Abstract
Background:
The microRNA-205 (miR-205) has been shown to be deregulated in prostate cancer (PCa). Here we continue to investigate the prognostic and therapeutic potential of this microRNA.
Methods:
The expression of miR-205 is measured by qRT–PCR and in situ hybridisation in a well-documented PCa cohort. An AGO2-based RIP-Chip assay is used to identify targets that are verified with western blots, luciferase reporter assay, ELISA and immunohistochemistry.
Results:
The expression of miR-205 is inversely correlated to the occurrence of metastases and shortened overall survival, and is lower in castration-resistant PCa patients. The miR-205 expression is mainly localised to the basal cells of benign prostate tissues. Genes regulated by miR-205 are enriched in, for example, the MAPK/ERK, Toll-like receptor and IL-6 signaling pathways. We demonstrate binding of miR-205 to the 3′UTR of androgen receptor (AR) and decrease of both AR transcript and protein levels. This finding was corroborated in the patient cohort were miR-205 expression inversely correlated to AR immunostaining in malignant prostate cells and to serum levels of prostate-specific antigen, an androgen-regulated protein.
Conclusion:
Taken together, these findings imply that miR-205 might have therapeutic potential, especially for the castration resistant and currently untreatable form of PCa.
Keywords: miRNA, miR-205, prostate cancer, androgen receptor, CRPC, metastases
Prostate cancer (PCa) is the most common cancer in men in the developed countries (Jemal et al, 2011). Most of these tumours are indolent and curable, but a few are aggressive. The focus of much of today's research is to find markers to determine at an early state which tumours will be life threatening and are in need of radical treatment. Androgen signaling through the androgen receptor (AR) is an important oncogenic pathway for PCa progression. Androgen deprivation therapy is a common treatment modality for advanced disease, but after initial regression of tumour burden, the disease will progress to a castration-resistant form (CRPC), for which there currently is no effective cure. Despite the low levels of androgens achieved by the androgen deprivation therapy, increased AR signaling is a key feature of CRPC, through, for example, increased expression of the AR, increased sensitivity of the AR to androgens, ligand promiscuity or ligand-independent activation of the AR (Feldman and Feldman, 2001). The prostate is composed of a stromal and glandular compartment. The glands of the prostate are predominantly made up of basal epithelial cells and luminal cells. The basal cells are generally undifferentiated and androgen independent, and they have a relatively high proliferative and a low apoptotic rate. The basal cells and progenitor cells continuously adds to the pool of the luminal layer that consists of differentiated cells that secrete various androgen-regulated proteins such as prostate-specific antigen (PSA).
The human genome encodes around 2000 microRNAs (miRNAs), each of which is capable of regulating hundreds of protein-coding genes (Farh et al, 2005; Stark et al, 2005; Baek et al, 2008; Selbach et al, 2008). In general, miRNAs bind the 3′UTR of mRNAs resulting in a block of translation or degradation of the mRNA (He and Hannon, 2004; Du and Zamore, 2005). However, there have been reports of direct miRNA binding resulting in increased translation of target mRNA (Vasudevan et al, 2007). MicroRNAs have emerged as important regulators of gene expression, and there have been several reports of individual deregulated miRNAs that have tumour suppressive or oncogenic properties in PCa (Fuse et al, 2011; Boll et al, 2012; Jalava et al, 2012). For example, the expression of microRNA-205 (miR-205) has been shown to be decreased in PCa, partly explained by hypermethylation of the CpG islands in the miR-205 promoter (Ke et al, 2009; Bhatnagar et al, 2010; Hulf et al, 2011; Wiklund et al, 2011), and to act as a tumour suppressor by affecting migration, invasion and growth (Gandellini et al, 2009; Majid et al, 2010; Schaefer et al, 2010; Boll et al, 2012). However, the mechanisms of action and individual targets have only limited been investigated. In this paper, we put our efforts to verify prognostic powers of miR-205 in PCa in a well-documented cohort with a long follow-up. The expression of miR-205 has been found to be significantly lower in PCa compared with the BPH samples in this cohort (Larne et al, 2012) and receiver operator characteristic curves for differentiation of patients with clinical localised PCa from those with BPH gave an area under the curve of 0.80 (95% CI: 0.70–0.90). In addition, we aim to determine the expression pattern of miR-205 in prostate tissue and identify novel targets of miR-205. These findings will provide new insight into PCa disease biology and could potentially result in novel therapeutic approaches.
Materials and methods
Tissue specimens and cell culture
The PCa cell lines were obtained from American Type Culture Collection (Manassas, VA, USA) (DU145, PC3, 22Rv1 and LNCaP clone FGC) and European Collection of Cell Cultures (PNT2 and VCaP Public Health England, Salisbury, UK). The cells were cultured according to the manufacturer's recommendations. The prostate cohort of 49 PCa and 25 patients without PCa has been described previously (Hagman et al, 2010). It is to be noted that this is a pre-PSA screening era, TURP (transurethral resection of the prostate) cohort. Consequently, the Gleason score/WHO grade is higher and the outcome is worse than in a random contemporary PCa cohort. The patients undergoing treatment at the time of TURP received either testicular ablation or chemical treatment (zoladex, decapeptyl or eulexine). CRPC was defined as biochemical recurrence; two consecutive PSA levels >0.2 ng ml−1, one single observation >1 ng ml−1 or clinical progression. The percentage of cancer cells in the samples (5–90%) was determined by a pathologist on the adjacent slide. Detailed information regarding the clinical parameters can be seen in Supplementary Table 1. Adjacent tissue section slides were used for the immunohistochemistry (IHC) detection of AR and RNA extraction, and qRT–PCR of miR-205. For external validation of the correlation between miR-205 and AR transcript levels, we analysed a microarray data set from Taylor et al (2010) constituting 110 PCa tissue samples and 28 non-malignant adjacent benign prostate tissue samples (GEO accession number GSE21032).
IL-6 concentrations in culture lysate were determined by a commercial ELISA (cat# 900-K16, Peprotech, Rocky Hill, NJ, USA). Cells were transiently transfected with either miRIDIAN microRNA Mimic (80 nℳ, Dharmacon, Lafayette, CO, USA) or miRCURY LNA Knockdown probes (100 nℳ, Exiqon, Vedbaek, Denmark) using Oligofectamin reagent (Invitrogen, Carlsbad, CA, USA). Control experiments were performed in parallel, transfecting cells with miRIDIAN microRNA Mimic Negative Control (Dharmacon) and scramble-miR (Exiqon), respectively.
qRT–PCR
RNA was extracted from the prostate cohort and the different cell lines as described previously (Hagman et al, 2010). We also analysed the expression of miR-205 in different human tissues, with RNA extracted as described previously (Lundwall et al, 2002). The miRNA levels were quantified by the TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol and as described previously (Hagman et al, 2010). For normalising the expression in the cell lines the geometric mean of RNU48, RNU66 and RNU24 was used. For normalising the expression of miR-205 in different human tissues, the geometric mean of RNU66, RNU48, RNU44 and RNU24 was used.
