Abstract
Background:
As epidermal growth factor receptor (EGFR) is involved in the pathogenesis of malignant pleural mesotheliomas (MPMs), the anti-EGFR drugs may be effective in treating MPM patients. Mutations of the EGFR gene or its downstream effectors may cause constitutive activation leading to cell proliferation, and the inhibition of apoptosis and metastases. Consequently, molecular profiling is essential for select patients with MPM who may respond to anti-EGFR therapies.
Methods:
After manual macrodissection, genomic DNA was extracted from 77 histological samples of MPM: 59 epithelioid, 10 biphasic, and 8 sarcomatoid. Epidermal growth factor receptor gene mutations were sought by means of real-time polymerase chain reaction (PCR) and direct sequencing, KRAS gene mutations by mutant-enriched PCR, and PIK3CA and BRAF gene mutations by direct sequencing.
Results:
Gene mutations were identified in nine cases (12%): five KRAS, three BRAF, and one PI3KCA mutation; no EGFR gene mutations were detected. There was no difference in disease-specific survival between the patients with or without gene mutations (P=0.552).
Conclusions:
Mutations in EGFR downstream pathways are not rare in MPM. Although none of those found in this study seemed to be prognostically significant, they may support a more specific selection of patients for future trials.
Keywords: malignant mesothelioma, EGFR, mutation analysis, target therapy
Malignant mesotheliomas are relatively rare, highly malignant tumours that arise from the mesothelial cells lining the serosal cavities of the body. The most frequent are malignant pleural mesotheliomas (MPMs) (Boutin et al, 1998), whose estimated annual incidence in Europe (15–33 cases per million inhabitants) is expected to increase further over the next 20 years because of their long period of latency (Peto et al, 1999). The main carcinogen associated with MPMs is asbestos, and occupational exposure to it has been considered the main risk factor (Wagner et al, 1960; Lanphear and Buncher, 1992; Carbone et al, 2002); however, other potential carcinogenic agents include infection by Simian Virus 40 and radiation exposure (Yang et al, 2008).
The median survival of patients with MPM is currently 12 months from the time of diagnosis despite treatment (Vogelzang et al, 2003). The European Organisation for Research and Treatment of Cancer has indicated that the main predictors of a negative prognosis are a poor performance status (PS), high white blood cell counts, male gender, a sarcomatoid subtype, anaemia, and thrombocytosis (Curran et al, 1998), but it has been recently suggested that short survival may also be related to a high nuclear tumour grade, a high MIB-1 labelling index (Comin et al, 2000), and high epidermal growth factor receptor (EGFR) expression in tumour cells (Rena et al, 2011).
There is no standard of care for MPM, but systemic chemotherapy is the only possible treatment option for most patients at the time of diagnosis. It has been found that pemetrexed combined with cisplatin is more effective than platinum alone in terms of overall survival (12.1 vs 9.3 months), time to progression (5.7 vs 3.9 months), and objective responses (41.3% vs 16.7% of tumour shrinkage) (Vogelzang et al, 2003), but, as most patients progress during or shortly after these first-line treatments, there is a clear need to develop more effective antitumour agents.
Epidermal growth factor receptor is a receptor tyrosine kinase that is overexpressed in 60–70% of MPM tissue specimens (mainly of the epithelial subtype), but not in the normal mesothelium (Dazzi et al, 1990; Destro et al, 2006; Okuda et al, 2008). Exposure to asbestos fibres causes EGFR aggregation, and the subsequent autophosphorylation and activation of EGFR activates both the RAS/RAF/MAPK pathway (which induces cell proliferation, metastasis, and invasion) (Pache et al, 1998) and the PI3KCA/AKT/mTOR pathway, which leads to the inhibition of apoptosis (Suzuki et al, 2009). Consequently, inhibiting EGFR pathways should have an antitumoral effect. Two classes of EGFR inhibitors have been developed for cancer therapy: tyrosine kinase inhibitors (TKIs), which block EGFR autophosphorylation by competing with ATP binding, and anti-EGFR monoclonal antibodies (mABs), which compete with the ligand binding the extracellular domain of EGFR. It has been reported that mutation analysis of the EGFR gene and of some of its downstream signal-transduction proteins predicts the response of lung adenocarcinomas to TKIs (Shepherd et al, 2005) and the response of colorectal cancer to anti-EGFR mABs (cetuximab, panitumumab) (Jonker et al, 2007).
Preclinical studies have shown that EGFR TKIs are highly efficacious (Barbieri et al, 2011), but two phase II studies of gefitinib and erlotinib used alone to treat malignant pleural and peritoneal mesotheliomas failed to demonstrate their clinical efficacy, although it needs to be pointed out that the patients in both trials were not selected on the basis of any molecular criteria (Govindan et al, 2005; Garland et al, 2007). One recent study has shown that cetuximab effectively blocks the growth of MPM cells in cell cultures and mouse models (Kurai et al, 2012) and, as in the case of colorectal cancer and lung adenocarcinomas, the potential efficacy of these TKIs in MPM may depend on the mutation status of EGFR gene and its downstream effectors (Lièvre et al, 2006; Pirker et al, 2011).
