Abstract
The single nucleotide polymorphism, rs2866164, in the MTP gene, has been associated with human longevity but has not been validated by subsequent longevity studies. Using our population of Ashkenazi Jews, we find that the MTP CC genotype is significantly overrepresented in centenarians and their offspring, as compared with controls (p < .05). However, when we examined MTP CC genotype frequency pattern with aging, we observed a monotonic decline between ages 55–85 years followed by a dramatic enrichment after age 90 years, forming a U-shape pattern (p < .05). Furthermore, the MTP CC genotype was buffered by three validated longevity genotypes (p < .05). This buffering effect was found to confer an enrichment of the MTP CC genotype in centenarians, whereas their absence in CC controls resulted in poorer survivorship (p < .05). Thus, we conclude that MTP CC is a buffered-deleterious genotype and that assessing genotype frequency across aging is essential for discerning longevity from buffered-deleterious genotypes.
Key Words: Genetics, Aging, Microsomal triglyceride transfer protein, Buffering mechanism
Microsomal triglyceride transfer protein (MTP) is a lipid transfer protein found in the liver and intestine and is necessary for the proper assembly of apolipoprotein B–containing lipoproteins, very low-density lipoproteins (VLDL), and chylomicrons (1). Gene association studies have linked haplotypes and single nucleotide polymorphisms (SNPs) in the MTP gene with several diseases and risk factors including fatty liver disease (2), atherosclerotic risk factors (3), type 2 diabetes (4), and blood pressure (5). In light of its pleiotropic effects and harmful associations with many diseases, it was not surprising that MTP also surfaced as one of the first genes linked to exceptional human longevity (6).
Using sibpair analysis on centenarians and their siblings, Puca and colleagues (7) performed the first genomic screening for exceptional human longevity and reported linkage with a region on chromosome 4. Fine mapping of the candidate region uncovered two SNPs, Q95H [rs61733139], and rs2866164 in the MTP gene (6). By comparing a population of exceptionally long-lived individuals residing in the United States (mean age 100.8 years) to a younger U.S. control cohort (mean age 38.6 years), it was found that the minor allele of SNP rs2866164 (G) was the risk allele, as it was underrepresented in the long-lived population. However, several studies in other long-lived populations have since failed to detect any association between MTP and longevity (8–12) and have led several to conclude that the findings of Geesaman and colleagues (6) were confounded by population stratification in control participants.
Because most humans die between ages 60 and 100 years, with fewer “survivors” populating each successive decade, one may assume that a monotonic trend (increase or decrease) in genotype prevalence with age suffices to discern potential beneficial longevity genes from deleterious genes. In the absence of other diseases, such a trend could provide a unique opportunity for genetic analysis of longevity. Indeed, a monotonic increase in genotype frequency with aging suggests that it may confer a survival advantage, as we and others have shown for four genotypes: CETP (VV) [rs5882] (13), APOC3 (CC) [rs2542052] (14), AdipoQ (del/del APM1+2019) [rs56354395] (15), and most recently, FOXO3a (16,17). Conversely, it is expected that higher prevalence of harmful genotypes in a cohort will result in increasing rates of mortality in that cohort, leading to a monotonic decline in deleterious genotype frequency over time. However, it has been observed that centenarian populations are carriers of some variants of deleterious genes, even at levels comparable to or greater than younger control groups (18,19).
Recently, we have addressed the limitations of comparing genotypic frequency between two groups (young controls and long-lived individuals) as well as monotonic trend analysis in a population containing participants at multiple ages (18). In order to aid in the discernment between longevity genotypes and deleterious genotypes, we proposed the Buffering Mechanisms in Aging hypothesis. This hypothesis states that an important property of many longevity genes (which are enriched in centenarians) is their ability to buffer against the harmful effects of deleterious genotypes (18). Specifically, in a subpopulation lacking longevity genotypes, the prevalence of a deleterious genotype will decline monotonically with age, whereas in a subpopulation endowed with a favorable longevity genotype(s), the prevalence of a buffered-deleterious genotype is expected not to vary or even increase with age. Indeed, we have shown that the deleterious variant of two age-related disease genes, Klotho and Lp(a), demonstrates a monotonic decline with age, but this is followed by a marked enrichment in prevalence among those surviving beyond 80 years, forming a U-shape pattern with aging (18). Importantly, this pattern in genotype frequency would have been missed if only a young and old cohort were compared.
To determine whether the MTP gene may be a longevity or buffered-deleterious genotype, we characterized the prevalence of SNP rs2866164 in our well-characterized population of Ashkenazi Jew centenarians, their offspring, and age-matched Ashkenazi controls. We then tested its relationship to the metabolic phenotype and performed a buffering analysis for MTP genotype on survivorship. Here, we show that the CC variant of SNP rs2866164, in the MTP gene, is indeed a deleterious genotype that is buffered by longevity genotypes.
METHODS
Study Population
A total of 205 Ashkenazi Jewish centenarians (probands, 141 women, median age = 97 years and 64 men, median age = 97 years), their offspring (n = 145 total, 80 women, median age = 69 years and 65 men, median age = 68 years), and a control group of 288 Ashkenazi Jews (167 women, median age = 74 years and 121 men, median age = 75 years), not family related to the earlier participants, but from the same geographic area, were similarly assessed. The study population was identified from the Longevity Genes Study at Albert Einstein College of Medicine, as described elsewhere (13,14), and the Einstein Aging Study, a community-based longitudinal study designed to identify predictive factors for cognitive decline and dementia (20,21). Participants were recruited through publicity, and stated age was verified by checking birth certificates or U.S. passports in all participants. Medical history, demographic characteristics, and clinical data were obtained uniformly using a structured questionnaire. All participants underwent a physical examination and provided a blood sample. We performed identity by descent analysis to exclude cryptic relatedness among control–probands and control–offspring. In addition, Affymetrix 6.0 was used to classify Ashkenazi origin (four grandparents) of all participants by principle component analysis. Informed written consent was obtained in accordance with the policy of the Committee on Clinical Investigation of the Albert Einstein College of Medicine.
