Abstract
Histone modifications are now well-established mediators of transcriptional programs that distinguish cell states. However, the kinetics of histone modification and their role in mediating rapid, signal-responsive gene expression changes has been little studied on a genome-wide scale. Vascular endothelial growth factor A (VEGFA), a major regulator of angiogenesis, triggers changes in transcriptional activity of human umbilical vein endothelial cells (HUVECs). Here, we used chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to measure genome-wide changes in histone H3 acetylation at lysine 27 (H3K27ac), a marker of active enhancers, in unstimulated HUVECs and HUVECs stimulated with VEGFA for 1, 4, and 12 h. We show that sites with the greatest H3K27ac change upon stimulation were associated tightly with EP300, a histone acetyltransferase. Using the variation of H3K27ac as a novel epigenetic signature, we identified transcriptional regulatory elements that are functionally linked to angiogenesis, participate in rapid VEGFA-stimulated changes in chromatin conformation, and mediate VEGFA-induced transcriptional responses. Dynamic H3K27ac deposition and associated changes in chromatin conformation required EP300 activity instead of altered nucleosome occupancy or changes in DNase I hypersensitivity. EP300 activity was also required for a subset of dynamic H3K27ac sites to loop into proximity of promoters. Our study identified thousands of endothelial, VEGFA-responsive enhancers, demonstrating that an epigenetic signature based on the variation of a chromatin feature is a productive approach to define signal-responsive genomic elements. Further, our study implicates global epigenetic modifications in rapid, signal-responsive transcriptional regulation.
Genome-wide profiling of chromatin components between different cell types has demonstrated that transcriptional regulatory elements are decorated by characteristic patterns of post-translational histone modifications and other chromatin features and that these features contribute to cell type-specific gene regulation (Heintzman et al. 2007; Ernst et al. 2011; Kharchenko et al. 2011; Bonn et al. 2012). Such epigenetic signatures have been used to functionally annotate transcriptional regulatory elements that distinguish different cell types. For example, active enhancers, genomic elements that stimulate gene transcription, are marked by acetylation of histone H3 at lysine 27 (H3K27ac) (Creyghton et al. 2010; Kharchenko et al. 2011; Rada-Iglesias et al. 2011; Zentner et al. 2011), the presence of the chromatin regulator EP300 (Visel et al. 2009; Creyghton et al. 2010), hypersensitivity to nuclease digestion (Boyle et al. 2008), and expression of RNA transcripts known as eRNAs (Kim et al. 2010; Wang et al. 2011). However, much less is known about how chromatin signatures change during rapid cellular responses to extracellular cues and the effectiveness of epigenetic profiles in identifying transcriptional elements that mediate signal-responsive changes in gene expression.
We studied rapid, signal-responsive changes in chromatin features using vascular endothelial growth factor A (VEGFA)-stimulated endothelial cells as a model system. Blood vessels nourish nearly every organ. Their growth is tightly regulated, and inadequate, excessive, or abnormal blood vessel growth is linked to a panoply of diseases, including ischemic heart disease, blinding eye diseases, and cancer (Carmeliet and Jain 2011). A central regulator of blood vessel growth is vascular endothelial growth factor A. In response to VEGFA signaling, endothelial cells dramatically change their phenotype and gene expression profile (Schweighofer et al. 2009). Intracellular signaling downstream from VEGFA has been studied in depth, but relatively less is known about the transcriptional regulatory elements that respond to VEGFA signaling.
We profiled activating chromatin epigenetic marks in a 12-h time course of endothelial cell stimulation by VEGFA. We show that temporal variation of H3K27ac is a novel epigenetic signature that identifies VEGFA-regulated enhancers and predicts VEGFA-responsive gene expression. Our work further shows that the catalytic activity of EP300 is required for dynamic changes in H3K27ac occupancy, altered chromatin architecture, and regulation of gene expression by VEGFA.
Results
VEGFA-induced changes in H3K27ac
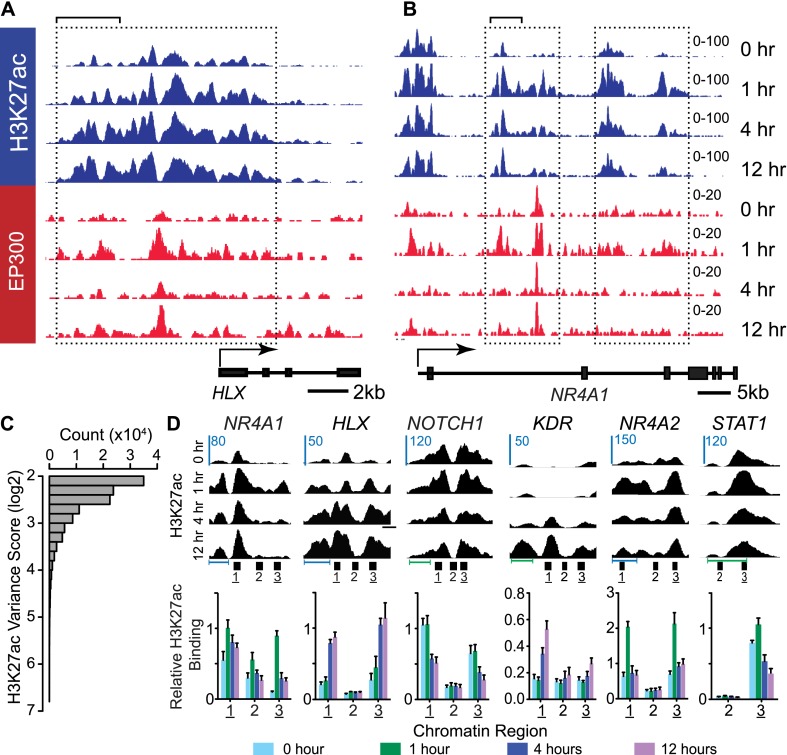
To study transcriptional and epigenetic regulation during angiogenesis, we measured H3K27ac chromatin occupancy genome-wide in human umbilical vein endothelial cells (HUVECs) treated for 0 (unstimulated), 1, 4, and 12 h with VEGFA (Supplemental Table 1). Overall, our ChIP-seq data correlated with H3K27ac in ENCODE data sets, with some differences likely attributable to differences in growth conditions (Supplemental Fig. 1). Inspection of the H3K27ac ChIP-seq data showed regions with substantial changes in H3K27ac as a result of VEGFA stimulation (boxed regions, Fig. 1A,B). To identify regions with VEGFA-induced variation in H3K27ac, we calculated the H3K27ac signal-normalized variance (variance score) for 200-bp sliding windows across the 12-h time course. This approach captured thousands of regions with substantial VEGFA-induced variation (Fig. 1C). Out of sites with a log2 variance score greater than 3, we selected those near eight genes implicated in angiogenesis for validation by ChIP-qPCR. Sites near six genes were successfully assayed, and, in all six cases, the ChIP-qPCR results were consistent with the ChIP-seq data (Fig. 1D).
Figure 1.
