Abstract
Individual cells from a genetically identical population exhibit substantial variation in gene expression. A significant part of this variation is due to noise in the process of transcription that is intrinsic to each gene, and is determined by factors such as the rate with which the promoter transitions between transcriptionally active and inactive states, and the number of transcripts produced during the active state. However, we have a limited understanding of how the DNA sequence affects such promoter dynamics. Here, we used single-cell time-lapse microscopy to compare the effect on transcriptional dynamics of two distinct types of sequence changes in the promoter that can each increase the mean expression of a cell population by similar amounts but through different mechanisms. We show that increasing expression by strengthening a transcription factor binding site results in slower promoter dynamics and higher noise as compared with increasing expression by adding nucleosome-disfavoring sequences. Our results suggest that when achieving the same mean expression, the strategy of using stronger binding sites results in a larger number of transcripts produced from the active state, whereas the strategy of adding nucleosome-disfavoring sequences results in a higher frequency of promoter transitions between active and inactive states. In the latter strategy, this increased sampling of the active state likely reduces the expression variability of the cell population. Our study thus demonstrates the effect of cis-regulatory elements on expression variability and points to concrete types of sequence changes that may allow partial decoupling of expression level and noise.
Fluctuations in the process of transcription can generate considerable cell-to-cell variability in the expression level of genes across isogenic cell populations (Blake et al. 2003; Kaern et al. 2005; Maheshri and O'Shea 2007; Raj and van Oudenaarden 2009). This variability can result in phenotypic diversity and thus lead to potentially important effects on many cellular and developmental processes. Consequently, unraveling the sources that underlie expression variability has been a topic of considerable interest. Several studies demonstrated that genes are transcribed in bursts (Ross et al. 1994; Blake et al. 2003; Golding et al. 2005; Larson et al. 2011; Suter et al. 2011), such that the expression variability due to transcription is determined by the frequency with which the bursts occur (burst frequency) and the number of transcripts produced per burst (burst size) (Cai et al. 2006). These parameters are determined in part by fluctuations in trans factors that are extrinsic to the promoter, such as the concentrations of the regulating transcription factors and RNA polymerases (Elowitz et al. 2002; Kaern et al. 2005). The remaining variability is gene-specific intrinsic noise that was shown to vary greatly across promoters (Raser and O'Shea 2004; Bar-Even et al. 2006), and which is at least partly encoded by cis-acting regulatory elements embedded within the DNA sequence of each promoter (Raser and O'Shea 2004; Blake et al. 2006).
Several types of regulatory elements were shown to affect transcriptional noise. Mutating a TATA-box in two yeast promoters reduces expression variability, in a manner that is consistent with an effect of TATA-boxes on the size of transcriptional bursts (Raser and O'Shea 2004; Blake et al. 2006; Hornung et al. 2012). The number and affinity of transcription factor binding sites were also shown to affect expression variability (Murphy et al. 2007; To and Maheshri 2010; Suter et al. 2011), with one study demonstrating that using two sites for the mammalian transcription factor NF-Y instead of one, or using a higher affinity NF-Y site, increased expression variability in an artificial promoter construct (Suter et al. 2011). Similar to the effect of TATA-boxes, the increase in expression variability observed with two NF-Y sites or with a higher-affinity site was mediated primarily by an increase in the average burst size, whereas the burst frequency was largely unaffected (Suter et al. 2011). Chromatin regulation has also been linked to expression variability (Blake et al. 2003; Raser and O'Shea 2004; Field et al. 2008; Choi and Kim 2009; Bai et al. 2010), with two studies showing that adding nucleosome-disfavoring sequences to a yeast promoter resulted in lower nucleosome occupancy and reduced expression variability (Bai et al. 2010; Raveh-Sadka et al. 2012). However, in contrast to the effect of TATA-boxes and TF sites, chromatin was suggested to mainly affect the frequency with which promoters transition between transcriptionally active and inactive states (Raser and O'Shea 2004). In yeast, statistically significant genome-wide associations were found between higher-expression variability and the presence of TATA-boxes, the number of TF binding sites, and the encoding of high nucleosome occupancy (Field et al. 2008; Tirosh et al. 2008).
A recent study showed that the quantitative increase in the mean expression over a cell population that results from adding nucleosome disfavoring sequences can be comparable to the increase in expression achieved when increasing the affinity of a transcription factor (TF) binding site (Raveh-Sadka et al. 2012). Several lines of evidence suggested that the effect of the nucleosome-disfavoring sequences was likely mediated by the lower nucleosome occupancy and thus increased accessibility that nucleosome-disfavoring sequences confer over the nearby promoter elements, such as TF sites (Raveh-Sadka et al. 2012). Thus, combined with the above studies, an intriguing hypothesis is that the two distinct types of sequence changes in either TF sites or in nucleosome-disfavoring sequences provide a genetic mechanism that may allow partial decoupling of mean expression level and transcriptional noise. Specifically, since mean expression is the product of burst frequency and burst size, and noise (under some assumptions) is the inverse of burst frequency (Paulsson and Ehrenberg 2000; Friedman et al. 2006), then increasing burst frequency is expected to result in lower noise compared with a similar increase in mean expression that is due to an increase in burst size.
Here, we set out to test the above hypothesis, as well as to directly compare the effects that changes to transcription factor binding sites have on expression variability with the effects induced by changes to sequences that affect nucleosome occupancy. To associate the observed changes in expression variability with one of these types of sequence changes, we always performed the two types of sequence changes being compared within the same promoter context. By monitoring fluorescent intensity in single cells using time-lapse microscopy, we compared the dynamics of promoters in which expression is increased by adding or lengthening nucleosome-disfavoring sequences with the dynamics induced by changes to the binding affinity of sites for two different transcription factors. Our results demonstrate that these two distinct DNA-encoded strategies for increasing expression indeed have opposing effects on promoter dynamics, allowing the same mean expression level to be achieved but with lower noise when the strategy of nucleosome-disfavoring sequences is used. These results hold for the two different transcription factors that we tested and are further supported by independent flow cytometry measurements that we performed.