Analysis of SNPs
The MTP gene spans 60 kb with 18 exons and is located on the long arm of chromosome 4. We selected the rs2866164 SNP of the MTP gene and genotyped it in probands, offspring, and controls, using the PSQ HS 96A Pyrosequencer according to the manufacturer’s recommendations (Pyrosequencing, Uppsala, Sweden; www.pyrosequencing.com). Briefly, a polymerase chain reaction product was generated from a primer pair that included one primer covalently coupled to biotin, the biotinylated template was bound to streptavidin-coated Sepharose High Performance beads, and this mixture was then annealed to a sequencing primer. Stepwise elongation of a sequencing primer strand upon sequential addition of a specified sequence of deoxynucleotide triphosphates and the degradation of nucleotides by apyrase were carried out simultaneously. As the sequencing reaction progressed, the DNA strand was extended, and the sequence was determined from the measured signal output of light upon nucleotide incorporation. The resulting peaks in the pyrogram were analyzed using Pyrosequencing software. Primer sequences are available from the authors upon request. Error rates based on blind replicates were estimated to be 1.5%.
Lipids and Lipoproteins
Total plasma cholesterol, high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein-cholesterol (LDL-C), and apolipoprotein A-I (ApoA-I) and B (Apo-B) concentrations for the participants were performed by standard automated methods at the clinical laboratories of Montefiore Medical Center, Bronx, NY. Plasma VLDL, LDL, triglycerides (TG) and HDL subclass levels, mean particle sizes, and chylomicrons were determined for all participants by nuclear magnetic resonance spectroscopy at LipoScience Inc. (Raleigh, NC) as previously described (22,23). The LDL and HDL subclass distributions and particle sizes determined by nuclear magnetic resonance are highly correlated with those measured by gradient gel electrophoresis and density gradient ultracentrifugation (24,25). The analytical reproducibility (given by the coefficient of variation) of LDL and HDL size is less than 0.5% (22), and the stability on repeated drawing for LDL size was 0.9% and for HDL size was 1.1%.
U-Shape Trend Analysis
In order to determine whether the MTP CC SNP is a buffered-deleterious genotype, we utilized the Buffering Mechanisms in Aging approach, whose statistical considerations are described in detail elsewhere (18). Briefly, controls and probands were grouped together, and a generalized linear model was used to fit the pattern of CC genotype prevalence with age. We confirmed the existence of a significant U-shape pattern with age using a binomial model incorporating both linear and quadratic terms for age and testing for the significant quadratic component using the equation: P(Y = 1) = b 0 + b 1age + b 2age2, where Y = 1 was indicative of an individual having the MTP CC genotype. Maximum likelihood estimates of the coefficients b 0, b 1, and b 2 were then obtained by the Fisher scoring method, and the significance of the quadratic term (ie, U-shape trend) was confirmed using the likelihood ratio test prior to performing interaction analysis on the MTP CC genotype.
Buffering Analysis
Upon confirmation of a significant U-shape trend for the CC genotype in controls and probands, we then combined all offspring and control participants (excluding probands) harboring the CC genotype and subgrouped them by presence or absence of one or more longevity genotypes, including CETP (homozygosity for the 405 V allele, VV) [rs5882] (13), APOC3 (homozygosity for the −641 C allele, CC) [rs2542052] (14), and ADIPOQ (del/del APM1+2019) [rs56354395] (15). We next performed logistic regression to test the interaction between the trend of the MTP CC genotype across age with or without the presence of favorable longevity genotypes. Because we did not detect any additive effect of harboring more than one favorable genotype, any carrier of one or more of the three longevity genotypes were combined as a single group. To test the significance of interaction effects between age and genotype, the model testing for main effect only and the model testing for the interaction effects were compared using the log-likelihood ratio test.
Statistical Analysis
We used JMP software, version 9 (SAS Institute Inc., Cary, NC) for data analysis. Plasma VLDL and chylomicron were tested for normality of distribution using Shapiro–Wilk test and D’Agostino’s K-squared test and were found to violate the normality assumption. Thus, a comparison between nonparametric variables was performed using Kruskal–Wallis one-way analysis of variance by ranks and Mann–Whitney rank sum test. Data are presented as means ± SE unless cited differently, and a p value less than or equal to .05 was considered statistically significant. The participants’ survival distribution was estimated by the Kaplan–Meier method, and the significance of the difference in survival distribution among the MTP genotypes was tested by means of a log-rank test. Wilcoxon statistics were calculated to test homogeneity between groups, and cox proportional hazard models were used for survival analysis. Average time of follow-up for participants was 8.5 ± 0.3 and 8.7 ± 0.3 years for the “buffered” and “unbuffered” groups, respectively. Approximately 70% of the buffered groups and 64% of the unbuffered groups were still alive at the time of the analysis.
RESULTS
Participant Characteristics for Aging Cohorts
Characteristics of Ashkenazi Jew centenarians, their offspring, and Ashkenazi controls are presented in Table 1. There were no significant differences observed between offspring and controls for glucose, insulin, apolipoproteins, LDL-C, or LDL particle size. Offspring did have greater levels of TG (p < .001) and tended to have elevated chylomicrons (p = .06), VLDL-TG (p = .06), and total cholesterol (p = .09), as compared with controls. However, offspring also had greater total unadjusted HDL-C (p < .01) and smaller HDL particle size (p < .001), though HDL-C was no longer significant after adjusting for several covariates.
Table 1.