VEGFA-stimulated dynamic changes in H3K27ac occupancy. (A,B) Browser views of H3K27ac and EP300 ChIP-seq enrichment at HLX and NR4A1 at four time points following VEGFA stimulation. Note areas with varying H3K27ac occupancy in response to VEGFA (boxed regions). Bracketed regions are enlarged in portions of panel C. (C) H3K27ac variance score histogram. VEGFA-stimulated significant changes in H3K27ac regions during the 12-h time course. The plot only includes regions with a log2 variance score greater than or equal to 2. (D) Chip-qPCR validation of VEGFA-induced changes in H3K27ac occupancy. Browser views show ChIP-seq enrichment, and lower plots indicate H3K27ac ChIP-qPCR using numbered amplicons indicated in the browser views. Underlined numbers indicate regions where ChIP-seq predicted VEGFA-stimulated H3K27ac occupancy change, while regions without underline are adjacent controls. Horizontal scale bars: (blue) 1 kb, (green) 0.5 kb.
A dynamic VEGFA-regulated H3K27ac signature is tightly linked to EP300 chromatin occupancy
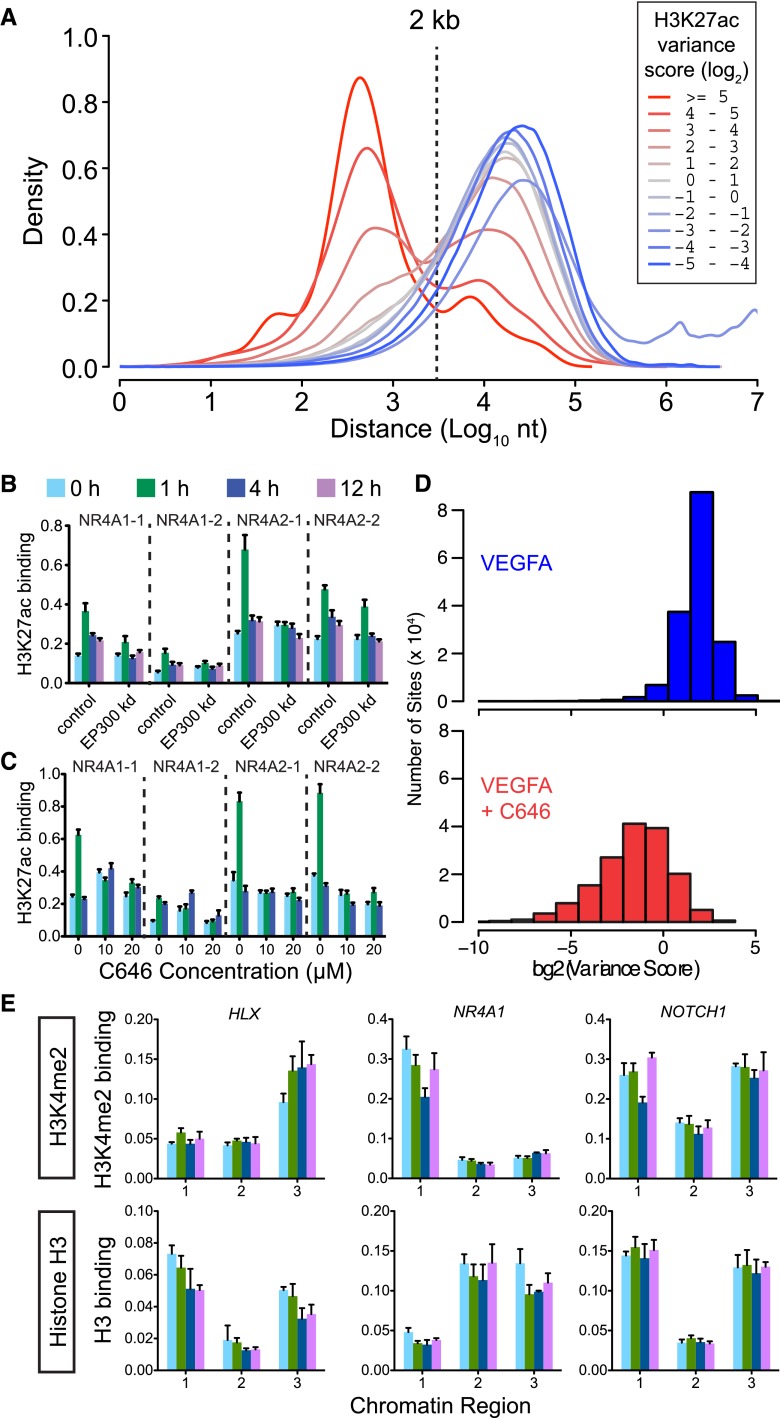
The transcriptional coactivator EP300 acetylates histones (Ogryzko et al. 1996) and occupies tissue-specific enhancers (Visel et al. 2009). To define the relationship of EP300 chromatin occupancy to dynamic H3K27ac sites, we measured EP300 chromatin occupancy during the VEGFA stimulation time course by ChIP-seq. The EP300 ChIP-seq data were reproducible between independent biological duplicates (Pearson r > 0.92) (Supplemental Fig. 2A). Comparison to publicly available ENCODE EP300 occupancy data for the immortalized B-cell cell line GM12878 (Reddy et al. 2012) indicated that EP300 binding was largely cell type-specific (Supplemental Fig. 2B). Next, from the VEGFA time course data, we calculated the distance from EP300 sites to neighboring H3K27ac sites, stratified by their H3K27ac variance score. Interestingly, most of the regions with the greatest H3K27ac variance scores occurred within 2 kb of EP300 sites, while less variant H3K27ac sites tended to be further from EP300 (Fig. 2A; Supplemental Fig. 3). This result supported a tight relationship between EP300 and dynamic but not static H3K27ac marks.
Figure 2.
Dynamic H3K27ac regions were associated with EP300. (A) Distance relationship of H3K27ac sites to nearest EP300-occupied region as a function of H3K27ac variance score. The H3K27ac regions with the highest variance score were predominantly located within 2 kb of EP300. (B,C) EP300 is required for dynamic H3K27 acetylation. ChIP-qPCR of H3K27ac in HUVEC cells treated with VEGFA and EP300 siRNA (B) or small molecule EP300 acetyltransferase inhibitor C646 (C). Two dynamic H3K27ac regions near each of the indicated four genes were tested (see also Supplemental Fig. 5A,B). (D) C646 pretreatment suppresses H3K27ac change at highly variant sites. We considered genomic windows with a log2 variance score of at least 2 and at least the sum of 10 reads across all four time points. We recalculated the variance score for each site under VEGFA stimulation in the absence or presence of C646 for hours 0, 1, and 4. The histogram shows that C646 reduced the variance score at the vast majority of sites. (E) Chip-qPCR of H3K4me2 and total histone H3 binding at three dynamic H3K27ac sites showed that altered nucleosome positioning is unlikely to account for VEGFA-stimulated changes in H3K27ac. Numbered chromatin regions are indicated in Fig. 1D.