Thus, our results show how mean expression and expression variability may be partially decoupled, providing a potentially useful tool for synthetic biology, and raising intriguing hypotheses regarding the extent to which evolution of native promoters has used these genetic mechanisms to partly decouple mean expression and noise levels.
Results
Experimental design and measurement of promoter dynamics
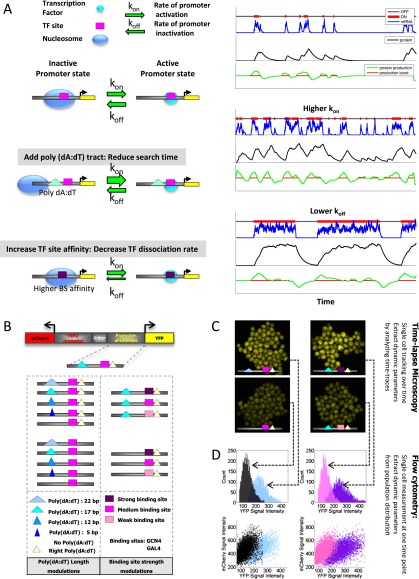
We sought to compare the effect on promoter dynamics of two distinct strategies for increasing expression, strengthening transcription factor binding sites, and adding or lengthening nucleosome-disfavoring sequences, since we hypothesized that each type of sequence change would affect different aspects of promoter dynamics. Specifically, in the classical scenario in which the binding of a transcription factor induces expression via its stabilizing effect on polymerase, we expect that strengthening the affinity of a TF site may increase the time that the factor remains bound at its site, resulting in more transcripts produced during the active state, and thus increasing expression by increasing the burst size (Fig. 1A, bottom). In contrast, since addition of poly(dA:dT) tracts, which disfavor nucleosome formation (Segal and Widom 2009), was shown to result in lower nucleosome occupancy and more accessible DNA in the vicinity of the tract (Iyer and Struhl 1995; Raveh-Sadka et al. 2012), we hypothesized that this would result in faster on/off binding dynamics of a TF to a nearby site, and thus increase expression by increasing the burst frequency (Fig. 1A, middle). Thus, we predicted that the strategy of increasing expression by lowering the encoded nucleosome occupancy of a TF binding site would result in faster promoter dynamics and thus lower expression variability across a cell population, as compared with the strategy of increasing expression by strengthening the TF binding site.
Figure 1.
Promoter variants and experimental setup. (A) Illustration of hypothesized promoter dynamics induced by two distinct DNA-encoded strategies for increasing expression and a matched stochastic simulation of promoter switching, transcription, and translation (right panel) for each strategy. For a simplified model in which promoters transition at some rate between transcriptionally inactive and active states (top), we hypothesize that addition of nucleosome disfavoring elements such as poly(dA:dT) tracts would increase the accessibility of the transcription factor binding site, thereby reducing the time that a factor molecule spends in search of its site (middle). In contrast, we hypothesize that increasing the affinity of a factor binding site would reduce the factor's dissociation rate. Note that both types of sequence changes result in a higher mean expression over the cell population, but with distinct hypothesized effects on promoter dynamics. Using the Gillespie algorithm, we simulate the kinetic scheme in three scenarios. In the simulation runs, we record, as a function of time, the promoter state (black–red line), mRNA levels (blue line), protein levels (black line), and the protein production rate (green line), which is the derivative of the protein levels with respect to time. In addition, we record protein production bursts (red line), when production is positive. The “normal” promoter (top) represents the reference point for the parameter changes. The fast promoter (middle) has an increased kon to simulate an increase in promoter accessibility [added poly(dA:dT)]. The “slow” promoter (bottom) has a decreased koff (with respect to the normal promoter) to simulate an increase in TF binding site affinity. The three example runs shown illustrate that although both parameters can increase the overall expression level, kon increases the frequency of production bursts, while koff increases the length of the bursts. (B) Illustration of promoter variants used in this study. All promoters are genomically integrated upstream of a yellow fluorescent protein (YFP) reporter and into a region that also contains an mCherry fluorescent protein driven by a constant TEF2 promoter. Promoter variants differ in the presence and length of two poly(dA:dT) tracts and in the affinity of the transcription factor binding site for either the Gcn4 or Gal4 transcriptional activators. (C) Representative YFP time-lapse microscopy images of four promoter variants, imaged in a microfluidic platform that supplies a continuous flow of medium. Each cell was followed over time, and its lineage, YFP, and mCherry signal intensity were extracted (Methods). (D) Single-cell flow cytometry data collected for the corresponding promoter variants from C, shown as a histogram of normalized YFP values (top) and as a scatterplot of YFP (x-axis) against mCherry fluorescence (bottom).
To test this hypothesis, we used a controlled setting in which only the elements hypothesized to have differential effects on promoter dynamics are altered. To this end, we used yeast strains in which different promoters are separately integrated into the same genomic location and upstream of the same yellow fluorescent protein (YFP) reporter (Zeevi et al. 2011). To control for experimental variability and as a marker for segmenting and tracking the cells in all promoter variants, an mCherry fluorescent reporter is also integrated into the same genomic location in all strains, downstream from a constant promoter (TEF2, a translation elongation factor). We used a set of synthetic variants that were generated in a recent study, in which poly(dA:dT) tracts with different lengths and binding sites with different affinities for two distinct transcription factors were integrated into the same promoter context (Fig. 1B; Raveh-Sadka et al. 2012). The promoter context is derived from the native yeast HIS3 promoter, and contains two poly(dA:dT) tracts flanking a single site for the transcriptional activator Gcn4. Measurements of mean expression levels of these promoter variants showed that variants with longer poly(dA:dT) tracts exhibit higher YFP expression. Consistent with an effect mediated by changes in nucleosome organization, variants with longer poly(dA:dT) had lower nucleosome occupancy over the nearby transcription factor binding site (Raveh-Sadka et al. 2012). Important for our purposes, this recent study showed that these two distinct strategies of increasing expression, once by modulating binding site affinity and once by modulating poly(dA:dT) length, can achieve comparable quantitative increases in the mean expression level of the cell population.
