Abstract
Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) provide powerful platforms for genome editing in plants and animals. Typically, a single nuclease is sufficient to disrupt the function of protein-coding genes through the introduction of microdeletions or insertions that cause frameshifts within an early coding exon. However, interrogating the function of cis-regulatory modules or noncoding RNAs in many instances requires the excision of this element from the genome. In human cell lines and invertebrates, two nucleases targeting the same chromosome can promote the deletion of intervening genomic segments with modest efficiencies. We have examined the feasibility of using this approach to delete chromosomal segments within the zebrafish genome, which would facilitate the functional study of large noncoding sequences in a vertebrate model of development. Herein, we demonstrate that segmental deletions within the zebrafish genome can be generated at multiple loci and are efficiently transmitted through the germline. Using two nucleases, we have successfully generated deletions of up to 69 kb at rates sufficient for germline transmission (1%–15%) and have excised an entire lincRNA gene and enhancer element. Larger deletions (5.5 Mb) can be generated in somatic cells, but at lower frequency (0.7%). Segmental inversions have also been generated, but the efficiency of these events is lower than the corresponding deletions. The ability to efficiently delete genomic segments in a vertebrate developmental system will facilitate the study of functional noncoding elements on an organismic level.
Sequence-specific nucleases (SSNs), such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), provide effective tools for engineering the genomes of plants and animals (Urnov et al. 2010; Carroll 2011; Joung and Sander 2013). These reagents are revolutionizing the application of reverse genetic approaches in a broad array of organisms by facilitating the generation of targeted double-strand breaks (DSBs) within the genome. These DSBs can disrupt gene function through imprecise break repair or promote tailor-made alterations to the genome through homologous recombination from an exogenously supplied donor DNA (Urnov et al. 2010; Carroll 2011; Joung and Sander 2013). Each SSN comprises a pair of programmable DNA-binding domains recognizing neighboring genomic sequences fused to the nuclease domain from FokI. Upon binding of both partners to the target locus, the appended FokI domains dimerize and generate a DSB in the spacer between the monomer binding sites.
The creation of two DSBs within the genome can stimulate the formation of larger genomic aberrations. In cultured cells, two nucleases that target nearby regions within the same chromosome can generate deletions of the intervening genomic segment (∼15 kb) with efficiencies approaching 10% of the treated genomes (Lee et al. 2010; Carlson et al. 2012; Kim et al. 2012). In addition, these tandem DSBs can generate segmental inversions, as well as segmental duplications through unequal exchange between homologous/sister chromosomes (Carlson et al. 2012; Lee et al. 2012). Likewise, the generation of DSBs on disparate chromosomes can promote translocations between the cleaved segments albeit at low frequency (Brunet et al. 2009; Simsek et al. 2011). Targeted genomic deletions have also been generated in invertebrate animals using two SSNs (Ma et al. 2012). In vertebrates, the description of segmental deletions using two SSNs has been limited to a single founder in rats harboring a 2.5-kb deletion (Menoret et al. 2010). However, this study lacks a description of the deletion rates induced in treated embryos and the transmission frequency of the associated deletion, which leaves undefined the efficiency of these processes in a complex vertebrate organism.
The ability to generate large segmental deletions reliably within a vertebrate genome would provide an avenue for the functional interrogation of noncoding elements, such as intergenic or intronic cis-regulatory modules, miRNA clusters, or lincRNAs. Due to the paucity of described functional sequences or conservation in such elements, they may be challenging to disrupt using a single nuclease. Targeted deletions would also facilitate the removal of alternately spliced exons to specifically disable the function of a single protein isoform. Here we examine the feasibility of creating segmental deletions and inversions within the zebrafish genome using two SSNs targeting three different protein-coding loci (apoea, flt4, slc24a5-lepa) and two noncoding sequences (the linc-birc6 [megamind] lincRNA gene and the globin locus control region). We find that two highly active SSNs can efficiently create segmental deletions of up to 69 kb at frequencies between 1% and 15% in somatic cells. Importantly, we demonstrate that these deletions can be transmitted through the germline of treated animals with comparable frequencies. Local segmental inversions can also be created through this approach, although these aberrations occur at a lower frequency than simple deletions. Given the simplicity with which SSNs can be constructed or purchased for any desired target sequence (Urnov et al. 2010; Carroll 2011; Joung and Sander 2013), large targeted deletions can be generated throughout the genome in vertebrates, allowing the functional analysis of noncoding sequence elements during development or in disease progression.