Demographic and Metabolic Characteristics of Ashkenazi Probands, Offspring, and Controls
| Trait | Probands, n = 205 | Offspring, n = 145 | Controls, n = 288 | p Value, Offspring vs Control | |
| * | † | ||||
| Female (%) | 69 | 55 | 58 | 0.58 | |
| Age, y | 98 ± 0.2 | 68.2 ± 0.6 | 74.0 ± 0.5 | <.0001 | <.0001 |
| Age range, y | 95–109 | 53–92 | 44–93 | ||
| Glucose, mg/dL | 101 ± 2 | 94 ± 3 | 93 ± 2 | .76 | .57 |
| Insulin, mg/dL | 27.9 ± 3.3 | 22.6 ± 3.2 | 17.8 ± 1.8 | .19 | .14 |
| CRP | 0.76 ± 0.18 | 0.44 ± 0.08 | 0.34 ± 0.03 | .16 | .11 |
| Cholesterol, mg/dL | 189 ± 3 | 209 ± 3 | 200 ± 3 | .03 | .09 |
| Triglycerides, mg/dL | 139 ± 5 | 149 ± 8 | 126 ± 4 | .002 | 2e-3 |
| Apo-A1, mg/dL | 141 ± 3 | 156 ± 3 | 156 ± 3 | .94 | .59 |
| Apo-B, mg/dL | 100 ± 2 | 104 ± 2 | 101 ± 2 | .38 | .52 |
| VLDL-TG, mg/dL | 85 ± 5 | 82 ± 7 | 74 ± 3 | .17 | .06 |
| VLDL particle size, nm | 49.4 ± 0.9 | 48.8 ± 1.3 | 47.1 ± 0.3 | .38 | .52 |
| LDL, mg/dL | 109 ± 5 | 116 ± 3 | 117 ± 2 | .8 | .76 |
| LDL1, mg/dL | 78.6 ± 4.3 | 69.6 ± 4.6 | 70.6 ± 3.1 | .85 | .79 |
| LDL3, mg/dL | 13.3 ± 2.6 | 29.3 ± 3.5 | 29.7 ± 2.4 | .93 | .94 |
| LDL particle size, nm | 21.4 ± 0.1 | 21.0 ± 0.1 | 21.1 ± 0.1 | .55 | .91 |
| HDL, mg/dL | 55.5 ± 1.2 | 63.8 ± 1.6 | 58.4 ± 1.1 | .005 | .13 |
| Large HDL, mg/dL | 32.2 ± 1.8 | 29.4 ± 1.8 | 32.1 ± 1.2 | .21 | .47 |
| Small HDL, mg/dL | 15.3 ± 0.6 | 20.3 ± 0.7 | 16.0 ± 0.5 | <.0001 | .18 |
| HDL particle size, nm | 9.47 ± 0.05 | 9.10 ± 0.05 | 9.30 ± 0.05 | .005 | .01 |
| Chylomicrons, mg/dL | 2.65 ± 0.56 | 2.27 ± 0.84 | 0.73 ± 0.16 | .01 | .06 |
Notes: Data are expressed as means ± SE. Crude means are shown. TG, which were not normally distributed, were log-transformed for analysis but presented as untransformed raw values. LDL = low-density lipoprotein; HDL = high-density lipoprotein; VLDL = very low-density lipoproteins.
p Values between offspring versus control unadjusted.
p Values between offspring versus control adjusted for age and sex (glucose [mg/dL], insulin [mg/dL], CRP, cholesterol [mg/dL], triglycerides [mg/dL], VLDL-TG [mg/dL], LDL [total levels, 1 and 3; mg/dL], HDL [total levels, large and small; mg/dL], VLDL particle size [nm], LDL particle size [nm], HDL particle size [nm], Chylomicron [mg/dL]).
MTP Genotype and Allele Frequency in Centenarians, Offspring, and Controls
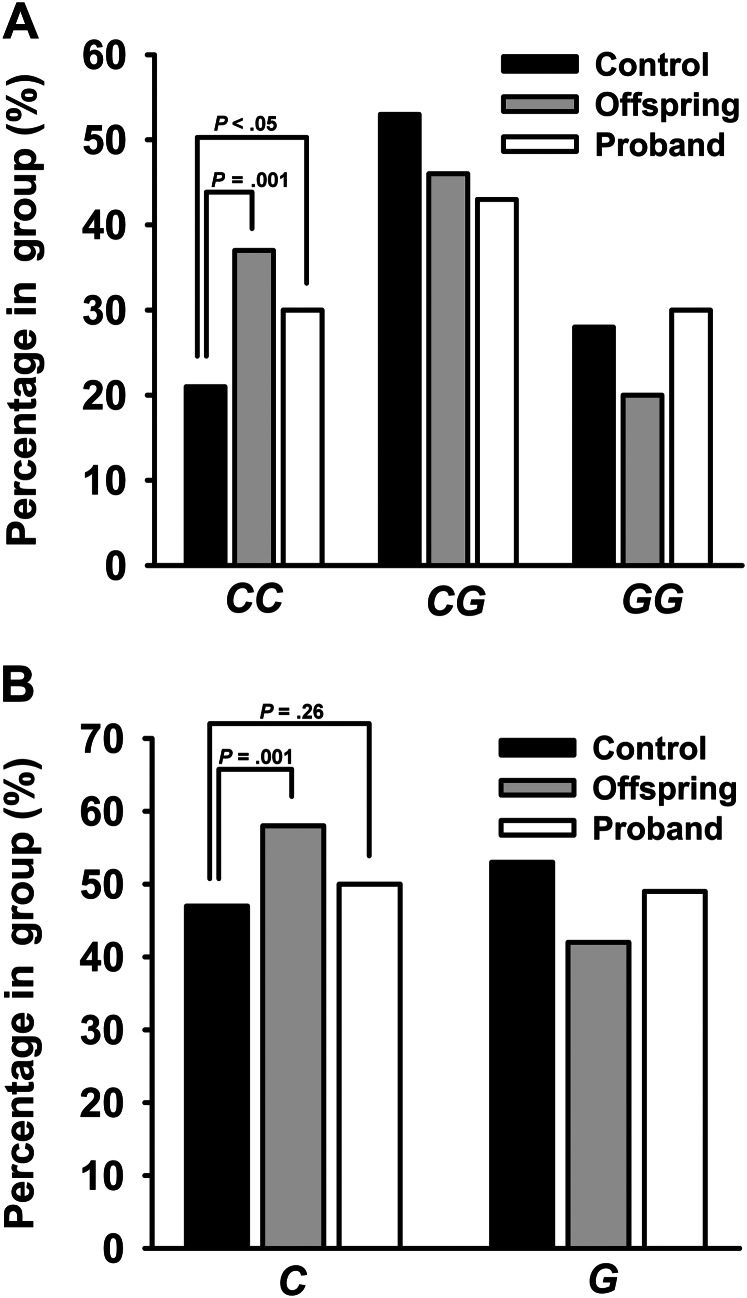
The prevalence of the MTP rs2866164 SNP and genotype frequency among centenarians, offspring, and controls are presented in Figure 1A and B, respectively. As compared with Ashkenazi controls, the CC genotype was more prevalent in both centenarians and their offspring (p < .05, Figure 1A), but no significant differences were observed in CG or GG genotype prevalence among groups. When assessing allele frequency in all three groups, the C allele was found to be overrepresented in offspring (p = .001), as compared with controls, but no significant difference was found between centenarians and controls (p = .26, Figure 1B).