To test our hypothesis that these distinct genetic strategies for increasing expression have opposing effects on promoter dynamics, we used two independent approaches that can each provide estimates of promoter dynamics (Fig. 1C,D). The first monitors YFP fluorescence in single cells over time and thus provides more direct measurements of promoter dynamics. To this end, we grew cells in a microfluidic plate that maintains continuous supply of media, and used an automated microscopy system to image fluorescence continuously over 15 h and at a resolution of 3 min (Fig. 2A; Supplemental Fig. S6). We developed analysis software that uses the mCherry fluorescence signal to segment the cells and track the segmented cells over time, resulting in time traces of mCherry and YFP signal intensities (Fig. 2A,B; Supplemental Methods). We followed each cell over multiple cell doublings, which was identified by the marked decrease in the fluorescence signal that is caused by diffusion of the fluorescent reporter into the daughter cell (Fig. 2B; Supplemental Methods). For each cell, we then computed the rates of its mCherry and YFP production over time, by differentiating the dilution-corrected signal tracks (Fig. 2B; Supplemental Methods). To compare promoter dynamics across different strains, we normalized the YFP signal by the mCherry signal (Fig. 2B), because this accounts for many extrinsic factors that are correlated with the measured protein production rates, such as variations in cell size, time within cell cycle, and abundance of general components of the transcriptional machinery (Supplemental Methods). As a second independent method from which indirect estimates of promoter dynamics can be extracted (Friedman et al. 2006), we used flow cytometry to measure the distribution of single-cell fluorescent intensity at a single time point (Fig. 1D). Notably, in both measurement methods, the mean expression of the cell population increases with both the length of the poly(dA:dT) tract and the affinity of the transcription factor binding sites, consistent with previous measurements (Supplemental Fig. S1; Raveh-Sadka et al. 2012).
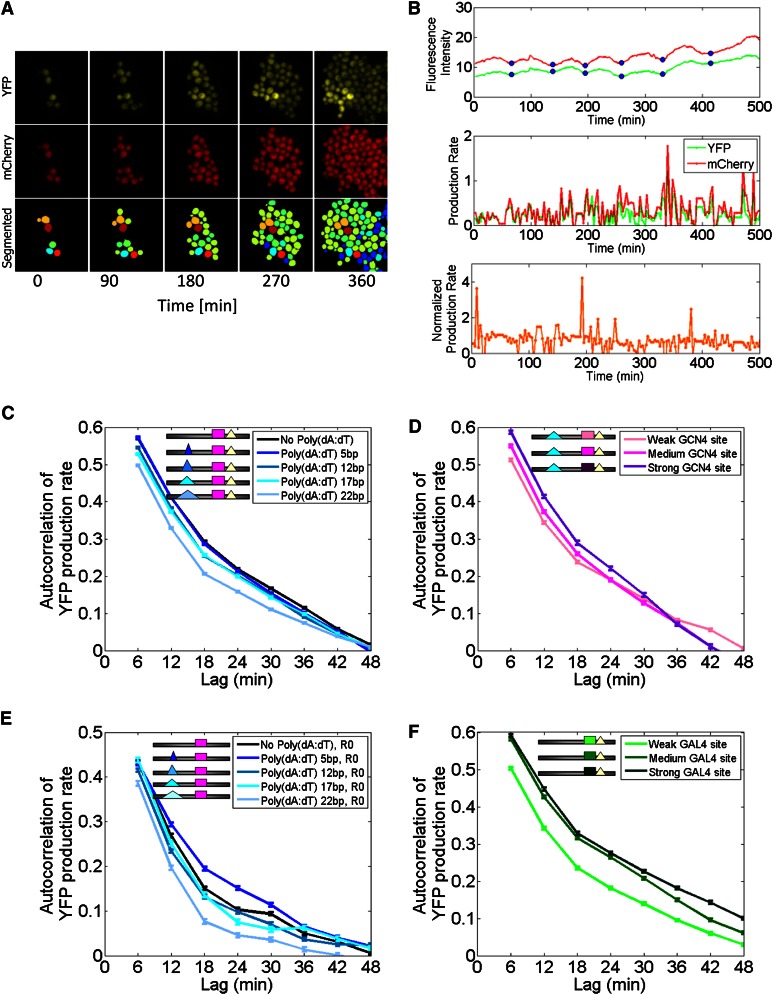
Figure 2.
Opposing effects on promoter dynamics for lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites. (A) Representative time-lapse microscopy images of one imaging area at five different time points, displaying YFP fluorescence (top), mCherry fluorescence (middle), and automatically segmented cells. (B) Representative time-lapse traces of YFP and mCherry fluorescence of a single cell over time (top), along with YFP and mCherry production rates (middle), and normalized YFP production rates (bottom; normalization done by mCherry; see Methods). Blue circles denote cell cycles. (C) Longer poly(dA:dT) tracts result in faster promoter dynamics. Shown is the average autocorrelation of normalized YFP production rates across thousands of different cell traces for each of five different promoter variants with poly(dA:dT) tracts of length 0, 5, 12, 17, or 22 bp. Bars denote standard error. (D) Higher-affinity binding sites result in lower promoter dynamics. Same as C, for three promoter variants that differ only in the affinity of the Gcn4 site. (E) Same as C, but where the poly(dA:dT) tract variants were inserted into a different genetic background in which the right poly(dA:dT) tract is deleted (R0). (F) Same as D, but for three Gal4 sites that differ in their affinity.
Lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites have opposing effects on the rate of promoter dynamics
As a direct and unbiased approach to compare dynamics of promoter activity across our various promoter variants, we first computed the autocorrelation function of the YFP production rates as measured by time-lapse microscopy of every variant (Sigal et al. 2006). Higher correlations indicate production rates that are maintained across longer time periods and thus suggest slower changes in dynamics of promoter activity. To perform the analysis, we extracted the time traces of YFP production of thousands of individual cells from each promoter variant, computed the autocorrelation of each time trace at different time lags in 6-min resolution, and, for every time lag, averaged the autocorrelation values of the cells of each variant.