To the best of our knowledge, only a few low-powered studies have investigated the presence and frequency of EGFR gene mutations in MPM (Cortese et al, 2006; Enomoto et al, 2012), and none has searched for mutations in the KRAS, BRAF, and PI3KCA downstream effectors. We searched a large series of histological MPM samples for mutations in EGFR gene and its main downstream signalling effectors to evaluate their frequency and possible prognostic significance, and their possible use as predictors of the response of MPMs to targeted therapies.
Materials and methods
Patients and samples
The study involved 77 consecutive MPM patients admitted to the Thoracic Unit of the University Hospital of Novara between January 2008 and December 2010, all of whom were diagnosed as having MPM on the basis of multiple pleural biopsies taken by means of video-assisted thoracoscopy. The tumour samples were immediately fixed in formalin for 24 h, embedded in paraffin, and routinely processed for histology and immunohistochemistry, and the diagnosis of MPM was based on standard histological and immunohistochemical criteria, including positivity to calretinin, vimentin, and cytokeratins 5 and 6, and negativity to carcinoembryonic antigen, thyroid transcription factor 1, and Ber Epy 4. The MPMs were classified on the basis of the WHO classification of pleural tumours (Travis et al, 2004), and clinically and pathologically staged on the basis of the TNM staging system (Sobin et al, 2009). The patients' PS at the time of diagnosis was graded using the Eastern Cooperative Oncology Group (ECOG) scale (Oken et al, 1982), and the patients with a PS of 0–2 underwent therapeutic protocols indicated by the referring oncologist.
Haematoxylin/eosin-stained slides of the pleural biopsies and corresponding formalin-fixed, paraffin-embedded blocks were reviewed by a pathologist (RB) to select the area with >50% of tumour cells.
DNA extraction
The tumoral areas of the formalin-fixed, paraffin-embedded sections were macrodissected manually, and then five 5-μm-thick sections were prepared and collected in a 1.5 ml tube. Genomic DNA was extracted using EDTA-SDS/proteinase K followed by phenol–chloroform, and resuspended with 30 μl of DEPC-treated and RNAse-free water (Promega, Madison, WI, USA).
Mutational analysis
Epidermal growth factor receptor gene
All of the samples were analysed using the TheraScreen EGFR29 Mutation Kit (Qiagen, Manchester, UK), which combines the two technologies of ARMS and Scorpion chemistry to detect mutations in a real-time polymerase chain reaction (PCR). This kit allows the detection of in-frame deletions on exon 19, insertions on exon 20, and G719X, S768I, T790M, L858R, and L861Q mutations against a background of WT genomic DNA with a sensitivity of 1%. Starting from 2 μl of genomic DNA, the analyses were made in accordance with the manufacturer's instruction using RotorGene Q (Qiagen), and the results were interpreted following the datasheet.
To determine the presence of other less common mutations, the samples underwent further PCRs to amplify the whole sequence of exons 18–21 of the EGFR gene as described previously by Paez et al (2004). Table 1 shows the primers and PCR conditions.
Table 1. Sequences of primers and PCR reaction protocol.
| Gene | Primer name | Sequence | Cycle | Length |
|---|---|---|---|---|
| KRAS codon 12 (outer) | 3F | 5′-ACTGAATATAAACTTGTGGTAGTTGGACCT–3′ | 95 °C × 10 min; (95 °C × 30 s, 50 °C × 1 min, 72 °C × 1 min) × 20 cycles; 72 °C × 3 min | 143 |
| |
10B |
5′-ACTCATGAAAATGGTCAGAGAAACCTTTAT-3′ |
|
|
| KRAS codon 13 (outer) | 9F | 5′-ACTGAATATAAACTTGTGGTAGTTGGCCCTGGT-3′ | 95 °C × 10 min; (95 °C × 30 s, 54 °C × 1 min, 72 °C × 1 min) × 20 cycles; 72 °C × 3 min | 113 |
| |
10B |
5′-ACTCATGAAAATGGTCAGAGAAACCTTTAT-3′ |
|
|
| KRAS codon 12 (inner) | 3F | 5′-ACTGAATATAAACTTGTGGTAGTTGGACCT-3′ | 95 °C × 10 min; (95 °C × 30 s, 54 °C × 1 min, 72 °C × 1 min) × 45 cycles; 72 °C × 3 min | 143 |
| 14B | 5′-TCAAAGAATGGTCCTGGACC-3′ | |||
| KRAS codon 13 (inner) | 9F | 5′-ACTGAATATAAACTTGTGGTAGTTGGCCCTGGT-3′ | 113 | |
| |
4B |
5′-TCAAAGAATGGTCCTGCACC-3′ |
|
|
| EGFR exon 18 | EGFR18F | 5′-TCCAGCATGGTGAGGGCTGAG-3′ | 50 °C × 2 min; 95 °C × 10 min; (95 °C × 40 s, 58 °C × 40 s, 72 °C × 35 s) × 40 cycles; 72 °C × 3 min | 242 |
| EGFR18R | 5′-GGCTCCCCACCAGACCATG-3′ | |||
| EGFR exon 19 | EGFR19F | 5′-TGGGCAGCATGTGGCACCATC-3′ | 217 | |
| EGFR19R | 5′-AGGTGGGCCTGAGGTTCAG-3′ | |||