Results
Targeted genomic deletions at the zebrafish apoea locus
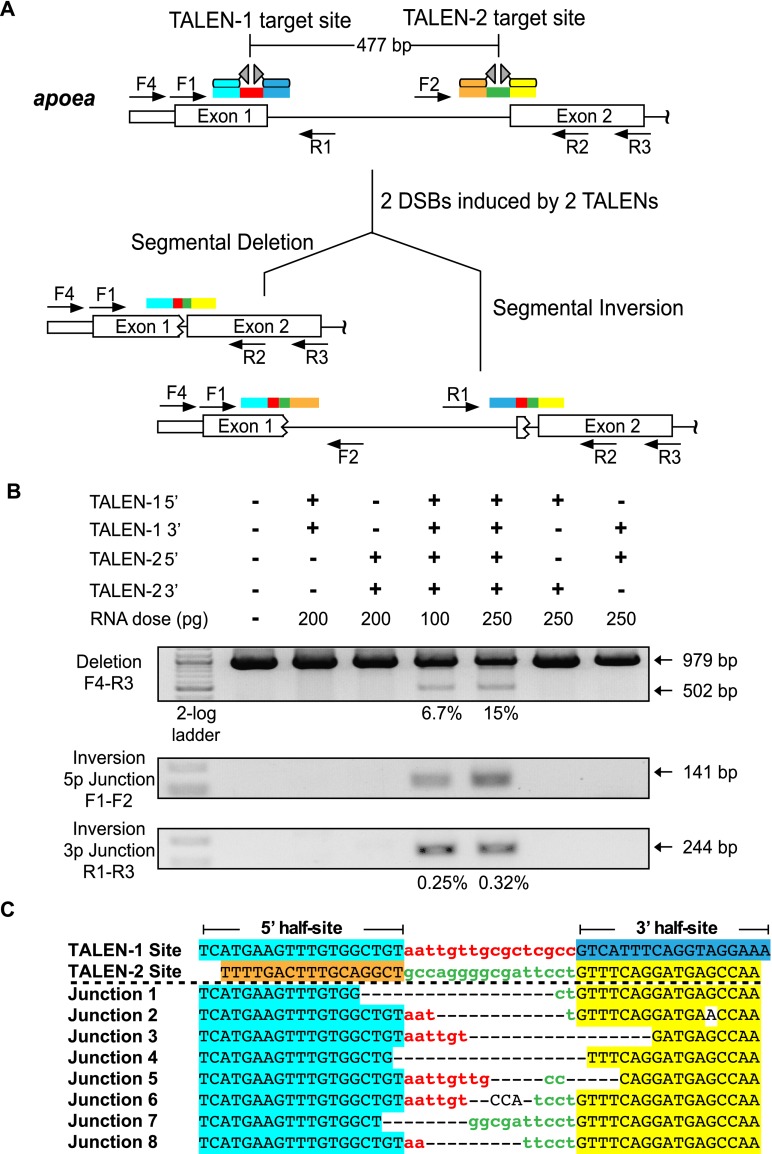
To examine the feasibility of generating targeted deletions within the zebrafish genome, we created four TALENs within the coding exons of apoea that span different segments of this locus. We tested the activity of each TALEN at its target site upon injecting its mRNAs into zebrafish embryos and verified that the lesion rates for each nuclease were in excess of 20% in normal embryos 24 h post-fertilization (hpf) (Supplemental Fig. 1; Meng et al. 2008; Gupta et al. 2012). We then examined whether coinjecting mRNAs encoding the two most proximal TALENs would delete the intervening genomic fragment, which spanned ∼500 bp (Fig. 1A). Consistent with prior results in human cell culture (Lee et al. 2010), segmental deletions were generated at appreciable frequencies only when mRNAs encoding both nucleases were coinjected (Fig. 1B). Based on a standard curve analysis, these segmental deletions occur in genomes of treated embryos at a rate of ∼15% (Supplemental Fig. 2). Sequence analysis of cloned PCR amplicons confirmed deletion of the intervening genomic fragment between the TALEN target sites and revealed a set of heterogeneous fusion sequences between the 5′ and 3′ spacer regions (Fig. 1C). The junction sequences display hallmarks of imprecise NHEJ repair with evidence of short microhomologies between some of the joined sequences (Simsek et al. 2011). In addition to the presence of segmental deletions, inversions were also detected within the genome of treated embryos (Fig. 1B), but at a lower frequency (∼0.3%) relative to segmental deletions, where the ends of the inverted segment are exchanged between the TALEN cleavage sites (Supplemental Fig. 3). The presence of segmental duplications created through unequal exchange between alternate break positions on sister/homologous chromosomes was also detected (Lee et al. 2012), but these events occur at a low frequency relative to deletions (Supplemental Fig. 4).
Figure 1.
TALEN-generated deletions within the apoea locus. (A) Schematic diagram of the zebrafish apoea locus displaying the positions of the two proximal TALEN binding sites separated by ∼500 bp. The half-sites recognized by each TALEN are differentially colored (cyan, blue, orange, yellow), as are the spacers between the half-sites (red, green). Examples of the possible alterations (segmental deletions or inversions) in a single chromosome in the context of two DSBs are indicated below with the expected rearrangement of the TALEN target sites. Each primer position and directionality is indicated by an arrow within the schematic. (B) Segmental deletions are readily detected in normal-appearing zebrafish embryos when two TALENs are coinjected, where the primer pair used for detection is listed to the left of the gel image. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each component. PCR products corresponding to the unmodified locus (979 bp) and the segmental deletion (∼502 bp) are present, where the latter occurs only in embryos injected with both TALENs. Estimated deletion rates are indicated below the positive lanes. Segmental inversions are also detected in embryos treated with two TALENs, where the inversion rates are lower than the corresponding segmental deletions. (C) Junction sequences for the 500-bp segmental deletions at the apoea locus. The TALEN-1 and TALEN-2 recognition sequences are indicated above the junction sequences, where the half-sites recognized by each TALEN are differentially colored. PCR products spanning this region were shotgun-cloned and sequenced to define the junctions at these deletions. This reveals a heterogeneous set of fusion sequences between the TALEN binding sites, where microhomologies are present at many of these fusion points.