Comparing the autocorrelations of five different variants with poly(dA:dT) tracts of length 0, 5, 12, 17, and 22 bp, we found significant gradual reductions in the autocorrelation of the variants as the length of the tract increases (Fig. 2C). In contrast, we found an opposite trend of an increase in the autocorrelation values of three different promoter variants in which the affinity of the binding site for Gcn4 is strengthened (Fig. 2D). In both cases, the differences between variants were significant across several lags (Student's t-test, P < 0.05), and the most significant lags correspond to lags that are <20 min and thus much shorter than the average cell cycle time (∼90 min).
To verify that these effects are not specific to the tested variants and to Gcn4, we repeated the analysis in three additional sets of variants consisting of a set of five variants in which the above poly(dA:dT) tracts were added to a different promoter background; a set of three variants with increasing binding site affinities for the transcriptional activator Gal4, which is not known to regulate the native promoter from which our variants were derived; and another set of the same three Gal4 sites added to a modified promoter background. Consistent with our above results, we found that variants with longer poly(dA:dT) tracts within the modified Gcn4 promoter background had lower autocorrelation values than shorter tracts (Fig. 2E), and variants with higher-affinity Gal4 sites in both promoter backgrounds had higher autocorrelations than lower-affinity sites (Fig. 2F; Supplemental Fig. S2). Moreover, comparing the two sets of Gal4 site variants between the two promoter backgrounds that differ in the presence of a poly(dA:dT) tract, we found that the variants within the poly(dA:dT)-containing promoter background had lower autocorrelation values, providing further support to the reduced autocorrelation that occurs upon addition of poly(dA:dT) tracts.
Our results suggest that these two distinct strategies for increasing expression, namely, lengthening poly(dA:dT) tracts or strengthening binding site affinity, do so with opposing effects on the time-dependent dynamics of promoter activity.
Longer poly(dA:dT) tracts induce faster promoter transition rates
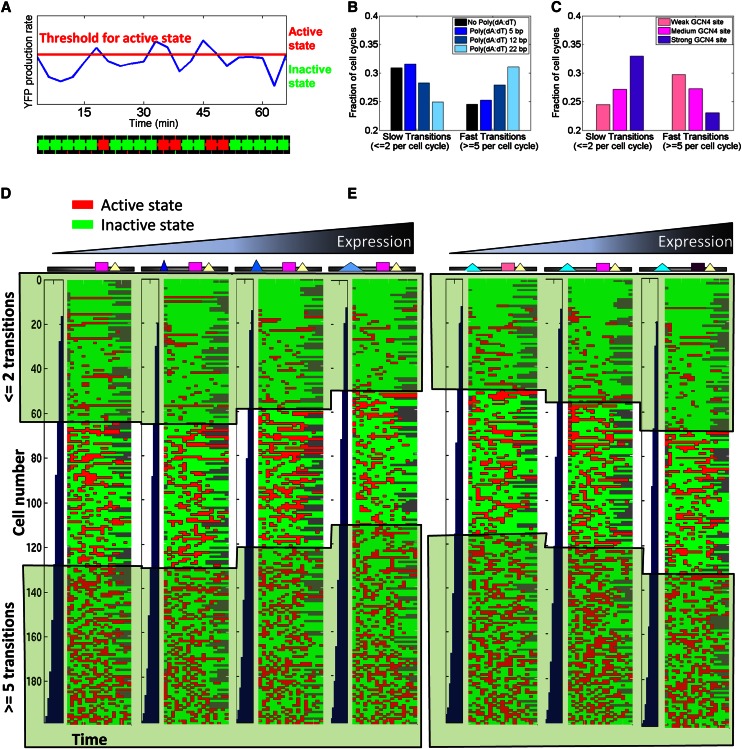
To obtain a more direct visual view of the above results at the single-cell level, we used our data to estimate the frequency with which each of the above promoter variants transitions between high-expressing and low-expressing states. To this end, we defined the high- and low-expressing states by whether the amount of YFP produced during the examined time window was above or below some arbitrary threshold, respectively. We again used the time traces of YFP production of thousands of individual cells from each promoter variant, and for a given YFP production threshold, classified every 3-min time window of each cell's YFP time trace into high- and low-expressing states (Fig. 3A). From these classifications, we then computed, for each promoter variant, its probability of transitioning between high- and low-expressing states, as well as the fraction of all of its cell cycle time traces that had k transitions, for all possible values of k. Thus, this analysis examines how the high- and low-expressing time windows are distributed across the time traces of each variant. For example, a fast-transitioning variant in which 50% of its time windows are high expressing would transition between high- and low-expressing states every 3 min, whereas an extremely slow variant would be continuously high expressing in half of all of the cell cycles examined and continuously low expressing in the other half. To ensure that the results are not sensitive to the choice of threshold, we performed the analysis across a wide range of threshold values.
Figure 3.
Lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites have opposing effects on the rate of promoter transitions between active and inactive states. (A) Illustration of our analysis of promoter transition rates. For each cell cycle of every cell, we classify its trace of normalized YFP production rate (blue trace illustrated here for one cell cycle of one cell) into active (red) and inactive (green) states according to whether they are above or below a predefined arbitrary threshold (red horizontal line), respectively. (B) Increasing the length of a poly(dA:dT) tract results in a higher rate of transitions between active and inactive states. For promoter variants that differ in the length of a poly(dA:dT) tract, shown is the fraction of all of its measured cell cycle traces in which the number of transitions between active and inactive states was at most two (slow transitions; left bar graph) or at least five (fast transitions; right bar graph). The comparison of these different promoter variants was done at a threshold in which the fraction of all inactive states in each variant was 70% (since absolute expression levels vary across variants, the absolute threshold value is different for each variant). See Supplemental Figure S4 for similar analyses at a range of thresholds from 50% to 90%. (C) Increasing the affinity of a transcription factor binding site results in a lower rate of transitions between active and inactive states. Same as B, but for variants that differ in the affinity of a Gcn4 binding site. (D) Visual illustration of cell cycle traces corresponding to the bar graphs from B in which the length of poly(dA:dT) tracts was varied. For each promoter variant, shown are 200 rows that each correspond to a time trace of one cell cycle of one cell with colored entries representing active (red) or inactive (green) states at a threshold in which 70% of all states were inactive. Rows are sorted according to the number of transitions between active and inactive states, and the 200 rows were sampled from all cell cycle traces such that they accurately represent the same probability distribution of the number of transitions across all cell cycle traces. (E) Same as D, but for the bar graphs from C in which the affinity of Gcn4 sites was altered.