| EGFR exon 20 | EGFR20F | 5′-CCTCCTTCTGGCCACCATGCG-3′ | 296 | |
| EGFR20R | 5′-CATGTGAGGATCCTGGCTCC-3′ | |||
| EGFR exon 21 | EGFR21F | 5′-CCTCACAGCAGGGTCTTCTC-3′ | 229 | |
| |
EGFR21R |
5′-CCTGGTGTCAGGAAAATGCT-3′ |
|
|
| BRAF exon 15 | BRAF15F | 5′-TCATAATGCTTGCTCTGATAGGA-3′ | 95°C × 10 min; (95 °C × 15 s, 52 °C × 30 s, 72 °C × 30 s) × 45 cycles; 72 °C × 3 min | 224 |
| |
BRAF15R |
5′-GGCCAAAAATTTAATCAGTGGA-3′ |
|
|
| PIK3CA exon 9 | PIK3CA9F | 5′-GGGAAAAATATGACAAAGAAAGC-3′ | 95 °C × 10 min; (95 °C × 35 s, | 251 |
| PIK3CA9R | 5′-CTGAGATCAGCCAAATTCAGTT-3′ | 56 °C × 30 s, 72 °C × 30 s) × 40 cycles; 72 °C × 10 min | ||
| PIK3CA exon 20 | PIK3CA20FPIK3CA20R | 5′-CTCAATGATGCTTGGCTCTG-3′5′-TGGAATCCAGAGTGAGCTTTC-3′ | 241 |
KRAS gene
The KRAS gene was analysed by means of a mutant-enriched PCR (ME-PCR) to detect the hotspots in codons 12 and 13 of exon 2 that include more than 95% of the known gene mutations. The ME-PCR consisted of two amplification steps (seminested PCR) in which artificial restriction sites were introduced into the wild-type amplicon using mismatched primers (Table 1). The restriction sites (BstNI for codon 12 and BglI for codon 13) introduced during the first PCR step were localised immediately next to the KRAS codon in the analysis to distinguish wild-type and mutant sequences. The wild-type amplicons were then digested by BstNI or BglI restriction enzymes, and the mutant products were enriched for a second round of amplification. The ME-PCR and digestion conditions have been described previously (Molinari et al, 2011). Mutant-enriched PCR has a sensitivity of up to 0.01%. All of the samples underwent automated sequencing by using an ABI PRISM 3130 (Applied Biosystems, Foster City; CA, USA) and reverse primers.
BRAF gene
Exon 15 of the BRAF gene (which contains the hotspot codon 600, where more than 90% of gene mutations occur) was analysed by means of direct sequencing in accordance with previously published protocols (Di Nicolantonio et al., 2008), starting from 50 ng of genomic DNA. The primers and PCR conditions are shown in Table 1.
PI3KCA gene
The analysis of the PIK3CA gene was concentrated on exons 9 and 20, which include all of the hotspot codons, using previously published protocols (Sartore-Bianchi et al., 2009). The primers and PCR conditions are shown in Table 1.
Sequence analysis
All of the PCR products and KRAS second enzymatic digestions were analysed by means of 3% agarose gel electrophoresis, and then purified using NucleoSpin Gel and the PCR clean-up kit (Macherey-Nagel, Düren, Germany). The sequence of each gene was analysed using an ABIPrism 3130 Genetic Analyzer (Applied Biosystems), and all of the mutated cases were confirmed two times starting from independent PCR reactions.
Statistical analysis
This examined the correlations between the presence of gene mutations and other demographic, clinical and pathological variables. The associations between categorical variables were determined using the χ2 or Fisher's exact test. The statistical differences of the average values were tested using a Student's t-test and analysis of variance, followed by Bonferroni's test.
The impact of the different variables on long-term outcomes was analysed using the Kaplan–Meier method of analysing disease-specific survival (DSS); the survival data were compared using the log-rank test.
P-values of <0.05, with a 95% confidence interval, were considered statistically significant.
Results
In all, 57 patients were male (74%) and 20 were female (26%); their average age at the time of diagnosis was 68 years (range 43–90, median 64.5 years). Of these, 50 patients (64.9%) had previously been exposed to asbestos at work. Histological examination showed that 59 MPMs (77%) were epithelioid, 10 (13%) biphasic, and 8 (10.4%) sarcomatoid. In total, 41 patients had stage II tumours, 30 stage III tumours, and 6 stage IV tumours. Eastern Cooperative Oncology Group PS was 0–2 in 68 patients, and >2 in nine patients. In all, 41 patients were treated with platinum plus pemetrexed and 22 with platinum alone; 14 received no treatment because their PS was >2 or because they refused.
Follow-up data were collected from 74 patients (three were lost to follow-up). In all, 15 patients were still alive at June 2012 with a median follow-up of 24.5 months (range 14–39 months). The median DSS of the cohort as a whole was 12.5 months (range 1–39 months).
