Abstract
FOXO3 is generally recognized as a “master” gene in aging since its association with longevity has been replicated in multiple organisms and human populations. A group of single nucleotide polymorphisms in linkage disequilibrium with a coding region has been associated with human longevity, but the actual functional variant is unidentified. Therefore, we sequenced the coding region in our long-lived Japanese American population in order to enhance resources for fine mapping this region. We demonstrate that of 38 published variants, 6 are misalignments with homologous nonallelic sequences from FOXO3B (ZNF286B), a pseudogene on a different chromosome; 2 are attributable to ZNF286B only, and the remaining 30 were unconfirmed, indicating that they are very rare and not likely involved in longevity. Furthermore, we identified a novel, unique, nonsynonymous coding variant in exon 3 (Gly566Ala; rs138174682) that is prevalent in multiple ethnic groups but appeared too rare for major longevity effects in our study populations.
Key Words: Aging, FOXO3, Genetic, Human longevity
The world’s population is rapidly aging and this has enormous implications for society. Therefore, it is vitally important to understand how and why we age in order to enhance our odds of aging with better health and less disability (1). Genetics may account for 10%–50% of the variability in human life span, depending upon the population, but little is known about the effector genes (2–6). Studies of model organisms have identified several evolutionarily conserved biological pathways that have major influence on aging and age-related traits (6–8). Daf-16, an evolutionarily conserved gene in nematodes was identified among early candidates as a potential modulator of human aging (8).
On this basis, we tested the hypothesis that the daf-16 human homologue, FOXO3, a gene on chromosome 6 that codes for a forkhead Box O transcription factor, might be important in human aging and longevity (9; see Figure 1). We first reported on associations of three single nucleotide polymorphisms (SNPs) of FOXO3 with healthy aging and longevity in male Americans of Japanese ancestry (rs2764264 [p value = .0002], rs13217795 [p = .0006], and rs2802292 [p < .0001]), associations that were initially confirmed (with effects pointing in the same direction) in German and French populations (10) and later in Italian, American (northern and western European ancestry), Chinese, and several other populations (11–15). Since the association of FOXO3 with longevity has been observed in genetically diverse populations of European and Asian descent, these results point to FOXO3 playing an important role in human aging. However, the “functional” SNP has yet to be identified.
Figure 1.
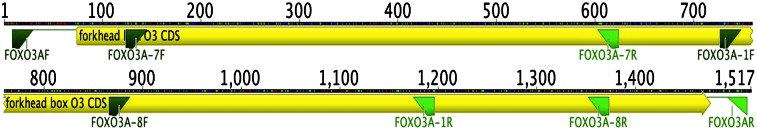
Sequencing strategy. Chromosome 6–specific primers were used to amplify a 1.5-Kb genomic fragment of exon 3. Sequences were generated from 95 individuals of various ethnicities living in Hawaii and screened for nucleotide substitutions. Forward primers are denoted as dark green, whereas reverse primers are light green. The yellow box denotes the protein-coding exon 3, whereas the solid line denotes the noncoding genomic sequence.
SNPs are gene variants that occur on average once every 300 bp. They frequently can be used to discover nearby functional variants that are associated with a disease or other phenotype (16). Once an association between a trait and a gene variant has been discovered, it can be difficult to find which variant is actually responsible for the biologic change that produced the phenotype because several closely linked variants may be in strong linkage disequilibrium (correlation) with the original gene variant. There are numerous SNPs within the FOXO3 locus that are associated with longevity, but they are all located in a haplotype block on the order of 100 kb in size on chromosome 6. Because there is a high degree of linkage disequilibrium, a “longevity-associated functional” variant could be anywhere in this 100+ kb region, which is centered on the second of four introns (known as “intron 2”; see Figure 2). This putative functional variant might act through any number of mechanisms. For example, it might, when translated, change an amino acid sequence. Or it could be a splice-site variant, a transcription enhancer element, a copy-number variant, or affect some other genetic structure. The quest to identify a functional variant (typically a coding variant) in FOXO3 will undoubtedly help us better understand the role that this gene plays in human longevity and healthy aging, but initial sequencing work is required before functional studies are undertaken.
Figure 2.
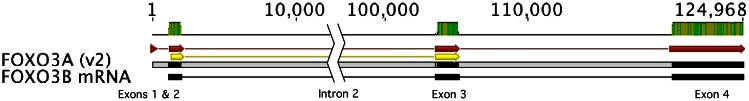
FOXO3 Genomic and FOXO3B mRNA Alignment. Shown is an alignment of the FOXO3 gene (GenBank NM_201559.2), also known as FOXO3A, with the homologous genomic sequences on chromosome 17 (FOXO3B [NR_026718, complement]). Homologies are noted in the top row with green bars. The red arrows refer to the FOXO3 transcript, version 2. The yellow arrows denote the protein-coding exons 2 and 3, and the black boxes denote the noncoding FOXO3B transcript. It is apparent from this alignment that the FOXO3B pseudogene is derived from the FOXO3 processed transcript variant 2, including exons 2–4. The present study assessed single nucleotide polymorphisms from exons 2–3, shown by the yellow arrows. The large intron 2 has been truncated to reduce the size of the figure.