Notably, we found that variants with longer poly(dA:dT) tracts had higher probabilities of transitioning between high- and low-expressing states and a higher fraction of cell cycles with more than five transitions (Fig. 3B,D). Conversely, variants with higher-affinity binding sites had lower transition probabilities and a higher fraction of cell cycles with at most one transition between high- and low-expressing states (Fig. 3C,E; Supplemental Fig. S3). These results were robust across all tested threshold choices (Supplemental Fig. S4). Thus, although our data do not directly measure high- and low-expressing states, they clearly demonstrate that across the broad range of threshold choices for the high-expressing states, increasing the length of poly(dA:dT) tracts results in faster transitions between the classified states, whereas increasing the affinity of a transcription factor binding site results in slower transitions. This suggests that these two distinct strategies for increasing the mean expression level of a cell population are mediated by opposing effects on promoter dynamics, consistent with the above autocorrelation analysis.
Agreement between time-lapse promoter dynamics and static expression measurements
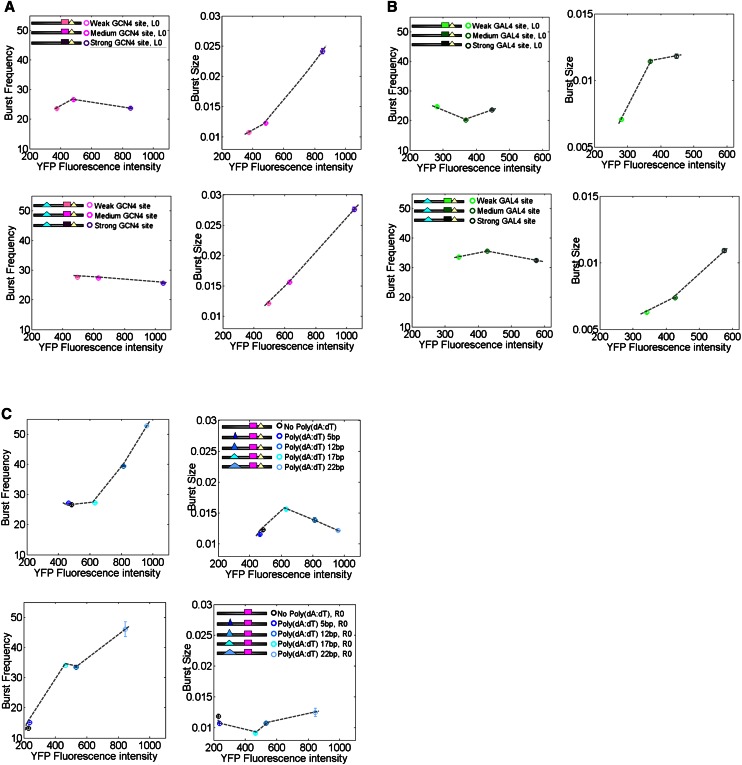
As another experimentally independent way to estimate promoter dynamics, we also measured the single-cell YFP and mCherry fluorescence intensity distributions of each promoter variant at a single time point of mid-log phase using flow cytometry. Under certain assumptions, previous studies have shown that by fitting the static distribution of such single cell fluorescence intensities to a gamma distribution, the two fitted parameters can be interpreted as the burst frequency with which genes are transcribed and the size of the bursts (Friedman et al. 2006; Taniguchi et al. 2010). To remove much of the extrinsic noise that is due to variations such as cell size, we normalized the YFP intensity of each cell by the measured mCherry intensity to get a pathway-specific measure of noise. Although this measure does not fully correspond to intrinsic noise in our experimental system, the distributions of normalized YFP fluorescent intensities were well fitted to a gamma distribution, thus supporting the use of this framework for estimating promoter dynamics (Supplemental Fig. S5).
Examining the fitted parameters of the gamma distribution for each promoter variant, we found that increasing the affinity of the transcription factor binding site resulted in higher values for the inferred burst size parameter, and in little effect on the inferred burst frequency parameter (Fig. 4A,B). We found this behavior in four different sets of variants that correspond to changes in the affinity of sites for both Gcn4 and Gal4, each in two different promoter backgrounds. In contrast, we found that increasing the length of a poly(dA:dT) tract resulted in higher values for the burst frequency parameter, and in little change in the burst size parameter (Fig. 4C). Here, too, we found similar behavior across two different promoter backgrounds in which the length of the poly(dA:dT) tract was increased. Thus, these results, obtained by an independent experimental system that measures the population-level distribution of fluorescent intensities, suggest that longer poly(dA:dT) tracts increase expression primarily by inducing a higher frequency of promoter transitions to the active state, whereas higher-affinity binding sites do so primarily by increasing the size of the bursts from the active state. These results are in accord with the more direct measurements of promoter dynamics that we obtained using time-lapse microscopy.
Figure 4.