Segmental deletions were also efficiently generated between two TALENs that were separated by either 750 bp or 4.2 kb within the apoea locus (Fig. 2A). A modest decrease in the deletion rate was observed with increasing separation, because 750-bp and 4.2-kb segmental deletions occurred at rates of 10% and 2.7%, respectively (Fig. 2B). Sequence analysis of cloned PCR amplicons spanning these regions confirmed deletion of the intervening genomic fragment between the TALEN target sites (Fig. 2C; Supplemental Fig. 5). Segmental inversions were also generated by these TALENs at a modest frequency (1%–2%) (Fig. 2B; Supplemental Fig. 5). Local duplications due to unequal exchange between homologous chromosomes were also evident (Supplemental Fig. 6). Thus, analysis at the apoea locus establishes that genomic segments on the kilobase scale can be effectively excised from the zebrafish genome in somatic cells using two TALENs.
Figure 2.
TALEN-generated 750-bp and 4.2-kb deletions within the zebrafish apoea locus. (A) Schematic diagram of the apoea locus demarcating the positions of all four TALEN binding sites and the distance separating them. (B) Segmental deletions of 750 bp and 4.2 kb are readily detected in normal-appearing TALEN-treated embryos when two TALENs are coinjected. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each component. PCR products corresponding to the desired 750-bp and 4.2-kb segmental deletion products (∼517 and ∼402 bp, respectively) are detected only in embryos injected with both TALENs. Estimated deletion rates are indicated below the positive lanes. Segmental inversions are detected in embryos treated with two TALENs for both the 750-bp and 4.2-kb segments. The calculated inversion rates are lower than the corresponding segmental deletions. (C) Junction sequences for the 4.2-kb segmental deletions at the apoea locus. The TALEN-1 and TALEN-4 recognition sequences are indicated above the junction sequences, where the half-sites recognized by each TALEN are differentially colored. The recovered junction sequences contain variable length deletions as well as evidence of insertions between some of the target sites, where the latter are indicated in plain text.
Targeted genomic deletions at other zebrafish loci
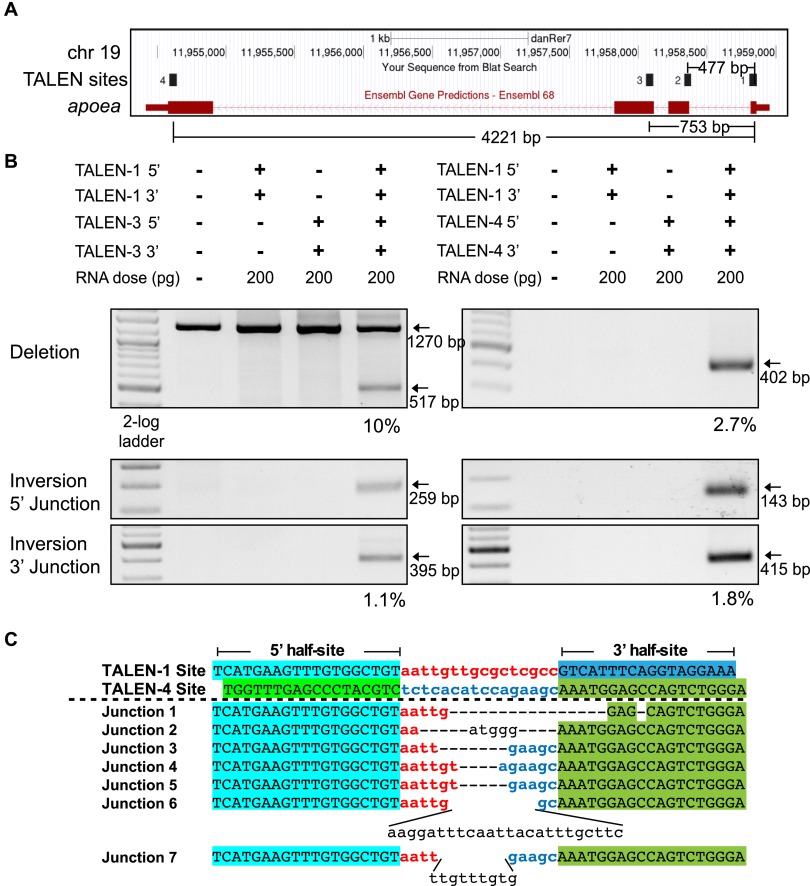
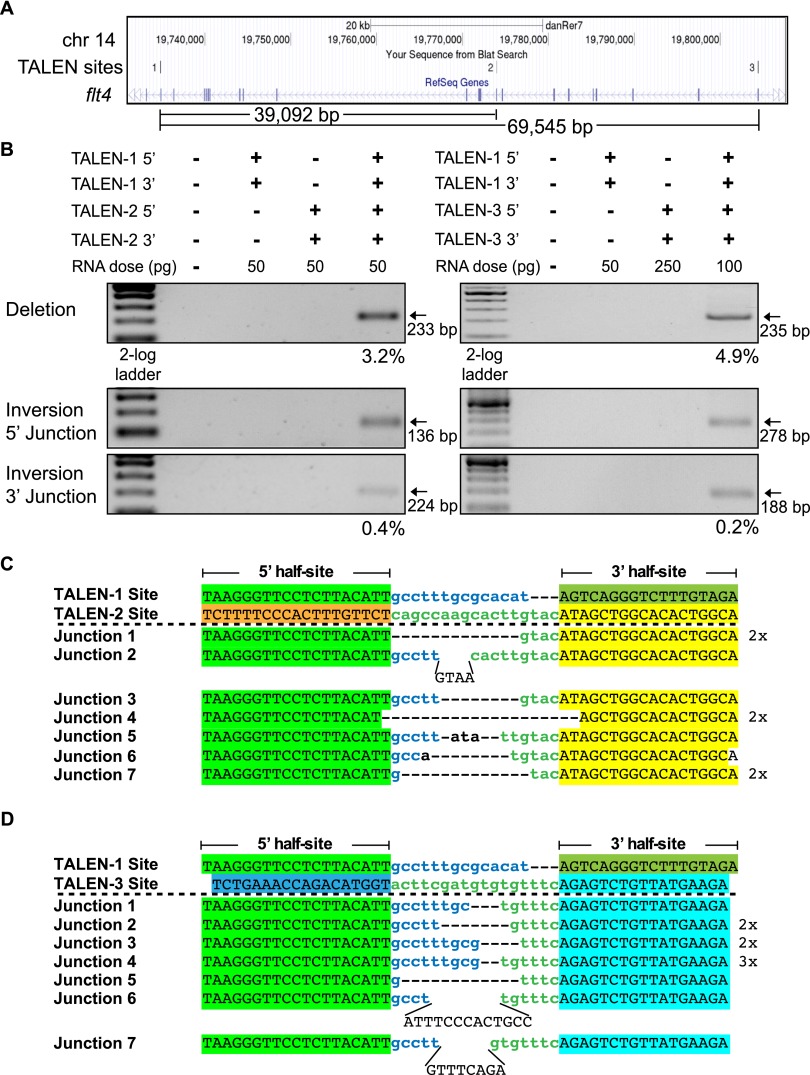