Androgen receptor expression levels
Total RNA was extracted with Trizol (Invitrogen), according to the manufacturer's protocol. A total of 500 ng RNA was reverse transcribed (random hexamers and Oligo(dT) primers) using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, Thermo Scientific, Wilmington, DE, USA). The qRT–PCR was performed using specific AR primers (Applied Biosystems, cat. #4331182), and the TaqMan master mix (Applied Biosystems, cat. #4369016).
Androgen regulation
The expression level of miR-205 was determined by qRT–PCR in LNCaP cell lines in the absence or presence of androgens. Cells were seeded in normal media and the day after, media was changed to androgen-free media with charcoal-stripped fetal bovine serum (Invitrogen). After 3 days, 0, 0.1 or 1 nℳ R1881 (a synthetic androgen) was added to the medium. Total RNA was extracted 2 days later with Trizol (Invitrogen). The miR-205 levels were quantified using the TaqMan MicroRNA Assays (Applied Biosystems) as described above. RNU48 was used as endogenous control. As a positive control for the experiment, concentrations of free and total PSA were measured with a dual label immunofluorometric assay (DELFIA Prostatus PSAF/T, Perkin-Elmer Life Sciences, Waltham, MA, USA. Mitrunen et al, 1995) and concentration of KLK2 was measured as described previously (Vaisanen et al, 2006). Both PSA and KLK2 levels increased with higher concentrations of R1881.
In situ hybridisation
In situ hybridisation was performed essentially as described previously (Hansson et al, 2002), with some modifications. Briefly, formalin-fixed paraffin-embedded tissue slides were deparaffinised, rehydrated and digested with 10 μg ml−1 proteinase K (Fermentas, Thermo Scientific) in proteinase K buffer (30 mℳ Tris–HCl pH 7.5 and 10 mℳ CaCl2). The slides were refixed with 4% paraformaldehyde (pH∼9), and thereafter fixed further with 1-methylimidazol buffer (0.13 ℳ 1-methylimidazol, 300 mℳ NaCl, pH 8) for 2 × 10 min and 0.16 ℳ 1-ethyl-3(3-dimethylaminopropyl) carbodiimide for 1 h in a humidified chamber. The slides were acetylated and pre-hybridised in hybridisation buffer (50% formamide, 5 × SSC, 5 × Denhardt's, 10% dextran, 10% CHAPS, 20% Tween, 0.4 mg ml−1 salmon sperm DNA and 20 mg ml−1 Roche blocking reagent (Roche, Basel, Switzerland) at the probes Tm minus 23 °C for 1 h and then O/N at the same temperature with hybridisation buffer containing 250 nℳ locked nucleic acid (LNA) double DIG (3′ and 5′) antisense probe (Exiqon). The slides were incubated with blocking reagent (TSA kit #T20915, Invitrogen) for 1 h and then with 1 : 100 anti-DIG-AP antibody (#11093274910, Roche) for 1 h. Slides were washed, equilibrated in AP buffer (100 mℳ Tris–HCl pH 9.5, 50 mℳ MgCl2, 100 mℳ NaCl) and developed with nitro-blue tetrazolium (NBT)/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP)/levamisol solution (75 mg ml−1 NBT, 50 mg ml−1 BCIP, 0.25 mg ml−1 levamisol, 0.05% Tween-20, in AP buffer). The reaction was stopped after colour development and the slides were mounted with ProLong Gold (P36930, Invitrogen). The specificity of the in situ hybridisation was determined by a negative scramble double DIG labeled LNA probe (Exiqon) that did not give any signal (Supplementary Figure 1). We also added mimic miRNA probes with 0, 1, 2 or 3 mismatches to the hybridisation buffer, in addition to the DIG-labeled detection probe, in a competition assay. The signal was abolished when adding a surplus of mimic miRNA. The signal increased stepwise by increasing the number of mismatches in the probe (Supplementary Figure 1).
In situ hybridisation combined with IHC
To further characterise the cellular expression pattern of miR-205 in prostatic tissue, we used tyramide signal amplification (TSA) and co-detection of cytokeratin 5, a basal cells marker. After hybridisation as described above, the slides were immersed in 3% H2O2 in PBS for 30 min. The slides were blocked (TSA kit, Invitrogen) for 1 h and then incubated with 1 : 100 diluted anti-DIG antibody (#11333062910, Roche) for 1 h. The slides were incubated with anti-mouse-HRP antibody (TSA kit) for 1 h and then incubated in the dark with tyramide diluted in amplification buffer (TSA kit) for 10 min. Immunohistochemistry was performed by incubating with an antibody against cytokeratin 5 (Ab776, clone 34bE12, Abcam, Cambridge, UK), diluted 1 : 500, for 1 h. The slides were washed and incubated with anti-rabbit-alexa (#A11008, Invitrogen, green emission) for 1 h. Finally, the slides were incubated with 4′,6-diamidino-2-phenylindole (D1306, Invitrogen) for 12 min. The slides were mounted with ProLong Gold (Invitrogen) and photos were taken with Olympus AX70 microscope (Center Valley, PA, USA) (filter from Semrock for fluorescence), equipped with a Nikon digital camera (Nikon, Tokyo, Japan).
