Abstract
Background:
Our recent studies of microRNA (miRNA) expression signature demonstrated that microRNA-874 (miR-874) was significantly downregulated in maxillary sinus squamous cell carcinoma (MSSCC), and a putative tumour-suppressive miRNA in human cancers. Our aim of this study was to investigate the functional significance of miR-874 in cancer cells and to identify novel miR-874-mediated cancer pathways and responsible genes in head and neck squamous cell carcinoma (HNSCC).
Methods:
Gain-of-function studies using mature miR-874 were performed to investigate cell proliferation and cell cycle distribution in HNSCC cell lines (SAS and FaDu). To identify miR-874-mediated molecular pathways and targets, we utilised gene expression analysis and in silico database analysis. Loss-of-function assays were performed to investigate the functional significance of miR-874 target genes.
Results:
Expression levels of miR-874 were significantly downregulated in HNSCC tissues (including oral, pharyngeal and laryngeal SCCs) compared with normal counterpart epithelia. Restoration of miR-874 in SAS and FaDu cell lines revealed significant inhibition of cell proliferation and induction of G2/M arrest and cell apoptosis. Our expression data and in silico analysis demonstrated that miR-874 modulated the cell cycle pathway. Moreover, histone deacetylase 1 (HDAC1) was a candidate target of miR-874 regulation. Luciferase reporter assays showed that miR-874 directly regulated HDAC1. Silencing of the HDAC1 gene significantly inhibited cell proliferation and induced G2/M arrest and cell apoptosis in SAS cells.
Conclusions:
Downregulation of miR-874 was a frequent event in HNSCC. miR-874 acted as a tumour suppressor and directly targeted HDAC1. Recognition of tumour-suppressive miRNA-mediated cancer pathways provides new insights into the potential mechanisms of HNSCC oncogenesis and suggests novel therapeutic strategies for the disease.
Keywords: miR-874, HDAC1, cell cycle arrest, apoptosis, HNSCC
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer in the world and approximately about 500 000 cases are diagnosed every year (Jemal et al, 2010). In spite of considerable advances in multimodality therapy, including surgery, radiotherapy and chemotherapy, the overall 5-year survival rate for patients with HNSCC is only 40%–50% (Haddad and Shin, 2008). Local tumour recurrence and distant metastasis after conventional therapy appear to be major contributing factors for restricted survival of HNSCC patients (Leemans et al, 2011). Therefore, understanding the molecular cancer pathways underlying HNSCC would help to improve diagnosis, approaches to therapy and prevention of the disease.
The discovery of non-coding RNA in the human genome was an important conceptual breakthrough in the post-genome sequencing era (Mattick, 2004). Improved understanding of non-coding RNA is necessary for continued progress in cancer research. MicroRNAs (miRNAs) constitute a class of small, non-coding RNA molecules, 19–22 nucleotides in length, which modulate gene expression. This is achieved through imperfect pairing with target messenger RNAs (mRNAs) of protein-coding genes and transcriptional or post-transcriptional regulation of their expression (Bartel, 2004). Currently, 2042 human mature miRNAs are registered at miRBase release 19.0 (http://microrna.sanger.ac.uk/).
A growing body of evidence indicates that miRNAs also contribute to the initiation and development of various types of cancers (Esquela-Kerscher and Slack, 2006). Many human cancers show aberrant expression of miRNAs that can function either as tumour suppressors or oncogenes (Esquela-Kerscher and Slack, 2006). In cancer pathways, normal regulatory mechanisms are disrupted by altered expression of tumour-suppressive or oncogenic miRNAs. Therefore, identification of differentially expressed miRNAs is an important step to understanding human oncogenesis.
Recently, a maxillary sinus squamous cell carcinoma (MSSCC) miRNA expression signature revealed that microRNA-874 (miR-874) was significantly downregulated. Moreover, restoration of miR-874 inhibited cell proliferation in a MSSCC cell line, IMC-3 (Nohata et al, 2011c). To the best of our knowledge, this is the only investigation of miR-874 in cancer biology. The aim of this study was to investigate the functional significance of miR-874 and identify the molecular pathways and responsible genes it regulated in HNSCC cells. Genome-wide gene expression analysis of miR-874 transfectants and in silico database analysis showed that the cell cycle was a promising candidate target of miR-874. Furthermore, histone deacetylase 1 (HDAC1) was a candidate target of miR-874 regulation.
Histone deacetylase 1 belongs to the family of HDACs and is a component of the HDAC complex. It also interacts with retinoblastoma tumour-suppressor protein and this complex is essential for cell proliferation and differentiation (Giacinti and Giordano, 2006). Several studies indicated that HDAC inhibitors (HDACis) are a new class of cytostatic agents that inhibit the proliferation of tumour cells in culture and in vivo by inducing cell cycle arrest, differentiation and/or apoptosis (Cruz and Matushansky, 2012; Popovic and Licht, 2012). We focused on HDAC1 as a putative miR-874 target and investigated the functional significance of HDAC1 in HNSCC. Tumour-suppressive miRNA-modulated cancer pathways provide new insights into the potential mechanisms of HNSCC oncogenesis and suggest novel therapeutic strategies for treatment of the disease.
Materials and methods
Clinical HNSCC specimens
Twenty-three pairs of primary HNSCC and corresponding normal epithelial samples were obtained from patients with HNSCC in Chiba University Hospital (Chiba, Japan) from 2005 to 2011. The samples considered normal were free of cancer cells by pathologic examination. The patient's backgrounds and clinicopathological characteristics are summarised in Table 1. The patients were classified according to the 2002 Union for International Cancer Control TNM staging criteria (Sobin and Wittekind, 2002) before treatment. Written consent of tissue donation for research purposes was obtained from each patient before tissue collection. The protocol was approved by the Institutional Review Board of Chiba University. The specimens were immersed in RNAlater (Qiagen, Valencia, CA, USA) and stored at −20 °C until RNA was extracted.
Table 1. Patient's characteristics.