To validate that segmental deletions can be generated at alternate protein-coding loci, we used two TALENs separated by 39 or 69 kb within the flt4 locus (Fig. 3A). As with the apoea TALENs, each flt4 TALEN displays high activity at its individual target site (Supplemental Fig. 7). When two flt4 TALENs were coinjected into zebrafish embryos, segmental deletions were observed for both the 39-kb and 69-kb segments with efficiencies that were similar (3.2% and 4.9%, respectively) to the shorter segmental deletions at the apoea locus (Fig. 3B). The junctions at these deletions are consistent with the segmental deletion of DNA between two DSBs generated by the TALENs (Fig. 3C). Segmental inversions (Fig. 3B) and duplications (Supplemental Fig. 8) could be detected as well. These results confirm that segmental deletions can be reliably generated within coding genes of sufficient length to span a vertebrate gene of moderate size.
Figure 3.
TALEN-generated 39-kb and 69-kb deletions within the zebrafish flt4 locus. (A) Schematic diagram of the flt4 locus demarcating the positions of all three TALEN binding sites and the distance separating them. (B) Segmental deletions of 39 and 69 kb are readily detected in normal-appearing TALEN-treated embryos when two TALENs are coinjected. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each TALEN component. PCR products corresponding to the desired 39-kb and 69-kb segmental deletion products are detected only in embryos injected with both TALENs. Estimated deletion rates are indicated below the positive lanes. Segmental inversions are detected in embryos treated with two TALENs for both segments, but at lower rates than the corresponding segmental deletions. Junction sequences for the 39-kb (C) and 69-kb (D) segmental deletions at the flt4 locus. The TALEN-1 and TALEN-2/3 recognition sequences are indicated above the junction sequences, where the half-sites recognized by each TALEN are differentially colored. In the recovered junction sequences, there are variable length deletions as well as evidence of insertions between some of the target sites, where the latter are indicated in plain text.
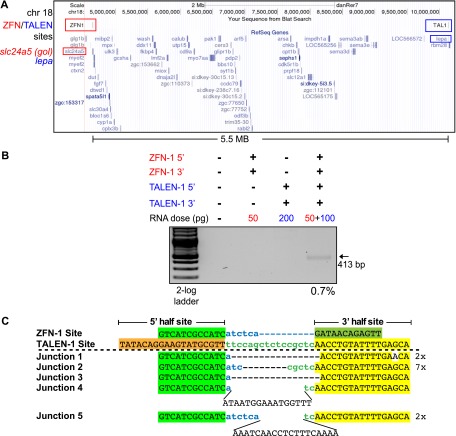
We examined the ability to generate even more expansive deletions on chromosome 18 using a ZFN that recognizes a target site within the slc24a5 (golden) locus and a TALEN that recognizes a target site within the lepa locus. The intervening segment between these target sites spans a 5.5-Mb gene-rich region of the genome (Fig. 4A). Both of these nucleases are efficient with the ZFN generating lesions at a 24% rate (Supplemental Fig. 9). Coinjection of mRNAs encoding both nucleases generated deletions of this entire genomic segment (Fig. 4B). Based on PCR analysis, we estimate the frequency of this large segmental deletion at 0.7%. Sequence analysis of cloned PCR amplicons spanning this region confirmed deletion of the intervening genomic fragment (Fig. 4C). Thus, sizeable deletions can be created using this dual nuclease approach within somatic cells of a vertebrate animal, albeit at a low frequency.
Figure 4.