Immunoprecipitation of human AGO2 complexes in PC3 cells
The immunopurification protocols were adapted from Easow et al (2007) and Peritz et al (2006). Cells were trypsinised 2 days after transfection with miR-205 or scramble mimics, and washed in cold PBS before lysis of the cells (20 mℳ Tris–HCl pH 7.5, 150 mℳ NaCl, 0.3% Nonidet P-40, 2 mℳ EDTA, 1 mℳ NaF, 1 mℳ DTT, Protease inhibitor (Pierce, Rockford, IL, USA), Superase In (Applied Biosystems)). The lysate was kept on ice for 30 min before centrifugation at 10 000 g for 30 min at 4 °C. To reduce background signal, the lysate was pre-cleared with protein G sepharose beads (4 fast flow, GE Healthcare, Waukesha, WI, USA) blocked with 1 mg ml−1 tRNA (Applied Biosystems) and BSA (1 mg ml−1, RNase free) for 30 min on rotation at 4 °C, before incubated with antibody against human AGO2 (11A9, IgG2a, rat, Ascenion, Germany (Rudel et al, 2008)) on rotation O/N at 4 °C. Next day, the mixture was incubated with blocked protein G sepharose beads on rotation at 4 °C for 2 h. The sample was washed four times with wash buffer (50 mℳ Tris–HCl pH 7.5, 500 mℳ NaCl, 0.3% Nonidet P-40, 1 mℳ MgCl2, Superase In) and one time with PBS. Thereafter, the protein/RNA complexes were digested with 40 μg Proteinase K (Qiagen, Germantown, MD, USA) in 30 mℳ Tris–HCl pH 8, 10 mℳ EDTA, 1% SDS buffer, for 30 min at 50 °C. The RNA was isolated by phenol/chloroform extraction and EtOH precipitation essentially as described earlier (Chomczynski and Sacchi, 1987). The RNA concentration was measured using a NanoDrop (ND-1000, Spectrophotometer, Thermo Scientific) and the RNA quality assessed using a bioanalyser (Agilent 2100 Bioanalyzer, Agilent, Santa Clara, CA, USA).
Microarray hybridisation and data analysis
Single-stranded cDNA was generated using the GeneChip Whole Transcript cDNA Synthesis and Amplification Kit (Affymetrix Inc, Santa Clara, CA, USA) using 100 ng of total RNA. Amplified cDNA was fragmented and labeled using the GeneChip WT Terminal Labelling Kit (Affymetrix). Subsequently, the biotinylated cDNA was hybridised to the GeneChip Human Gene 1.0 ST Arrays (Affymetrix). The arrays were washed and stained on a GeneChip Fluidics Station 450 (Affymetrix) according to the manufacturer's recommendations. Arrays were scanned using the GeneChip Scanner 3000 and image analysis was performed using GeneChip Operating Software (Affymetrix) and in Genepix 4.0 (Axon Instruments, Molecular Devices, Sunnyvale, CA, USA). The data were normalised, background corrected and summarised using the Robust Multichip Average algorithm implemented in the Expression Console version 1.1.2 software (Affymetrix). The data were analysed using SAM analysis to identify significantly differentially expressed genes between the groups. The list of differentially expressed genes (thresh hold >1.5; and q=0) was analysed using Ingenuity Pathway Analyses (Ingenuity, Montain View, CA, USA). A score was computed for each network according to the fit of the original set of significant genes. This score reflects the negative logarithm of the P-value, which indicated the likelihood of the focus genes in a network being enriched in the data set by chance.
Western blot
Cells were lysed with M-PER (Pierce) supplemented with Halt protease inhibitor cocktail EDTA-free (Pierce) and 13.4 mℳ EDTA. Protein concentration was measured and equal amount of proteins were separated on a NuPAGE (Invitrogen) and transferred onto an Immobilon PVDF membrane (Millipore Corporation, Bedford, MA, USA). The membrane was incubated with antibodies directed against E-cadherin 1 : 2000 (#610181, BD Biosciences, San Jose, CA, USA), EPCaM 1 : 100 (sc25308, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), AR 1 : 100 (N-20, Santa Cruz Biotechnology, Inc.), GAPDH 1 : 20 000 (a-GAPDH, MAB374, Chemicon, Millipore, Temecula, CA, USA) and α-actinin (sc-17829, Santa Cruz Biotechnology, Inc.). Signals from HRP-coupled polyclonal secondary antibodies (mouse or rabbit, Dako, Glostrup, Denmark) were generated by ECL (ECL plus, GE Healthcare) and recorded using a CCD camera (LAS-3000, Fujifilm, Tokyo, Japan). Band intensities were quantified and normalised to GAPDH, using the ImageJ software.
AR 3′UTR construct and luciferase assay
Due to the length of the AR 3′UTR region (almost 7 kb) it has been cloned in several pieces, seven fragments have been described earlier (Ostling et al, 2011), but the missing part (fragment 8, located 4344–5364 bp upstream of the stop codon in the AR 3′UTR; Ostling et al, 2011) was synthesised and cloned into the pMIR-REPORT Luciferase vector (Ambion, Austin, TX, USA) by DNA2.0 (Menlo Park, CA, USA). For luciferase assays, PC3 cells were seeded 24 h before transfection. The cells were co-transfected with 1 μg of AR 3′-UTR reported plasmid, 40 ng of Renilla luciferase plasmid and with 80 nℳ of mimic with Lipofectamine 2000 (Invitrogen). Firefly luciferase and Renilla activity was assayed 48 h after transfection with a Dual-Glo Luciferase Assay System (Promega, Fitchburg, WI, USA).
In vitro attachment assay
Three days after PC3 and 22Rv1 cells were transfected, 1 × 105 cells were seeded in a 12-well plate. Five hours later, the cells were fixed and stained with sulforhodamine B (SRB), to determine cell density, LNCaP cells were allowed to attached for 24 h. Cells were fixed in 10% trichloroacetic acid and stained with 0.4% SRB (Sigma-Aldrich, St Louis, MO, USA) in 1% acetic acid for 15 min. Bound SRB was dissolved in 10 mℳ Tris base and absorbance was read at 490 nm using a microplate reader (El808, BioTek Instruments, Winooski, VT, USA).
Statistical analysis
The real-time qRT–PCR data are presented as median and analysed by the Mann–Whitney test. GraphPad Prism version 5 was used for statistical analysis (GraphPad, La Jolla, CA, USA). For in vitro assay results, we used Student's t-test, where P<0.05 was considered significant.