| No. | Location | Age | Sex | T | N | M | Differentiation | Stage |
|---|---|---|---|---|---|---|---|---|
| 1 |
Tongue |
68 |
M |
2 |
0 |
0 |
Well |
II |
| 2 |
Tongue |
66 |
M |
2 |
1 |
0 |
Moderate |
III |
| 3 |
Tongue |
76 |
F |
1 |
0 |
0 |
Well |
I |
| 4 |
Tongue |
69 |
M |
1 |
0 |
0 |
Well |
I |
| 5 |
Tongue |
76 |
F |
1 |
0 |
0 |
Well |
I |
| 6 |
Tongue |
67 |
M |
4a |
2c |
0 |
Moderate |
IVa |
| 7 |
Tongue |
36 |
F |
3 |
1 |
0 |
Moderate |
III |
| 8 |
Tongue |
64 |
M |
1 |
0 |
0 |
Well |
I |
| 9 |
Oral floor |
65 |
M |
2 |
2c |
0 |
Moderate |
IVa |
| 10 |
Oropharynx |
67 |
M |
3 |
2c |
0 |
Poor |
IVa |
| 11 |
Oropharynx |
52 |
M |
3 |
2a |
0 |
Moderate |
IVa |
| 12 |
Oropharynx |
76 |
M |
2 |
0 |
0 |
Poor |
II |
| 13 |
Larynx |
63 |
M |
3 |
0 |
0 |
Moderate |
III |
| 14 |
Larynx |
69 |
M |
3 |
0 |
0 |
Well |
III |
| 15 |
Larynx |
66 |
M |
4a |
0 |
0 |
Moderate |
IVa |
| 16 |
Larynx |
82 |
M |
3 |
0 |
0 |
Poor |
III |
| 17 |
Hypopharynx |
68 |
M |
4a |
1 |
0 |
Moderate |
IVa |
| 18 |
Hypopharynx |
73 |
M |
3 |
1 |
0 |
Well |
III |
| 19 |
Hypopharynx |
66 |
M |
2 |
2c |
0 |
Moderate |
IVa |
| 20 |
Hypopharynx |
68 |
M |
2 |
2b |
0 |
Poor |
IVa |
| 21 |
Hypopharynx |
65 |
M |
1 |
2b |
0 |
Moderate |
IVa |
| 22 |
Hypopharynx |
64 |
M |
3 |
0 |
0 |
Moderate |
III |
| 23 | Hypopharynx | 55 | M | 3 | 2b | 0 | Poor | IVa |
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RNA concentrations were determined spectrophotometrically, and molecular integrity was checked by gel electrophoresis. RNA quality was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Cell lines and cell culture
The following cell lines were used: human HNSCC; SAS (derived from a primary lesion of tongue SCC), FaDu (derived from a primary lesion of hypopharyngeal SCC), HSC3 (derived from a metastatic lymph node of tongue SCC), IMC-3 (derived from a primary lesion of maxillary sinus SCC), human fibroblast; MRC-5. All cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37 °C.
5-Aza-2′-deoxycytidine (5-Aza-dC) treatment
We investigated the effects of the demethylation agent (5-Aza-dC) treatment (Sigma-Aldrich, St Louis, MO, USA) on HNSCC and fibroblast cell lines (SAS, FaDu, HSC3, IMC-3 and MRC-5). Cells were treated with 5 μℳ of the 5-Aza-dC, and cells harvested after 72- or 120-h treatment. Experiment manner was as described previously (Enokida et al, 2004). The difference expression status of miR-874 and HDAC1 was evaluated by real-time PCR methods as follows using before and after 5-Aza-dC treatments.
Mature miRNA transfection and small interfering RNA treatment
The following mature miRNAs species were used in this study: hsa-miR-874, Ambion Pre-miR, PM12355 (Applied Biosystems, Foster City, CA, USA) and miRIDIAN Mimics, MIMAT0004911 (Thermo Scientific Dharmacon, Waltham, MA, USA). The following siRNAs were used: Stealth Select RNAi siRNA; si-HDAC1_1 Cat# HSS104726 and si-HDAC1_2 Cat# HSS179192 (Invitrogen), and negative control miRNA/siRNA (Applied Biosystems, P/N: AM17111). RNAs were incubated with OPTI-MEM (Invitrogen) and LipofectamineTM RNAiMax reagent (Invitrogen) as described previously (Ichimi et al, 2009). Transfection efficiency of miRNA in cell lines was confirmed based on downregulation of TWF1 (PTK9) mRNA following transfection with miR-1 as previously reported (Ichimi et al, 2009).
Cell proliferation assays
Cells were transfected with 10 nℳ miRNA and siRNA by reverse transfection and plated in 96-well plates at 3 × 103 cells per well. After 72 h, cell proliferation was determined with the XTT assay, using the Cell Proliferation Kit II (Roche Molecular Biochemicals, Mannheim, Germany) as previously reported (Chiyomaru et al, 2010). Triplicate wells were measured for cell viability in each treatment group.
Flow cytometry
Cell cycle status and apoptosis were examined using an APC BrdU Flow kit (BD Bioscience, San Jose, CA, USA) according to the manufacturer's protocol. SAS and FaDu cells transiently transfected with miR-control, miR-874, si-control or si-HDAC1 were harvested 72 h after transfection by trypsinisation. Experiments were done in triplicate.
Genome-wide gene expression analysis and in silico analysis for miR-874-modulated pathways
To gain further insight into which genes were affected by miR-874, we performed genome-wide gene expression analysis using miR-874 transfectants of SAS and FaDu cells. Oligo-microarray human 4 × 44K (GPL10332) (Agilent Technologies) was used for gene expression analysis; the experimental procedure was described previously (Sugimoto et al, 2009). To identify the biological processes or pathways potentially regulated by mi-874, we performed GENECODIS analysis (Carmona-Saez et al, 2007; Nogales-Cadenas et al, 2009; http://genecodis.cnb.csic.es/) with our microarray results. Then, to assess the networks regulated by miR-874 and their target genes, we analysed and characterised those genes in KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway Database (Kanehisa et al, 2012).
Quantitative real-time RT–PCR
First-strand cDNA was synthesised from 1 μg of total RNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene-specific PCR products were assayed continuously using a 7900-HT Real-Time PCR System according to the manufacturer's protocol. The initial PCR step consisted of a 10-min hold at 95 °C, followed by 40 cycles consisting of a 15-s denaturation at 95 °C and a 1-min annealing/extension at 63 °C. TaqMan probes and primers for, BUB3 (P/N: Hs00190920_m1), CCNE1 (P/N: Hs01026536_m1), CDC23 (P/N: Hs00946641_m1), CDC25B (P/N: Hs01550934_m1), HDAC1 (P/N: Hs02621185_s1) and GAPDH (P/N: Hs02758991_g1) as an internal control were obtained from Applied Biosystems (Assay-On-Demand Gene Expression Products). The expression levels of miR-874 (assay ID: 002268) were analysed by TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay; Applied Biosystems) and normalised to RNU48 (assay ID: 001006). The delta delta Ct method was adopted and applied to calculate the relative quantities of subject genes. All reactions were performed in triplicate, and included negative control reactions that lacked cDNA.
Western blots
Cells were harvested 72 h after transfection and lysates were prepared. In all, 50 μg of protein lysate was separated by Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA) and transferred to PVDF membranes. Immunoblotting was performed with diluted (1 : 1000) rabbit HDAC1 antibody (600-401-879; Rockland, Gilbertsville, PA, USA), with GAPDH antibody (ab8245, Abcam, Cambridge, UK) used as an internal control. The membrane was washed and incubated with a goat anti-rabbit IgG (H+L)–HRP conjugate (Bio-Rad). Complexes were visualised with an Immun-Star WesternC Chemiluminescence Kit (Bio-Rad), and the expression levels of these genes were evaluated by ImageJ software (ver.1.44; http://rsbweb.nih.gov/ij/).