The enrichment of EP300 near dynamic H3K27ac sites suggested that EP300 is functionally involved in deposition of H3K27ac in response to VEGFA stimulation. To test the functional requirement for EP300 in VEGFA-stimulated deposition of H3K27ac, we measured the effect of EP300 knockdown on H3K27ac chromatin occupancy. siRNA EP300 knockdown (Supplemental Fig. 4) blocked VEGFA-stimulated deposition of H3K27ac at NR4A1, HLX, and KDR (Fig. 2B; Supplemental Fig. 5A). To determine if EP300 histone acetyltransferase catalytic activity, as opposed to other functions mediated by EP300 (e.g., coactivator complex formation through protein-protein interactions), is required for VEGFA-stimulated H3K27ac, we inhibited EP300 enzymatic activity using the small molecule C646 (Bowers et al. 2010). Pretreatment of HUVEC cells for 30 min with C646 blocked VEGFA-stimulated deposition of H3K27ac (Fig. 2C; Supplemental Fig. 5B).
We next interrogated the extent to which EP300 activity is required for dynamic H3K27ac deposition genome-wide by performing H3K27ac ChIP-seq on cells pretreated with C646 and then stimulated with VEGFA for 0, 1, and 4 h. The cells became unhealthy by 12 h, precluding analysis at this time point. This experiment demonstrated that EP300 inhibition caused widespread reduction in H3K27ac variation induced by VEGFA (Fig. 2D; Supplemental Fig. 5C,D). However, some VEGFA-stimulated changes in H3K27ac persisted, indicating that additional mechanisms also contribute to H3K27ac changes induced by VEGFA. Overall, these data indicate a key role of EP300 in contributing to H3K27 acetylation induced by VEGFA.
Changes in nucleosome positioning were previously reported to underlie rapid changes in the occupancy profile of histone H3 dimethylated at lysine 4 (H3K4me2) (He et al. 2010). We tested the hypothesis that changes in nucleosome occupancy contribute to the observed dynamic changes in H3K27ac by measuring total histone H3 and H3K4me2 occupancy at six dynamic H3K27ac sites (Fig. 2E; Supplemental Fig. 6). We did not observe significant changes in histone H3 or H3K4me2 occupancy at any of these sites, indicating that acetylation of histone H3 rather than shifts in its position cause altered H3K27ac occupancy.
Temporal clustering of H3K27ac variation defined groups of chromatin regions with distinct function annotations and enriched transcription factor motifs
To investigate the significance of EP300-associated variation in H3K27ac, we focused on the subset of H3K27ac sites within 2 kb of EP300 with the highest variance scores (upper 20th percentile) (see Methods; Supplemental Table 2; Supplemental Fig. 3). Hierarchical clustering showed that H3K27ac enrichment at these sites followed three predominant temporal patterns (Fig. 3A). We labeled these clusters as H1 (peak H3K27ac signal at 1 h; 4689 regions), H4-12 (peak H3K27ac signal at 4–12 h; 3947 regions), and H0 (decreased H3K27ac signal at 4–12 h; 3601 regions). Plotting H3K27ac signal intensity for each region illustrated the significant dynamic changes of H3K27ac binding in each temporal cluster (Fig. 3B; Supplemental Fig. 7A). Cluster H4-12 was particularly interesting, because it showed initial depletion of H3K27ac signal at the peak center at 0 and 1 h and subsequent “filling-in” of the depleted region at 4 and 12 h (Fig. 3B; Supplemental Fig. 7A).
Figure 3.
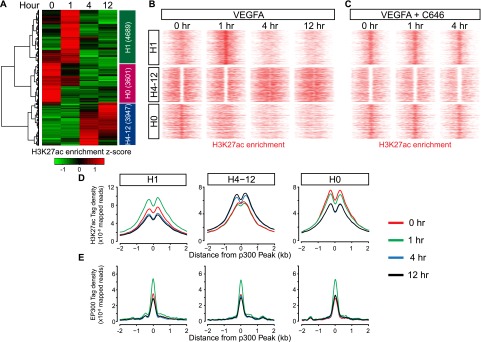
Dynamic, EP300-associated H3K27ac regions grouped into three temporal clusters. (A) Hierarchical clustering heat map of dynamic EP300-associated H3K27ac variants. Cluster names and number of regions per cluster are indicated in the adjacent colored bar. (B,C) Tag density map of dynamic H3K27ac clusters during VEGFA stimulation time course, without (B) or with (C) the EP300 inhibitor C646. Each row represents a 4-kb region centered on a dynamic H3K27ac site. The same color scale was applied within each cluster. Note the loss of VEGFA-induced changes of H3K27 acetylation. (D) H3K27ac aggregation plots, centered on nearby EP300 peaks. Maximal H3K27ac density occurred adjacent to EP300 peak centers. (E) EP300 aggregation plots, centered on EP300 peaks within 2 kb of dynamic H3K27ac regions.
Our genome-wide analysis of the effect of the EP300 inhibitor C646 on VEGFA-stimulated H3K27 acetylation showed a mechanistic requirement for EP300 at the majority of sites. We, therefore, examined the C646 effect on the H1, H4-12, and H0 clusters in detail. Consistent with its essential role in VEGFA-stimulated deposition of H3K27ac, C646 strongly blunted H3K27ac accumulation in the H1 and H4-12 clusters (Fig. 3C; Supplemental Fig. 7B). Interestingly, the down-regulation of H3K27ac seen in the H0 cluster was also blunted by C646, suggesting secondary effects on counter-regulatory mechanisms that remove H3K27ac marks.
When the regions were centered on EP300 enrichment, aggregation plots of H3K27ac signal showed that maximal H3K27ac signal variation occurred adjacent to, rather than overlapping, EP300 (Fig. 3D). Prior work showed that the chromatin landscape at most transcription factor binding sites is asymmetric. When we applied an algorithm for function strand segregation (Kundaje et al. 2012), we found that H3K27ac and EP300 occupancy were both asymmetric in the H1, H4-12, and H0 clusters (Supplemental Fig. 7C). Interestingly, the distribution of H3K27ac and EP300 with respect to the peak center was largely concordant, consistent with a mechanistic role of EP300 in establishing the H3K27ac marks.
EP300 aggregation plots showed that EP300 binding also changed during the VEGFA-stimulation time course (Fig. 3E). We confirmed VEGFA-stimulated enrichment of EP300 by ChIP-qPCR (Supplemental Fig. 7D). For cluster H4-12, on average, EP300 binding increased before H3K27ac occupancy. At cluster H1, these events appear to occur concurrently, suggesting that either the sequence of events differs at 1 h or that the data did not contain sufficient temporal resolution to order events peaking at this early time point. For cluster H0 with decreasing H3K27ac signal, EP300 signal increased at 1 h, but H3K27ac signal did not (Fig. 3B–E; Supplemental Fig. 7D), suggesting that other factors, such as increased HDAC activity, impeded H3K27ac deposition at these regions.