Figure 1.
MTP genotype frequencies in Ashkenazi centenarians, their offspring, and Ashkenazi controls. (A) The frequency of homozygosity (CC) for the MTP rs2866164 SNP of MTP in probands (n = 126), offspring (n = 145), and Ashkenazi controls (n = 288). The frequency of the CC genotype is 1.5- and 1.6-fold increased in offspring (p < .05) and probands (p = .001), respectively, compared with controls. (B) The frequency of the C allele for the MTP rs2866164 SNP in probands (n = 126), offspring (n = 145), and controls (n = 288). The frequency of the C allele is significantly increased in offspring (p = .001), but not in probands (p = .26), as compared with controls.
MTP CC Genotype Is Associated With an Unfavorable Lipid Profile
Characteristics of offspring and controls by MTP genotype (CC vs CG/GG) are presented in Table 2. There were no significant differences observed for glucose, insulin, cholesterol, apolipoproteins, VLDL particle size, HDL-C, or LDL-C (Table 2). However, individuals with the CC genotype had significantly greater levels of total TG (p < .05, Figure 2A), VLDL (p < .05, Figure 2B), and chylomicrons (p < .05, Figure 2C), even after adjustment for confounders.
Table 2.
Demographic and Metabolic Characteristics of Ashkenazi Offspring and Controls Categorized by MTP Genotype
| Trait | CC, n = 113 | CG/GG, n = 320 | p Value, CC vs CG/GG | |
| * | † | |||
| Female (%) | 51 | 59 | .15 | .14 |
| Age, y | 70.2 ± 0.8 | 73.1 ± 0.5 | .004 | .004 |
| Age range, y | 44–92 | 49–93 | ||
| Glucose, mg/dL | 90 ± 4 | 95 ± 2 | .25 | .26 |
| Insulin, mg/dL | 19.2 ± 3.2 | 20.8 ± 2.3 | .68 | .33 |
| CRP | 0.39 ± 0.07 | 0.37 ± 0.04 | .73 | .53 |
| Cholesterol, mg/dL | 204 ± 4 | 203 ± 2 | .78 | .61 |
| Apo-A1, mg/dL | 154 ± 3 | 157 ± 2 | .45 | .42 |
| Apo-B, mg/dL | 105 ± 3 | 102 ± 2 | .37 | .40 |
| VLDL particle size, nm | 48.9 ± 1.7 | 47.6 ± 0.8 | .45 | .54 |
| LDL, mg/dL | 117 ± 4 | 116 ± 2 | .85 | .54 |
| LDL1, mg/dL | 67.2 ± 5.6 | 71.4 ± 2.9 | .47 | .72 |
| LDL3, mg/dL | 32.8 ± 4.2 | 28.2 ± 2.3 | .3 | .46 |
| LDL particle size, nm | 20.9 ± 0.1 | 21.0 ± 0.1 | .34 | .77 |
| HDL, mg/dL | 59.5 ± 1.7 | 60.8 ± 1.1 | .53 | .4 |
| Large HDL, mg/dL | 28.8 ± 2.1 | 32.1 ± 1.2 | .17 | .72 |
| Small HDL, mg/dL | 18.2 ± 0.7 | 17.5 ± 0.5 | .66 | .92 |
| HDL particle size, nm | 9.06 ± 0.05 | 9.16 ± 0.03 | .15 | .48 |
Notes: Data are expressed as means ± SE. Crude means are shown. TG, which were not normally distributed, were log-transformed for analysis but presented as untransformed raw values. HDL = high-density lipoprotein; LDL = low-density lipoprotein; VLDL = very low-density lipoproteins.
p Values between CC carriers and CG/GG carriers unadjusted.
p Values between CC carriers and CG/GG carriers adjusted for age and sex (glucose [mg/dL], insulin [mg/dL], CRP, cholesterol [mg/dL], triglycerides [mg/dL], Apo-A1 [mg/dL], ApoB [mg/dL], VLDL particle size [nm], LDL [total levels, 1 and 3; mg/dL], LDL particle size [nm], HDL [total levels, large, and small; mg/dL], HDL particle size [nm]).
Figure 2.