Therefore, we conducted an initial long-range sequencing effort with two purposes: (i) validate known coding variants and (ii) identify novel variants that might be responsible for the association with the longevity phenotype.
MATERIALS AND METHODS
Study Populations
Ninety-five individuals of various ethnicities (Caucasians, Filipinos, Japanese, Chinese, and multiracial) living in Hawaii contributed DNA for this sequencing study, which yielded 85 intact, complete, high quality sequences for further study; an additional 282 persons with Japanese surnames living in Hawaii were used for allele frequency determination within the Japanese American population, and 675 study participants (Japanese American men) from the Kuakini Honolulu Heart Program were used for a longevity case–control study. Procedures were performed according to institutional guidelines and approved by the Institutional Review Board of Kuakini Medical Center.
Sequencing Strategy
Chromosome 6–specific primers (FOXO3AF and FOXO3AR) were designed using the Primer3 (17) component of Geneious and were used to amplify a 1.5 kb chromosome 6–specific genomic fragment containing exon 3, the location of the majority of the previously reported coding variants in FOXO3. Sequences were generated and screened for nucleotide substitutions using an ABI3100 Genetic Analyzer and Geneious software, using eight primers (shown in Table 1 and the triangles in Figure 1).
Table 1.
Primers Used for DNA Amplification and Sequencing
| Primer | Sequence | Tm (°C) | Length (bp) |
| FOXO3AF: | ATCATCTGGGTGCTCGGTTT | 62 | 1,503 |
| FOXO3AR: | GACCTGCTTTGCCCACTTC | 61 | |
| FOXO3A-1F: | CCCAACCAGCTCCTTTAACA | 60 | |
| FOXO3A-1R: | TGCATAGACTGGCTGACAGG | 60 | |
| FOXO3A-3F: | ACCAATTCTAACGCCAGCAC | 60 | |
| FOXO3A-3R: | GAGTCCGAAGTGAGCAGGTC | 60 | |
| FOXO3A-7F: | GAATGAGGGAACTGGCAAGA | 60 | |
| FOXO3A-7R: | CAGGTCGTCCATGAGGTTTT | 60 | |
| FOXO3A-8F: | GACCTGCTCACTTCGGACTC | 60 | |
| FOXO3A-8R: | AAATCCAACCCATCAGCATC | 60 |
Note: DNA sequencing primers used in the current study, which amplifies a 1,503 bp genomic chromosome 6–specific fragment of FOXO3 just outside the region of homology with FOXO3B.
FOXO3/FOXO3B Sequence Alignments
FOXO3 is known to have a pseudogene on chromosome 17, FOXO3B (shown in Figure 1 and 2). FOXO3 exon 3 was used in a BLAST search (18) to examine this homology in more detail. AF032887 was chosen to represent the homology of FOXO3B with FOXO3 (NM_201559), with 1425/1436 identity (99%).
Mapping of FOXO3 Coding Variants
Coding variants for FOXO3, published in dbSNP, were used in BLAST queries in an attempt to explore the origins of the sequence variations. For this, a portion of the DNA adjacent to the SNP was used in the NCBI website (megablast) as well as local alignments using the Geneious software (Biomatters Ltd, Auckland, New Zealand).
RESULTS
FOXO3/FOXO3B Sequence Alignment
FOXO3 is 125 kb in size, located on chromosome 6q21, and is composed of four exons (variant 2), of which exons 2 and 3 code for the translated protein. A variant of FOXO3 (variant 1) is missing the first exon, but this does not result in an amino acid change during translation. FOXO3 (also referred to as FOXO3A) is known to have a pseudogene on chromosome 17, referred to as FOXO3B. FOXO3 exons 2–4 appear to represent the entire FOXO3B pseudogene and support the finding that the latter is an integrated processed messenger RNA from FOXO3. Figure 2 demonstrates this homology. We observed that a portion of the FOXO3B pseudogene is annotated as being contained in exon 4 of the ZNF286B gene (Gene ID: 729288 [not shown]).
No Validated Coding SNPs in FOXO3 in the Most Recent dbSNP Build
A recent version of the Genome Reference Consortium (GRCh37, NCBI dbSNP Build 134), released on August 8, 2011 lists FOXO3 as having 39 polymorphic variants in the coding region (later revised down to 38 polymorphic variants in the November 8 build). This included 21 synonymous, 16 nonsynonymous (including our newly found variant), and 2 frameshift variants. These variants are of great potential interest as one of them might theoretically be the actual causal variant for the longevity and healthy aging phenotypes. We will refer to “transcript variant 2” (NM_201559.2) which describes all four exons for FOXO3, but transcript variant 1 (NM_001455.3) describes two exons. Both transcript variants code for the same protein, as exons 1 and 4 are noncoding but may play roles in gene regulation.