Results
Expression of miR-205 in a PCa cohort
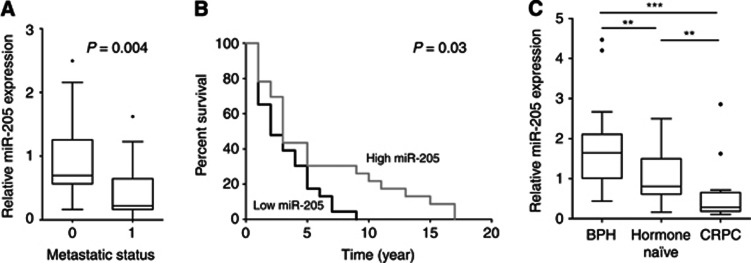
The prognostic properties of miR-205 expression levels, as measured by qRT–PCR on RNA extracted from FFPE prostatic tissues, were investigated. The clinical characteristics of the cohort have been thoroughly described previously (Hagman et al, 2010), but a shorter version can be seen in Supplementary Table 1. We found that miR-205 levels were significantly lower in patients with metastases compared with patients without metastasis (P=0.004, Mann–Whitney test), see Figure 1A. A Kaplan–Meier analysis of patient overall survival based on miR-205 expression levels divided into low (<median) and high (>median) expression, significantly divides the PCa patients into high-risk (median survival of 2 years) and low-risk patients (median survival of 3 years; P=0.025, log-rank test), with a hazard ratio of 2.33 (95% CI: 1.11–4.88), see Figure 1B. The same result was obtained if only the men that have PSA values from the diagnosis are tested. We found miR-205 level to be highest in BPH and decrease with higher WHO grade, the median miR-205 expression in BPH=1.67, WHO I=0.77, WHO II=1.03, WHO III=0.64 (Cuzick's trend test P<0.0001). The expression of miR-205 was also measured in six prostate cell lines: 22Rv1, DU145, LNCaP, PC3, VCaP and PNT2. In concordance with the findings in the primary tumour cohort, the expression was very low to undetectable in all PCa cell lines, whereas the expression was high in the immortalised normal cell line PNT2 (Supplementary Figure 2).
Figure 1.
(A) The miR-205 levels in prostate cancer patients with metastasis (denoted 1, n=18) and without metastases (denoted 0, n=17). (B) Kaplan–Meier analysis of patient survival based on miR-205 expression levels divided into low (<median) and high (>median) expression. (C) The miR-205 levels in benign hyperplasia (n=25), hormone naive (n=22) and castration-resistant prostate cancer (failed androgen ablation treatment; n=14). ** P < 0.01, *** P < 0.001.
Expression of miR-205 in the epithelial cells of prostate glands
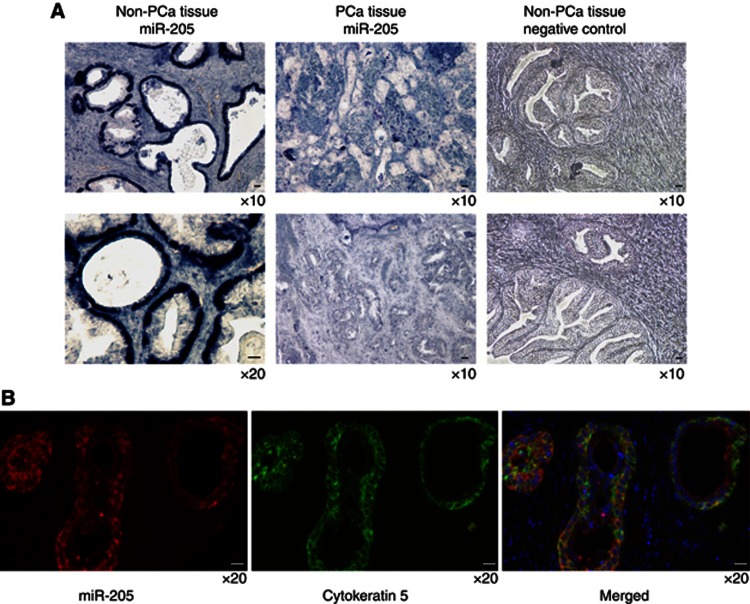
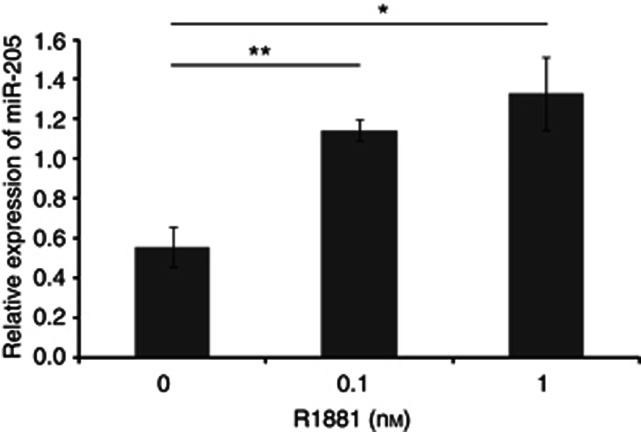
To determine the histological expression pattern of miR-205 in prostatic tissues, we performed in situ hybridisation on prostatic tissues from 10 patients using double DIG-labeled LNA antisense-miR-205. NBT/BCIP detection of miR-205 demonstrated a strong signal in the epithelial cells of the prostate glands, whereas the stromal cells lacked miR-205 expression (Figure 2A). Consistent with the qRT–PCR results we could see a lower expression of miR-205 in cancer foci. To be able to more specifically verify the cellular location, TSA amplification for detection of miR-205 expression in prostate tissue was performed. This detection method gave the same pattern as with NBT/BCIP detection; cytoplasmic within the epithelial cells. By a combined IHC and in situ hybridisation assay, to detect the expression of the prostate basal marker cytokeratin 5 and miR-205, miR-205 was found to be expressed in the basal cells (Figure 2B). The expression of miR-205 was also analysed in different human tissues using qRT–PCR (Supplementary Figure 3). Among the tissues with high miR-205 expression was breast, vas deferens, urethra, seminal vesicles, prostate and epididymis; tissues that are regulated by steroid hormones. This suggested that miR-205 might be hormonally regulated. Supporting this notion, in silico analyses of the promoter region of the miR-205 gene revealed potential androgen responsive elements. To investigate this LNCaP cells were grown in charcoal-stripped media with (0.1 or 1 nℳ R1881) or without androgen and the expression of miR-205 was measured by qRT–PCR. Expression of miR-205 was detected in androgen-depleted conditions, but increased significantly in the presence of R1881. Adding 0.1 nℳ R1881 gave a 105% increase and adding 1 nℳ R1881 gave a 139% increase; see Figure 3, indicating that androgens are not essential, but can incite miR-205 expression.
Figure 2.