Plasmid construction and dual-luciferase reporter assay
Partial wild-type sequences of HDAC1 3′-UTR and those with deleted miR-874 target sites (positions 149–155) were inserted between the XhoI–PmeI restriction sites in the 3′-UTR of the hRluc gene in psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). Sequences of oligonucleotides were described in Supplementary document 1. The synthesised DNA was cloned into the psiCHECK-2 vector. SAS cells were transfected with 15 ng of vector, 10 nℳ of miR-874 (Applied Biosystems), and 1 μl of Lipofectamine 2000 (Invitrogen) in 100 μl of Opti-MEM (Invitrogen). The activities of firefly and Renilla luciferases in cell lysates were determined with a dual-luciferase assay system (E1910; Promega). Normalised data were calculated as the quotient of Renilla/firefly luciferase activities.
Statistical analysis
The relationships between two groups and the numerical values obtained by real-time RT–PCR were analysed using the paired t-test. The relationship among three variables and numerical values was analysed using the Bonferroni adjusted Mann–Whitney U-test. All analyses were performed using Expert StatView (version 4, SAS Institute Inc., Cary, NC, USA).
Results
Expression levels of miR-874 in HNSCC clinical specimens and cell lines
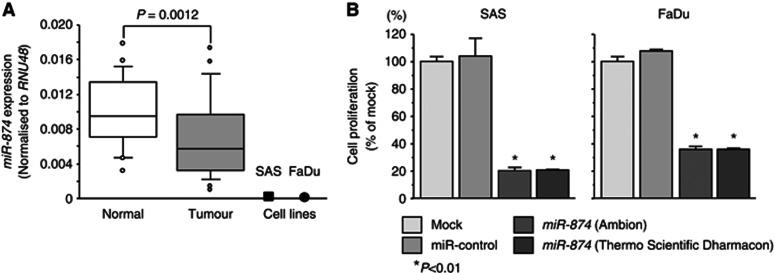
The expression levels of miR-874 were significantly lower in 23 clinical HNSCC specimens than those in corresponding adjacent non-cancerous specimens (P=0.0012). We also evaluated the expression levels of miR-874 in two HNSCC cell lines (SAS and FaDu). The expression levels of miR-874 in these two cell lines were much lower than those in non-cancerous epithelial cells (Figure 1A).
Figure 1.
Expression levels of miR-874 in HNSCC clinical specimens and effect of miR-874 transfection on SAS and FaDu cells. (A) Real-time RT–PCR showed that miR-874 was significantly downregulated in HNSCC clinical specimens compared with normal specimens (n=23). (B) Cell proliferation activity after transfection of miR-874 was determined by XTT assay. Suppression of cell proliferation was similarly observed in both Ambion and Thermo Scientific Dharmacon miR-874, taking into consideration the off-target effects of dsRNAs. Trials were independently conducted three times. *P<0.01.
Effects of miR-874 restoration on cell proliferation in HNSCC cell lines
To investigate the functional effects of miR-874, we performed gain-of-function studies using miRNA transfection of SAS and FaDu cell lines. We utilised two sources of mature miR-874 (Ambion and Thermo Scientific Dharmacon) to ensure reproducibility of the data. The XTT assay demonstrated that cell proliferation was significantly inhibited in miR-874 transfectants in comparison with the mock or miR-control transfectant cells. Specifically, we observed the following growth, expressed as a percentage of the control: (1) SAS – mock, 100.0±1.5; miR-control, 103.5±1.5; miR-874 (Ambion), 20.2±2.6; miR-874 (Thermo Scientific Dharmacon), 20.5±0.6 and (2) FaDu – mock, 100.0±1.7; miR-control, 107.3±1.1; miR-874, 36.8±2.5; miR-874 (Thermo Scientific Dharmacon), 36.6±2.3, with P<0.01 for both (Figure 1B).
Effect of miR-874 restoration on cell cycle and apoptosis
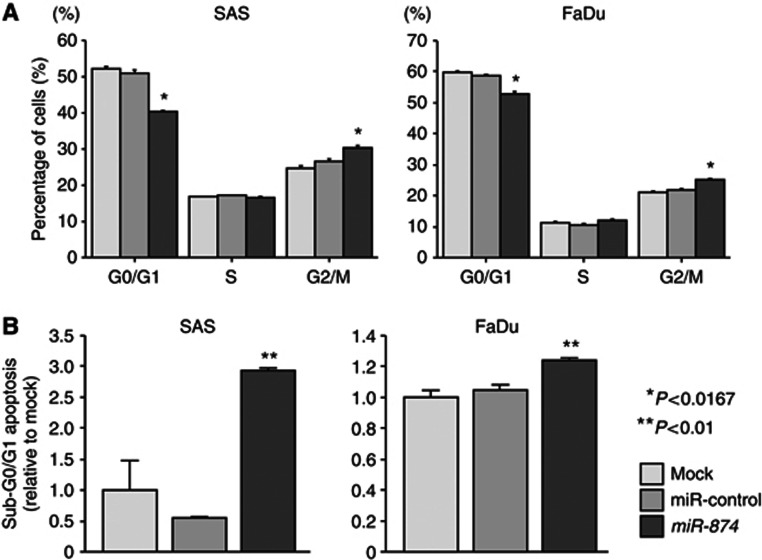
As miR-874 restoration significantly inhibited cell proliferation in SAS and FaDu cells, we hypothesised that their restoration induced cell cycle arrest and/or apoptosis. This hypothesis was tested by flow cytometry. With regard to the cell cycle distribution, the fraction of cells in the G2/M phase was significantly larger in miR-874 transfectants in comparison with the mock (both P<0.0167, Figure 2A). These results suggested that miR-874 restoration induced G2/M arrest in both cell lines. The sub-G0/G1 apoptotic cell fraction was significantly larger in miR-874 transfectants (2.9-fold in SAS and 1.3-fold in FaDu) than in the mock and miR-control (both P<0.01, Figure 2B). Flow cytometric data are shown in Supplementary Figure 1.
Figure 2.
Effect of miR-874 transfection on cell cycle and apoptosis in SAS and FaDu cells determined by flow cytometry. (A) Typical results of cell cycle analysis of mock, miR-control and miR-874 transfectants. The bar chart represents the percentage of the cells in the G0/G1, S or G2/M phase of the cell cycle. (B) Typical results of sub-G0/G1 apoptosis analysis of mock, miR-control and miR-874 transfectants. Induction of cell apoptosis was shown after transfection of miR-874. Trials were independently conducted three times. *P<0.0167, **P<0.01.