We suspected that some of these variants might not be true variants but rather result from misalignment between the coding region of the FOXO3 gene with its highly homologous pseudogene, FOXO3B (Gene ID = 2310). In an attempt to validate these SNPs, we designed long-range primers that were outside the region of FOXO3/FOXO3B homology that would amplify a 1,503 bp chromosome 6–specific fragment, including all of exon 3 that would serve as a template for DNA sequencing. We found a paucity of variation in this region after sequencing DNA from 95 persons in a random sample of persons living in Hawaii. These persons included primarily Caucasians, Filipinos, Japanese, and Chinese, but some were multiracial.
After completely sequencing the region of interest, we were not able to confirm the existence of any (see later) of the 38 sequence variants that were reported in the most recent version (version 134, GRCh37.p2 of November 8, 2011) of the SNP public database accessible through the NCBI website: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?geneId=2309 (see Table 2.). One variant that was previously reported in dbSNP (August update) is no longer listed. This variant is a T to C transition at position 108985893 (rs113367269), a serine at codon 619 previously described by Watkins and colleagues (19), which does not result in an amino acid change (ie, a synonymous variant). In an attempt to determine the reasons for this failure in confirming the remaining 38 variants, we performed BLAST searches of sequences adjacent to these published FOXO3 SNPs and found that many of them mapped to chromosome 17 with 100% homologies or were the result of misalignment with the location of the highly homologous FOXO3 pseudogene (20), known as FOXO3B (Figure 2). FOXO3B is homologous to exons 2, 3, and 4 of FOXO3, and there are a number of nonallelic nucleotides that align with published SNPs. None of the exon 3 SNPs was verified by our long-range chromosome 6–specific sequencing effort. In sum, none of the coding SNPs listed in dbSNP could be verified. Eight of the 38 previously published coding SNPs were found to result from misalignment of sequences between chromosomes 6 and 17, the remaining 30 could not be verified (not found in our multiethnic populations; Table 2). FOXO3B itself lies within another gene referred to as “ZNF286B,” a putative zinc finger protein 286B (Gene ID: 729288). Collectively, these results indicate that these SNPs are either extremely rare in our multiethnic populations, are the result of misalignment with other highly homologous sequences, or the result of sequencing and/or assembly errors. They are clearly not responsible for the longevity association in our prior work (9) and are unlikely to be important to human longevity.
Table 2.
Evaluation of Variants in FOXO3 From the dbSNP Public Database
| Chromosome Position | mRNA Position | dbSNP rs# Cluster ID | MAF | Function | dbSNP Allele | Protein Residue | Codon Position | Amino Acid Position | Present Results |
| 108882570 | 502 | rs11757217 | 0.1283 | Synonymous | T | Ala [A] | 3 | 53 | ZNF286B |
| Contig reference | C | Ala [A] | 3 | 53 | |||||
| 108882706 | 638 | rs79884776 | Missense | T | Trp [W] | 1 | 99 | Not conf. | |
| Contig reference | C | Arg [R] | 1 | 99 | |||||
| 108882830 | 762 | rs111556510 | 0.0345 | Missense | T | Val [V] | 2 | 140 | Not conf. |
| Contig reference | C | Ala [A] | 2 | 140 | |||||
| 108882855 | 787 | rs13204476 | Missense | T | Ser [S] | 3 | 148 | Not conf. | |
| Contig reference | G | Arg [R] | 3 | 148 | |||||
| 108882873 | 805 | rs146186567 | Synonymous | A | Arg [R] | 3 | 154 | Not conf. | |
| Contig reference | G | Arg [R] | 3 | 154 | |||||
| 108882888 | 820 | rs61758963 | Synonymous | T | Asn [N] | 3 | 159 | Not conf. | |
| Contig reference | C | Asn [N] | 3 | 159 | |||||
| 108882891 | 823 | rs139172563 | Synonymous | A | Leu [L] | 3 | 160 | Not conf. | |
| Contig reference | G | Leu [L] | 3 | 160 | |||||
| 108882906 | 838 | rs149906214 | Synonymous | A | Leu [L] | 3 | 165 | Not conf. | |
| Contig reference | G | Leu [L] | 3 | 165 | |||||
| 108882915 | 847 | rs150320900 | Synonymous | T | Arg [R] | 3 | 168 | Not conf. | |
| Contig reference | C | Arg [R] | 3 | 168 | |||||
| 108882933 | 865 | rs149189425 | Synonymous | A | Pro [P] | 3 | 174 | Not conf. | |
| Contig reference | G | Pro [P] | 3 | 174 | |||||
| 108882987 | 919 | rs142429317 | Synonymous | G | Pro [P] | 3 | 192 | Not conf. | |
| Contig reference | C | Pro [P] | 3 | 192 | |||||