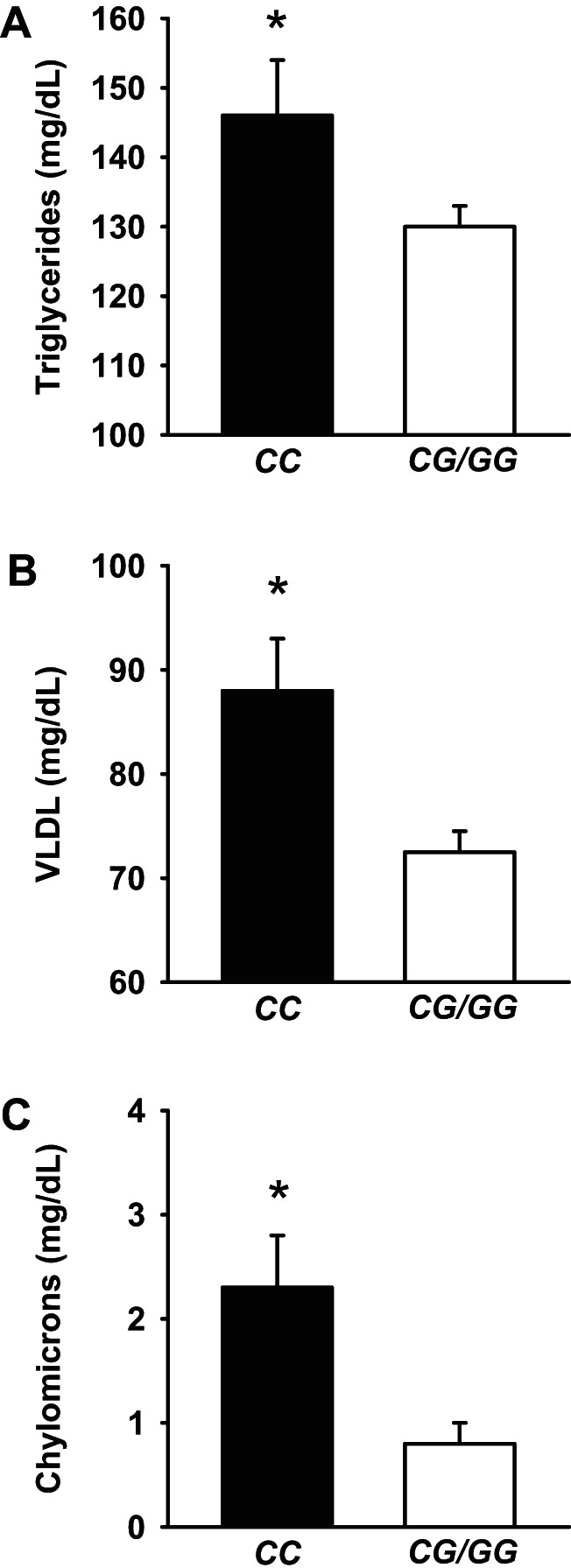
Concentration of triglycerides (TG) and two lipoproteins, VLDL and chylomicrons, which are under the physiologic control of MTP, according to MTP rs2866164 genotype (CC vs CG/GG) in offspring and controls. Results are adjusted for age and gender. (A) Total plasma TG, (B) plasma VLDL, and (C) chylomicrons. Values are means ± SE. *p < .05.
Effect of Buffering Genotypes on MTP CC Genotype Frequency During Normal Aging
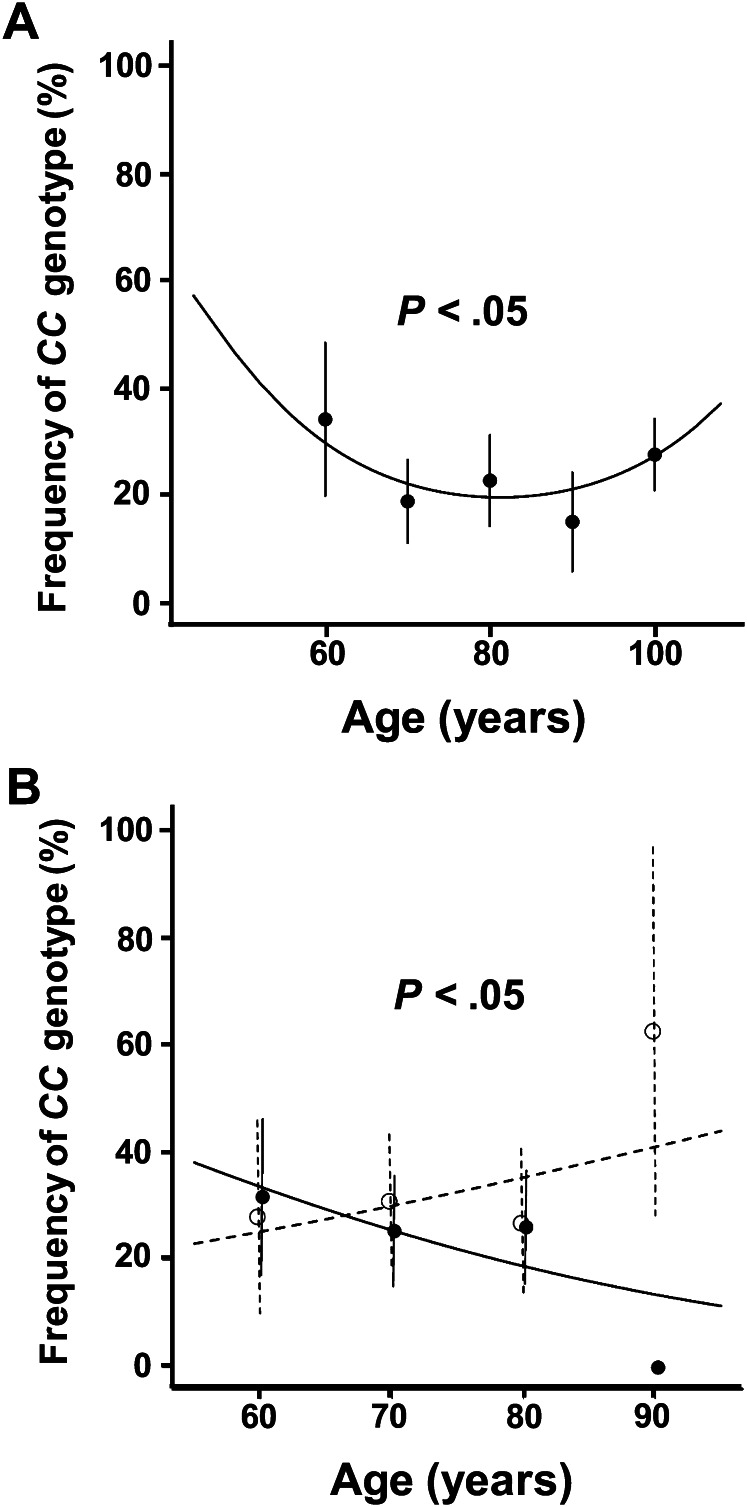
The frequency of the MTP CC genotype with normal aging in controls and probands is shown in Figure 3A. Treating age as a continuous variable to describe the distribution pattern of CC over the life course, we found that the prevalence of the CC genotype demonstrated a monotonic decline of ∼16%, from 38% to 22% between 60 and 80 years of age, respectively. However, between 80 and 100 years of age, the prevalence markedly increased by nearly 10% to near 30% prevalence, resulting in a U-shape frequency pattern with aging (p < .05). Using interaction analysis, when the frequency of the MTP CC genotype was assessed in the absence of favorable genotypes (AdipoQ [rs56354395], CETP [rs5882], and ApoC3 [rs2542052]) in offspring and controls, we observed a monotonic decline with aging from ∼33% at 60 years to near 0% survivorship by age 90 years (Figure 3B, p < .05). However, in individuals possessing one or more favorable longevity genotypes, the prevalence of the MTP CC genotype was relatively unchanged from the sixth to eighth decade of life but was followed by a marked enrichment to near 60% by 90 years of age (Figure 3B), as predicted by the buffering hypothesis (18).