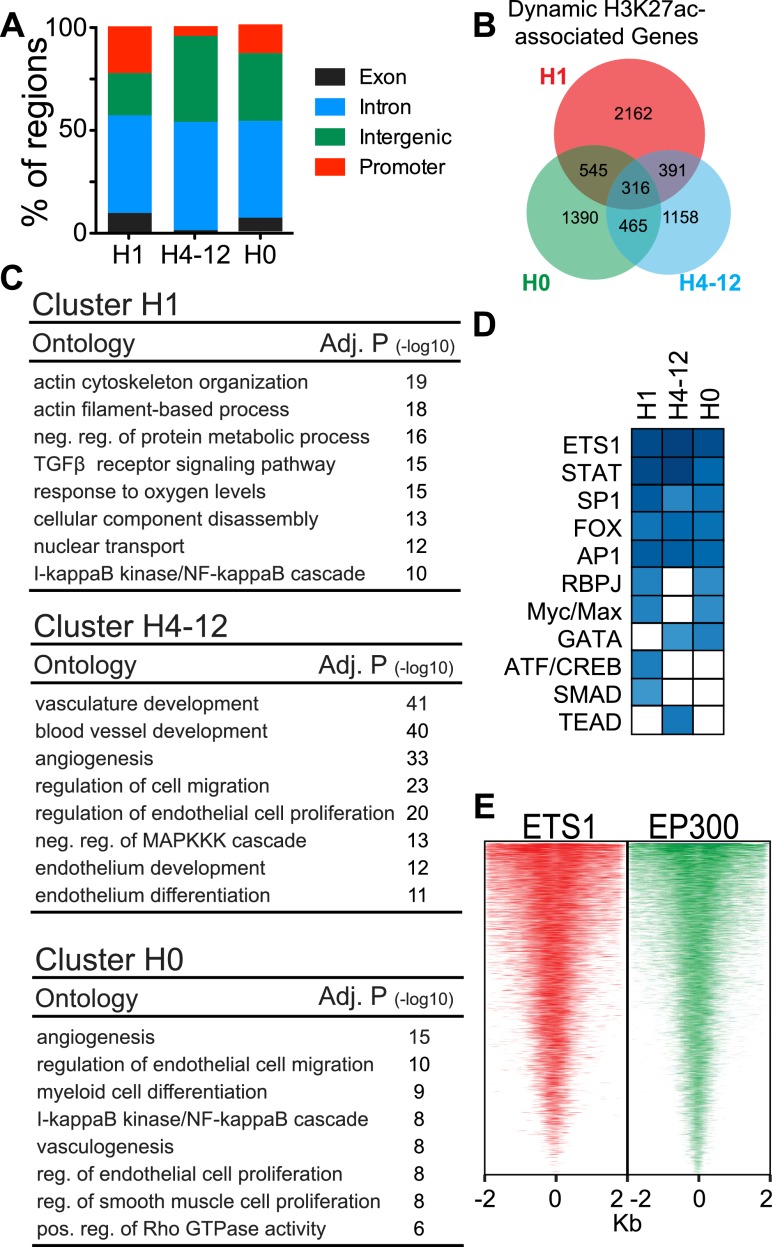
We further characterized the location and function of the dynamic, EP300-associated H3K27ac sites. Most of these sites were located distal to transcriptional start sites (TSSs) of genes, consistent with the reported predominant location of EP300 (Visel et al. 2009; Creyghton et al. 2010). However, a significantly greater proportion of sites in cluster H1 were located in promoters, near gene TSSs (P-value < 10−46), while a significantly greater proportion of sites in cluster H4-12 were located in intergenic regions (P-value < 10−10) (Fig. 4A). Dynamic, EP300-associated H3K27ac sites were associated with 8454 adjacent genes. The majority of these genes did not overlap between temporal H3K27ac clusters (Fig. 4B). Gene Ontology (GO) analysis showed that these different gene sets have distinct functional properties (Fig. 4C). Both of the late-responding clusters, H4-12 and H0, were strongly enriched for terms related to vascular development, endothelial differentiation, and angiogenesis. In contrast, the early-responding H1 cluster was enriched for terms related to cell morphology, protein metabolism, response to oxygen levels, and TGFbeta receptor signaling, which are relevant to cellular stress responses in many cells types, including endothelial cells. Collectively, our data indicate that each temporal cluster of dynamic EP300-associated H3K27ac sites was linked to regulation of varying aspects of cellular function.
Figure 4.
Characterization of dynamic, EP300-associated H3K27ac regions. (A) Genomic distribution of dynamic H3K27ac regions. (B) Venn diagram of genes associated with the three clusters of dynamic H3K27ac regions. (C) Biological process terms were selected from the 20 most significantly enriched for dynamic H3K27ac regions. (D) Transcription factor families with enriched motifs found under EP300 peaks at dynamic H3K27ac regions. Color intensity indicates significance score (see Supplemental Table 3 for full table). (E) ETS1 and EP300 colocalized near dynamic H3K27ac regions. ETS1 and EP300 tag densities are plotted in EP300-bound regions, sorted by ETS1 tag enrichment.
To identify transcription factors that regulate EP300 recruitment and VEGFA-regulated H3K27 acetylation, we searched for overrepresented transcription factor binding motifs among sequences at EP300 peaks in each cluster. De novo motif discovery revealed highly significant enrichment of ETS, FOX, AP1, STAT, and SP1 families in all three clusters. To further validate the motif discovery results, we performed ChIP-seq for ETS1 and found that 51% of EP300-bound regions were co-occupied by ETS1 (Fig. 4E).
Our analysis also identified transcription factor motifs that occurred in one or two of the H3K27ac clusters. The motif of ATF and CREB1, mediators of immediate early responses in multiple cell types (Altarejos and Montminy 2011), was significantly enriched in the H1 cluster. Also notable was overrepresentation of the SMAD binding motif in the H1 cluster, where we observed enrichment for the TGFbeta receptor signaling pathway (Fig. 4D). We found overrepresentation of GATA and TEAD binding motifs (Fig. 4D; Supplemental Table 3) in the H0 and H4-12 clusters, while the binding motif of RBP/J, the nuclear target of Notch signaling, was enriched in the H0 and H1 clusters (Fig. 4D; Supplemental Table 3). These results suggest that members of these transcription factor families are important in orchestrating EP300 recruitment and H3K27ac deposition in response to VEGFA stimulation.
Enrichment of TF motifs at dynamic H3K27ac sites suggested that these TFs recruit EP300 and thereby contribute to changes in H3K27ac. To test this hypothesis, we knocked down ETS1 or C-JUN (a component of the AP1 heterodimer) and measured the effect on dynamic H3K27ac sites directly bound by these factors. Validation experiments demonstrated efficient ETS1 or C-JUN knockdown in HUVECs after siRNA transfection (Supplemental Fig. 8A,B,E,F) and corresponding reduction of ETS1 or JUN (also known as c-Jun) occupancy of tested dynamic H3K27ac sites (two sites tested per factor) (Supplemental Fig. 8C,G). This reduction of ETS1 or JUN binding attenuated H3K27ac changes at these sites in response to VEGFA (Supplemental Fig. 8D,H). These results suggest that TFs with enriched motifs are functionally important in mediating dynamic H3K27ac changes in response to VEGFA.