(A) In situ hybridisation with double DIG-labeled LNA probe and NBT/BCIP detection. Expression of miR-205 was detected in the epithelial cells in non-cancerous prostate tissue, but not in prostate cancer foci. No signal was seen for negative control in non-cancerous prostate tissue. (B) In situ hybridisation with double DIG-labeled LNA probe and tyramide signal amplification (miR-205, red), was combined with IHC on the same tissue section for the basal cell marker cytokeratin 5 (green). The hybridisation signals for miR-205 and cytokeratin 5 co-localised suggesting that miR-205 is expressed in the basal cells in non-cancerous prostate tissue. We used DAPI for nuclear staining (blue). Scale bar=20 μm.
Figure 3.
The expression level of miR-205 increases with increasing androgen concentrations (0, 0.1 or 1 nℳ R1881). The expression of miR-205 was determined by real-time PCR and the expression of RNU48 was used as an endogenous control. *P < 0.05, **P < 0.01.
Regulation of the AR
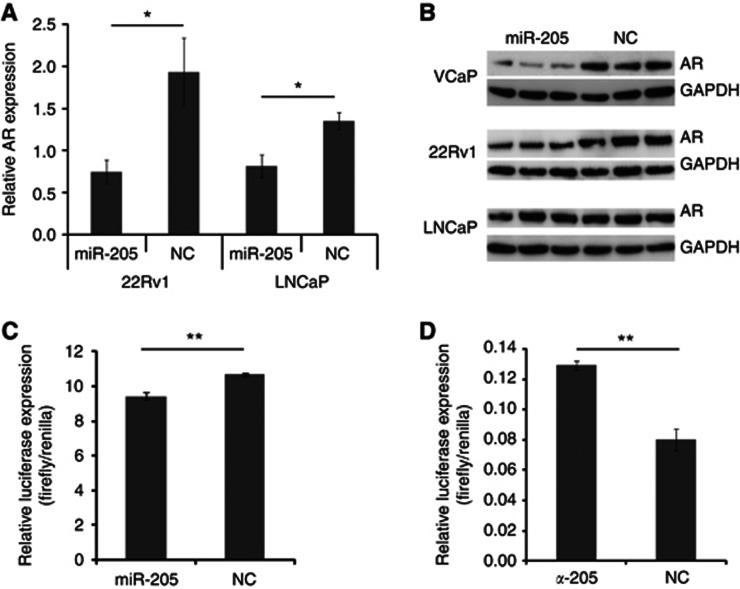
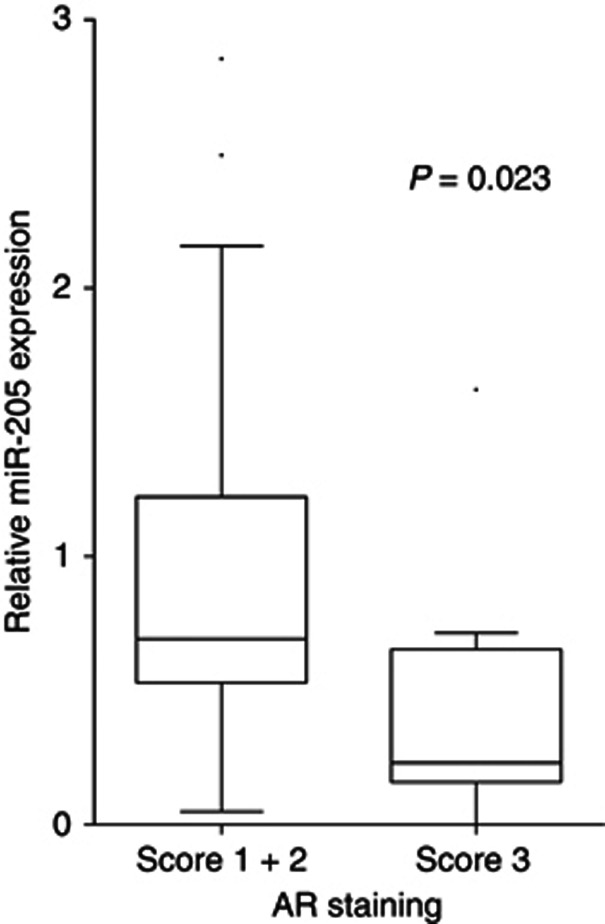
The miR-205 levels in castration-resistant patients (patients who have failed androgen ablation treatment) were found to be significantly lower than in hormone naive patients (P=0.0045; Mann–Whitney test), see Figure 1C. In addition, an inverse correlation between miR-205 levels and pre-TURP total PSA was found (P=0.019; Spearman test). As PSA is regulated by androgens through the AR, and castration resistance often is linked to increased AR levels, these findings raised the question whether miR-205 might be involved in regulation of the AR. To investigate this, we made in vitro expression studies. Ectopic expression of miR-205 resulted in decreased AR transcript levels in 22Rv1 (62% decrease, P=0.018) and in LNCaP cells (40% decrease, P=0.034), see Figure 4A. In concordance with this, ectopic expression of miR-205 also resulted in a decrease of AR protein levels in VCaP (80% decrease, P=0.015) and 22Rv1 (38% decrease, P=0.018), see Figure 4B. In the LNCaP cells, the result was spread and although there was a trend (27% less AR proteins 4 days after transfection, and 37% less AR proteins 5 days after transfection) this was not statistically significant (results not shown). Using the target prediction programme RNA22, we found miR-205-binding site in the AR 3′UTR, both in fragment 2, 4 and 8. This was tested by luciferase reporter assays in the AR-negative cell line to avoid binding to the endogenous protein. Vectors containing fragment 1–8 of the AR 3′UTR were co-transfected with miR-205 or scramble mimics. A significant reduction in luciferase signal was seen for construct 8 in PC3 (P=0.005), indicating that miR-205 can regulate the AR expression level by binding to this part of the 3′UTR. Further, blocking endogenous levels of miR-205 in PNT2 (the only cell line with high miR-205 levels as shown in Supplementary Figure 2) gave an increased binding of miR-205 to construct 8 of the AR 3′UTR, indicating that this interaction can occur in prostate cells. Next, we analysed whether miR-205 inversely correlate with AR levels in PCa patients. The AR content has previously been determined by IHC on PCa tissue in the investigated cohort (Ostling et al, 2011), and we now correlated this to miR-205 expression levels as measured by qRT–PCR on adjacent slides. We found a statistically significant inverse correlation in the malignant epithelial cells (P=0.023 for intensity score 1+2 vs 3; Mann–Whitney test), but no correlation in the benign epithelium, see Figure 5. We confirmed an inverse correlation of AR and miR-205 level in an external microarray data set of 138 men (P=0.0009, Spearman, r=−0.2788; Taylor et al, 2010).
Figure 4.