Figure 3.
Prevalence of homozygosity (CC) for the MTP rs2866164 SNP according to age and adjustment for buffering genotypes in Ashkenazi Jews. (A) Frequency of MTP CC genotype in controls and probands (range 55–104 years) by the specified age ranges with the indicated number of subjects for each age range: 55–64 years = 44, 65–74 years = 101, 75–84 years = 97, 85–94 years = 60, and 95–104 years = 179. (B) The frequency of homozygosity for the CC genotype of the MTP rs2866164 SNP according to age with or without buffering genes in offspring and controls (see Methods for description of CETP [rs5882], ApoC3 [rs2542052], AdipoQ [rs56354395]). Filled circles (and continuous line) indicate prevalence of the CC homozygosity in those that did not harbor buffering genes. Open circles (and dashed line) indicate prevalence of the CC homozygosity in participants that harbor buffering genes. Frequency of MTP CC genotype either with or without buffering genes are given by the specified age ranges with the indicated number of subjects for each age range (buffered/unbuffered): 55–64 years = 32/51, 65–74 years = 50/72, 75–84 years = 42/58, 85–92 years = 9/22.
MTP CC Genotype and Survivorship in Ashkenazi Offspring and Controls
Kaplan–Meier curves comparing survivorship between MTP genotypes are presented in Figure 4A and B. As compared with Ashkenazi offspring and controls with either CG/GG genotypes, those with the CC genotype demonstrated significantly worse survival (p < .05, Figure 4A). However, after adjustment for the presence of favorable longevity genotypes, this difference in mortality no longer remained (p = .53, Figure 4B).
Figure 4.
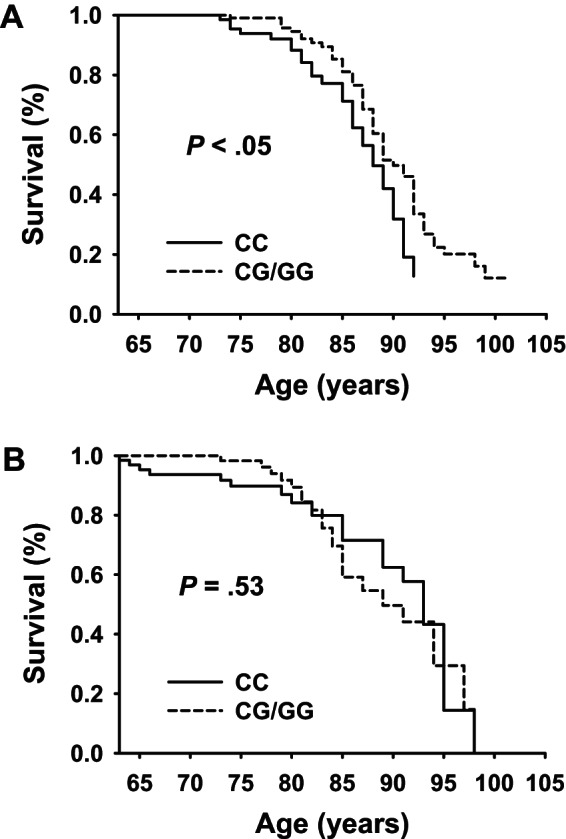
Survival Analysis according to MTP genotype in Ashkenazi offspring and controls. To describe the relationship between genotype and death, we plotted the Kaplan–Meier survival function estimates of controls by MTP genotype based upon whether or not individuals harbored a buffering gene (see Methods for description of CETP [rs5882], ApoC3 [rs2542052], and AdipoQ [rs56354395]). (A) Kaplan–Meier curve of offspring and control Ashkenazi’s without buffering genes; CC genotype (n = 43) was associated with worse survivorship than CG/GG genotypes (n = 126) in offspring and controls without buffering genes (Wilcoxon, p < .05), (B) Kaplan–Meier curve of offspring and control Ashkenazi’s with buffering genes (CC genotype, n = 32; CG/GG genotype, n = 71). There was no significant difference in survivorship between genotypes in individuals with buffering genes (Wilcoxon, p = .53).
DISCUSSION
Here, we show that the prevalence of the MTP CC genotype is enriched in centenarians and their offspring, as compared with Ashkenazi Jew controls. Paradoxically, this genotype was associated with an unfavorable lipid profile, including greater concentrations of TG, VLDL, and chylomicrons, all known risk factors for cardiovascular and metabolic diseases. This paradox between phenotype and genotype is explained by the fact that the MTP CC genotype demonstrated a monotonic decline with aging until ∼80 years of age, followed by a marked increase in centenarians, forming a U-shape pattern with aging.
Offspring and individuals belonging to the control group with the MTP CC genotype across ages were then categorized by presence or absence of one or more known longevity genotypes {CETP (VV) [rs5882] (13), APOC3 (CC) [rs2542052] (14), and AdipoQ (del/del APM1+2019) [rs56354395] (15)}. Absence of these genotypes resulted in a monotonic decline in CC prevalence to near 0% by 90 years, whereas a sharp rise in prevalence was observed in those harboring longevity genotypes beyond 80 years old. Finally, we show that control and offspring without any of the three favorable longevity genotype variants, but possessing the MTP CC genotype, demonstrated a poorer survivorship than those with the MTP CG/GG genotype. These findings lend support to the hypothesis that MTP CC is in fact a deleterious genotype, whose effects can be buffered by longevity genes. However, the exact biological mechanism(s) whereby these favorable genotypes attenuate the harmful effect(s) of the MTP CC genotype are unclear.