The dynamic H3K27ac signature defines VEGFA-responsive transcriptional regulatory elements
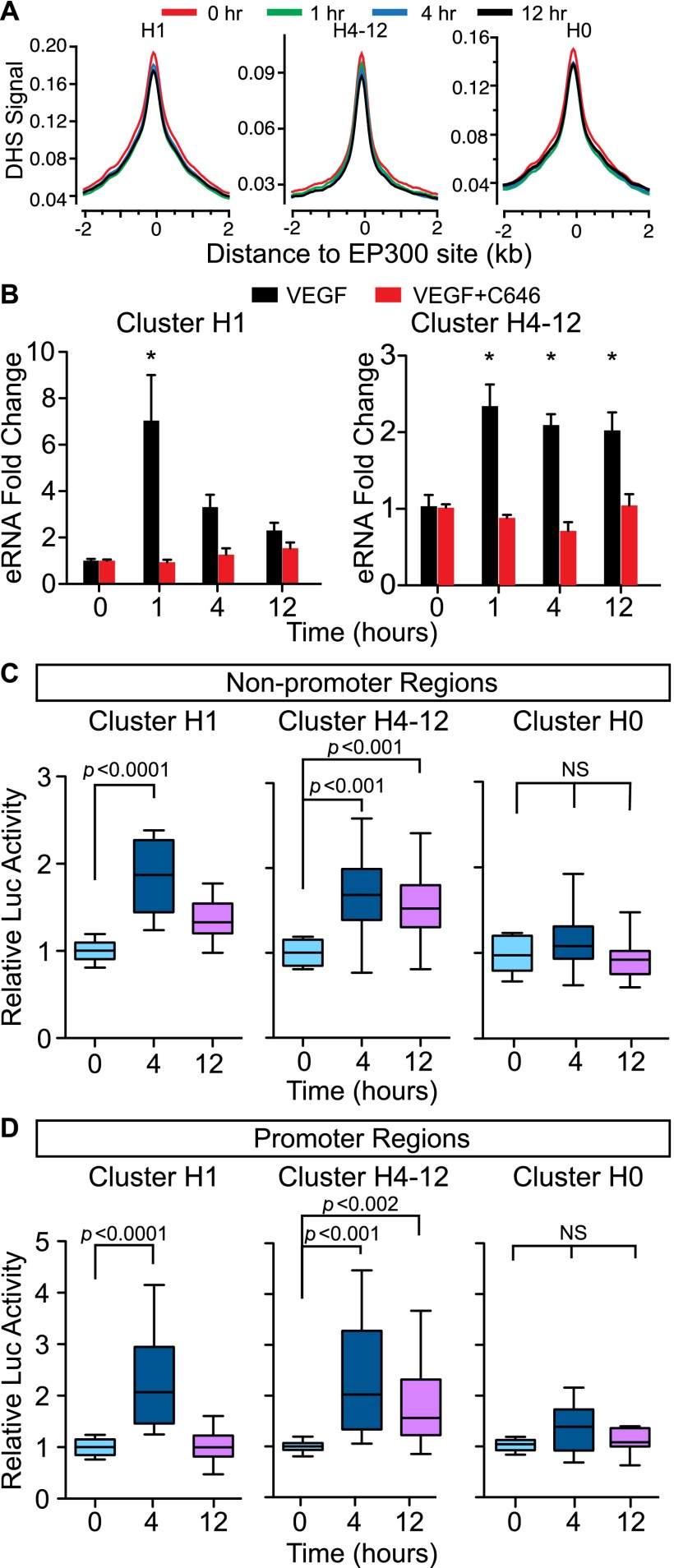
Active transcriptional regulatory elements are characterized by hypersensitivity to digestion by DNase I (Xi et al. 2007). To further investigate whether dynamic, EP300-associated H3K27ac sites are activating transcriptional regulatory elements, we performed genome-wide measurement of DNase I hypersensitivity during the VEGFA-stimulation time course using DNase-seq (Boyle et al. 2008). Biological duplicate samples showed that the technique is highly reproducible (Supplemental Fig. 9A). Dynamic, EP300-associated H3K27ac loci were DNase I hypersensitive (Fig. 5A), consistent with their function as active transcriptional regulatory elements. Interestingly, on average, these regions did not change significantly in their sensitivity to DNase I digestion during the VEGFA-stimulation time course (Fig. 5A; Supplemental Fig. 9B–D), suggesting that most of these sites are already “open” and poised to respond to VEGFA stimulation. These data are consistent with our observation that H3K27 acetylation was not associated with changes in nucleosome occupancy (Fig. 2D; Supplemental Fig. 6).
Figure 5.
Dynamic H3K27ac regions had functional properties of transcriptional regulatory regions. (A) DNase-seq showed that dynamic, EP300-associated sites are hypersensitive to DNase I digestion, but the sensitivity did not change substantially during the VEGFA stimulation time course. DNase signal is plotted as reads per 10−6 mapped reads. (B) eRNA expression from dynamic H3K27ac regions with or without EP300 inhibition by C646. Each bar represents the average of 5–6 different regions (individual data are shown in Supplemental Fig. 7). (*) P < 0.05. (C,D) Luciferase activity of dynamic H3K27ac regions at promoter and nonpromoter sites. Luciferase activity was expressed relative to the activity measured at time 0. n = 5–8 per group. Line, bar, and whiskers represent median, quartiles, and min-max values, respectively.
Active enhancers are also characterized by production of transcripts known as eRNAs (Kim et al. 2010). To further characterize the dynamic, EP300-associated H3K27ac loci and confirm their enhancer activity, we measured eRNA transcript levels from the activating clusters (H1 and H4-12) by qRT-PCR (Fig. 5B; Supplemental Fig. 10A), testing 11 regions distal to gene bodies. In regions belonging to cluster H1, eRNA was strongly up-regulated at 1 h and then decreased at 4 and 12 h. In regions belonging to cluster H4-12, eRNA was up-regulated to maximal levels by 1 h and was sustained through hours 4 and 12. As controls, we measured eRNA transcripts from nearby regions with H3K27ac enrichment that did not change during the VEGFA time course. Although some control regions also showed VEGFA-stimulated changes in transcript level, their number and overall fold increase were less than at H3K27ac dynamic regions (Supplemental Fig. 10A).
H4-12 cluster regions showed increased EP300 recruitment and eRNA activity at 1 h, whereas H3K27ac did not increase until 4–12 h. To determine if EP300 activity was required for VEGFA-stimulated increases in eRNA, we blocked EP300 activity with C646 and measured eRNA levels. We found that acute EP300 inhibition blocked VEGFA-stimulated up-regulation of eRNA (Fig. 5B; Supplemental Fig. 10B). In the H1 cluster, our experiments were unable to resolve temporal differences in EP300 recruitment, eRNA activity, and H3K27ac binding, which concurrently peaked at 1 h. Nevertheless, EP300 inhibition also blocked eRNA up-regulation at H1 regions, suggesting a similar role of EP300 in this cluster. Our results suggest that EP300 recruitment and acetylase activity are required for eRNA synthesis and precedes H3K27 acetylation.