Flow cytometry validation of opposing effects of lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites. (A) Strengthening the affinity of a transcription factor binding site mainly affects burst size. For two different sets of promoter variants, each with three different binding site affinities for Gcn4, shown are the values of the two parameters from the Gamma function when fitted to the normalized YFP intensities of each promoter variant measured over the cell population at a single time point using a flow cytometer. Under certain assumptions (Friedman et al. 2006; Taniguchi et al. 2010), these two parameters correspond to the burst frequency (left graph) and burst size (right graph). Note the larger variation in burst size across these variants. (B) Same as A, for variants in which the affinities of Gal4 binding sites were varied. (C) Lengthening a poly(dA:dT) tract mainly affects burst frequency. Same as A, for variants in which the length of a poly(dA:dT) tract was varied. In contrast to A and B, note the larger variation in burst frequency across these variants. [R0, right poly(dA:dT) deleted; L0, left poly(dA:dT) deleted.]
Achieving similar mean expression levels with predictably different noise levels
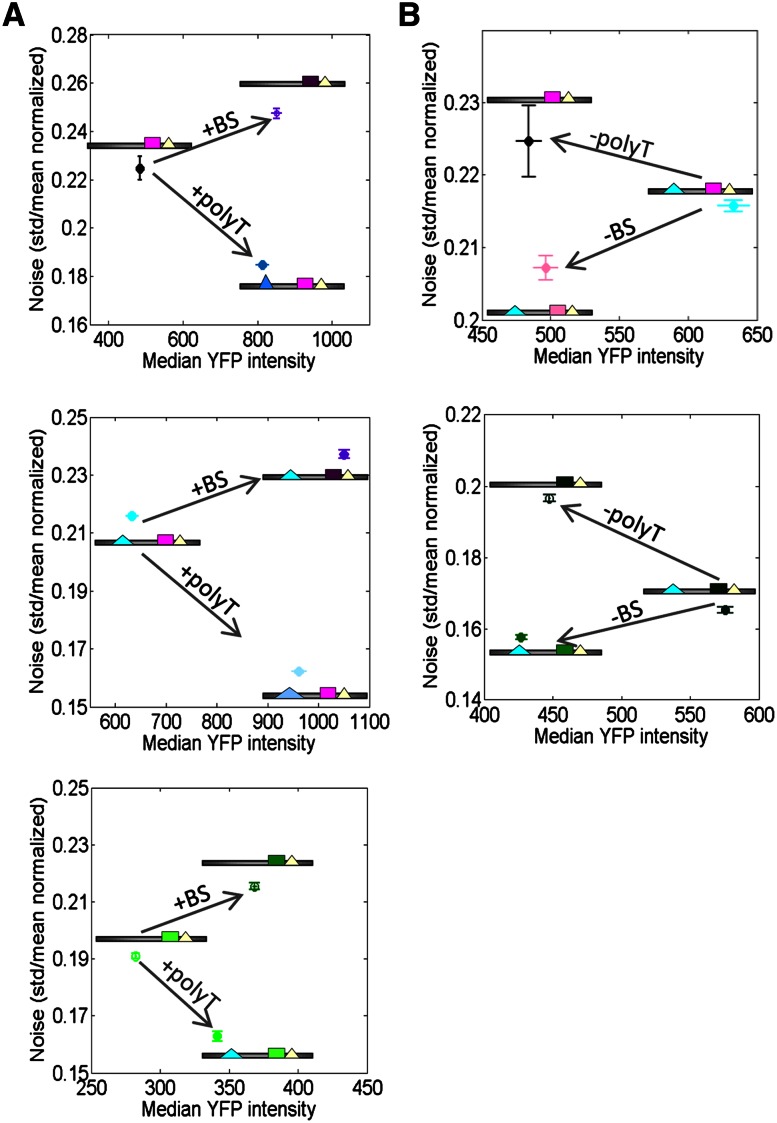
Finally, analytical models predict that when two promoters achieve the same mean expression level but with different burst frequency and burst size, then the promoter with the higher burst frequency will exhibit less noise, defined as the standard deviation of the expression across single cells divided by the mean expression level of the population (Elowitz et al. 2002). We used our promoter variants to test this intriguing prediction, since according to our above estimates of promoter dynamics, our variants represent a case in which the same increase in mean expression can be achieved through different effects on burst frequency and burst size. To this end, we examined promoter triplets that each consist of three promoter variants in which promoters are modified by either adding/lengthening a poly(dA:dT) tract or strengthening a transcription factor binding site such that these modifications increase the mean expression level to similar levels. For each variant in every set, we then used our above flow cytometry measurements to compute its noise level. In all cases, the promoter in which a poly(dA:dT) tract was added had lower noise than the promoter in which the similar expression level was reached by increasing the binding site strength. This result is consistent with the prediction we set out to test, if, indeed, the same mean expression levels were achieved with the poly(dA:dT)-containing promoter exhibiting a higher burst frequency (Fig. 5A). Notably, these sets included sites for both Gcn4 and Gal4, suggesting that the results may hold generally for more transcription factors.
Figure 5.
Adding poly(dA:dT) tracts and strengthening transcription factor binding sites have opposing effects on transcriptional noise. (A) Adding a poly(dA:dT) tract results in a similar increase in mean expression level but in lower noise compared with strengthening the affinity of a transcription factor binding site. For three sets of promoter variants (three different graphs), shown are the median YFP expression (x-axis) and expression noise (standard deviation of expression divided by the mean expression) of a starting promoter (leftmost promoter in each graph with lowest median YFP expression) and two promoters representing modifications to the starting promoter, in which either a poly(dA:dT) tract was added (bottom right promoter in each plot) or the binding site was strengthened (top right promoter in each plot). Note that both promoter modifications result in similar mean expression levels, but the promoter in which the poly(dA:dT) tract was added always has lower noise. Bars denote standard error. (B) Same as A, but where the starting promoter contained a poly(dA:dT) tract and a strong transcription factor binding site, and the modifications either deleted the poly(dA:dT) tract (top left promoter in each plot) or weakened the binding site (bottom left promoter in each plot). As in A, both promoter modifications result in similar mean expression levels, but the promoter with the poly(dA:dT) tract always has lower noise.