Identification of miR-874-modulated molecular pathways and putative target genes in HNSCC
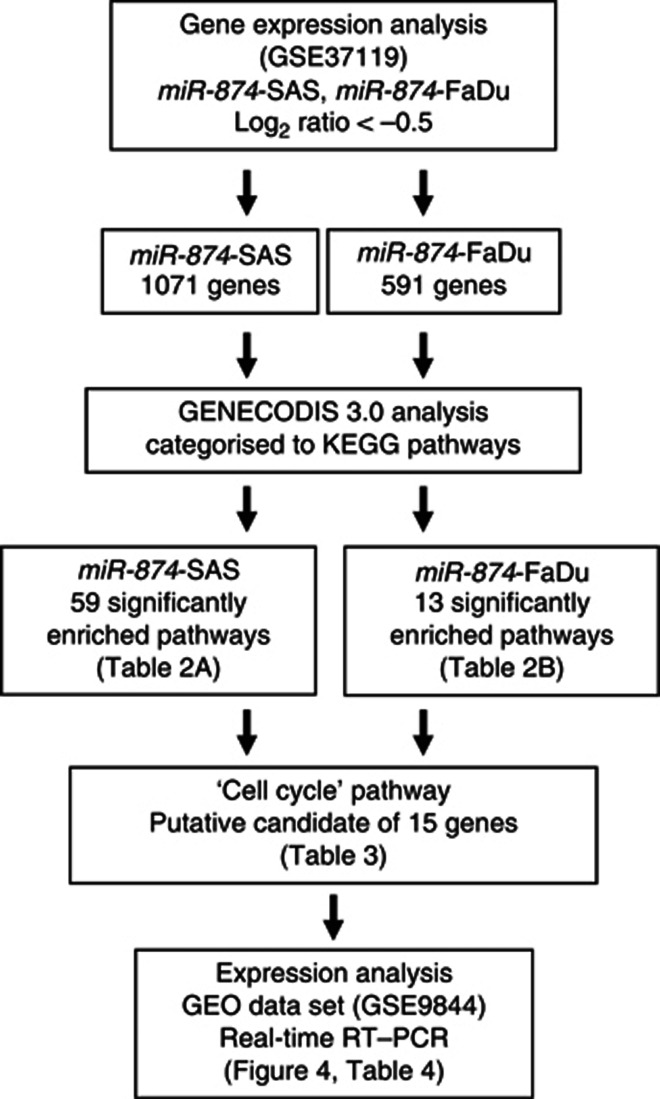
To gain further insight into molecular mechanisms and pathways regulated by tumour-suppressive miR-874 in HNSCC, we performed genome-wide gene expression analysis using miR-874 transfected cells, miR-874-SAS and miR-874-FaDu. Entries from the microarray data were approved by the Gene Expression Omnibus (GEO) and were assigned GEO accession number GSE37119. A total of 1071 and 591 genes were downregulated <−0.5 (log2 ratio) in miR-874-SAS and miR-874-FaDu transfectants, respectively, compared with control cells. These genes were analysed and characterised in KEGG pathway categories by the GENECODIS 3.0 program.
The GENECODIS analysis categorised 59 and 13 significantly enriched signalling pathways in the miR-874-SAS and miR-874-FaDu transfectants, respectively (Table 2a and b). We focused on the ‘cell cycle' pathway and the 15 genes listed within this pathway (Table 3). Of those 15 genes, five (budding uninhibited by benzimidazoles 3 homolog: BUB3, cyclin E1: CCNE1, cell division cycle protein 23 homolog: CDC23, cell division cycle protein 25 homolog B: CDC25B and HDAC 1: HDAC1) were downregulated by transfection of miR-874 in both SAS and FaDu cells, and contained putative binding sites of miR-874 in their 3′-UTR by TargetScan database (Table 3). Our strategy for selection of miR-874 target genes is shown in Figure 3.
Table 2. (a) Fifty-nine significantly enriched pathways in SAS; (b) Thirteen significantly enriched pathways in FaDu.
| KEGG pathway | Genes | P-value |
|---|---|---|
|
(a) SAS | ||
| Protein processing in endoplasmic reticulum |
30 |
4.67E–13 |
| Pathways in cancer |
40 |
1.75E–11 |
| Endocytosis |
29 |
1.90E–10 |
| Focal adhesion |
29 |
2.40E–10 |
| Small cell lung cancer |
19 |
4.29E–10 |
| Regulation of actin cytoskeleton |
29 |
7.09E–10 |
| Lysosome |
21 |
5.25E–09 |
| Axon guidance |
21 |
1.35E–08 |
| ECM–receptor interaction |
15 |
1.07E–06 |
| Insulin signalling pathway |
17 |
1.91E–05 |
| Amoebiasis |
14 |
6.64E–05 |
| p53 signalling pathway |
11 |
1.15E–04 |
| Valine, leucine and isoleucine degradation |
9 |
1.05E–04 |
| Vasopressin-regulated water reabsorption |
9 |
1.05E–04 |
| Prostate cancer |
12 |
2.55E–04 |
| Bacterial invasion of epithelial cells |
10 |
7.80E–04 |
| Notch signalling pathway |
8 |
9.55E–04 |
| ErbB signalling pathway |
11 |
9.49E–04 |
| Phagosome |
14 |
1.11E–03 |
| Cell cycle |
13 |
1.31E–03 |
| Glioma |
9 |
1.37E–03 |
| mTOR signalling pathway |
8 |
1.80E–03 |
| Tight junction |
13 |
1.99E–03 |
| HCM |
10 |
2.00E–03 |
| Pancreatic cancer |
9 |
2.61E–03 |
| Chronic myeloid leukaemia |
9 |
3.45E–03 |
| Dilated cardiomyopathy |
10 |
3.47E–03 |
| Prion diseases |
6 |
4.19E–03 |
| MAPK signalling pathway |
19 |
4.51E–03 |
| Melanogenesis |
10 |
6.69E–03 |
| Calcium signalling pathway |
14 |
8.25E–03 |
| Galactose metabolism |
5 |
8.43E–03 |
| Nucleotide excision repair |
6 |
9.87E–03 |
| Bladder cancer |
6 |
1.09E–02 |
| NOD-like receptor signalling pathway |
7 |
1.08E–02 |
| Inositol phosphate metabolism |
7 |
1.08E–02 |
| ARVC |
8 |
1.08E–02 |
| Glycosaminoglycan degradation |
4 |
1.33E–02 |
| Vascular smooth muscle contraction |
10 |
1.50E–02 |
| Ubiquitin-mediated proteolysis |
11 |
1.77E–02 |
| Amino sugar and nucleotide sugar metabolism |
6 |
1.80E–02 |
| SNARE interactions in vesicular transport |
5 |
2.10E–02 |
| RNA degradation |
7 |
2.09E–02 |
| Starch and sucrose metabolism |
6 |
2.28E–02 |
| Long-term potentiation |
7 |
2.36E–02 |
| Endometrial cancer |
6 |
2.40E–02 |
| CAMs |
10 |
2.55E–02 |
| Non-small cell lung cancer |
6 |
2.47E–02 |
| Vibrio cholerae infection |
6 |
2.47E–02 |
| Melanoma |
7 |
2.44E–02 |
| PPAR signalling pathway |
7 |
2.44E–02 |
| Adherens junction |
7 |
2.58E–02 |
| Oocyte meiosis |
9 |
2.91E–02 |
| Sphingolipid metabolism |
5 |
2.91E–02 |
| Fatty acid metabolism |
5 |
3.18E–02 |
| B-cell receptor signalling pathway |
7 |
3.22E–02 |
| Phosphatidylinositol signalling system |
7 |
3.39E–02 |
| Glycosphingolipid biosynthesis – ganglio series |
3 |
3.60E–02 |
| Protein digestion and absorption |
7 |
3.75E–02 |
|
(b) FaDu | ||
| Protein processing in endoplasmic reticulum |
17 |
4.67E–07 |
| Systemic lupus erythematosus |
13 |
3.33E–07 |
| Pathways in cancer |
17 |
3.24E–03 |
| Lysosome |
9 |
1.11E–02 |
| Endocytosis |
11 |
1.94E–02 |
| Valine, leucine and isoleucine degradation |
5 |
2.65E–02 |
| Cell cycle |
8 |
3.32E–02 |
| Bile secretion |
6 |
3.03E–02 |
| Neurotrophin signalling pathway |
8 |
2.72E–02 |
| Insulin signalling pathway |
8 |
3.80E–02 |
| Ubiquitin-mediated proteolysis |
8 |
3.79E–02 |
| Inositol phosphate metabolism |
5 |
4.26E–02 |
| Oocyte meiosis | 7 | 3.97E–02 |
Abbreviations: ARVC=arrhythmogenic right ventricular cardiomyopathy; CAMs=cell adhesion molecules; ECM=extracellular matrix; HCM=hypertrophic cardiomyopathy; KEGG=Kyoto Encyclopedia of Genes and Genomes; MAPK= mitogen-activated protein kinase; mTOR= mammalian target of rapamycin; NOD=nucleotide-binding oligomerization domain; PPAR=peroxisome proliferator-activated receptor; SNARE=soluble NSF attachment protein receptor..