| 108984783 | 1090 | rs61756661 | Synonymous | A | Arg [R] | 3 | 249 | Not conf. | |
| Contig reference | G | Arg [R] | 3 | 249 | |||||
| 108984823 | 1130 | rs141893794 | Missense | G | Gly [G] | 1 | 263 | Not conf. | |
| Contig reference | A | Ser [S] | 1 | 263 | |||||
| 108984834 | 1141 | rs147028825 | Synonymous | T | Arg [R] | 3 | 266 | Not conf. | |
| Contig reference | C | Arg [R] | 3 | 266 | |||||
| 108984867 | 1174 | rs112124249 | Synonymous | T | Ala [A] | 3 | 277 | Not conf. | |
| Contig reference | C | Ala [A] | 3 | 277 | |||||
| 108984882 | 1189 | rs138297794 | Synonymous | T | Asp [D] | 3 | 282 | Not conf. | |
| Contig reference | C | Asp [D] | 3 | 282 | |||||
| 108984883 | 1190 | rs140968061 | Missense | A | Asn [N] | 1 | 283 | Not conf. | |
| Contig reference | G | Asp [D] | 1 | 283 | |||||
| 108984951 | 1258 | rs150216371 | Synonymous | T | Ala [A] | 3 | 305 | Not conf. | |
| Contig reference | G | Ala [A] | 3 | 305 | |||||
| 108985003 | 1310 | rs145756480 | Missense | T | Cys [C] | 1 | 323 | Not conf. | |
| Contig reference | C | Arg [R] | 1 | 323 | |||||
| 108985057 | 1364 | rs145259784 | Missense | A | Thr [T] | 1 | 341 | Not conf. | |
| Contig reference | G | Ala [A] | 1 | 341 | |||||
| 108985080 | 1387 | rs149503832 | Synonymous | T | Tyr [Y] | 3 | 348 | Not conf. | |
| Contig reference | C | Tyr [Y] | 3 | 348 | |||||
| 108985092 | 1399 | rs149158541 | Synonymous | G | Ala [A] | 3 | 352 | Misalign | |
| Contig reference | C | Ala [A] | 3 | 352 | |||||
| 108985094 | 1401 | rs11551770 | Missense | T | Ile [I] | 2 | 353 | Not conf. | |
| Contig reference | G | Ser [S] | 2 | 353 | |||||
| 108985119 | 1426 | rs146009555 | Synonymous | C | Pro [P] | 3 | 361 | Not conf. | |
| Contig reference | G | Pro [P] | 3 | 361 | |||||
| 108985136 | 1443 | rs34223850 | Missense | T | Leu [L] | 2 | 367 | Misalign | |
| Contig reference | C | Pro [P] | 2 | 367 | |||||
| 108985149 | 1456 | rs34754045 | Synonymous | C | Asp [D] | 3 | 371 | Misalign | |
| Contig reference | T | Asp [D] | 3 | 371 | |||||
| 108985176 | 1483 | rs34133353 | Frameshift | G | Glu [E] | 3 | 380 | Misalign | |
| Contig reference | Asp [D] | 3 | 380 | ||||||
| 108985269 | 1576 | rs34079373 | 0.0147 | Synonymous | T | Ser [S] | 3 | 411 | Misalign |
| Contig reference | C | Ser [S] | 3 | 411 | |||||
| 108985274 | 1581 | rs138742093 | Missense | C | Thr [T] | 2 | 413 | Not conf. | |
| Contig reference | G | Ser [S] | 2 | 413 | |||||
| 108985277 | 1584 | rs148296241 | Missense | A | Tyr [Y] | 2 | 414 | Not conf. | |
| Contig reference | T | Phe [F] | 2 | 414 | |||||
| 108985280 | 1587 | rs141876866 | Missense | T | Leu [L] | 2 | 415 | Not conf. | |
| Contig reference | C | Pro [P] | 2 | 415 | |||||
| 108985306 | 1613 | rs34488332 | 0.0090 | Missense | A | Ser [S] | 1 | 424 | Misalign |
| Contig reference | G | Gly [G] | 1 | 424 | |||||
| 108985326 | 1633 | rs146169955 | Synonymous | C | Phe [F] | 3 | 430 | Not conf. | |
| Contig reference | T | Phe [F] | 3 | 430 | |||||
| 108985552 | 1859 | rs148405845 | Missense | T | Cys [C] | 1 | 506 | Not conf. | |
| Contig reference | C | Arg [R] | 1 | 506 | |||||
| 108985557 | 1864 | rs142533192 | Synonymous | C | Arg [R] | 3 | 507 | Not conf. | |
| Contig reference | G | Arg [R] | 3 | 507 | |||||
| 108985581 | 1888 | rs150535671 | Synonymous | A | Pro [P] | 3 | 515 | Not conf. | |
| Contig reference | G | Pro [P] | 3 | 515 | |||||
| 108985618 | 1925 | rs147010831 | Missense | G | Val [V] | 1 | 528 | ZNF286B | |
| Contig reference | T | Leu [L] | 1 | 528 | |||||
| 108985733 | 2040 | rs138174682 | Missense | C | Ala [A] | 2 | 566 | This study | |
| Contig reference | G | Gly [G] | 2 | 566 | |||||
| 108985856 | 2163 | rs34600091 | Frameshift | C | Ser [S] | 2 | 607 | Not conf. | |
| Contig reference | Phe [F] | 2 | 607 |
Note: Chromosome position is in reference to the Genome Reference Consortium (GRCh37.2, NCBI dbSNP Build 132 [NT_025741]) released November 2010. The mRNA position is in reference to the “A” in the AUG start codon, and codon refers to the position in the amino acid codon at which the variant occurs. “ZNF286B” indicates that the SNP has been assigned to the ZNF286B gene on chromosome 17; “not conf.” indicates that there is no evidence for this SNP in the present study; “misalign” indicates that the SNP appears to result from misalignment of FOXO3 and FOXO3B sequences. Variant “rs138174682” was identified in the present study and is now listed in dbSNP. MAF = minor allele frequency, mRNA = messenger RNA, and SNP = single nucleotide polymorphism.
