Abstract
Background:
Few studies have attempted to characterise genomic changes occurring in hereditary epithelial ovarian carcinomas (EOCs) and inconsistent results have been obtained. Given the relevance of DNA copy number alterations in ovarian oncogenesis and growing clinical implications of the BRCA-gene status, we aimed to characterise the genomic profiles of hereditary and sporadic ovarian tumours.
Methods:
High-resolution array Comparative Genomic Hybridisation profiling of 53 familial (21 BRCA1, 6 BRCA2 and 26 non-BRCA1/2) and 15 sporadic tumours in combination with supervised and unsupervised analysis was used to define common and/or specific copy number features.
Results:
Unsupervised hierarchical clustering did not stratify tumours according to their familial or sporadic condition or to their BRCA1/2 mutation status. Common recurrent changes, spanning genes potentially fundamental for ovarian carcinogenesis, regardless of BRCA mutations, and several candidate subtype-specific events were defined. Despite similarities, greater contribution of losses was revealed to be a hallmark of BRCA1 and BRCA2 tumours.
Conclusion:
Somatic alterations occurring in the development of familial EOCs do not differ substantially from the ones occurring in sporadic carcinomas. However, some specific features like extensive genomic loss observed in BRCA1/2 tumours may be of clinical relevance helping to identify BRCA-related patients likely to respond to PARP inhibitors.
Keywords: ovarian cancer, hereditary, BRCA1, BRCA2, genomic alteration
Epithelial ovarian cancer is the most lethal gynaecological malignancy and the fifth leading cause of cancer-related death in women in western countries. Due to lack of specific symptoms and effective screening methods, the majority of cases are diagnosed at advanced stages. Since survival probability drops significantly with increasing stage, epithelial ovarian carcinoma (EOC) patients in general face a poor prognosis, with an estimated 5-year survival rate of around 27% (Siegel et al, 2012).
Epithelial ovarian cancer is a very heterogeneous disease that can be stratified using different criteria. Based on molecular and developmental features, EOCs can be divided into type I and type II tumours. Type I tumours arise in a progressive manner from benign, through borderline to low-malignant potential neoplasm. Type II tumours grow rapidly without well-defined premalignant lesions and are typically diagnosed as high-grade serous, high-grade endometroid, or undifferentiated carcinomas depending on the dominant pattern (Kurman and Shih Ie, 2011). Tumours can also be classified as hereditary or sporadic when they arise in patients with or without a family history of the disease, respectively. Overall, around 10–15% of invasive EOCs are estimated to involve hereditary susceptibility (Bast et al, 2009). The majority of these cases are explained by germline mutations in the BRCA1 or BRCA2 tumour suppressor genes (Bast et al, 2009; Pennington and Swisher, 2012) although additional genes such as BRIP1, RAD51C and RAD51D have been recently shown to confer ovarian cancer susceptibility (Pennington and Swisher, 2012). Clinical and histopathological differences between BRCA1- and BRCA2-related tumours and those arising in non-mutation carriers have been reported (Soslow et al, 2012). Importantly, germline BRCA1/2 mutations have been associated with improved survival and chemotherapy response (Alsop et al, 2012; Bolton et al, 2012; Pennington and Swisher, 2012).
It has been suggested that hereditary and sporadic EOCs might evolve in distinct ways, especially due to early homologous recombination (HR) impairment in carriers of BRCA1 or BRCA2 mutations (Patael-Karasik et al, 2000; Israeli et al, 2003; Walsh et al, 2008). However, it is still unclear exactly which mechanisms are involved in cancer development in BRCA1/2 mutation carriers, and whether they differ from those taking place in sporadic cases. One way to get insight into this issue is to compare the rate and pattern of DNA copy number changes exhibited by these tumour types. This approach is particularly relevant in a view of the results from the recently published Cancer Genome Atlas (TCGA) study (TCGA, 2011). This the most comprehensive analysis of high-grade EOCs carried out so far revealed that these tumours present a relatively simple mutational spectrum, but are characterised by a large degree of genome instability.