(A) Androgen receptor (AR) transcript levels, as measured by qRT–PCR, after ectopic expression of miR-205 in 22Rv1 and LNCaP cells. PGK1 and HPRT1 transcripts were used as endogenous controls. (B) Western blot on the protein levels of AR after ectopically expressing miR-205 in VCaP, 22Rv1 and LNCaP cells. GAPDH is used as a loading control. (C) Luciferase reporter assay of AR 3′UTR construct 8. PC3 cells were co-transfected with reporter construct and miRNA mimics. (D) Luciferase reporter assay of AR 3′UTR construct 8. PNT2 cells were co-transfected with reporter construct and LNA antisense. The luciferase signal was measured 24 h after transfection and the firefly luciferase was normalised to Renilla. *P < 0.05, **P < 0.01.
Figure 5.
Inverse correlation of miR-205 and AR levels in prostate malignant epithelium. miR-205 was detected with qRT–PCR in 47 prostatic cancer tissues obtained by TURP. The AR protein content in malignant epithelium was determined by immunostaining and scored by overall intensity (score: 1 weak; 2 moderate to strong; and 3 intense). Significant inverse correlation was observed for miR-205 when compared with AR intensity score 1+2 vs 3 (Mann–Whitney test).
Identification of potential miR-205 targets in PC3 cells
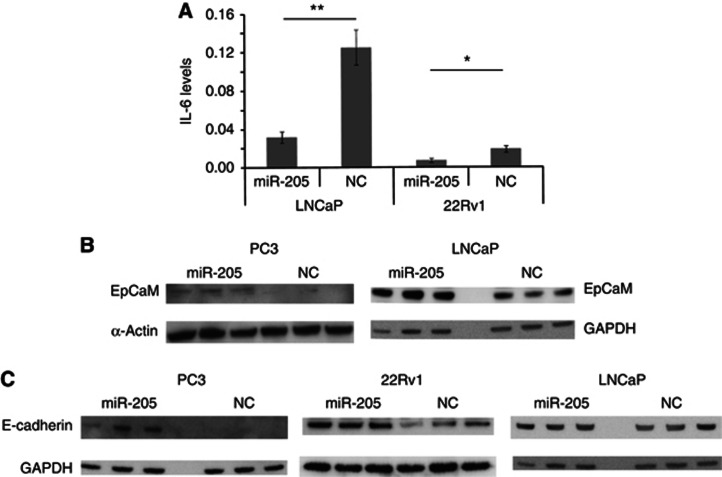
To further investigate what proteins are directly regulated by miR-205, a RIP-Chip assay was performed, and the enriched mRNAs was analysed by an Affymetrix microarray. The complete set of targets identified is provided in Supplementary Table 2, and the data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002), accession number GSE39735. Pathways in which the identified targets are enriched in were identified by Ingenuity Pathway Analysis, and pathways that are important in cancer came out at the top; for example, the MAPK/ERK, Toll-like receptor and IL-6 signaling pathways (Supplementary Table 3). We confirmed an effect on IL-6 levels by ELISA after overexpressing miR-205 (in 22Rv1 a 63% decrease, P=0.044; in LNCaP a 74% decrease P=0.008), see Figure 6A. We also identified interesting individual miR-205 targets with relevance for tumour progression, for example, EGR1, EPCaM, CCL20, FGFBP1, CDH1, FOSB and JUNB, see Table 1. We could confirm an increased level of EPCaM upon miR-205 expression in LNCaP and PC3 cells (Figure 6B). We also confirmed increased levels of E-cadherin (CDH1) upon miR-205 ectopic expression in PC3 and 22Rv1 cells, in accordance with previous studies done in the androgen-independent cell lines DU145 and PC3 cells (Gandellini et al, 2009) and in non-prostatic tissues (Gregory et al, 2008; Wiklund et al, 2011), see Figure 6C. Interestingly, we did not see this effect in the androgen-dependent LNCaP cells. We also found individual targets known to enhance the expression of AR, for example, IL-8 (fold change 3.7) and EDN1 (fold change 3.5); and targets known to inhibit AR, such as ATF3 (fold change 2.3). Furthermore, other members of the nuclear receptor family (NR4A1, NR4A2, and NR4A3) were among the miR-205 target.
Figure 6.
(A) The effect of miR-205 on the levels of IL-6 levels in LNCaP and 22Rv1 cells. (B) Western blot on the levels of EPCaM after ectopically expressing miR-205 in 22Rv1 and PC3 cells. (C) Western blot on the levels of E-cadherin after ectopically expressing miR-205 in PC3, 22Rv1, and LNCaP cells. GAPDH or α-actinin was used as loading controls. *P < 0.05, **P < 0.01.
Table 1. Molecules identified by RIP-ChiP, fold change in miR-205 expressing cells compared with control cells.
| Gene symbol | Gene description | Fold change |
|---|---|---|
| NR4A2 |
Nuclear receptor subfamily 4, group A, member 2 |
5.6 |
| EGR1 |
Early growth response 1 |
5.1 |
| NR4A1 |
Nuclear receptor subfamily 4, group A, member 1 |
5.1 |
| EPCAM |
Epithelial cell adhesion molecule |
4.9 |
| CCL20 |
Chemokine (C–C motif) ligand 20 |
4.5 |
| C1orf116 |
Chromosome 1 open reading frame 116, SARG |
3.9 |
| FGFBP1 |
Fibroblast growth factor-binding protein 1 |
3.9 |
| CDH1 |
Cadherin 1, type 1, E-cadherin (epithelial) |
3.7 |
| IL8 |
Interleukin 8 |
3.7 |
| EDN1 |
Endothelin-1 |
3.5 |
| FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
2.9 |
| JUNB | Jun B proto-oncogene | 2.3 |
miR-205 affects adhesion of PCa cells
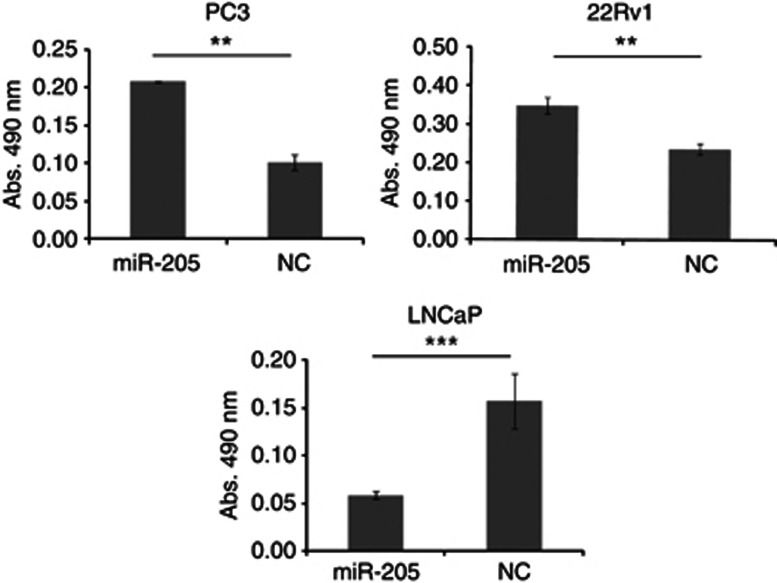
miR-205 has previously been reported to affect cell proliferation, apoptosis, colony formation, migration and invasion of PCa cells (Gandellini et al, 2009; Majid et al, 2010). We assessed the effect of miR-205 on cell adhesion using a gain-of-function approach. Ectopic expression of miR-205 in androgen-independent PC3 and 22Rv1 cells gave a 103% and 47% increase in adhesion, respectively (P=0.003 resp. P=0.001). Transfection of androgen-dependent LNCaP cells on the contrary, resulted in 73% less adhesion compared with negative control-transfected cells (P=0.0004), see Figure 7. Ectopic expression of miR-205 or blocking of miR-205 did not affect adhesion in PNT2 (results not shown).