Table 3. Cell cycle-related genes regulated by miR-874.
| No. | Symbol | SAS | FaDu | miR-874 site |
|---|---|---|---|---|
| 1 |
ANAPC11 |
− |
+ |
− |
| 2 |
BUB3 |
+ |
+ |
2 |
| 3 |
CCNE1 |
+ |
+ |
1 |
| 4 |
CDC23 |
+ |
+ |
2 |
| 5 |
CDC25B |
+ |
+ |
1 |
| 6 |
CDK6 |
+ |
− |
1 |
| 7 |
CDKN1B |
+ |
− |
− |
| 8 |
CDKN2D |
+ |
+ |
− |
| 9 |
GSK3B |
+ |
− |
1 |
| 10 |
HDAC1 |
+ |
+ |
1 |
| 11 |
RAD21 |
− |
+ |
1 |
| 12 |
RB1 |
+ |
− |
− |
| 13 |
RBL2 |
+ |
− |
− |
| 14 |
SMAD3 |
+ |
− |
4 |
| 15 | TGFB2 | + | − | − |
Abbreviation: miR-874=microRNA-874.
Figure 3.
Workflow for the in silico analysis of miR-874-regulated pathways. miR-control transfectants that produced raw signal values of <1000 were excluded before comparisons were made. Downregulated genes in miR-874 transfectants with <−0.5 (log2 ratio) downregulation compared with the control transfectants were chosen from the results of microarray analysis (GSE37119). A total of 1071 and 591 genes were identified as genes downregulated by miR-874 in SAS and FaDu cells, respectively. The gene sets were then analysed and categorised with the KEGG Pathway Database using the GENECODIS 3.0 program. In all, 59 and 13 significantly enriched signalling pathways were derived from the gene sets in SAS and FaDu cells, respectively. In total, 13 and 8 genes were included in the ‘cell cycle' pathway in SAS and FaDu cells, respectively.
Expression levels of five candidate genes regulated by miR-874 in HNSCC clinical specimens
In cancer pathways, normal regulatory mechanisms can be disrupted by altered expression of tumour-suppressive or oncogenic miRNAs. We hypothesised that downregulation of miR-874 led to overexpression of oncogenic genes in HNSCC clinical specimens. To support this hypothesis, we investigated the two independent gene expression analyses, one is deposited in GEO database, accession number is GSE9844, and the other is our own expression data by using real-time RT–PCR.
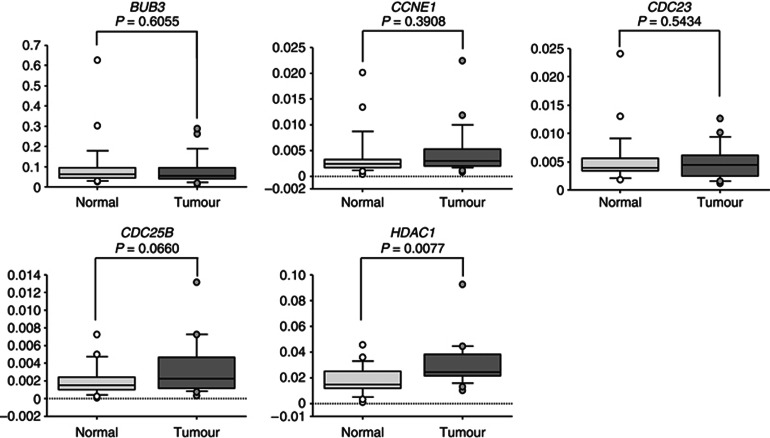
First, we screened the expression of those 5 candidates and other 10 genes in HNSCC clinical samples using data deposited in GEO (accession number GSE9844) by another research group. We noted that CCNE1, HDAC1, CDC25B, CDC23 and BUB3 were upregulated in cancer tissues compared with normal epithelia (Table 4 and Supplementary Figure 2). Next, we used real-time RT–PCR to measure the mRNA expression levels of five candidate genes in HNSCC clinical specimens. Four genes (BUB3, CCNE1, CDC23 and CDC25B) were not upregulated in cancer tissues (Figure 4). In contrast, HDAC1 was significantly upregulated in cancer tissues compared with adjacent normal epithelia (P=0.0077, Figure 4). Therefore, we focused on the HDAC1 gene as a possible target gene regulated by miR-874 in HNSCC and subjected it to further experiments.
Table 4. Expression levels of 15 genes downregulated by miR-874 in ‘cell cycle' pathway (GSE9844).
| Entrez gene | Symbol | Fold change (tumour/normal) | P-value |
|---|---|---|---|
| 898 |
CCNE1 |
2.262 |
0.0012 |
| 3065 |
HDAC1 |
1.539 |
0.0012 |
| 994 |
CDC25B |
1.782 |
0.0023 |
| 8697 |
CDC23 |
1.331 |
0.0095 |
| 1021 |
CDK6 |
1.932 |
0.0118 |
| 1027 |
CDKN1B |
−1.569 |
0.0644 |
| 5885 |
RAD21 |
1.567 |
0.0897 |
| 5934 |
RBL2 |
−1.622 |
0.0945 |
| 2932 |
GSK3B |
1.276 |
0.1100 |
| 7042 |
TGFB2 |
1.405 |
0.1158 |
| 9184 |
BUB3 |
1.162 |
0.1239 |
| 51 529 |
ANAPC11 |
1.264 |
0.2215 |
| 5925 |
RB1 |
1.076 |
0.7581 |
| 4088 |
SMAD3 |
−1.076 |
0.8110 |
| 1032 | CDKN2D | 1.089 | 0.9277 |
Abbreviation: miR-874=microRNA-874.