Since the dynamic EP300-associated H3K27ac loci had chromatin features of transcriptional regulatory regions, we used luciferase reporter assays to measure the transcriptional activity of 1- to 2-kb regions centered on 38 dynamic EP300-associated H3K27ac loci. An equal number of loci were arbitrarily selected from each cluster, and tested regions were further subdivided into those located in promoter or nonpromoter regions. After reporter plasmid transfection, HUVECs were treated with VEGFA or vehicle. Luciferase activity measurements showed that dynamic, EP300-associated H3K27ac regions belonging to H1 and H4-12 clusters activated transcription in response to VEGFA, while regions from the H0 cluster did not (Fig. 5C,D). Regions belonging to the H1 cluster activated luciferase expression by 4 h, and expression then returned to baseline levels at 12 h. In contrast, regions from the H4-12 cluster increased luciferase activity at 4 h and maintained this through 12 h. Regions from promoter and nonpromoter regions behaved similarly (Fig. 5C,D). These data indicate that regions in H1 and H4-12 clusters function as VEGFA-responsive transcriptional enhancers, while those in the H0 cluster with decreasing H3K27ac signal did not. Together, the DNase hypersensitivity, eRNA, and luciferase assays support VEGFA-responsive transcriptional enhancer activity of dynamic, EP300-associated H3K27ac regions.
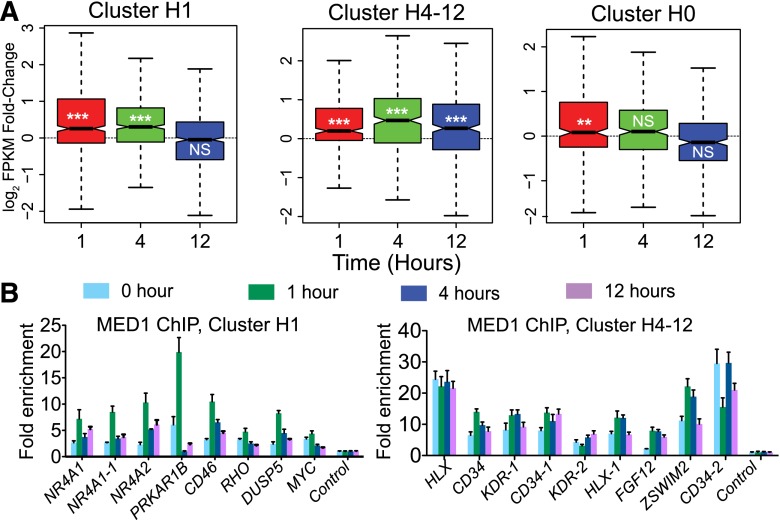
We next investigated the relationship of dynamic, EP300-associated H3K27ac loci to gene expression. We profiled gene transcript levels at 0, 1, 4, and 12 h after VEGFA stimulation by RNA-seq (Supplemental Fig. 11A). As expected, gene expression was highly dynamic following VEGFA stimulation, with 495 genes differentially expressed in at least one time point (Q-value < 0.05) (Supplemental Fig. 11A; Supplemental Table 4.). The RNA-seq data was validated by qRT-PCR and was generally concordant with previously reported microarray gene expression profiling data for HUVECs stimulated with VEGFA for 0 and 1 h (Supplemental Fig. 11B,C; Schweighofer et al. 2009) and ENCODE HUVEC RNA-seq data (Supplemental Fig. 12). To evaluate the effect of the dynamic H3K27ac loci on VEGFA-regulated gene expression, we examined expression of genes that were differentially expressed and within 100 kb of dynamic, EP300-associated H3K27ac sites (Fig. 6A). For genes associated with the H1 cluster, transcript levels were significantly higher at 1 and 4 h compared to 0 h, and returned to baseline by 12 h. For genes associated with the H4-12 cluster, expression increased by 1 h, became further increased by 4 h, and was sustained through 12 h. Expression of H0-associated genes was slightly but significantly increased at 1 h but returned to baseline levels at 4 and 12 h. Thus, each cluster of H3K27ac variation was associated with a corresponding temporal pattern of altered gene expression. These data further support the activity of the dynamic H3K27ac loci in the H1 and H4-12 clusters as transcriptional enhancers.
Figure 6.
VEGFA-increased gene expression and mediator binding associated with dynamic H3K27ac loci in H1 and H4-12 clusters. (A) Fold change of differentially expressed genes within 100 kb of dynamic H3K27ac loci belonging to the indicated clusters, compared to 0 h. Line, boxes, and whiskers are as in Fig. 5A,B. Notches are a function of the interquartile difference and inversely related to the square root of the sample size. (***) P < 0.0001, (**) P < 0.001. (NS) Not significant. (B) MED1 ChIP-qPCR of dynamic H3K27ac loci belonging to H1 or H4-12 clusters.
Dynamic H3K27sites and EP300 participate in VEGFA-stimulated chromatin looping
Enhancers are thought to stimulate transcription from promoters by forming chromatin loops (Tolhuis et al. 2002). We investigated whether VEGFA rapidly stimulated chromatin looping at dynamic, EP300-associated H3K27ac loci. The Mediator complex has been implicated in the formation of chromatin loops (Kagey et al. 2010). MED1 and MED12, encoding Mediator complex subunits, were highly expressed in HUVECs (Supplemental Fig. 13A). We measured MED1 and MED12 occupancy of dynamic, EP300-associated H3K27ac sites by ChIP-qPCR (Fig. 6B; Supplemental Fig. 13B–E). At seven of eight loci belonging to cluster H1, MED1 and MED12 enrichment strongly increased at 1 h of VEGFA treatment, then declined to basal levels at 4 to 12 h. In cluster H4-12, MED1 and MED12 were enriched at most loci, but the degree of enrichment did not change in a consistent temporal pattern following VEGFA treatment. These results indicate that the Mediator complex is bound to dynamic, EP300-associated H3K27ac sites and suggest that these sites may undergo VEGFA-stimulated looping.
To directly test the hypothesis that dynamic, EP300-associated H3K27ac sites loop into proximity with promoters after VEGFA stimulation, we used chromatin conformation capture (Dekker et al. 2002) to study temporal changes in chromatin conformation involving three loci with VEGFA-stimulated increases in H3K27ac. Upstream of DUSP5, a dynamic H3K27ac site belonging to cluster H1 became transiently associated with the promoter at 1 h (Fig. 7), when it was maximally occupied by H3K27ac, EP300, and MED1/12, and maximally transcribed as eRNA. At later time points, the association of these regions declined, coincident with decreased EP300 and MED1/12 occupancy, and decreased eRNA transcription. Upstream of KDR (encoding VEGFR2, a VEGFA receptor), dynamic H3K27ac sites belonging to cluster H4-12 became associated with the promoter within 1 h of VEGFA stimulation (Fig. 7). This correlated with its time course of EP300 and MED1/12 occupancy and eRNA transcription but preceded its maximal occupancy by H3K27ac. Similar observations were made at a second dynamic H3K27ac site from cluster H4-12 located upstream of the endothelial gene CD34 (Fig. 7). Thus, at these sites, VEGFA stimulation rapidly altered chromatin conformation and stimulated eRNA transcription, and these events preceded deposition of H3K27ac.