Newly Identified Coding SNP
One novel variant that we discovered in the current sequencing effort is shown in Table 3; it was a nonsynonymous SNP located at position 108985733 on chromosome 6 which results in a G to C transversion and an alanine being substituted for a glycine at amino acid 566 (SNP ID: rs138174682).
Table 3.
Validated Variants in FOXO3 exon 3
| Chromosome Position | mRNA Position | SNP ID | MAF | Type | Allele | Amino Acid | Codon | Amino Acid |
| 108985733 | 1697 | n/a | 0.082 | Missense | G | Gly [G] | 2 | 566 |
| C | Ala [A] | 2 | 566 | |||||
| 108985893 | 1857 | n/a | 0.012 | Synonymous | C | Ser [S] | 3 | 619 |
| T | Ser [S] | 3 | 619 |
Note: Chromosome position is in reference to the Genome Reference Consortium (GRCh37, NCBI dbSNP Build 132 [NT_025741]) released October 9, 2010. The mRNA position is in reference to the “A” in the AUG start codon, SNP IDs are those designated in dbSNP, and codon refers to the position in the amino acid codon at which the variant occurs. A multiethnic group of 85 participants were sequenced for a total of 170 chromosomes. MAF = minor allele frequency, mRNA = messenger RNA, and SNP = single nucleotide polymorphism.
Allele Frequencies of G1697C (Gly566Ala)
A collection of 85 random multiethnic (Caucasian, Filipino, Japanese, Chinese, and mixed ethnicity) genetic samples was chosen for allele frequency determination from the original 95 sequenced samples on the basis of intact, complete, high quality sequences. The ages of these participants ranged from 10 to 78 years. We found that of 85 individuals, 10 were heterozygous and 2 were homozygous for the G1697C variant (rs138174682; see Table 4). To determine if this variant might be useful to study longevity in our Kuakini Honolulu Heart Program cohort (American men of Japanese ancestry), we selected 282 persons with Japanese surnames for a further analysis. Of these 282, only 8 were heterozygous and none was homozygous for this variant. Because the allele frequency was not extremely rare, we decided to assess this SNP with our original Hawaii Lifespan Study case–control study sample (9) to assess whether it might be an important contributor to the “longevity phenotype.” The cases had a longevity phenotype defined as survival to at least 95 years of age (mean attained age 97.9 years), and the controls had an “average-lived phenotype” defined as death by age 81 years (mean attained age 78.5 years). There were 675 study participants. For these men, the allele frequency was 0.01, with only 14 heterozygotes. The small number of carriers precluded any meaningful statistical power for association studies.
Table 4.
Allele Frequencies of G1697C in Different Populations
| Random Multiethnic in Hawaii (n = 85) | Japanese Surnames in Hawaii (n = 282) | KHHP (n = 675) |
| 14/170 alleles (0.082) | 8/564 (0.014) | 14/1,350 (0.0104) |
Note: Allele frequencies for the G566A variant at chromosome position 108985733 were determined in (1) a random sample of 85 participants living in Honolulu, HI, including Filipinos, Japanese, Chinese, and Caucasian (Random Hawaii); (2) 282 random samples from participants with Japanese surnames living in Honolulu, HI; and (3) 675 participants from the Kuakini Honolulu Heart Program (KHHP).
Regarding the C1857T variant (chromosome position 108985893), at codon 619, only 2 of 85 individuals in the random Hawaii sample were heterozygous—for an allele frequency of 0.012, also precluding any meaningful power for association studies.
DISCUSSION
FOXO3 is generally recognized as a “master” gene for human aging because it represents one of only two genes with strong associations with aging-related phenotypes and longevity that have had replications in multiple human populations (9–15). The other—ApoE—is a lipid transport gene, which, while also pleiotropic, appears to act principally via cardiovascular mechanisms on several age-related diseases (15). The E4 allele is the principal risk allele (15). Conversely, the functional SNP in FOXO3 is not known—nor is the mechanism(s) of influence over human aging.
The functions of FOXO3 are highly pleiotropic, ranging from tumor suppression to energy metabolism and thus could affect specific age-related diseases, such as cancer and cardiovascular diseases and/or influence organismal processes, such as oxidative stress, that have wider implications for aging and its related phenotypes (6,21). Important downstream targets of FOXO3 include autophagy, apoptosis, glycolysis and gluconeogenesis, cell proliferation/differentiation, and stress resistance. Therefore, finding the functional variant and its mechanism would be an important step in understanding human aging.