Mutation analysis
Mutations in the EGFR downstream pathway were identified in nine patients (12%): five in the KRAS gene, three in the BRAF gene, and one in the PIK3CA gene. No mutations were detected in the EGFR gene by direct sequencing or the Scorpions-ARMS assay, even though the latter has a sensitivity of 1% (vs the 10–20% of direct sequencing).
KRAS and BRAF gene mutational profiling
The KRAS gene was successfully amplified in all of the samples, five of which showed mutations: two patients had the GGT→GtT point mutation in codon 12 leading to a glycine-to-valine amino-acid substitution (G12V); two had the GGC→GaC point mutation in codon 13 leading to a glycine-to-aspartic acid substitution (G13D); and one had the rare GGC→aGC mutation in codon 13 leading to a glycine-to-serine substitution (G13S). As shown in Table 2, three of the five mutations occurred in patients with epithelioid MPMs (G12V, G13D, and G13S), one in a patient with a biphasic MPM (G13D), and one in a patient with a sarcomatoid subtype (G12V). All five patients with KRAS mutations reported previous occupational asbestos exposure.
Table 2. Characteristics of patients with gene mutations.
| Patient | Gene | Amino-acid substitution | Gender | Age | Histotype | Asbestos exposure | DSS |
|---|---|---|---|---|---|---|---|
| 1 | KRAS | G12V | Male | 81 | Sarcomatoid | Yes | 4 |
| 2 | G12V | Male | 55 | Epithelioid | Yes | 14 | |
| 3 | G13D | Male | 82 | Epithelioid | Yes | 4 | |
| 4 | G13S | Male | 60 | Epithelioid | Yes | 5 | |
| 5 |
|
G13D |
Male |
77 |
Biphasic |
Yes |
19 |
| 6 | BRAF | V600E | Female | 51 | Epithelioid | None | 9 |
| 7 | V600E | Male | 57 | Biphasic | None | 19 | |
| 8 |
|
V600E |
Male |
73 |
Epithelioid |
None |
33 |
| 9 | PIK3CA | M1040I | Male | 68 | Biphasic | None | 7 |
Abbreviation: DSS=disease-specific survival.
The BRAF gene mutational analysis showed the classical valine-to-glutamic amino-acid substitution in codon 600 (V600E) in three patients: two with epithelioid MPMs and one with a biphasic tumour (Table 2). None of them reported previous occupational asbestos exposure.
PI3KCA gene mutational profiling
The DNA of exons 9 and 20 of the PIK3CA gene was successfully amplified from 75 of the 77 specimens. A point mutation was detected in only one case: it occurred in exon 20, and led to a methionine-to-isoleucine substitution in position 1040 (M1040I). The patient had a biphasic mesothelioma and no previous occupational asbestos exposure (Table 2).
Statistical analysis
The correlations between the presence/absence of gene mutations and demographic, clinical, and pathologic features (gender, age, occupational asbestos exposure, history of previous cancer, histological type, ECOG PS, treatment) were investigated, without finding any significant differences (Table 3).
Table 3. Statistical correlation between demographic, clinical, pathological data, and gene mutations.
| Wild type (n=68) | Mutations (n=9) | P-value | |
|---|---|---|---|
| Age, mean±s.d. (years) | 66±21 | 67±12 | 0.89 |
| Gender (male proportion) | 49/68 | 8/9 | 0.491 |
| Previous cancer | 5/68 | 2/9 | 0.385 |
| Asbestos exposure |
44/68 |
6/9 |
0.799 |
|
Histological subtype | |||
| Epithelial | 54/68 | 5/9 | 0.241 |
| Biphasic | 7/68 | 3/9 | 0.135 |
| Sarcomatoid |
7/68 |
1/9 |
0.623 |
|
ECOG score | |||
| 0–2 | 60/68 | 8/9 | 0.892 |
| >2 |
8/68 |
1/9 |
0.617 |
|
Clinical stage | |||
| II | 37/68 | 4/9 | 0.689 |
| III–IV |
31/68 |
5/9 |
0.576 |
|
Treatment type | |||
| None | 13/68 | 1/9 | 0.623 |
| Platinum | 19/68 | 3/9 | 0.876 |
| Platinum+pemetrexed | 36/68 | 5/9 | 0.776 |
Abbreviations: ECOG=Eastern Cooperative Oncology Group; s.d.=standard deviation.
The Kaplan–Meier analysis of the influence of some variables on long-term outcomes revealed no difference in DSS between the patients with and without gene mutations (P=0.552). Moreover, separate evaluation of the patients with KRAS and BRAF mutations did not indicate any advantage in terms of DSS (P=0.363 and 0.752) and, within the mutated group, no mutation significantly correlated with DSS (KRAS, P=0.363; BRAF, P=0.187).