We next examined two additional sets that also consist of three promoter variants each in which starting from a promoter with a poly(dA:dT) tract and a high-affinity transcription factor binding site, deleting the poly(dA:dT) tract, or lowering the site affinity reduced expression to similar mean levels. In both sets, the promoter in which the poly(dA:dT) was deleted had higher noise, which is again consistent with the original prediction, if, indeed, the promoter without the tract achieved the same mean expression with a lower burst frequency (Fig. 5B). Here too, the two sets represent sites for two different transcription factors (Gcn4 and Gal4).
Taken together, our results suggest that we can achieve similar quantitative effects on the mean expression of a cell population by either adding poly(dA:dT) tracts or strengthening the affinity of a transcription factor binding site, but the promoter with the poly(dA:dT) will have a higher burst frequency, lower burst size, and, as predicted by analytical models (Paulsson and Ehrenberg 2000; Kepler and Elston 2001; Raser and O'Shea 2004; Friedman et al. 2006), lower noise, as compared with the promoter with the stronger site. Since the effects of these manipulations are predictable, these two distinct strategies may allow for partially decoupling mean expression level and noise.
Discussion
Understanding the dynamical process by which promoters transition between active and inactive states is central to an understanding of transcriptional regulation. Here, we focused on the role of promoter DNA sequence in this process and compared the effect on single-cell expression of two different DNA-encoded strategies that are capable of increasing the mean expression level of a cell population by similar magnitudes. Compared with strengthening transcription factor binding sites, we found that increasing expression by adding nucleosome-disfavoring sequences results in faster promoter dynamics and lower expression variability across an isogenic cell population, suggesting that mean expression level and transcriptional noise can be partially decoupled. Notably, these results held in several different contexts and for two distinct transcription factors. Previous studies showed that gene expression and noise can be decoupled by mutating the TATA-box (Murphy et al. 2010) or by introducing negative autoregulation (Nevozhay et al. 2009). Our work suggests that such decoupling can also be achieved through chromatin.
Although our experimental system does not directly measure the frequency with which promoters transition between active and inactive states or the number of transcripts produced from each transcriptional burst, both our time-lapse microscopy and flow cytometry measurements suggest that the expression increase of each of the two types of sequence changes that we tested is mediated by distinct mechanisms. In the case of adding poly(dA:dT) tracts, our results suggest that the increase in expression is primarily achieved by a higher frequency of transitions between the inactive and active states. We propose that the lower nucleosome occupancy induced by the poly(dA:dT) tracts (Raveh-Sadka et al. 2012) increases the accessibility of the nearby promoter region, thereby reducing the time that it takes the cognate transcription factor to find the site and leading to a higher frequency of successful binding events and promoter activation. This suggestion is supported by a study showing that promoter firing rate is dictated by the time required for a transcription factor to find its gene within the nucleus (Larson et al. 2011). In contrast, when strengthening a transcription factor binding site, our results suggest that the increase in expression is primarily achieved by an increase in the average number of transcripts produced during the active state. We propose that a stronger binding site reduces the dissociation rate between the binding site and its cognate factor, resulting in more stable binding of the transcriptional machinery and thus a higher probability of transcription reinitiation and longer and more sustained bursts, similar to the effects suggested for promoter TATA-boxes. This suggestion is supported by recent findings that measured bursting kinetics in mammalian genes and found increased mean burst sizes for artificial promoters designed with a higher-affinity binding site for a transcriptional activator (Suter et al. 2011).
Finally, comparing the expression of five pairs of promoters in which the promoters in each pair have similar expression levels, we found that in every pair, the promoter with the longer poly(dA:dT) tract and lower-affinity transcription factor binding site always had lower noise. Since noise is the inverse of burst frequency (allowing for some assumptions) (Friedman et al. 2006; Taniguchi et al. 2010), and mean expression is the product of burst frequency and burst size, these results are also consistent with longer poly(dA:dT) tracts causing a higher burst frequency. We note that our experimental system provides a measure of pathway-specific noise rather than intrinsic noise. However, stringent filtering gives us a measure that correlates well with intrinsic noise. Our results are consistent with a model in which the promoter elements affect promoter state switching (see the Supplemental Methods). Although we cannot rule out that other mechanisms may affect the observed transcription rates, our results suggest that the frequency and length of transcriptional activity change more than the magnitude of the activity itself (Supplemental Figs. S21, S22).
Taken together, our results suggest genetic mechanisms by which partial decoupling of mean expression and noise can be achieved. It will be interesting to test the effect of other types of sequence changes on promoter dynamics and to identify cases in which these signals may have been used in evolution for achieving biologically important single-cell behaviors.
Methods
Yeast strains
The set of promoters analyzed consists of 22 promoters that were each genomically integrated into a shared master strain upstream of the yellow fluorescence protein (YFP) reporter and the HIS3 proximal promoter (100 bp upstream of the ATG). The integrated region also contains a fixed control promoter for the TEF2 gene (a translation elongation factor) upstream of an mCherry fluorescent protein. The mCherry reporter serves as an internal control for normalization and cell segmentation. Promoter variants consist of different binding sites for the Gcn4 and Gal4 transcriptional activators and different lengths of poly(dA:dT) tracts upstream of the factor binding sites. Variants of the Gcn4 binding site are as described (Raveh-Sadka et al. 2012). Variants of the Gal4 binding sites were designed to span a range of weak medium and strong affinity as follows (strong site, CGGAAGACTCTCCTCCG; medium, AGGAAGACTCTCCTCCG taken from the GAL1–GAL10 UASg site 3; and weak, CGGATTAGAAGCCGCCG, taken from the GAL1–GAL10 UASg site 1). Full details regarding the strains and the fluorescent proteins are available in the Supplemental Material.
Growth conditions
Yeast strains were grown at 30°C in synthetic complete medium supplemented with 2% glucose, in a 96-well plate for 48 h to saturation. For fluorescence microscopy of single cells, cells were diluted to reach an optical density of 1 prior to the experiment in the desired medium and then diluted and loaded into the microfluidic imaging plate (The ONIX Microfluidic Perfusion System, CellASIC Corporation). Cells were loaded into a 4-μm trap region that ensures monolayer growth of the cells for optimal maintenance of focal plane. A constant flow of media was kept, by applying a stable pressure of 1 psi that enabled constant flow at a rate of 2.5 μL/h. For the flow cytometry measurements, cells were grown to saturation and then diluted to reach mid-log phase (optical density of ∼0.2) in the specified conditions.