TALEN/ZFN-generated 5.5-Mb deletions on chromosome 18. (A) Schematic diagram of the target region within chromosome 18 demarcating the position of the ZFN target site in the slc24a5 (golden) locus and the TALEN target site in the lepa locus. (B) Segmental deletions of 5.5 Mb are detected in normal-appearing nuclease-treated embryos when the ZFN and TALEN RNAs are coinjected. The presence or absence of each nuclease component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each component. PCR products corresponding to the desired 5.5-Mb segmental deletion (413 bp) are detected only in embryos injected with both nucleases. The estimated deletion rate is indicated below the positive lane. (C) Junction sequences for the 5.5-Mb segmental deletions on chromosome 18. The nuclease recognition sequences are indicated above the junction sequences, where the half-sites are differentially colored. In the recovered junction sequences, there are variable length deletions as well as evidence of insertions between some of the target sites, where the latter are indicated in plain text.
Targeted genomic deletions of noncoding sequence elements
The creation of segmental deletions would also have broad application in the study of regulatory elements or noncoding RNAs within the genome, because they may not be readily disrupted by InDels generated by a single SSN. As a proof-of-principle, we targeted the locus control region (LCR) for the major globin locus on chromosome 3 (Ganis et al. 2012). The LCR spans ∼6 kb within an intron of the neighboring nprl3 gene. An ∼200-bp region within the LCR contains a cluster of recognition sites for transcription factors that regulated the globin locus and is sufficient to drive expression of a GFP transgene in zebrafish erythrocytes (Ganis et al. 2012).
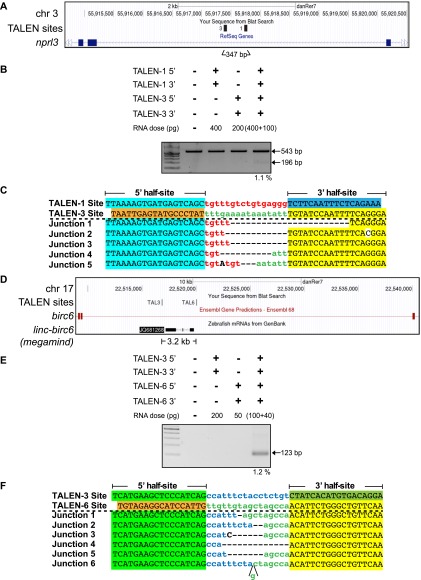
We created four TALENs that flank this 200-bp element within the LCR to facilitate its removal (Fig. 5A). To our surprise, two of these TALENs were inactive, and one displayed low activity (Supplemental Fig. 10). We believe that this reflects an inherent difficulty in targeting noncoding sequence elements that are more prone to contain repetitive sequences and have low complexity relative to protein-coding exons. We sequenced the target region from these embryos to confirm that the failure of a TALEN was not due to a difference between the genomic sequence of our zebrafish lines and those from the latest zebrafish genome assembly (Zv9, Ensembl) (Collins et al. 2012; data not shown). Using our most promising TALENs, we were able to achieve targeted deletion of this LCR regulatory element within the genome (Fig. 5B), although the efficiency of this deletion was modest (1.1%). As anticipated, the sequences of the PCR products that span this deletion indicate the excision of the genomic segment between the TALEN target sites (Fig. 5C).
Figure 5.
TALEN-generated deletions of a segment of the globin LCR on chromosome 3 and the linc-birc6 (megamind) on chromosome 17. (A) Schematic diagram of the nprl3 locus demarcating the positions of the two TALEN binding sites flanking the core LCR regulatory element within this intron and the distance separating them. (B) Segmental deletion of the 300-bp element is detected in normal-appearing TALEN-treated embryos when two TALENs are coinjected. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each TALEN component. A PCR product corresponding to the desired segmental deletion product is detected only in embryos injected with both TALENs. Estimated deletion rates are indicated below the positive lanes. (C) Junction sequences for the segmental deletion of this element are shown below, where there are variable length deletions between the distal TALEN recognition sites. (D) Schematic diagram of the birc6 locus demarcating the positions of the two TALEN binding sites flanking the linc-birc6 (megamind) lincRNA gene and the distance separating them. (E) Segmental deletion of the 3.2-kb element is readily detected in normal-appearing TALEN-treated embryos when two TALENs are coinjected. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each TALEN component. A PCR product corresponding to the desired segmental deletion product is detected only in embryos injected with both TALENs. The estimated deletion rate is indicated below the positive lane. (F) Junction sequences for the 3.2-kb segmental deletions of the linc-birc6 (megamind). The TALEN-3 and TALEN-6 recognition sequences are indicated above the junction sequences, where the TALEN half-sites are differentially colored. In the recovered junction sequences, there are variable length deletions.
We examined the feasibility of deleting an entire lincRNA by creating TALENs flanking the linc-birc6 (megamind) locus (Ulitsky et al. 2011), which spans 3.2 kb on chromosome 17 within an intron of birc6 (Fig. 5D). As with the globin LCR TALENs, the activity of TALENs targeting this locus was more variable; nonetheless, we identified two nucleases flanking the locus that displayed moderate activity (Supplemental Fig. 11). Coinjection of these nucleases produced deletions at a moderate frequency (1.2%) within zebrafish embryos (Fig. 5E). The sequences of the PCR products that span this deletion indicate excision of the genomic segment between the TALEN target sites (Fig. 5F). Thus, deletions can be readily created that excise noncoding sequence elements in their entirety using two TALENs, but the generation of active nucleases within noncoding sequence elements is more challenging.