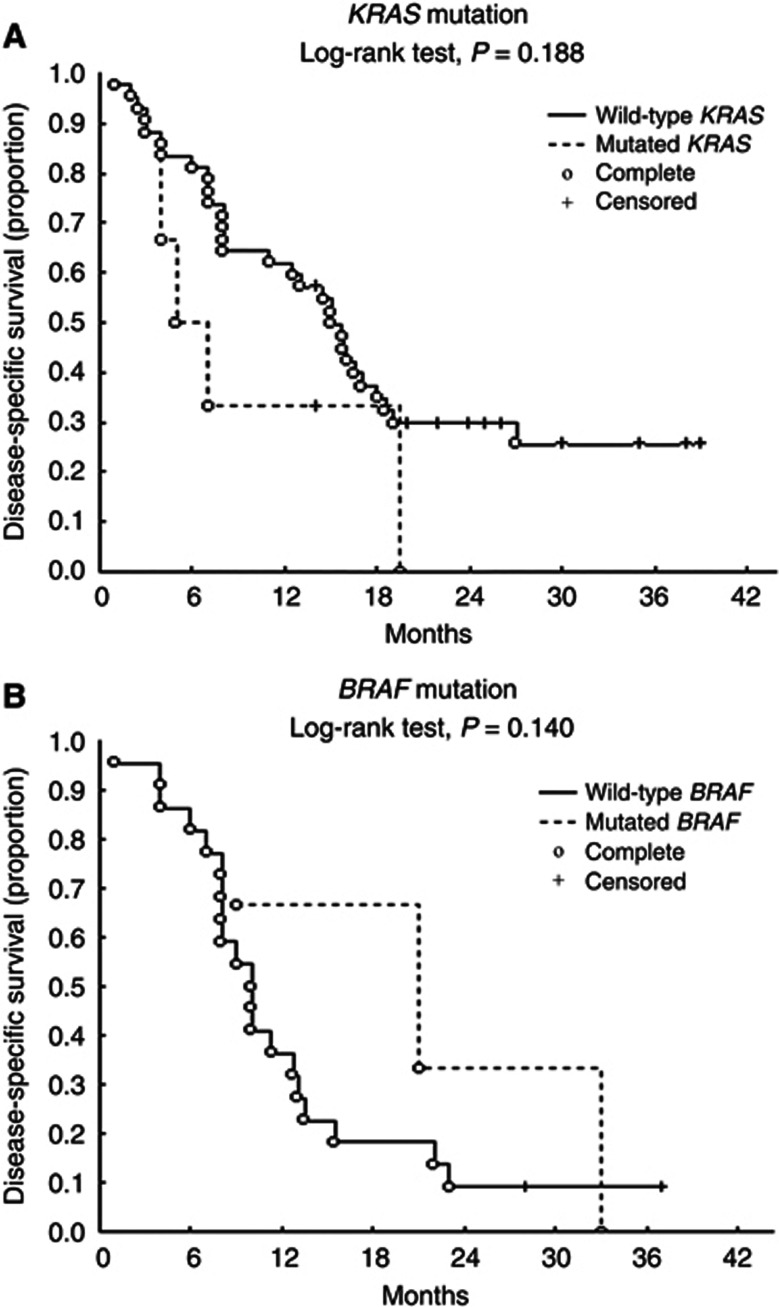
Interestingly, the patients with KRAS gene mutations reported occupational asbestos exposure, whereas those with BRAF and PI3KCA gene mutations did not. When the DSS of the patients with reported asbestos exposure was considered, the five KRAS gene-mutated patients had a worse prognosis than those with wild-type KRAS (n=42), although the difference was not statistically significant (mean survival, 9.20±6.91 vs 15.6±10.39 months; P=0.188) (Figure 1A). On the contrary, the DSS of the patients without reported occupational asbestos exposure was better in the BRAF gene-mutated patients (n=3) than in those without BRAF mutations (n=22), although, once again, the difference was not statistically significant (mean survival, 20.33±12.06 vs 12.1±8.37 months; P=0.140) (Figure 1B).
Figure 1.
Kaplan–Meier DSS curves for MPM patients with KRAS mutation vs wild-type (A) and with BRAF mutation vs wild-type (B).
Discussion
As EGFR is involved in the carcinogenesis of MPM, it is possible that EGFR-targeted therapies may be efficacious in MPM patients (Barbieri et al, 2011). Epidermal growth factor receptor TKI inhibitors, such as gefitinib and erlotinib, inhibit MPM cell migration and proliferation, enhance the response to radiation of human MPM cell lines, and reduce motility and invasion in MPM cell lines (Kurai et al, 2012). However, the promising results obtained in in vitro studies were not reproduced in two phase II trials involving patients with pleural and peritoneal mesotheliomas, although it should be noted that neither study evaluated the mutation status of the EGFR gene and its downstream signalling transduction pathway (Govindan et al, 2005; Garland et al, 2007). As in the case of colorectal cancer and lung adenocarcinoma, this lack of molecular selection could explain the therapeutic failure.