Figure 7.
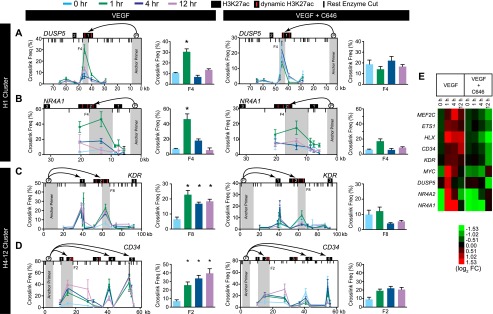
Promoter contact with dynamic H3K27ac loci in H1 and H4-12 clusters was stimulated by VEGFA and required EP300 activity. (A–D) Chromatin confirmation capture of dynamic H3K27ac sites (red lines) near DUSP5, NR4A1, KDR, and CD34. (Black boxes) Regions of H3K27ac enrichment; chromatin looping was attenuated by pretreatment of HUVEC cells with C646 for 30 min. (Red lines) Dynamic H3K27ac sites. (Gray bars) Promoter anchor primers and fragments graphed to the right. (Vertical black lines) Cutting sites of restriction enzyme used for chromatin conformation capture. (*) P < 0.05. (E) EP300 acetyltransferase activity was required for activation of gene expression downstream from VEGFA. Gene expression was measured by qRT-PCR during VEGFA stimulation in the presence or absence of the EP300 inhibitor C646.
To probe the requirement of EP300 in chromatin looping, we repeated the chromatin conformation capture experiments in the presence of the EP300 inhibitor C646 (Fig. 7). C646 blocked VEGFA-stimulated chromatin looping, thereby establishing the importance of EP300 in establishing chromatin loops. Consistent with a key role of EP300 acetyltransferase activity in mediating VEGFA-stimulated chromatin changes and activation of gene transcription, C646 potently blocked up-regulation of genes normally induced by VEGFA, including DUSP5, KDR, NR4A1, and CD34 (Fig. 7E).
Discussion
Epigenetic signatures define transcriptional regulatory elements that underlie the distinct gene expression programs of different cell types, and these signatures have been used to annotate cell type-specific functional elements (Heintzman et al. 2007; Ernst et al. 2011; Kharchenko et al. 2011; Bonn et al. 2012). However, less is known about how the chromatin landscape responds to transient environmental cues. To gain insights into this area, we studied changes in H3K27 acetylation that occur within 12 h of endothelial cell stimulation with VEGFA, a major regulator of angiogenesis. We showed that VEGFA induces rapid changes in H3K27ac at thousands of genomic loci. We demonstrated that dynamic changes in H3K27ac define VEGFA-regulated transcriptional regulatory elements. These regions had characteristics of activity-regulated enhancers: they were tightly linked to EP300 chromatin occupancy, had functional annotations linked to blood vessel development, were transcribed as VEGFA-stimulated eRNAs, and engaged in VEGFA-regulated chromatin looping and gene expression. These regions with dynamic H3K27ac exhibited VEGFA-stimulated transcriptional activity in both luciferase assays and in HUVEC gene expression profiles, and EP300 inhibition blocked VEGFA-induced changes in H3K27ac and gene expression. Thus, our study indicates that the epigenome is an integral participant in signal-induced transcriptional responses.
We developed a novel epigenetic signature based on the signal-induced variation of H3K27ac chromatin occupancy. Using this signature, we identified thousands of novel endothelial, VEGFA-responsive transcriptional regulatory elements and the transcription factor families that are likely to regulate them. These regulatory elements were associated with genes that regulate angiogenesis and could be separated into distinct functional groups based upon their temporal variation of H3K27 acetylation. These data will provide an important resource for future studies of the transcriptional regulation of angiogenesis, although we note that this study was performed in cultured venous endothelial cells and that other VEGFA-responsive endothelial enhancers active in other endothelial cell types or active in vivo were likely not detected in this system. More broadly, we anticipate that application of our epigenetic signature based upon signal-induced chromatin feature variation to other biological systems will enhance annotation of activity-regulated functional elements genome-wide.
Previous studies suggested that nucleosome dynamics define activity-regulated transcriptional enhancers (He et al. 2010; Bonn et al. 2012). However, our data suggest that rapid changes in H3K27ac were not due to changes in chromatin accessibility/nucleosome occupancy. Rather, dynamic H3K27 acetylation was closely associated with EP300, and, indeed, EP300 and its acetyltransferase activity were required to write these marks. These data indicate that epigenetic enzymatic activity is also an important factor that establishes activity-regulated transcriptional enhancers.
Our experiments highlight the crucial role of EP300 in mediating signal-responsive changes in H3K27ac and gene expression. EP300 is a histone acetyltransferase that was previously reported to occupy tissue-specific transcriptional enhancers (Visel et al. 2009). Our data show the proximity of regions occupied by EP300 and regions with VEGFA-stimulated variation in H3K27ac. Inhibition of VEGFA-induced H3K27ac accumulation by EP300 antagonists supports the causal role of EP300 in dynamic variation of H3K27ac occupancy. Furthermore, EP300 inhibition dramatically blocked gene expression changes induced by VEGFA. These data directly demonstrate the key role of EP300 in executing VEGFA-induced transcriptional responses and suggest more broadly that EP300 is required for signal-induced changes in histone acetylation and gene expression.
To identify transcription factors that participate in the VEGFA transcriptional response and recruit EP300 to dynamic H3K27ac sites, we found transcription factor motifs enriched in EP300-bound regions. ETS, FOX, AP1, and STAT transcription factor motifs were enriched in all three clusters, suggesting that members of these transcription factor families broadly participate in VEGFA-driven transcriptional changes. The key role of several ETS factors in angiogenesis was reviewed recently (Randi et al. 2009). We directly confirmed ETS1 occupancy of most EP300-bound regions, validating the motif analysis and providing a resource for further study of the role of ETS1 in angiogenesis and VEGFA-induced gene expression changes. The extensive overlap between EP300 and ETS1 binding suggests that ETS1 may contribute to EP300 recruitment. Consistent with this hypothesis, ETS1 knockdown blocked VEGFA-induced H3K27ac changes at ETS1-bound loci. Compared to ETS, relatively less is known about the role of FOX, AP1, and STAT transcription factor family members as effectors of VEGFA signaling. Our data identify regions potentially regulated by these factors downstream from VEGFA. Recently, FOX transcription factors were reported to interact with ETS factors to regulate vasculogenesis, and similar interactions may also contribute to angiogenesis (De Val et al. 2008). Our data also indicate that AP1 is an important transcriptional effector of VEGFA. Although this role of AP1 has not been studied, AP1 is well-positioned in intracellular signaling pathways to act in this capacity: AP1 is a major nuclear target of MAPK signaling, which is robustly activated downstream from VEGFA (D'Angelo et al. 1995).