Germline transmission rates for genomic deletions and inversions
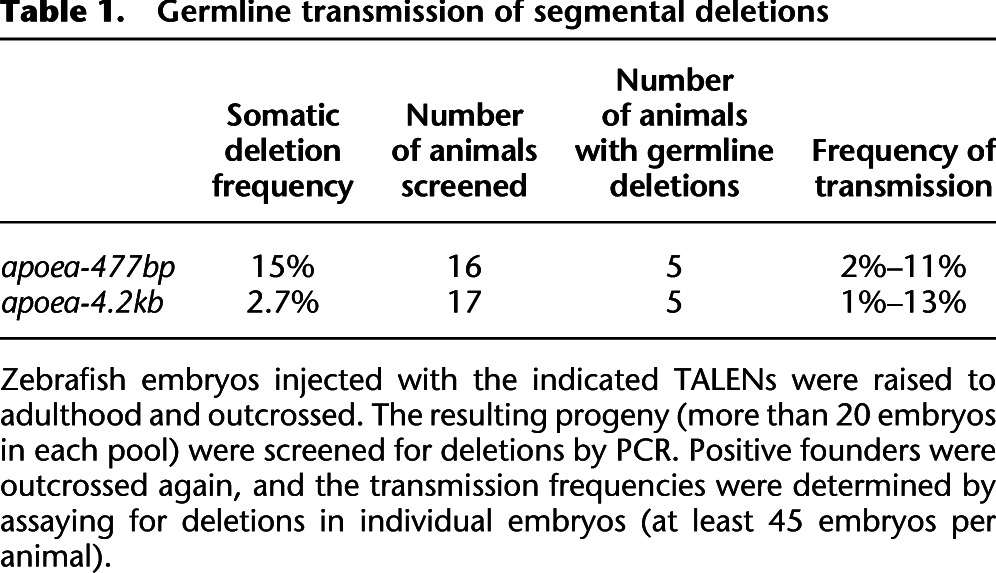
Our analysis of SSN-mediated segmental deletions has focused on assessments in somatic cells of injected embryos. While this provides a simple framework for defining the activity of these nucleases, the value of these segmental deletions will only be realized if they can be stably transmitted to offspring for subsequent phenotypic analysis. To our knowledge, there are currently no published data on the germline transmission rates of nuclease-generated segmental deletions in vertebrates. Consequently, we raised animals from our 500-bp and 4.2-kb apoea TALEN injections to adulthood and determined the rate of germline transmission of segmental deletions by treated adults. Consistent with our observed lesion rates in somatic cells, we were successful in achieving transmission of segmental deletions through the zebrafish germline. For animals treated with TALENs to induce a 500-bp deletion, five of 16 adults spawned progeny carrying deletions with transmission rates varying from 2% to 11% (Table 1; Supplemental Fig. 12). Similar rates of transmission were observed for the adults treated with nucleases to generate 4.2-kb deletions: Five of 17 adults spawned progeny harboring deletions with transmission rates varying from 1% to 13% (Table 1; Supplemental Fig. 13). The transmitted alleles displayed variation at the junctions of the deletion, where three founders transmitted two different deletion alleles (Supplemental Figs. 12, 13). One 500-bp inversion was also transmitted by a single founder among the 33 screened animals between the 500-bp and 4.2-kb TALEN-treated sets (Supplemental Figs. 12, 13). We found no evidence of transmission of duplications within this population. Thus, moderately sized TALEN-generated segmental deletions are readily transmitted through the zebrafish genome with efficiencies that allow the rapid generation of families of heterozygous carriers.
Table 1.
Germline transmission of segmental deletions
Discussion
Recent reports from the ENCODE Consortium suggest that upward of 20% of the genome may contain regulatory function (The ENCODE Project Consortium 2012). In addition, analysis of transcripts of protein-coding genes across multiple cell lines indicates that there are on average about four alternately spliced isoforms per locus that result in the production of different protein products (The ENCODE Project Consortium 2012). Thus, there is a wealth of genomic features and coding region complexity to deconvolute in vertebrate model systems, which will be facilitated by the ability to create heritable segmental deletions.
Herein we provide the first detailed analysis of the generation and transmission rates of nuclease-induced segmental deletions in a vertebrate. We find that the generation and transmission of deletions for nucleases targeting protein-coding regions is quite facile with transmission rates sufficient to allow the generation of carriers (>25% of the animals are founders with transmission rates between 1% and 13%) (Table 1). Segmental deletions could be generated with either ZFNs or TALENs, but the incredible targeting capacity and efficiency of the TALEN platform facilitate the creation of nucleases with the high efficiencies that are necessary to generate these deletions (Christian et al. 2010; Huang et al. 2011; Miller et al. 2011; Sander et al. 2011; Bedell et al. 2012; Dahlem et al. 2012; Reyon et al. 2012b).
Efficiently generating deletions at noncoding sequence elements proved slightly more challenging, since only five of 10 TALENs generated local lesions at a modest frequency (>5%) (Supplemental Table 1). The fundamental composition of the noncoding TALEN target sites is not significantly different from TALENs targeting protein-coding sequence (Supplemental Table 2). However, the presence of similar sequences at other sites in the genome was more prevalent for the TALENs targeting noncoding sequence elements, which could act as a sink reducing their on-target activity. Consequently, end-users should evaluate the presence of repeat elements within their target regions when designing constructs for noncoding sequence elements using existing online tools (Sander et al. 2011; Doyle et al. 2012; Sanjana et al. 2012).