Figure 7.
The effect of miR-205 ectopic expression on the attachment potential of PC3, 22Rv1 and LNCaP cells, as measured by a sulforhodamine B assay. **P < 0.01, ***P < 0.001.
Discussion
In the present study, we examined the expression pattern and potential targets of miR-205 in PCa. We and others have seen that miR-205 is significantly downregulated in PCa specimens and the same was found to be true in prostatic cancer cell lines consistent with previous reports (Porkka et al, 2007; Gandellini et al, 2009; Majid et al, 2010; Schaefer et al, 2010). Furthermore, miR-205 expression was higher in patients with localised disease compared with patients with distant metastases, confirming earlier results (Gandellini et al, 2009; Tucci et al, 2012). Overall survival was 1 year longer in the group with high levels of miR-205. Furthermore, the expression of miR-205 was found to be localised to the basal cells of the prostate, in agreement with a report published while this paper was prepared (Gandellini et al, 2012).
Transient expression of miR-205 was found to affect the levels of AR in androgen-dependent VCaP and 22Rv1 cells. We have data indicating binding to the AR 3′UTR by a luciferase assay, both when overexpressing ectopic miR-205 in PC3 and when blocking endogenous levels in PNT2. In LNCaP, there is an effect on mRNA level, but no statistically significant effect on protein levels, although there is a trend. The lack of difference on AR protein level in LNCaP is in accordance with an earlier report (Boll et al, 2012). Potentially, miR-205 has other targets in LNCaP that repress AR protein levels and thereby counteract the effect of miR-205 on AR protein levels. One possibility is degradation of phosphorylated AR by the PI3K–AKT pathway (Lin et al, 2002), which is induced by growth factors such as PDGFA/B and executed via growth factor receptors such as IGF1R, these are proteins that was present in the IP from LNCaP cells (Boll et al, 2012), but not in the PC3 cells.
In the PCa patient cohort, the expression of miR-205 was found to be inversely correlated to AR. This was also verified at transcript level in an external PCa cohort (Taylor et al, 2010). In addition, we found that the level of miR-205 inversely correlated to serum levels of the androgen-regulated PSA in our cohort, corroborating the targeting of AR signaling. Moreover, miR-205 was lower in castration-resistant patients where AR is frequently upregulated (Feldman and Feldman, 2001), compared with hormone naive patients. It has been observed that many miRNAs and their targets are expressed in a mutually exclusive manner, either temporally or spatially (Stark et al, 2005). Targets are typically present in foci adjacent to the miRNA-expressing cells. We found that miR-205 is expressed mainly in the basal cells of prostatic glands. The AR expression is mainly localised to the luminal cells, and we did not detect AR in the basal cells, in concordance with earlier results showing that AR expression is undetectable (Prins et al, 1991; Uzgare et al, 2004) or very low (Bonkhoff et al, 1998) in basal cells. In the RIP-Chip assay, miR-205 was found to target pathways important for cancer progression, for example, the IL-6 signaling that was verified in two PCa cell lines. There were also several previously reported miR-205 targets from other tissues identified by the RIP-Chip assay, such as PKP3, IL6, CDH3, EDN1 and FOS (Gandellini et al, 2009; Supplementary Table 5); IL-24 (Majid et al, 2010); CYR61 (Xie et al, 2012); EGR1 and IL-18 (Greene et al, 2010; Supplementary Table 1). However, when comparing our data from PC3 cells to a similar RIP-Chip assay performed in LNCaP cells (Boll et al, 2012) the results are very different. Only 4% of the miR-205 targets (with a thresh hold of >1.5 fold) we found in PC3 cells overlapped with the miR-205 targets reported in LNCaP (Boll et al, 2012). The discrepancy could be due to differences in methodology, the available transcriptomes (e.g., the androgen-regulated KLK2 identified in the LNCaP cells is not expressed in PC3 cells) or target sequences variations, but also that miR-205 might be acting by other mechanisms in an androgen-independent setting compared with an androgen dependent. It is possible that cofactors essential for miR-205 action are repressed by androgen signaling. This hypothesis is strengthen by a resent paper that show that miR-205 negatively affect viability only in the absence of androgen (Hulf et al, 2012). This is an interesting concept as the natural setting of miR-205 is in the AR-devoid basal cells, hence, the augmenting effect of androgens on miR-205 expression could be counteracted in AR-containing cells by an alternative mechanism ensuring that miR-205 action is not occurring outside the original site of expression. This would imply that if endogenous miR-205 was used therapeutically, it would decrease the AR in the androgen-independent malignant epithelial, luminar and possibly stem cells, but without affecting the benign epithelial cells. As we continued to investigate other targets of miR-205, we found more examples of differential regulation in the androgen-dependent and -independent cell lines. Ectopic expression of miR-205 increases E-cadherin protein levels in 22Rv1 and PC3 cells (also seen in DU145; Gandellini et al, 2009), but not in LNCaP cells. We also find differences between the androgen-dependent and -independent cell lines when studying adhesion. Ectopic expression of miR-205 induces increased adhesion in PC3 and 22RV1 cells, but decreased adhesion in LNCaP cells (in PNT2 adhesion was not affected by miR-205). These results indicate that a decrease of miR-205 in PCa would give less adhesion in an androgen-independent setting, with concomitant increase in migration and invasion. This could possibly accounts for parts of the increased risk of metastasis that was seen in the patient cohort.