On the surface, our results seem to be in agreement with those of an initial fine-mapping experiment which first linked an overrepresentation of the major allele (C) of SNP rs2866164 in the MTP gene with longevity (6). However, our data clearly suggest that in spite of its enrichment in centenarians and their offspring, harboring two copies of this allele without buffering genes is not “good” for aging, but rather is detrimental. Furthermore, our results provide a “proof of concept” regarding the necessity to obtain genotype prevalence data over the life course coupled with functional analysis, in order to definitively distinguish beneficial and deleterious genotypes. Indeed, we have previously shown that the hazards posed by a deleterious variant of the Lp(a) gene is buffered by the favorable CETP VV genotype [rs5882] (18). Likewise, here, we demonstrate that the deleterious effect of the MTP CC genotype can be mitigated by any of the following favorable longevity genotypes, CETP (VV) [rs5882], APOC3 (CC) [rs2542052], and AdipoQ (del/del APM1+2019) [rs56354395].
Although our results were able to somewhat substantiate those of Geesaman and colleagues (6), albeit with a different interpretation, the fact that five separate follow-up studies, utilizing unique populations of long-lived individuals and controls (8–12), failed to detect any difference among groups deserves further clarification. First, consistent with these follow-up studies, we also fail to see any significant difference in allele frequency between younger controls and probands. However, our results demonstrate that a stark enrichment of the CC genotype exists in Ashkenazi centenarians and offspring, as compared with controls. We attribute this finding in part to the unique risk posed by the CC genotype, as opposed to those harboring one or two copies of the G allele, to mortality in participants without longevity genotypes.
A second important consideration that deserves further explanation is the issue of comparing two data points for the purpose of identifying either deleterious or longevity genotypes. Most human longevity studies are structured as case–control analyses, with only a small range around the two selected ages. Thus, these populations are unable to perform trend analysis, which is only possible in our population, due to the distribution of participants across this entire segment of the life span, where age-related mortality rate is greatest (ie, 55–100+ years). Indeed, as we have already mentioned, some deleterious genotype frequencies show a propensity to form a U-shape pattern with aging, and this can be a major concern in regards to distinguishing longevity and deleterious genotypes.
For example, comparing only case–control analysis between 60-year-old individuals and centenarians in our study, which are on opposite ends of the “U shape”, would have led to the conclusion that genotype frequency changed very little with aging (Figure 3A). In contrast, comparing 70 years old to centenarians could have led us to conclude that the CC genotype is a longevity gene due to its relative enrichment in Ashkenazi probands. On the contrary, as is evident from the pattern in genotype frequency during aging (Figure 3A), CC genotype prevalence begins to decrease at ages where the mortality rate is expected to increase. However, as individuals approach exceptional old age, there is a marked enrichment in the CC genotype, not because this genotype suddenly becomes beneficial but rather due to the buffering effect of longevity genotypes, which are also becoming enriched (Figure 3B).
The observation that the MTP CC genotype is associated with a risky lipid profile and adversely affects survival in control Ashkenazi’s without longevity genes is in agreement with our prior studies. Indeed, we first reported that a hallmark feature of individuals with exceptional longevity was a healthy lipoprotein profile, and that this phenotype was heritable (13). Furthermore, this phenotype was associated with increased homozygosity for the I405V variant in CETP [rs5882] (13), an enzyme involved in reverse cholesterol transport. Likewise, we have observed that a variant in the APOC3 gene [rs2542052], which expresses an apolipoprotein involved in TG metabolism, is associated with lower APOC3 levels, larger HDL and LDL particle sizes, and longevity (14). Moreover, we have implicated the VV variant for CETP [rs5882] as a modulator of age-related cognitive function (26). Taken together, these findings suggest that gene variants involved in lipoprotein metabolism play an important role in healthy aging and longevity.
In summary, we show that enrichment of the MTP CC genotype in centenarians and their offspring, relative to controls, is misleading unless the pattern in genotype frequency during aging is observed. Indeed, as opposed to prior observations, which had conflicting conclusions regarding the role of this genotype in human longevity, we show that the CC genotype demonstrates all the hallmarks of a buffered-deleterious genotype. We also demonstrate that a functional consequence of the CC genotype in controls without longevity genes includes poorer survivorship. Finally, this study is a vital proof of concept for the importance of populating the “age axis” as well as accounting for buffering mechanisms when attempting to discern longevity genotypes from deleterious genotypes.
FUNDING
This work has been supported by grants from the Paul Beeson Physician Faculty Scholar in Aging Award, the Ellison Medical Foundation Senior Scholar Award, the General Clinical Research Center (M01-RR12248), and the Diabetes Research and Training Center (DK20541) at the Albert Einstein College of Medicine and grants AG027734, AG028872, RR12248, and M01RR12248 from the National Institutes of Health. D.M.H. is supported by an National Institute of Aging-sponsored K99 award (AG037574).
Acknowledgments
We are indebted to all participants and their families for their commitment and enthusiasm. We are also grateful to the following institutions that assisted in recruitment: The Hebrew Home for the Aging, Riverdale, NY; Kittay House, Bronx, NY; the Hebrew Home Hospital, West Hartford, CT; and the Jewish Home for the Aged, New Haven, CT.