Although our newly discovered Gly566Ala variant (rs138174682) does not appear to be involved in any known modification sites at amino acid 566, such as acetylation, phosphorylation, or ubiquitination, it is three bases away from a cyclic-AMP responsive element binding (CREB-binding protein) site at K569 (22,23). While this is a conservative amino acid substitution, it might change the conformation of the FOXO3 protein so could theoretically have functional implications. This variant and others identified from further sequencing efforts will require validation in other populations. Unfortunately, the variant was too infrequent in our study population of Japanese Americans to account for our previous study findings (9), but it might theoretically contribute to longevity in other populations.
While this variant might appear promising, there is a plethora of human studies that have found one or more genetic variants associated with longevity, but the vast majority has not been replicated. This has historically been the rule rather than the exception. For example, in 1987, in the first candidate gene studies of human longevity, several HLA genes found in Okinawans (24) were not replicated until over a decade later when one of the original findings (DR alleles) were replicated in a European population (25). The second major genetic finding, the APOE gene (26) was widely replicated over the next several years (for review, see [15]) and inspired a plethora of studies of new candidate longevity genes and genome-wide scans (27). Over the next decade, multiple novel genetic findings were reported with inconsistent or no replications until the FOXO3 gene finding (9). Thereafter, multiple replications occurred in less than 2 years (for review, see [15]).
The fact that only two genes (APOE and FOXO3) have consistent, and widespread replication is puzzling. What is the reason for the general dearth of replications for most preliminary genetic findings? One potential cause that can lead to apparent lack of replication is population-specific variants. Similar phenotypes may result from completely different variants that are common in one population but simply do not exist in other populations, such as population-specific variation within the CETP gene. A relation between variation within this gene and human longevity was first reported by Barzilai and colleagues (28) in Ashkenazi Jews but was never replicated until Koropatnick and colleagues (29) tested a population-specific variant in American men of Japanese ancestry from the Honolulu Heart Program and Hawaii Lifespan Study. The particular SNPs thought to be responsible for the shared phenotypes (elevated high density lipoprotein and exceptional longevity) do not exist in the other population. Had Koropatnick and colleagues (29) focused on the specific SNPs found in the initial study of Ashkenazi Jews (and/or those in LD), this may have led to another uninformative study. Other potentially more common reasons for lack of replication include the existence of rare variants, misalignments with other highly homologous sequences, sequencing/assembly errors, among other causes.
Regarding the current study, rapid progress in genomic methodology has made sequencing a viable and cost-effective approach to uncover variants that can then be assessed for functional significance. SNPs found within a coding sequence have generally been of most interest because they are more likely to alter the biological function of a protein than those in noncoding sequences. Therefore, the current investigation focused on all putative coding sequence variants (n = 38) that were reported in the most recent version of the SNP public database (see earlier). Surprisingly, only one of the SNPs (located in exon 3) was verified by our long-range chromosome 6–specific sequencing (a noncoding SNP). Further investigation revealed that, in fact, 8 of the 38 previously published coding SNPs resulted from misalignment of sequences between chromosomes 6 and 17 (Table 2), the remaining 30 are from exon 3 and could not be verified (not found at all in our populations).
This highlights the aforementioned replication issue and points to an important additional challenge for future studies. That is, a significant proportion of SNPs in public databases is, in fact, not well curated and is composed of paralogous sequence variants. Paralogous sequence variants can arise by segmental duplications of the human genome sequence as well as by homologies with pseudogenes (30). Musumeci and colleagues (31) provide evidence that as many as 8.32% of the biallelic coding SNPs listed in the NCBI’s dbSNP database are not reliable and appear to be artifacts due to the presence of highly similar genes in the human genome. This problem arises when there is extensive DNA sequence similarity between two or more genomic regions and their sequence differences masquerade as normal SNPs. Care must be taken to reduce this problem in DNA sequencing studies so that primer selection includes unique-copy regions. In the present study, we report that all 38 of the previously reported coding SNPs in FOXO3 are either invalid or too rare to be useful for phenotype–genotype association and other genetic mapping studies.
The plethora of large-scale genotyping efforts, such as genome-wide association studies that utilize nonvalidated variants from short “shotgun” sequencing efforts, will no doubt add many invalid variants to public available data sets that may lead to wasted efforts and unrealistic expectations when mapping complex genetic traits (32). Through the use of longer range DNA sequencing and careful annotation of the genome, primarily through efforts like the ENCODE Project (33), there should be an improvement in the resources that can help elucidate functional variants that are associated with complex phenotypes.
With regard to discovering the longevity-associated functional variant for FOXO3, this is as yet unresolved. It appears from the current study that it is unlikely (although not impossible) that a coding SNP will be identified as the functional variant responsible for human longevity. Interestingly, Banasik and colleagues (34) did find that one variant in intron 2, rs2802292, a noncoding SNP, demonstrates a small but significant difference in messenger RNA levels in functional studies. This same variant had the strongest association in our previous case–control study of healthy aging and longevity (9). Whether this SNP is the functional variant remains to be proven and more functional studies are required. For example, cell physiology experiments measuring messenger RNA levels need to be performed, keeping all genetic information the same, except for this variant, as well as aforementioned model organism and human studies.