Figure 4.
Expression levels of five candidate genes of miR-874 target. BUB3, CCNE1, CDC23, CDC25B and HDAC1 mRNA expression levels in HNSCC clinical specimens were measured by quantitative real-time RT–PCR. GAPDH was used for normalisation. All reactions were performed in triplicate.
HDAC1 is a direct target of miR-874
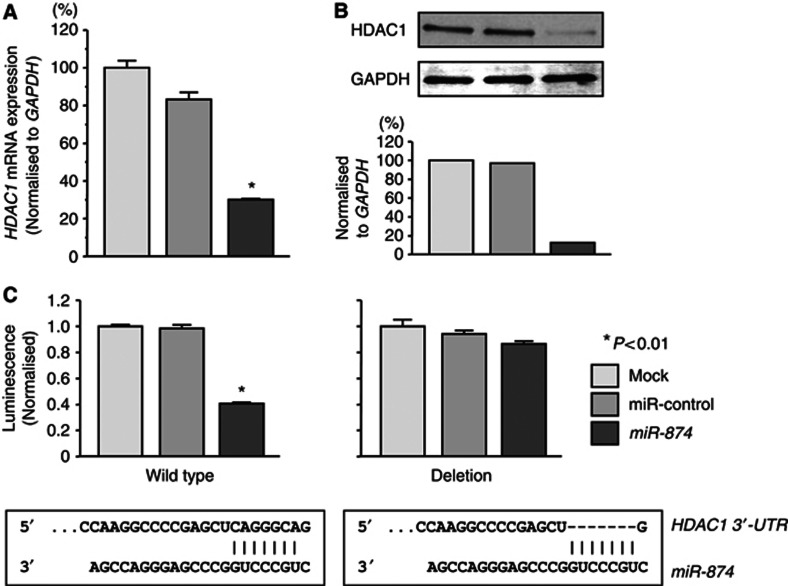
We performed quantitative real-time RT–PCR and western blotting to investigate whether HDAC1 expression was downregulated by restoration of miR-874. The mRNA and protein expression levels of HDAC1 were significantly repressed in miR-874-transfectants in comparison with mock or miR-control transfectants (P<0.01, Figures 5A and B).
Figure 5.
Effect of miR-874 on HDAC1 mRNA and protein expression in SAS and FaDu cells. (A) HDAC1 mRNA expression 72 h after transfection with 10 nℳ miR-874. HDAC1 mRNA expression was significantly repressed in miR-874 transfectants. GAPDH was used as an internal control. (B) HDAC1 protein expression 72 h after transfection with miR-874. GAPDH was used as a loading control. The protein expression level of HDAC1 was also repressed in miR-874 transfectants. (C) Interaction of miR-874 with HDAC1 3′-UTR. After 24-h transfection with 10 nℳ miR-874, miR-control or mocks, a reporter plasmid containing HDAC1 wild type-3′-UTR or deletion-3′-UTR and a plasmid expressing Renilla luciferase (hRluc) were co-transfected into SAS cells. Firefly luciferase activity was normalised to Renilla luciferase activity. Relative luciferase activity in miR-874 transfectants was compared with that in mock cultures, which was set at 1, in cells transfected with wild type-3′-UTR or deletion-3′-UTR. Trials were independently conducted three times. *P<0.01.
We performed a luciferase reporter assay to determine whether HDAC1 mRNA had a target site for miR-874. We used a vector encoding either the partial sequence of the 3′-UTR of HDAC1 mRNA, including the predicted miR-874 target site (positions 149–155) or a vector lacking the miR-874 target site. We found that the luminescence intensity was significantly reduced by transfection of the wild type 3′-UTR of HDAC1 while deletion of positions 149–155 blocked the decrease in luminescence (P<0.01, Figure 5C).
HNSCC and fibroblast cells were subjected to real-time RT–PCR to evaluate the expression levels of miR-874 and HDAC1. At baseline, expression levels of the miR-874 were negative in all cell lines. However, after 5-Aza-dC treatment, the expression levels of miR-874 were significantly increased in all cell lines. Conversely, the expression levels of HDAC1 mRNA were significantly decreased by 5-Aza-dC treatment. Inverse correlation was recognised between miR-874 and HDAC1 expression after 5-Aza-dC treatment (Supplementary Figure 3). This data suggested that demethylation restored the expression of miR-874 in HNSCC and fibroblast cells and that reduction of the HDAC1 expression was caused by recovery for expression of miR-874 in cancer cells.
Effects of HDAC1 silencing on cell proliferation, cell cycle and apoptosis in HNSCC cell lines
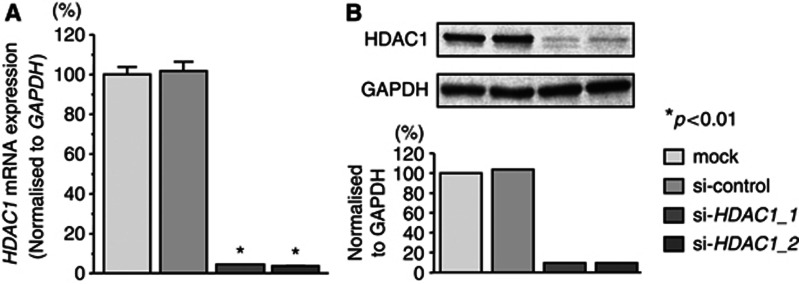
To investigate the functional role of HDAC1, we performed loss-of-function studies using si-HDAC1 transfectants. First, we evaluated the knockdown efficiency of si-HDAC1 treatments. The expression levels of HDAC1 mRNA were repressed in two si-HDAC1 transfectants in comparison with mock and si-control transfectants (P<0.01, Figure 6A). Moreover, the expression levels of HDAC1 protein were repressed in si-HDAC1-1 and the si-HDAC1-2 transfectants in comparison with mock and si-control transfectants (Figure 6B). These results showed that the two siRNAs were effective for loss-of-function assays in this study.
Figure 6.
Silencing of HDAC1 in HNSCC cell lines by siRNAs. (A) HDAC1 mRNA expression 72 h after transfection with 10 nℳ si-HDAC1_1, si-HDAC1_2 or si-control. HDAC1 mRNA expression was repressed in si-HDAC1_1 and si-HDAC1_2 transfectants. GAPDH was used as an internal control. (B) HDAC1 protein expression 72 h after transfection of the siRNAs. GAPDH was used a loading control. The protein expression level of HDAC1 was also repressed in si-HDAC1_1 and si-HDAC1_2 transfectants. Trials were independently conducted three times. *P<0.01.