References
- 1.Gordon DA, Jamil H. Progress towards understanding the role of microsomal triglyceride transfer protein in apolipoprotein-B lipoprotein assembly. Biochim Biophys Acta. 2000;1486:72–83. doi: 10.1016/s1388-1981(00)00049-4. [DOI] [PubMed] [Google Scholar]
- 2.Jun DW, Han JH, Jang EC, et al. Polymorphisms of microsomal triglyceride transfer protein gene and phosphatidylethanolamine N-methyltransferase gene in alcoholic and nonalcoholic fatty liver disease in Koreans. Eur J Gastroenterol Hepatol. 2009;21:667–672. doi: 10.1097/MEG.0b013e3283196adc. [DOI] [PubMed] [Google Scholar]
- 3.Gastaldi M, Diziere S, Defoort C, et al. Sex-specific association of fatty acid binding protein 2 and microsomal triacylglycerol transfer protein variants with response to dietary lipid changes in the 3-mo Medi-RIVAGE primary intervention study. Am J Clin Nutr. 2007;86:1633–1641. doi: 10.1093/ajcn/86.5.1633. [DOI] [PubMed] [Google Scholar]
- 4.Rubin D, Helwig U, Pfeuffer M, et al. A common functional exon polymorphism in the microsomal triglyceride transfer protein gene is associated with type 2 diabetes, impaired glucose metabolism and insulin levels. J Hum Genet. 2006;51:567–574. doi: 10.1007/s10038-006-0400-y. [DOI] [PubMed] [Google Scholar]
- 5.Yamada Y, Ando F, Shimokata H. Association of a microsomal triglyceride transfer protein gene polymorphism with blood pressure in Japanese women. Int J Mol Med. 2006;17:83–88. [PubMed] [Google Scholar]
- 6.Geesaman BJ, Benson E, Brewster SJ, et al. Haplotype-based identification of a microsomal transfer protein marker associated with the human lifespan. Proc Natl Acad Sci U S A. 2003;100:14115–14120. doi: 10.1073/pnas.1936249100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puca AA, Daly MJ, Brewster SJ, et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci U S A. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bathum L, Christiansen L, Tan Q, Vaupel J, Jeune B, Christensen K. No evidence for an association between extreme longevity and microsomal transfer protein polymorphisms in a longitudinal study of 1651 nonagenarians. Eur J Hum Genet. 2005;13:1154–1158. doi: 10.1038/sj.ejhg.5201468. [DOI] [PubMed] [Google Scholar]
- 9.Beekman M, Blauw GJ, Houwing-Duistermaat JJ, Brandt BW, Westendorp RG, Slagboom PE. Chromosome 4q25, microsomal transfer protein gene, and human longevity: novel data and a meta-analysis of association studies. J Gerontol A Biol Sci Med Sci. 2006;61:355–362. doi: 10.1093/gerona/61.4.355. [DOI] [PubMed] [Google Scholar]
- 10.Nebel A, Croucher PJ, Stiegeler R, Nikolaus S, Krawczak M, Schreiber S. No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc Natl Acad Sci U S A. 2005;102:7906–7909. doi: 10.1073/pnas.0408670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neville MJ, Clarke R, Evans JG, Rubinsztein DC, Karpe F. Absence of relationship between MTTP haplotypes and longevity. J Gerontol A Biol Sci Med Sci. 2007;62:202–205. doi: 10.1093/gerona/62.2.202. [DOI] [PubMed] [Google Scholar]
- 12.Novelli V, Viviani Anselmi C, Roncarati R, et al. Lack of replication of genetic associations with human longevity. Biogerontology. 2008;9:85–92. doi: 10.1007/s10522-007-9116-4. [DOI] [PubMed] [Google Scholar]
- 13.Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 14.Atzmon G, Rincon M, Schechter CB, et al. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS Biol. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atzmon G, Pollin TI, Crandall J, et al. Adiponectin levels and genotype: a potential regulator of life span in humans. J Gerontol A Biol Sci Med Sci. 2008;63:447–453. doi: 10.1093/gerona/63.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pawlikowska L, Hu D, Huntsman S, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8(4):460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging. PLoS Comput Biol. 2007;3:e170. doi: 10.1371/journal.pcbi.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijg J, Perls T, Franceschi C, van Orsouw NJ. BRCA1 gene sequence variation in centenarians. Ann N Y Acad Sci. 2001;928:85–96. doi: 10.1111/j.1749-6632.2001.tb05639.x. [DOI] [PubMed] [Google Scholar]
- 20.Verghese J, Lipton RB, Hall CB, Kuslansky G, Katz MJ, Buschke H. Abnormality of gait as a predictor of non-Alzheimer's dementia. N Engl J Med. 2002;347:1761–1768. doi: 10.1056/NEJMoa020441. [DOI] [PubMed] [Google Scholar]
- 21.Verghese J, Lipton RB, Katz MJ, et al. Leisure activities and the risk of dementia in the elderly. N Engl J Med. 2003;348:2508–2516. doi: 10.1056/NEJMoa022252. [DOI] [PubMed] [Google Scholar]
- 22.Otvos JD. Measurement of lipoprotein subclass profiles by nuclear magnetic resonance spectroscopy. Clin Lab. 2002;48:171–180. [PubMed] [Google Scholar]
- 23.Otvos JD, Jeyarajah EJ, Bennett DW, Krauss RM. Development of a proton nuclear magnetic resonance spectroscopic method for determining plasma lipoprotein concentrations and subspecies distributions from a single, rapid measurement. Clin Chem. 1992;38:1632–1638. [PubMed] [Google Scholar]
- 24.Blake GJ, Otvos JD, Rifai N, Ridker PM. Low-density lipoprotein particle concentration and size as determined by nuclear magnetic resonance spectroscopy as predictors of cardiovascular disease in women. Circulation. 2002;106:1930–1937. doi: 10.1161/01.cir.0000033222.75187.b9. [DOI] [PubMed] [Google Scholar]
- 25.Grundy SM, Vega GL, Otvos JD, Rainwater DL, Cohen JC. Hepatic lipase activity influences high density lipoprotein subclass distribution in normotriglyceridemic men. Genetic and pharmacological evidence. J Lipid Res. 1999;40:229–234. [PubMed] [Google Scholar]
- 26.Barzilai N, Atzmon G, Derby CA, Bauman JM, Lipton RB. A genotype of exceptional longevity is associated with preservation of cognitive function. Neurology. 2006;67:2170–2175. doi: 10.1212/01.wnl.0000249116.50854.65. [DOI] [PubMed] [Google Scholar]