In conclusion, all published nonsynonymous (coding and result in amino acid change) SNPs identified in the FOXO3 gene, to date, are either invalid or too rare to be of consequence to human longevity. The novel variant that we identified through detailed sequencing appears to be too low in frequency (0.082) to be responsible for our previously reported association of FOXO3 variants with human longevity (9), although it is conceivable that this novel SNP is associated with a disease phenotype at a younger age or is important in other populations. This suggests that the functional SNP is most likely a noncoding SNP from intron 2, where all previously reported associations have occurred. Future research should include complete sequencing of intron 2 and annotation of regulatory elements, in order to identify candidates for functional study of the FOXO3 gene and human aging.
In the future, when more evidence accumulates as to a functional SNP(s), rigorous assessment with model organisms and human studies will be needed. Work with model organisms has already provided some clues that might be helpful for mechanistic studies of the effects of FOXO3 variation in humans. For example, oxidative stress, still a leading theory of how we age (35,36), is one potential mechanism. Indeed, the PI3K/Akt signaling cascade, a downstream target of insulin signaling, is an important inhibitor of FOXO function. Important antioxidant enzyme expression, including manganese superoxide dismutase (MnSOD) and catalase, depend on PI3K/Akt/FOXO3 activity (37,38), and oxidative stress appears linked to human longevity (39). Yet, work to date shows that mice deficient in the FOXO target MnSOD do not exhibit changes in life span (40). Nor do mice overexpressing the FOXO target MnSOD exhibit changes in life span (41). Interestingly, Ferber and colleagues (42) recently found that FOXO3 activation results in the repression of nuclear-encoded genes with mitochondrial function, this mediated by inhibition of c-Myc (independent of SOD2). FOXO3 activation also reduced mitochondrial DNA copy number, mitochondrial protein expression, respiratory complexes, and mitochondrial respiratory complexes and activity. More work is needed to assess potential relations between FOXO3, oxidative stress, and longevity, such as whether expression of common antioxidant enzymes correlates with genetic variation in FOXO3 or with other factors that influence FOXO3 expression.
Some of the most basic groundwork in humans could begin with assessment of the potential relation between mortality from the common diseases of aging, including coronary heart disease, stroke, and cancer, which account for the majority of human deaths, and variation in FOXO3. Already, there is growing evidence for FOXO3 as a tumor suppressor. For example, downregulation of FOXO3 activity is seen in various cancers (43,44). Indeed, the Hawaii Lifespan Study men who had the longevity phenotype (and were more likely to have a protective FOXO3 genotype) had fewer cancers in their medical history (9). Ongoing studies with prospectively collected data can address whether and how cause of death is modified by variation in the FOXO3 gene. This will provide further clues as to mechanisms for FOXO3’s effects on human aging and longevity. Such information may enhance translational efforts that link model organism and human research and could have important implications for human health span (45).
FUNDING
We appreciate the support of contract N01-AG-4-2149, grants 5 U01 AG019349-05 and 2R01 AG027060-05A2 (Hawaii Lifespan Study) from the National Institute on Aging and contract N01-HC-05102 from the National Heart, Lung, and Blood Institute.
Acknowledgments
Author contributions: T.A.D., B.J.W., Q.H., J.S.G., K.H.M., B.R., and J.D.C. designed research; T.A.D., B.J.W., Q.H., R.C., J.S.G., K.H.M., B.R., and J.D.C. performed research; T.A.D. contributed new reagents and analytic tools; Q.H. and J.S.G. analyzed data; and T.A.D., B.J.W., Q.H., J.S.G., K.H.M., B.R., D.C.W., and J.D.C. wrote the article.