Time-lapse microscopy
Time-lapse experiments were conducted using a commercial fully automated inverted fluorescence microscope (Nikon TiE) equipped with a motorized stage (TI-S-ER, Nikon), hardware-based focus maintenance system (PFS; Nikon), fast external shutters (SUTTER), 60× objective lens, and a cage incubator. Commercial filter sets were optimized for detection of YFP and mCherry (YFP ex500/20 em535/30 mCherry ex572/35 em632/60, Chroma). High-resolution images (effective pixel size of 0.216 μm) were acquired for 15 h at a resolution of 3 min using a cooled charge-coupled device camera (EMCCD; DU-888E Andor). Approximately 3000 cell traces were obtained for each imaging area.
Flow cytometry
Flow cytometry experiments were conducted using the Becton-Dickinson LSRII machine and standard protocols. Four channels were acquired: forward-scatter, side-scatter, YFP and mCherry (excitation wavelength was 350 nm for YFP and 740 nm for mCherry). About 150,000 cells were collected from each well at a flow rate of 1 μL/sec. To reduce cell variability, cells were gated in the forward-scatter and side-scatter channels. Analysis of the flow cytometry data is described in the Supplemental Material.
Image analysis
Automated image analysis and cell tracking was performed using a modified version of CellProfiler (Carpenter et al. 2006) and MATLAB to analyze and process single-cell data. We devised an automated and robust framework to analyze and filter the cell data tracks, leaving only high-quality cell data tracks. The framework included six modules that deal with image corrections, cell segmentation and tracking of the cells, segmentation and tracking post-processing, cell lineage analysis, data masking and filtering, correction for cell cycle dilution that stems from diffusion from the mother to the daughter cell, and calculation of production rates. The full description of all modules for image analysis is available in the Supplemental Material. The algorithm for calculating the cell lineage is also described in full in the Supplemental Material. In brief, the basic idea of the algorithm is to compute scores for each of the possible parents and then assign the best scoring parent for each of the buds, based on several parameters obtained from the florescent time-lapse data.
Data analysis
We normalized the data according to the mCherry distribution to account for changes across different well and experiments. Autocorrelation of promoter production rates was calculated for each cell, and then all autocorrelations across all cells were averaged for each resolution (see the Supplemental Material).
Analytical model and stochastic simulations
To analyze the temporal dynamics of gene expression, we used a stochastic simulation of a two-state kinetic scheme in which the promoter switches between an active and inactive promoter state, and has transcription, translation, mRNA degradation and protein degradation. The equations and rates used in the model, as well as a full description of all simulations we performed to support our data are specified in the Supplemental Material.
Acknowledgments
This work was supported by grants from the European Research Council (ERC) and the U.S. National Institutes of Health (NIH) to E.S. E.S. is the incumbent of the Soretta and Henry Shapiro Career Development Chair. D.D. was supported by FP7 FET Open project “DynaNets” (EU Grant Agreement no. 233847), “ViroLab” (EU project no. IST-027446), and by an EMBO Short-Term Fellowship and a travel grant from the BioRange program of the Netherlands Bioinformatics Centre (NBIC) and Netherlands Genomics Initiative (NGI). We thank Ilya Soifer and Gil Hornung from Naama Barkai's laboratory for their assistance.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.149096.112.
References
- Bai L, Charvin G, Siggia ED, Cross FR 2010. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell 18: 544–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N 2006. Noise in protein expression scales with natural protein abundance. Nat Genet 38: 636–643 [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kaern M, Cantor CR, Collins JJ 2003. Noise in eukaryotic gene expression. Nature 422: 633–637 [DOI] [PubMed] [Google Scholar]
- Blake WJ, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS 2006. Stochastic protein expression in individual cells at the single molecule level. Nature 440: 358–362 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim Y-J 2009. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat Genet 41: 498–503 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie X 2006. Linking stochastic dynamics to population distribution: An analytical framework of gene expression. Phys Rev Lett 97: 1–4 [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Hornung G, Bar-Ziv R, Rosin D, Tokuriki N, Tawfik DS, Oren M, Barkai N 2012. Noise–mean relationship in mutated promoters. Genome Res 22: 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ 2005. Stochasticity in gene expression: From theories to phenotypes. Nat Rev Genet 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Kepler TB, Elston TC 2001. Stochasticity in transcriptional regulation: Origins, consequences, and mathematical representations. Biophys J 81: 3116–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri N, O'Shea EK 2007. Living with noisy genes: How cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct 36: 413–434 [DOI] [PubMed] [Google Scholar]
- Murphy KF, Balazsi G, Collins JJ 2007. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci 104: 12726–12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KF, Adams RM, Wang X, Balázsi G, Collins JJ 2010. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res 38: 2712–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G 2009. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci 106: 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J, Ehrenberg M 2000. Random signal fluctuations can reduce random fluctuations in regulated components of chemical regulatory networks. Phys Rev Lett 84: 5447–5450 [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A 2009. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys 38: 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E 2012. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet 44: 743–750 [DOI] [PubMed] [Google Scholar]
- Ross IL, Browne CM, Hume DA 1994. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol Cell Biol 72: 177–185 [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U 2006. Variability and memory of protein levels in human cells. Nature 444: 643–646 [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science 332: 472–474 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Valouev A, Ichikawa J, Tonthat T, Song JS, Liu X, Liu XS, Mavrich TN, Ioshikhes IP, et al. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T-L, Maheshri N 2010. Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science 327: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Zeevi D, Sharon E, Lotan-pompan M, Lubling Y, Shipony Z, Raveh-sadka T, Keren L, Levo M, Weinberger A, Segal E 2011. Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Res 21: 2114–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]