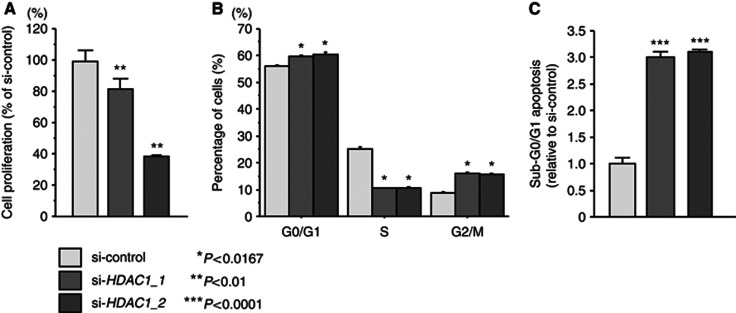
The XTT assay revealed significant inhibition of cell proliferation in the two different si-HDAC1 transfectants in comparison with growth of the mock and si-control transfectants. The following was observed, expressed as a percentage of control proliferation: mock, 100±4.2; si-control, 93.7±5.6; si-HDAC1_1, 74.7±6.4; si-HDAC1_2, 38.4±0.9, P<0.01; Figure 7A. As for the cell cycle distribution, the fractions of cells in the G1/G0 and G2/M phases were significantly larger in si-HDAC1 transfectants in comparison with the si-control (P<0.0167, Figure 7B), and the sub-G0/G1 apoptotic cell fraction was significantly larger (3.0-fold) in si-HDAC1 transfectants than in the si-control in SAS cells (P<0.01, Figure 7C). Flow cytometric data are shown in Supplementary Figure 4.
Figure 7.
Effect of si-HDAC1 transfectants on cell proliferation, cell cycle and apoptosis in SAS cells. (A) Suppression of cell proliferation after transfection of si-HDAC1_1 and si-HDAC1_2 into SAS cells as determined by XTT assay. (B) Typical results of cell cycle analysis of si-control, si-HDAC1_1 and si-HDAC1_2 transfectants. The bar chart represents the percentage of the cells in the G0/G1, S or G2/M phase. (C) Typical results of cell apoptosis analysis of mock, si-control, si-HDAC1_1 and si-HDAC1_2 transfectants. Trials were independently conducted three times. *P<0.0167, **P<0.01, ***P<0.0001.
Discussion
Despite considerable advances in cancer treatment, the overall survival rate of HNSCC patients has not markedly improved. Therefore, it is necessary to pursue new concepts to enhance treatment results and to develop new therapeutic strategies. For elucidation of the molecular mechanisms underlying HNSCC, we examined miRNA expression, focusing on its regulated molecular targets and novel cancer pathways based on HNSCC expression signatures (Kikkawa et al, 2010; Nohata et al, 2011c). Our recent studies demonstrated that miR-1, miR-133a, miR-218, miR-375 and miR-489 were significantly reduced in HNSCC clinical specimens. Moreover, they functioned as tumour suppressors through their targeting of several oncogenic genes in HNSCC cells (Kikkawa et al, 2010; Mutallip et al, 2011; Nohata et al, 2011a, 2011b, 2011c, 2011d, 2011e; Kinoshita et al, 2012a, 2012b, 2012c, 2012d). Our previous study of MSSCC miRNA expression signature showed that miR-874 was the most downregulated miRNA and significantly inhibited cancer cell proliferation in IMC-3 cells derived from MSSCC. We also identified putative targets of miR-874 by genome-wide gene expression analysis and PPP1CA was a direct target of miR-874 in MSSCC cells (Nohata et al, 2011c). The overexpression of PPP1CA was observed in oral cancer cell lines (Hsu et al, 2006) and this protein may act as an oncogene since dephosphorylated BRCA1 as breast and ovarian tumour suppressor is considered to be inactive form (Liu et al, 2002). The identification of tumour-suppressive miRNA-mediated novel cancer pathways is the first step to consider the effective and promising strategy for miRNA-based therapeutics for the treatment of human cancers. Except for our previous report, there are no data to suggest a relationship between miR-874 and human cancers. Thus, we believed that analysis of miR-874 could lead to discoveries of new molecular mechanisms in HNSCC.
First, we evaluated the expression level of miR-874 in HNSCC clinical specimens that were obtained from the oral cavity, larynx and pharynx. We confirmed that miR-874 was significantly downregulated in tumour tissues. The elucidation of silencing mechanisms of tumour-suppressive miRNAs will contribute to miRNA-based cancer therapies in near future. We performed preliminary analysis of molecular mechanism of miR-874 silencing in HNSCC cells. Our data demonstrated that miR-874 was re-expressed by treatment of 5-Aza-dC, suggesting that silencing of miR-874 in cancer cells was caused by epigenetic modification especially methylation of CpG island located on the upstream of miR-874 genomic locus (Supplementary Figure 3). Further detailed analysis is necessary to understand the molecular mechanism of miR-874 silencing in cancer cells. Next, we investigated the functional significance of miR-874 in HNSCC using two cell lines, SAS and FaDu. In this study, we used two kinds of miRNA to evaluate the function of miR-874. Restoration of mature miR-874 in cancer cells revealed significant inhibition of cancer cell proliferation, suggesting miR-874 is indeed a new tumour suppressor in HNSCC. It is likely that further analysis will provide added insight into HNSCC oncogenesis.
miRNAs are unique in their ability to regulate many protein-coding genes. Bioinformatic predictions indicate that miRNAs regulate >30% of protein-coding genes (Filipowicz et al, 2008). We have taken the position that identification of novel cancer pathways and responsible genes regulated by tumour-suppressive miR-874 is an important first step in understanding HNSCC oncogenesis. Based on this view, we performed genome-wide gene expression analysis of miR-874 transfectants (miR-874-SAS and miR-874-FaDu) and in silico analysis. A single miRNA is capable of targeting a number of genes to regulate biological processes globally. In fact, 1060 and 587 genes were downregulated in miR-874-SAS and miR-874-FaDu transfectants, respectively. We used GENECODIS analysis to reveal the functional significance of the genes potentially regulated by miR-874 in HNSCC. GENECODIS analysis applies many genes to known pathways in the KEGG Pathway Database and these data facilitate understanding of tumour-suppressive miRNA-mediated molecular pathways in human cancer. We devised this method of analysis and found that tumour-suppressive miRNA could efficiently regulate cancer-associated pathways (Fuse et al, 2012; Kinoshita et al, 2012a). In this study, we focused on the ‘cell cycle' pathway because restoration of miR-874 inhibited cancer cell proliferation in HNSCC cell lines.
The expression signatures of miR-874 transfectants in HNSCC cells and in silico analysis revealed that five genes (BUB3, CCNE1, CDC23, CDC25B and HDAC1) were involved in the ‘cell cycle' pathway and that they were candidate targets of miR-874 regulation. Out of the five genes, HDAC1 was significantly upregulated in HNSCC clinical specimens as assessed by both our real-time RT–PCR analysis and HNSCC gene expression data deposited in GEO. We further focused on HDAC1 as a responsible gene in HNSCC and investigated the functional significance of the gene.