References
- 1.Willcox BJ, Willcox DC, Ferrucci L. Secrets of healthy aging and longevity from exceptional survivors around the globe: lessons from octogenarians to supercentenarians. J Gerontol A Biol Sci Med Sci. 2008;63:1181–1185. doi: 10.1093/gerona/63.11.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yashin AI, Iachine IA, Harris JR. Half of variation in susceptibility to mortality is genetic: findings from Swedish twin survival data. Behav Genet. 1999;29:11–19. doi: 10.1023/a:1021481620934. [DOI] [PubMed] [Google Scholar]
- 3.Yashin AI, Debenedictis G, Vaupel JW, et al. Genes and longevity: lessons from studies of centenarians. J Gerontol Biol Sci. 2000;55:B319–B328. doi: 10.1093/gerona/55.7.b319. [DOI] [PubMed] [Google Scholar]
- 4.Melzer D, Hurst AJ, Frayling T. Genetic variation and human aging: progress and prospects. J Gerontol A Biol Sci Med Sci. 2007;62:301–307. doi: 10.1093/gerona/62.3.301. [DOI] [PubMed] [Google Scholar]
- 5.Gögele M, Pattaro C, Fuchsberger C, Minelli C, Pramstaller PP, Wjst M. Heritability analysis of life span in a semi-isolated population followed across four centuries reveals the presence of pleiotropy between life span and reproduction. J Gerontol A Biol Sci Med Sci. 2011;66(1):26–37. doi: 10.1093/gerona/glq163. [DOI] [PubMed] [Google Scholar]
- 6.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 7.Tatar M. Can we develop genetically tractable models to assess healthspan (rather than life span) animal models? J Gerontol A Biol Sci Med Sci. 2009;64:161–163. doi: 10.1093/gerona/gln067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 9.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flachsbart F, Caliebe A, Kleindorp R, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anselmi CV, Malovini A, Roncarati R, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Wang WJ, Cao H, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pawlikowska L, Hu D, Huntsman S, et al. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Study of osteoporotic fractures. Aging Cell. 2009;18:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Y, Cheng L, Chen H, et al. Effects of FOXO genotypes on longevity: a biodemographic analysis. J Gerontol A Biol Sci Med Sci. 2010;65:1285–1299. doi: 10.1093/gerona/glq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung WH, Dao RL, Chen LK, Hung SI. The role of genetic variants in human longevity. Ageing Res Rev. 2010;9(suppl 1):S67–S78. doi: 10.1016/j.arr.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson GC, Esposito L, Barratt BJ, et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Watkins WJ, Umbers AJ, Woad KJ, et al. Mutational screening of FOXO3A and FOXO1A in women with premature ovarian failure. Fertil Steril. 2006;86:1518–1521. doi: 10.1016/j.fertnstert.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 20.Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 21.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 23.Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27:2263–2275. doi: 10.1038/onc.2008.20. [DOI] [PubMed] [Google Scholar]
- 24.Takata H, Suzuki M, Ishii T, Sekiguchi S, Iri H. Influence of major histocompatibility complex region genes on human longevity among Okinawan-Japanese centenarians and nonagenarians. Lancet. 1987;2(8563):824–826. doi: 10.1016/s0140-6736(87)91015-4. [DOI] [PubMed] [Google Scholar]
- 25.Caruso C, Candore G, Romano GC, et al. Mech Ageing Dev. 2001;122(5):445–462. doi: 10.1016/s0047-6374(00)00255-4. [DOI] [PubMed] [Google Scholar]
- 26.Schächter F, Faure-Delanef L, Guénot F, et al. Nat Genet. 1994;6(1):29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 27.Willcox DC, Willcox BJ, Poon LW. Centenarian studies: important contributors to our understanding of the aging process and longevity. Curr Gerontol Geriatr Res. 2010;2010:484529. doi: 10.1155/2010/484529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290(15):2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 29.Koropatnick TA, Kimbell J, Chen R, et al. A prospective study of high-density lipoprotein cholesterol, cholesteryl ester transfer protein gene variants, and healthy aging in very old Japanese-American men. J Gerontol A Biol Sci Med Sci. 2008;63(11):1235–1240. doi: 10.1093/gerona/63.11.1235. [DOI] [PubMed] [Google Scholar]
- 30.Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum Mol Genet. 2002;11:1987–1995. doi: 10.1093/hmg/11.17.1987. [DOI] [PubMed] [Google Scholar]
- 31.Musumeci L, Arthur JW, Cheung FS, Hoque A, Lippman S, Reichardt JK. Single nucleotide differences (SNDs) in the dbSNP database may lead to errors in genotyping and haplotyping studies. Hum Mutat. 2010;31:67–73. doi: 10.1002/humu.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 34.Banasik K, Ribel-Madsen R, Gjesing AP, et al. The FOXO3A rs2802292 G-allele associates with improved peripheral and hepatic insulin sensitivity and increased skeletal muscle-FOXO3A mRNA expression in twins. J Clin Endocrinol Metab. 2011;96:E119–E124. doi: 10.1210/jc.2010-0881. [DOI] [PubMed] [Google Scholar]
- 35.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78(2):547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 36.Salmon AB, Richardson A, Pérez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic Biol Med. 2010;48(5):642–655. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 38.Kops GJ, Dansen TB, Polderman PE, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki M, Willcox DC, Rosenbaum MW, Willcox BJ. Oxidative stress and longevity in Okinawa: an investigation of blood lipid peroxidation and tocopherol in Okinawan centenarians. Curr Gerontol Geriatr Res. 2010;2010:380460. doi: 10.1155/2010/380460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Ikeno Y, Qi W, et al. Mice deficient in both Mn superoxide dismutase and glutathione peroxidase-1 have increased oxidative damage and a greater incidence of pathology but no reduction in longevity. J Gerontol A Biol Sci Med Sci. 2009;64:1212–1220. doi: 10.1093/gerona/glp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang YC, Perez VI, Song W. Overexpression of Mn superoxide dismutase does not increase life span in mice. J Gerontol A Biol Sci Med Sci. 2009;64(11):1114–1125. doi: 10.1093/gerona/glp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. doi: 10.1038/cdd.2011.179. [published online ahead of print December 2, 2011]. doi:10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15(3):752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greer E, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 45.Lebel M, Picard F, Ferland G, Gaudreau P. Drugs, nutrients, and phytoactive principles improving the health span of rodent models of human age-related diseases. J Gerontol A Biol Sci Med Sci. 2012;67(2):140–151. doi: 10.1093/gerona/glr038. [DOI] [PubMed] [Google Scholar]