The ability to generate inversions or duplications within zebrafish somatic cells was also readily apparent, although our efforts to recover these aberrations from injected animals yielded only one animal transmitting a small inversion. The difficulty in transmitting inversions or duplications through the germline, when compared with the highly successful transmission of segmental deletions, implies that the rate of inversions is lower than the segmental deletions, at least at the loci that we evaluated (Supplemental Table 1). This result is consistent with studies in transformed mammalian cells in which the rate of inversions or duplications in some cases was >10-fold lower than the rate of segmental deletions (Lee et al. 2012). The difficulty in recovering inversions or duplications generated via unequal exchange of sister/homologous chromosomes implies that SSN-mediated segmental deletions are primarily occurring at a single chromosome and that the loss of a genomic fragment is more favorable than the swapping of broken ends be they intra- or interchromosomal. Consequently, the generation of translocations via this approach may be inefficient.
In conjunction with methods to introduce small DNA segments in a targeted manner in zebrafish using single-stranded oligonucleotides (ssODNs) (Bedell et al. 2012), this methodology potentially provides a complementary approach for the functional analysis of coding and noncoding elements in zebrafish. Analysis of the transmitted alleles for animals treated with two TALENs indicates that both of these target sites can be independently cleaved and imprecisely repaired within the same allele without segmental deletion (Supplemental Fig. 14). Thus, it should be possible to capture two different ssODNs simultaneously at different positions on the same chromosome (e.g., for the introduction of two loxP sites flanking a set of exons) depending on the efficiency of this process. Furthermore, combination of the segmental deletions with the application of ssODNs (Chen et al. 2011; Bedell et al. 2012) or the introduction of nuclease-stimulated ligation of an exogenous donor DNA (Cristea et al. 2013; Maresca et al. 2013) could facilitate the targeted replacement of a desired genomic segment with a specific sequence variant for a more complete deconvolution of function.
Methods
Zebrafish lines
The use of zebrafish was in accordance with established protocols (Westerfield 1993) and in conformity with Institutional Animal Care and Use Committee guidelines of the University of Massachusetts Medical School.
slc24a5 locus zinc finger nuclease (ZFN) construction
Zinc finger arrays (ZFAs) targeting the slc24a5 locus (target site: cGTCATCGCCATCatctcaGATAACAGAGTTt) were constructed using a combination of modular assembly and selection (Supplemental Fig. 15). The 5′ ZFA was assembled from our archive of single finger modules (Zhu et al. 2011), whereas the 3′ ZFA was created using a combination of single finger modules (Zhu et al. 2011) and selection of a two-finger module for the AACAGA subsite (Gupta et al. 2012). These ZFAs were coupled to the DD/RR versions of FokI endonuclease (Miller et al. 2007; Szczepek et al. 2007) in a pCS2 backbone (Meng et al. 2008).
TALEN construction
TALE module arrays were constructed using the Voytas Golden Gate System (Cermak et al. 2011), where the constructed monomers contained between 16.5 and 19.5 modules with a 14- to 17-bp gap separating the TALEN target sites (Supplemental Fig. 16; Supplemental Tables 2, 3; Reyon et al. 2012b). All TALEN recognition sites incorporate a 5′ T, and standard modules were used (repeat-variable di-residues [RVDs]: Ade = NI; Cyt = HD; Gua = NN; Thy = NG) for base recognition in the remaining sequence (Boch et al. 2009; Moscou and Bogdanove 2009; Cermak et al. 2011). We changed the overall architecture of the final backbone of the Golden Gate System, since the original TALEN constructs contained longer N- and C-terminal elements than those used in zebrafish (Huang et al. 2011; Sander et al. 2011). Since pJDS vectors (Sander et al. 2011) from the Joung Laboratory REAL Assembly TALEN Kit (Addgene) (Reyon et al. 2012a) were highly active in zebrafish (Sander et al. 2011), we used the architecture from pJDS vectors for the incorporation of the Golden Gate assembled TALE arrays. To make pJDS vectors compatible with the Voytas laboratory Golden Gate Kit, the overhangs generated by Esp3I cleavage sites in the pJDS vectors were adjusted to GGGA/CTAT to ensure proper complementarity between intermediate vectors and the final vector. Moreover, since pJDS vectors already have the last half-repeat incorporated into the backbone (there are four different vectors, one for each base), we modified each pJDS vector to match the reading frame of the Golden Gate–assembled intermediate arrays. These vectors use the wild-type FokI nuclease domain.
ZFN and TALEN mRNA injection into zebrafish embryos
Each pJDS-TALEN clone was linearized with PmeI, and mRNA was transcribed and poly(A)-tailed using the mMessage mMachine T7 Ultra Kit (Ambion). Each pCS2-ZFN construct was linearized with NotI, and mRNA was transcribed using the mMessage mMachine SP6 Kit (Ambion). Injections of TALEN or ZFN mRNAs into the blastomere of one-cell-stage zebrafish embryos were carried out as previously described (Meng et al. 2008; Gupta et al. 2011). For analysis of lesion rates at individual target sites, or segmental deletions or inversions, injected embryos with normal appearance (20–30 embryos) and uninjected embryos were collected at 24 hpf, and incubated in 50 mM NaOH (8 μL/embryo) for 30 min at 95°C to isolate genomic DNA. Lysis reactions were subsequently neutralized with 0.5 M Tris-HCl (pH 7.5) (2 μL/embryo) and centrifuged at 13,000 rpm for 1 min, after which the supernatant containing genomic DNA was used in PCRs for analysis. For single embryo analysis, each embryo was lysed in 40 μL of 50 mM NaOH and neutralized with 10 μL of 0.5 M Tris-HCl (pH 7.5).