One of the few pathways that were affected by miR-205 both in PC3 and LNCaP cells was the MAPK pathway, however, targeting different transcripts in the pathway. MAPK signaling is involved in AR phosphorylation, which has been suggested to increase the ligand-independent activation of the AR under low androgen conditions such as castration. The MAPK pathway is also an oncogenic pathway that can promote growth and survival.
In conclusion, we find that miR-205 is correlated to overall survival and inversely correlated with negative prognostic parameters such as occurrence of distant metastases and castration resistance. This could be explained by the cellular location of miR-205 and the androgen-influenced effects on individual oncogenes and oncogenic pathways. These findings might have therapeutic implications especially for the castration resistant and currently untreatable form of PCa.
Acknowledgments
We thank Elise Nilsson for preparing prostatic tissue samples and Margareta Persson for technical assistance. AR 3′UTR constructs was a kind gift from Päivi Östling (Medical Biotechnology, VTT Technical Research Centre of Finland and Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, Turku, Finland). The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement no. 201438, the Swedish Research council, Gunnar Nilssons Cancer foundation, Jeanssons foundation and Gyllenstiernska Krapperups foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. doi: 10.1038/cddis.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Verhaegh G, Schalken J, von Bergen M, Horn F, Hackermuller J. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2012;32 (3:277–285. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H, Fixemer T, Remberger K. Relation between Bcl-2, cell proliferation, and the androgen receptor status in prostate tissue and precursors of prostate cancer. Prostate. 1998;34:251–258. doi: 10.1002/(sici)1097-0045(19980301)34:4<251::aid-pros2>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- Fuse M, Nohata N, Kojima S, Sakamoto S, Chiyomaru T, Kawakami K, Enokida H, Nakagawa M, Naya Y, Ichikawa T, Seki N. Restoration of miR-145 expression suppresses cell proliferation, migration and invasion in prostate cancer by targeting FSCN1. Int J Oncol. 2011;38:1093–1101. doi: 10.3892/ijo.2011.919. [DOI] [PubMed] [Google Scholar]
- Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, Valdagni R, Daidone MG, Zaffaroni N. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- Gandellini P, Profumo V, Casamichele A, Fenderico N, Borrelli S, Petrovich G, Santilli G, Callari M, Colecchia M, Pozzi S, De Cesare M, Folini M, Valdagni R, Mantovani R, Zaffaroni N. miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell Death Differ. 2012;19 (11:1750–1760. doi: 10.1038/cdd.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123:606–618. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- Hansson J, Bjartell A, Gadaleanu V, Dizeyi N, Abrahamsson PA. Expression of somatostatin receptor subtypes 2 and 4 in human benign prostatic hyperplasia and prostatic cancer. Prostate. 2002;53:50–59. doi: 10.1002/pros.10121. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hulf T, Sibbritt T, Wiklund ED, Bert S, Strbenac D, Statham AL, Robinson MD, Clark SJ. Discovery pipeline for epigenetically deregulated miRNAs in cancer: integration of primary miRNA transcription. BMC Genomics. 2011;12:54. doi: 10.1186/1471-2164-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulf T, Sibbritt T, Wiklund ED, Patterson K, Song JZ, Stirzaker C, Qu W, Nair S, Horvath LG, Armstrong NJ, Kench JG, Sutherland RL, Clark SJ.2012Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer Oncogenee-pub ahead of print 6 August 2012; doi: 10.1038/onc.2012.300 [DOI] [PubMed]
- Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, Seppala J, Lahdesmaki H, Tammela TL, Visakorpi T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 2012;31 (41:4460–4471. doi: 10.1038/onc.2011.624. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, Rotter V, Akslen LA, Oyan AM, Kalland KH. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larne O, Martens-Uzunova E, Hagman Z, Edsjo A, Lippolis G, Vredenbregt-van den Berg MS, Bjartell A, Jenster G, Ceder Y.2012miQ—a novel microRNA based diagnostic and prognostic tool for prostate cancer Int J Cancerdoi: 10.1002/ijc.27973 [DOI] [PubMed]
- Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundwall A, Bjartell A, Olsson AY, Malm J. Semenogelin I and II, the predominant human seminal plasma proteins, are also expressed in non-genital tissues. Mol Hum Reprod. 2002;8:805–810. doi: 10.1093/molehr/8.9.805. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrunen K, Pettersson K, Piironen T, Bjork T, Lilja H, Lovgren T. Dual-label one-step immunoassay for simultaneous measurement of free and total prostate-specific antigen concentrations and ratios in serum. Clin Chem. 1995;41:1115–1120. [PubMed] [Google Scholar]
- Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjo A, Kangaspeska S, Edgren H, Nicorici D, Bjartell A, Ceder Y, Perala M, Kallioniemi O. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Prins GS, Birch L, Greene GL. Androgen receptor localization in different cell types of the adult rat prostate. Endocrinology. 1991;129:3187–3199. doi: 10.1210/endo-129-6-3187. [DOI] [PubMed] [Google Scholar]
- Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Muller PA, Dotsch V, Kehrloesser S, Sayan BS, Giaccone G, Lowe SW, Takahashi N, Vandenabeele P, Knight RA, Levine AJ, Melino G. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzgare AR, Xu Y, Isaacs JT. In vitro culturing and characteristics of transit amplifying epithelial cells from human prostate tissue. J Cell Biochem. 2004;91:196–205. doi: 10.1002/jcb.10764. [DOI] [PubMed] [Google Scholar]
- Vaisanen V, Peltola MT, Lilja H, Nurmi M, Pettersson K. Intact free prostate-specific antigen and free and total human glandular kallikrein 2. Elimination of assay interference by enzymatic digestion of antibodies to F(ab')2 fragments. Anal Chem. 2006;78:7809–7815. doi: 10.1021/ac061201+. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS, Borre M, Peter ME, Orntoft TF, Kjems J, Clark SJ. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- Xie H, Zhao Y, Caramuta S, Larsson C, Lui WO. miR-205 expression promotes cell proliferation and migration of human cervical cancer cells. PLoS One. 2012;7:e46990. doi: 10.1371/journal.pone.0046990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.