The few studies that have sought mutations in the tyrosine kinase domain of the EGFR gene in patients with malignant mesotheliomas involved small populations and used a relatively insensitive method (the direct sequencing of exons 18–21) (Cortese et al, 2006; Velcheti et al, 2009; Enomoto et al, 2012). The primary objective of our study was to look for EGFR gene mutations in a larger series of patients (n=77) using two molecular methods: all of the cases were first screened using Scorpion-ARMS technology, which is capable of detecting 1% of mutated cells against a 99% background of wild-type cells, followed by direct sequencing to find rarer mutations or mutations that cannot be detected using the first method. However, despite this, we did not find any mutations in the TK domain of EGFR: in addition to confirming previous findings (Cortese et al, 2006; Velcheti et al, 2009), this also indicates that, unlike in the case of lung adenocarcinomas, mutations cannot be detected even when real-time PCR is used to increase sensitivity (Allegrini et al, 2012).
On the contrary, Enomoto et al (2012) have recently studied 38 patients and found EGFR missense mutations in exons 18 (n=1), 20 (n=3), and 21 (n=1) in six (16%) patients with pleural (n=3) or peritoneal mesotheliomas (n=3). Epidermal growth factor receptor gene mutations have been previously found in peritoneal mesotheliomas (Foster et al, 2009, 2010), but this is the only published report of EGFR gene mutations in MPM. However, the study involved Japanese patients, who are characterised by more frequent EGFR gene mutations in lung adenocarcinoma than Western patients (Endo et al, 2005). Furthermore, some of the detected mutations had never been reported before, and their biological and clinical significance is still unknown.
An alternative method of blocking EGFR is to use mABs, which may be extremely useful as it has been demonstrated that MPM patients show EGFR gene amplification (Dazzi et al, 1990; Destro et al, 2006; Okuda et al, 2008). No published studies have assessed the in vivo effects of anti-EGFR mAbs on MPMs, although one recent study has found that cetuximab is highly efficacious in cultured MPM cell lines (Kurai et al, 2012). It has been demonstrated that mutations in EGFR downstream pathways can affect the efficacy of EGFR mABs in other tumours such as colorectal adenocarcinoma (Jonker et al, 2007), and we found nine patients (11.7%) with missense mutations involving the KRAS (n=5), BRAF (n=3), and PIK3CA genes (n=1). Few other studies have separately investigated the presence of mutations in KRAS, BRAF, and PIK3CA genes in MPM samples and mesothelioma cell lines without success (see review by Agarwal et al, 2011), but, to the best of our knowledge, ours is the first to investigate these alterations systematically in a large series of MPM patients.
Various reasons may explain these discrepant results. We screened a large number of samples (n=77), whereas the other studies were based on smaller series and may have underestimated the real frequency of such mutations. Furthermore, we analysed KRAS gene mutations using an ME-PCR technology whose sensitivity is 0.1% (Molinari et al, 2011), and so it is possible that the percentage of KRAS gene-mutated cells is very low in MPM and that more widely used sequencing methods are unable to detect small clones.
Our findings show that, although infrequent, mutations in EGFR downstream pathways can be found in MPMs, thus supporting the hypothesis that EGFR mABs may be clinically effective in the majority of patients. On the other hand, patients with a molecular profile indicating putative resistance to EGFR mABs (because of the presence of KRAS or BRAF or PIK3CA mutations) may be directed towards new targeted therapies. One recent study has shown that vemurafenib is promising not only in patients with metastatic melanoma but also in patients with non-small-cell lung cancer carrying a BRAF mutation (Gautschi et al, 2012), and selumetinib and BYL-719, which target KRAS and PIK3CA mutations, are currently being evaluated in several clinical trials (http://ctmagnifier.org/). Our data therefore underline the importance of the molecular characterisation of patients with MPM.
The clinical implications of the gene mutations detected in our study are not clear. DSS was no different in the patients with or without gene mutations (whether analysed together or separately). Interestingly, all of the patients with KRAS gene mutations reported occupational asbestos exposure, but none of those with BRAF or PIK3CA gene mutations. Comparison of mean DSS in the KRAS and BRAF gene-mutated patients vs wild-type patients previously exposed to asbestos or not, showed that the KRAS gene-mutated patients (n=5) tended to have a worse prognosis than the wild-type patients (9.20±6.91 vs 15.6±10.39 months), and the BRAF gene-mutated patients (n=3) tended to have a better prognosis (20.33±12.06 vs 12.1±8.37 months). However, the differences were not statistically significant and our findings need to be confirmed in larger series of MPM patients.
