] [Google Scholar]
- Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Thiele DJ. Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 2001;276:20529–20535. doi: 10.1074/jbc.M102004200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.