We also identified transcription factor motifs that were enriched in a subset of dynamic H3K27ac clusters, suggesting a link to specific temporal patterns of H3K27 acetylation and to specific functional pathways. In the early-responding H1 cluster, we detected significant enrichment for the ATF1/CREB1 (activating transcription factor 1 and cyclic-AMP response element binding protein 1) motifs (Fig. 4D; Supplemental Table 3). These transcription factors mediate immediate early responses, which predominate the functional terms linked to the H1 cluster. GATA and TEAD motifs were overrepresented in the H4-12 and H0 clusters. GATA2 has been implicated as a key regulator of endothelial gene transcription (Linnemann et al. 2011), and Tead4 (also known as RTEF-1 and TEF-3) was recently reported to be required for VEGFA-stimulated angiogenesis (Liu et al. 2011). GATA2 and Tead4 likely contribute to endothelial cell-specific functional term enrichment in the H4-12 and H0 clusters. GATA factors are also crucial in regulating hematopoiesis. However, GO terms related to blood development were not overrepresented in the H4-12 or H0 cluster, suggesting that the GATA motifs identified by our analysis are selectively active in endothelial cells. The H0 and H1 clusters, which share a decline of H3K27ac at 4–12 h, were both enriched for the binding motif of RBP/J, the nuclear target of Notch signaling. Interestingly, VEGFA signaling activates the Notch pathway, which, in turn, antagonizes VEGFA action in an auto-regulatory loop (Hellstrom et al. 2007; Holderfield and Hughes 2008). Collectively, these data suggest that distinct transcription factor families contribute to the different temporal and functional properties of the H0, H1, and H4-12 clusters.
In addition to its role in depositing H3K27ac, EP300 catalytic activity was required for VEGFA-induced chromatin looping. To our knowledge, the requirement of EP300 in chromatin looping has not been reported previously. Furthermore, the time course data, surprisingly, suggest that, for members of the H4-12 cluster, eRNA expression and chromatin looping occur prior to up-regulation of H3K27ac, yet are dependent on EP300 catalytic activity. Further work is required to establish the mechanism(s) through which EP300 acetyltransferase activity promotes eRNA expression and chromatin looping and how the interplay between these chromatin properties regulates gene transcription.
Methods
Detailed descriptions of cell culture, ChIP-seq, RNA-seq, DNase-seq, chromatin conformation capture, luciferase assays, and their integrative analysis are provided in the Supplemental Material. Primary HUVEC cells were cultured overnight in EBM2 with 0.5% FBS, then treated with 50 μg/mL VEGFA for 0, 1, 4, and 12 h. Chromatin occupancy analysis was performed by ChIP-seq and ChIP-qPCR following established protocols (Lee et al. 2006), with minor modifications. Antibodies are listed in Supplemental Table 5. Polyadenylated RNA expression was profiled by mRNA-seq as described (Christodoulou et al. 2011), with minor modifications. DNase-seq was performed as described (Song and Crawford 2010), with modifications to library construction to permit multiplex sequencing. Chromatin conformation capture was performed as described (Hagege et al. 2007). Luciferase assays were performed using 1–2 kb genomic regions cloned upstream of luciferase. Primers used in this study are listed in Supplemental Table 6.
ChIP-seq and DNase-seq reads were mapped with Bowtie (Langmead et al. 2009) (summarized in Supplemental Table 5). EP300 peaks were called using SPP (Kharchenko et al. 2008). DNase-seq peaks were called using F-seq as described (Boyle et al. 2008). H3K27ac ChIP-seq enrichment over input in 200-bp windows, tiled at 50-bp intervals, were calculated across the genome. The H3K27ac variance score for each window was calculated as log2 of the variance of H3K27ac in the window over time, divided by the mean of H3K27ac in the window. Windows with low mean signal were discarded. For each EP300 site, we identified the H3K27ac region within 2 kb with greatest variance score. Of these regions, the top 20th percentile was defined as dynamic, EP300-associate H3K27ac regions. RNA-seq and differential expression analysis was performed using Cufflinks and Cuffdiff (Trapnell et al. 2010). Motif discovery was performed using DREME (Bailey 2011), and motifs were annotated with TomTom (Gupta et al. 2007).
Data access
All sequencing data were deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41166. The data are also available through the Cardiovascular Development Consortium at https://b2b.hci.utah.edu/gnomex/.
Acknowledgments
W.T.P. was supported by an American Heart Association Established Investigator Award and National Heart, Lung, and Blood Institute award U01HL098166, and by charitable contributions from Karen Carpenter, Edward Marram, and Gail Federici Smith. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Author contributions: B.Z. and W.T.P. designed the experiments. B.Z. and J.C. executed the experiments. L.S. and G.E.C. performed DNase-seq experiments. D.C. and J.G.S. assisted with RNase-seq experiments. B.Z., D.S.D., J.H., P.J.P., and W.T.P. analyzed the data. B.Z., D.S.D., and W.T.P. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.149674.112.
References
- Altarejos JY, Montminy M 2011. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL 2011. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. 2010. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem Biol 17: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK 2011. Molecular mechanisms and clinical applications of angiogenesis. Nature 473: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou DC, Gorham JM, Herman DS, Seidman JG 2011. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol 94: 4.12.1–4.12.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, Struman I, Martial J, Weiner RI 1995. Activation of mitogen-activated protein kinases by vascular endothelial growth factor and basic fibroblast growth factor in capillary endothelial cells is inhibited by the antiangiogenic factor 16-kDa N-terminal fragment of prolactin. Proc Natl Acad Sci 92: 6374–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, et al. 2008. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TJ, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS 2007. Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Phng LK, Gerhardt H 2007. VEGF and Notch signaling: The yin and yang of angiogenic sprouting. Cell Adhes Migr 1: 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holderfield MT, Hughes CC 2008. Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-β in vascular morphogenesis. Circ Res 102: 637–652 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A 2012. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res 22: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann AK, O'Geen H, Keles S, Farnham PJ, Bresnick EH 2011. Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci 108: 13641–13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao D, James L, Li J, Zeng H 2011. Requirement of the nuclear localization of transcription enhancer factor 3 for proliferation, migration, tube formation, and angiogenesis induced by vascular endothelial growth factor. FASEB J 25: 1188–1197 [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi AM, Sperone A, Dryden NH, Birdsey GM 2009. Regulation of angiogenesis by ETS transcription factors. Biochem Soc Trans 37: 1248–1253 [DOI] [PubMed] [Google Scholar]
- Reddy TE, Gertz J, Pauli F, Kucera KS, Varley KE, Newberry KM, Marinov GK, Mortazavi A, Williams BA, Song L, et al. 2012. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res 22: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer B, Testori J, Sturtzel C, Sattler S, Mayer H, Wagner O, Bilban M, Hofer E 2009. The VEGF-induced transcriptional response comprises gene clusters at the crossroad of angiogenesis and inflammation. Thromb Haemost 102: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE 2010. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RDG, Chenoweth JG, Tesar PJ, Furey TS, et al. 2007. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]