In conclusion, our extensive molecular characterisation of EGFR pathways may explain the failure of TKI administration and may open up the possibility of developing new targeted therapies.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Agarwal V, Lind M, Cawkwell L. Targeted epidermal growth factor receptor therapy in malignant pleural mesothelioma: where do we stand. Cancer Treat Rev. 2011;37:533–542. doi: 10.1016/j.ctrv.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Allegrini S, Antona J, Mezzapelle R, Miglio U, Paganotti A, Veggiani C, Frattini M, Monga G, Balbo P, Boldorini R. Epidermal growth factor receptor gene analysis with a highly sensitivity molecular assay in routine cytologic specimens of lung adenocarcinoma. Am J Clin Pathol. 2012;138:377–381. doi: 10.1309/AJCPVAGIUC1AHC3Y. [DOI] [PubMed] [Google Scholar]
- Barbieri F, Wurth R, Favoni RE, Pattarozzi A, Gatti M, Ratto A, Ferrari A, Bajetto A, Florio T. Receptor tyrosine kinase inhibitors and cytotoxic drugs affect pleural mesotelioma cell proliferation: insight into EGFR and ERK1/2 as antitumor targets. Biochem Pharmacol. 2011;82:1467–1477. doi: 10.1016/j.bcp.2011.07.073. [DOI] [PubMed] [Google Scholar]
- Boutin C, Schlesser M, Frenay C, Astoul P. Malignant pleural mesothelioma. Eur Respir J. 1998;12:972–981. doi: 10.1183/09031936.98.12040972. [DOI] [PubMed] [Google Scholar]
- Carbone M, Kratzke RA, Testa JR. The pathogenesis of mesothelioma. Semin Oncol. 2002;29:2–17. doi: 10.1053/sonc.2002.30227. [DOI] [PubMed] [Google Scholar]
- Comin CE, Anichini C, Boddi V, Novelli L, Dini S. MIB-1 proliferation index correlates with survival in pleural malignant mesothelioma. Histopathology. 2000;36:26–31. doi: 10.1046/j.1365-2559.2000.00793.x. [DOI] [PubMed] [Google Scholar]
- Cortese JF, Gowda AL, Wali A, Eliason JF, Pass HI, Everson RB. Common EGFR mutations conferring sensitivity to gefitinib in lung adenocarcinoma are not prevalent in human malignant mesothelioma. Int J Cancer. 2006;118:521–522. doi: 10.1002/ijc.21271. [DOI] [PubMed] [Google Scholar]
- Curran D, Sahmoud T, Therasse P, van Meerbeeck J, Postmus PE, Giaccone G. Prognostic factors in patients with pleural mesothelioma: The European Organization for Research and Treatment of Cancer experience. J Clin Oncol. 1998;16:145–152. doi: 10.1200/JCO.1998.16.1.145. [DOI] [PubMed] [Google Scholar]
- Dazzi H, Hasleton PS, Thatcher N, Wilkes S, Swindell R, Chatterjee AK. Malignant pleural mesothelioma and epidermal growth factor receptor (EGF-R). Relationship of EGF-R with histology and survival using fixed paraffin embedded tissue and the F4, monoclonal antibody. Br J Cancer. 1990;61:924–926. doi: 10.1038/bjc.1990.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destro A, Ceresoli GL, Falleni M, Zucali PA, Morenghi E, Bianchi P, Pellegrini C, Cordani N, Vaira V, Alloisio M, Rizzi A, Bosari S, Roncalli M. EGFR overexpression in malignant pleural mesothelioma. An immunohistochemical and molecular study with clinico-pathological correlations. Lung Cancer. 2006;51:207–215. doi: 10.1016/j.lungcan.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Endo K, Konishi A, Sasaki H, Takada M, Tanaka H, Okumura M, Kawahara M, Sugiura H, Kuwabara Y, Fukai I, Matsumura A, Yano M, Kobayashi Y, Mizuno K, Haneda H, Suzuki E, Iuchi K, Fujii Y. Epidermal growth factor receptor gene mutation in non-small cell lung cancer using highly sensitive and fast TaqMan PCR assay. Lung Cancer. 2005;50:375–384. doi: 10.1016/j.lungcan.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Enomoto Y, Kasai T, Takeda M, Takano M, Morita K, Kadota E, Iizuka N, Maruyama H, Haratake J, Kojima Y, Ikeda N, Inatsugi N, Nonomura A. Epidermal growth factor receptor mutations in malignant pleural and peritoneal mesothelioma. J Clin Pathol. 2012;65:522–527. doi: 10.1136/jclinpath-2011-200631. [DOI] [PubMed] [Google Scholar]
- Foster JM, Gatalica Z, Lilleberg S, Haynatzki G, Loggie BW. Novel and existing mutations in the tyrosine kinase domain of the epidermal growth factor receptor are predictors of optimal respectability in malignant peritoneal mesothelioma. Ann Surg Oncol. 2009;16:152–158. doi: 10.1245/s10434-008-0206-6. [DOI] [PubMed] [Google Scholar]
- Foster JM, Radhakrishna U, Govindarajan V, Carreau JH, Gatalica Z, Sharma P, Nath SK, Loggie BW. Clinical implications of novel activating EGFR mutations in malignant peritoneal mesothelioma. World J Surg Oncol. 2010;8:88. doi: 10.1186/1477-7819-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland LL, Rankin C, Gandara DR, Rivkin SE, Scott KM, Nagle RB, Klein-Szanto AJ, Testa JR, Altomare DA, Borden EC. Phase II study of erlotinib in patients with malignant pleural mesothelioma: a Southwest Oncology Group Study. J Clin Oncol. 2007;25:2406–2413. doi: 10.1200/JCO.2006.09.7634. [DOI] [PubMed] [Google Scholar]