ZFN or TALEN activity analysis at individual target sites
PCR primers were designed to amplify an ∼200-bp region bordering the ZFN or TALEN target site (Supplemental Table 4). PCR was run with 1 μL of the extracted genomic zebrafish DNA in a total reaction volume of 20 μL using the Phire Hot Start DNA polymerase (Finnzymes). Lesion rates were determined using T7 Endonuclease I (New England Biolabs) (Kim et al. 2009; Reyon et al. 2012b). In this analysis, 10 μL of PCR product was submitted to the following protocol on a thermocycler: 5 min at 95°C; 95°C–85°C at −2°C/sec; 85°C–25°C at −0.1°C/sec; hold at 4°C. Reannealed PCR products from this step were incubated with 10 units of T7 Endonuclease I in a 15-μL reaction for 45 min at 37°C in 1× NEB Buffer 2. The digestion products were run on a 3.5% 0.5× TBE UltraPure Agarose (Invitrogen) gel at 200 V for 15–20 min. Band intensities for the cleaved PCR product relative to the entire PCR product were used to estimate the lesion frequency at the ZFN and TALEN target sites using ImageJ (Schneider et al. 2012). Lesion rate calculations were performed as described (Guschin et al. 2010).
Determining segmental deletion and inversion rates
PCR primers spanning the anticipated junctions for segmental deletions or inversions (Supplemental Tables 4, 5) were used to amplify the desired regions. The intensity of the deletion, inversion, or duplication band was compared either with the full-length genomic amplicon (for detecting deletions at the apoea-500bp, apoea-750bp, and LCR loci) or with an unmodified genomic standard sequence of comparable length using a third primer in the PCR assay. Where possible, a PCR amplification standard curve was established as a benchmark for this analysis using genomic DNA from an individual embryo (obtained from founder outcross) that was heterozygous for the lesion of interest diluted in various quantities of wild-type genomic DNA. The intensity of bands for the lesion of interest and the control region (full-length amplicon or unmodified genomic DNA) was quantified by ImageJ analysis (Schneider et al. 2012) from an ethidium bromide–stained agarose gel.
Germline transmission analysis
TALENs and ZFNs were injected at optimal doses in wild-type zebrafish embryos. Injected embryos were grown to maturity and crossed with wild-type zebrafish to identify founders. Initially, pools of embryos (at least 20 embryos) from each nuclease-treated animal were analyzed by PCR (described above) to identify potential founders. Subsequently, individual embryos from each putative founder were characterized to assess the transmission rate of deletions or inversions. Deletion products from individual embryos were directly sequenced to identify the junction sequences that were transmitted by each founder animal. PCR products for inversions were shotgun-cloned and sequenced to identify the junction sequences that were transmitted by the founder animal.
Data access
Sequences of the primers are listed in Supplemental Tables 4 and 5. The target sites of the TALENs are listed in Supplemental Table 1, and information on the ZFNs is listed in Supplemental Figure 15. The TALEN scaffold sequence and RVD sequences are provided in Supplemental Figure 16 and Supplemental Table 3, respectively.
Acknowledgments
We gratefully acknowledge the other members of the Lawson and Wolfe laboratories for insightful comments and discussions. This work was supported by the U.S. National Institutes of Health (NIH) grants HL093766 (to N.D.L. and S.A.W.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.154070.112.
References
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG II, Tan W, Penheiter SG, Ma AC, Leung AY, et al. 2012. In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D 2011. Genome engineering with zinc-finger nucleases. Genetics 188: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8: 753–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, White S, Searle SM, Stemple DL 2012. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res 22: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD, Gregory PD, Cost GJ 2013. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng 110: 871–880 [DOI] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8: e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ 2012. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, Jia Z, Sabo PJ, Weaver M, Sandstrom R, et al. 2012. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol 366: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA 2011. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res 39: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA 2012. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods 9: 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ 2010. A rapid and general assay for monitoring endogenous gene modification. In Engineered zinc finger proteins (ed. Mackay JP, Segal DJ), pp. 247–256. Humana Press, Totowa, NJ [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B 2011. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29: 699–700 [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD 2013. TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS 2012. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22: 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS 2012. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res 22: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zhang S, Wang F, Liu Y, Liu Y, Xu H, Liu C, Lin Y, Zhao P, Xia Q 2012. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS ONE 7: e45035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Lin VG, Guo N, Yang Y 2013. Obligate Ligation-Gated Recombination (ObLiGaRe): Custom designed nucleases mediated targeted integration through non-homologous end joining. Genome Res 23: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA 2008. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menoret S, Iscache AL, Tesson L, Remy S, Usal C, Osborn MJ, Cost GJ, Bruggemann M, Buelow R, Anegon I 2010. Characterization of immunoglobulin heavy chain knockout rats. Eur J Immunol 40: 2932–2941 [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Reyon D, Khayter C, Regan MR, Joung JK, Sander JD 2012a. Engineering designer transcription activator-like effector nucleases (TALENs) by REAL or REAL-Fast assembly. Curr Protoc Mol Biol 100: 12.15.1–12.15.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK 2012b. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR 2011. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29: 697–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F 2012. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7: 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. 2011. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T 2007. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786–793 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 1993. The zebrafish book. University of Oregon Press, Eugene, OR. [Google Scholar]
- Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. 2011. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development 138: 4555–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]