Our luciferase reporter assay demonstrated that miR-874 directly regulated HDAC1 in HNSCC cells. It has been shown that HDAC1 is directly regulated by miR-449a in prostate cancer (Noonan et al, 2009) and lung cancer (Jeon et al, 2012). The seed sequence of mature miR-449a is identical to that of miR-34 families, which are well known as p53-responsive genes and tumour suppressors through regulating HDAC SIRT1, cyclin-dependent kinases, cyclins and E2Fs (Hermeking, 2010). Recent miRNA researches suggest that one of the reasons why HDACs is overexpressed in various cancers might be downregulation of miRNAs acting as endogenous HDACis (Datta et al, 2008; Nasser et al, 2008; Noonan et al, 2009; Jeon et al, 2012).
Histone deacetylases remove acetyl groups from histones and other nuclear proteins that induce chromatin condensation and transcriptional repression (Haberland et al, 2009). HDACs have important roles in aberrant epigenetic changes related to human malignant diseases (Marks, 2010). HDAC1 is upregulated in several types of cancer, such as lung (Sasaki et al, 2004), gastric (Choi et al, 2001), breast (Zhang et al, 2005), prostate (Weichert et al, 2008a), liver (Rikimaru et al, 2007) and colorectal cancers(Weichert et al, 2008b), and it correlates with poor prognosis. HDACis have been watched with keen interest in cancer therapy because aberrant expressions of HDACs are frequent events (Glozak and Seto, 2007; Marks, 2010). Vorinostat and romidepsin have been approved recently for the treatment of refractory cutaneous T-cell lymphoma, and clinical trials of other HDACis are in progress in other lymphomas (Grant et al, 2007).
This is the first report that upregulation of HDAC1 is caused by downregulation of tumour-suppressive miR-874 in HNSCC cells. Future work on miR-874 involving its targeting of oncogenic pathways and its targeting of oncogenes may lead to better understanding of HNSCC oncogenesis and development of new therapeutic strategies for HNSCC.
Conclusions
Downregulation of miR-874 was a frequent event in HNSCC. miR-874 inhibited cancer cell proliferation through direct targeting of HDAC1, suggesting that miR-874 is a new tumour-suppressive miRNA. Tumour-suppressive miR-874 appears to modulate cancer-associated pathways. The recognition that miR-874 target oncogenes may lead to better understanding of HNSCC oncogenesis and the development of new therapeutic strategies for HNSCC.
Acknowledgments
This study was supported by KAKENHI (C), 23592505 and 24592590.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883–891. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Kwon HJ, Yoon BI, Kim JH, Han SU, Joo HJ, Kim DY. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Cancer Res. 2001;92:1300–1304. doi: 10.1111/j.1349-7006.2001.tb02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FD, Matushansky I. Solid tumor differentiation therapy - is it possible. Oncotarget. 2012;3:559–567. doi: 10.18632/oncotarget.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Enokida H, Shiina H, Igawa M, Ogishima T, Kawakami T, Bassett WW, Anast JW, Li LC, Urakami S, Terashima M, Verma M, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. CpG hypermethylation of MDR1 gene contributes to the pathogenesis and progression of human prostate cancer. Cancer Res. 2004;64:5956–5962. doi: 10.1158/0008-5472.CAN-04-0081. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, Ichikawa T, Seki N. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Grant S, Easley C, Kirkpatrick P. Vorinostat. Nat Rev Drug Discov. 2007;6:21–22. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Huang X, Seasholtz S, Potter DM, Gollin SM. Gene amplification and overexpression of protein phosphatase 1alpha in oral squamous cell carcinoma cell lines. Oncogene. 2006;25:5517–5526. doi: 10.1038/sj.onc.1209563. [DOI] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, Na MJ, Park JY, Kang J, Son JW. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012;76:171–176. doi: 10.1016/j.lungcan.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Hanazawa T, Nohata N, Kikkawa N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012a;3:1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Fuse M, Hanazawa T, Kikkawa N, Fujimura L, Watanabe-Takano H, Yamada Y, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-133a regulates novel targets: moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2012b;418:378–383. doi: 10.1016/j.bbrc.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Watanabe-Takano H, Yoshino H, Hidaka H, Fujimura L, Fuse M, Yamasaki T, Enokida H, Nakagawa M, Hanazawa T, Okamoto Y, Seki N. Actin-related protein 2/3 complex subunit 5 (ARPC5) contributes to cell migration and invasion and is directly regulated by tumor-suppressive microRNA-133a in head and neck squamous cell carcinoma. Int J Oncol. 2012c;40:1770–1778. doi: 10.3892/ijo.2012.1390. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Yoshino H, Hanazawa T, Kikkawa N, Fujimura L, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int J Oncol. 2012d;40:185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- Liu Y, Virshup DM, White RL, Hsu LC. Regulation of BRCA1 phosphorylation by interaction with protein phosphatase 1alpha. Cancer Res. 2002;62:6357–6361. [PubMed] [Google Scholar]
- Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19:1049–1066. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: a new genetics. Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mutallip M, Nohata N, Hanazawa T, Kikkawa N, Horiguchi S, Fujimura L, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N. Glutathione S-transferase P1 (GSTP1) suppresses cell apoptosis and its regulation by miR-133a in head and neck squamous cell carcinoma (HNSCC) Int J Mol Med. 2011;27:345–352. doi: 10.3892/ijmm.2010.589. [DOI] [PubMed] [Google Scholar]
- Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Fujimura L, Yoshino H, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N. Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol. 2011a;38:209–217. [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Sakurai D, Fujimura L, Kawakami K, Chiyomaru T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC) J Hum Genet. 2011b;56:595–601. doi: 10.1038/jhg.2011.66. [DOI] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, Katayama A, Harabuchi Y, Okamoto Y, Seki N. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011c;105:833–841. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Sasaki K, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol. 2011d;39:1099–1107. doi: 10.3892/ijo.2011.1096. [DOI] [PubMed] [Google Scholar]
- Nohata N, Sone Y, Hanazawa T, Fuse M, Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Shozu M, Okamoto T, Seki N. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget. 2011e;2:29–44. doi: 10.18632/oncotarget.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–413. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikimaru T, Taketomi A, Yamashita Y, Shirabe K, Hamatsu T, Shimada M, Maehara Y. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72:69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Moriyama S, Nakashima Y, Kobayashi Y, Kiriyama M, Fukai I, Yamakawa Y, Fujii Y. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer. 2004;46:171–178. doi: 10.1016/j.lungcan.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Sobin L, Wittekind C.2002UICC TNM Classification of Malignant Tumors6th edn.Wiley-Liss: New York [Google Scholar]
- Sugimoto T, Seki N, Shimizu S, Kikkawa N, Tsukada J, Shimada H, Sasaki K, Hanazawa T, Okamoto Y, Hata A. The galanin signaling cascade is a candidate pathway regulating oncogenesis in human squamous cell carcinoma. Genes Chromosomes Cancer. 2009;48:132–142. doi: 10.1002/gcc.20626. [DOI] [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008a;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008b;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.