- Gautschi O, Pauli C, Strobel K, Hirschmann A, Printzen G, Aebi S, Diebold J. A patient with BRAF V600E lung adenocarcinoma responding to Vemurafenib. J Thorac Oncol. 2012;7:e23–e24. doi: 10.1097/JTO.0b013e3182629903. [DOI] [PubMed] [Google Scholar]
- Govindan R, Kratzke RA, Herndon JE, Niehans GA, Vollmer R, Watson D, Green MR, Kindler HL. Gefitinib in patients with malignant mesothelioma: a phase II study by the Cancer and Leukemia Group B. Clin Cancer Res. 2005;11:2300–2304. doi: 10.1158/1078-0432.CCR-04-1940. [DOI] [PubMed] [Google Scholar]
- Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, van Hazel G, Wierzbicki R, Langer C, Moore MJ. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- Kurai J, Chikumi H, Hashimoto K, Takata M, Sako T, Yamaguchi K, Kinoshita N, Watanabe M, Touge H, Makino H, Igishi T, Hamada H, Yano S, Shimizu E. Therapeutic antitumor efficacy of anti-epidermal growth factor receptor antibody, cetuximab, against malignant pleural mesothelioma. Int J Oncol. 2012;41:1610–1618. doi: 10.3892/ijo.2012.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanphear BP, Buncher CR. Latent period for malignant mesothelioma of occupational origin. J Occup Med. 1992;34:718–721. [PubMed] [Google Scholar]
- Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive to response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O, Girlando S, Soini B, Spitale A, Di Nicolantonio F, Saletti P, Crippa S, Mazzucchelli L, Marchetti A, Bardelli A, Frattini M. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res. 2011;17:4901–4914. doi: 10.1158/1078-0432.CCR-10-3137. [DOI] [PubMed] [Google Scholar]
- Oken MM, Creech RH, Tormey DC, Horton J, Davis TE, McFadden ET, Carbone PP. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. [PubMed] [Google Scholar]
- Okuda K, Sasaki H, Kawano O, Yukiue H, Yokoyama T, Yano M, Fujii Y. Epidermal growth factor receptor gene mutation, amplification and protein expression in malignant pleural mesothelioma. J Cancer Res Clin Oncol. 2008;134:1105–1111. doi: 10.1007/s00432-008-0384-4. [DOI] [PubMed] [Google Scholar]
- Pache JC, Janssen YM, Walsh ES, Quinlan TR, Zanella CL, Low RB, Taatjes DJ, Mossman BT. Increased epidermal growth-factor receptor protein in a human mesothelial cell line in response to long asbestos fibers. Am J Pathol. 1998;152:333–340. [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutation in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Peto J, Decarli A, La Vecchia C, Levi F, Negri E. The European mesothelioma epidemic. Br J Cancer. 1999;79:666–672. doi: 10.1038/sj.bjc.6690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker R, Herth FJF, Kerr KM, Filipits M, Taron M, Gandara D, Hirsch FR, Grunenwald D, Popper H, Smit E, Dietel M, Marchetti A, Manegold C, Schirmacher P, Thomas M, Rosell R, Cappuzzo F, Stahel R. Consensus of EGFR mutation testing in non-small cell lung cancer: result from a European workshop. J Thorac Oncol. 2011;5:1706–1713. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- Rena O, Boldorini LR, Gaudino E, Casadio C. Epidermal growth factor receptor overexpression in malignant pleural mesothelioma: prognostic correlations. J Surg Oncol. 2011;104:701–705. doi: 10.1002/jso.21901. [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P, Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Sobin LH, Gospodarowic MK, Wittekind2009TNM Classification of Malignant Tumours7th edn. pp142–145.Wiley-Blackwell: Oxford, UK [Google Scholar]
- Suzuki Y, Murakami H, Kawaguchi K, Tanigushi T, Fujii M, Shinjo K, Kondo Y, Osada H, Shimokata K, Horio Y, Hasegawa Y, Hida T, Sekido Y. Activation of the PI3K-AKT pathway in human malignant mesothelioma cells. Mol Med Report. 2009;2:181–188. doi: 10.3892/mmr_00000081. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Mueller-Hermelink HK, Harris CC. World Health Organization Classification of tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. IARC Press: Lyon, France; 2004. pp. 125–136. [Google Scholar]
- Velcheti V, Kasai Y, Viswanathan AK, Ritter J, Govindan R. Absence of mutations in the epidermal growth factor receptor (EGFR) kinase domain in patients with mesothelioma. J Thorac Oncol. 2009;4:559. doi: 10.1097/JTO.0b013e31819c8661. [DOI] [PubMed] [Google Scholar]
- Vogelzang NJ, Rusthoven JJ, Symanowski J, Denham C, Kaukel E, Ruffie P, Gatzemeier U, Boyer M, Emri S, Manegold C, Niyikiza C, Paoletti P. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- Wagner JC, Sleggs P, Marchand P. Diffuse pleural mesothelioma and asbestos exposure in the North Western Cape Province. Br J Ind Med. 1960;17:260–271. doi: 10.1136/oem.17.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Opt Oncol. 2008;9:147–157. doi: 10.1007/s11864-008-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]