So far few studies have specifically analysed the DNA copy number changes that characterise the different groups of hereditary ovarian tumours (BRCA1, BRCA2 and those from non-BRCA1/2-mutation carriers, also called ‘BRCAX') or have compared these changes with those observed in sporadic neoplasms (Patael-Karasik et al, 2000; Zweemer et al, 2001; Israeli et al, 2003; Ramus et al, 2003; Leunen et al, 2009). Moreover, the few studies conducted have yielded contradictory results, which might be due to the limited number of tumours included (Patael-Karasik et al, 2000; Israeli et al, 2003; Leunen et al, 2009), the use of low-resolution techniques (Patael-Karasik et al, 2000; Ramus et al, 2003) or the application of different algorithms. Interestingly, the TCGA ovarian study reported that tumours with BRCA1/2 alterations do not exhibit increased genomic instability compared with wild-type tumours (TCGA, 2011). However, that comparison was only made of the total level of DNA copy number alterations with no distinction between gained or lost events. In addition, the TCGA (TCGA, 2011) and other studies (Koul et al, 2000) jointly describe the DNA copy number alterations that occur in tumours carrying germline, somatic and/or epigenetic inactivation of the BRCA1 or BRCA2 genes. Nevertheless, it is still not clear whether the mechanisms by which these genes are rendered non-functional might be relevant to the natural history of the tumours and to the changes that arise and are selected throughout the oncogenic process.
Our study addresses some of these limitations by using a homogeneous series of ovarian tumours from patients of well-characterised high-risk breast and ovarian cancer families. Moreover, this series includes not only cases from carriers of germline BRCA1/2 mutations, but also from hereditary BRCAX cases. In addition, we use high-resolution array Comparative Genomic Hybridisation (aCGH) and pay special attention to the separate analysis of gain and loss events. This approach, although still limited, allowed us to obtain further insight into this poorly explored field and to define potential differences and similarities in genomic instability between these tumour groups.
Materials and methods
Additional information can be found in Supplementary Methods.
Patients and tumours
At total of 72 formalin-fixed paraffin-embedded (FFPE) epithelial ovarian tumours were analysed. Fifty seven corresponded to patients from high-risk breast and ovarian cancer families and fifteen to sporadic patients. Families selected for this study fulfilled one of the following criteria: (a) at least two cases of ovarian cancer in the same family line; (b) at least one case of ovarian cancer and at least one case of breast cancer in the same family line; (c) at least one woman with both breast and ovarian cancer; (d) at least one woman with bilateral ovarian cancer. Mutation testing of BRCA1 and BRCA2 genes was carried out using previously described methods (Milne et al, 2008). In total, paraffin blocks from 21 BRCA1, 6 BRCA2 and 30 BRCAX tumours were obtained from different hospitals throughout Spain. Sporadic cases (with no reported first or second degree relative with breast or ovarian cancer), used for comparison purposes, were obtained from a single institution (Hospital Virgen del Rocio, Seville) and were selected to match the distribution of histological subtypes in familial series.
All tumours were blindly reviewed by two pathologists (IMR and JP) and classified histopathologically. Immunohistochemical expression of markers such as Wilms Tumour protein (WT1), tumour protein p53 (TP53), oestrogen receptor (ESR), progesterone receptor (PGR) and cyclin-dependent kinase inhibitor 2A (p16) (CDKN2A) was performed to assist in the differential diagnosis (Kobel et al, 2009; Kalloger et al, 2011). Grading of serous tumours was performed according to two-tier MD Anderson Cancer Center (MDACC) system (Malpica et al, 2004; Gilks et al, 2008) while the rest of the histological types was graded according to World Health Organisation criteria (Silverberg, 2000; World Health Organization, 2004). A subgroup of tumours within the type II carcinomas was defined to allow for comparisons between more homogenous groups of high-grade neoplasms. This subgroup consisted of high-grade serous tumours of solid growth pattern and undifferentiated carcinomas (hereafter referred as to ‘subgroup of type II tumours'). Detailed information is shown in Supplementary Table 1. The study was approved by the research ethics committees from each of the participating centres and all patients gave informed consent.
DNA isolation and labelling
Genomic DNA was extracted from three 10-μm-thick FFPE tissue sections per tumour. After deparaffination and rehydration, sections were hematoxylin and eosin (H&E) stained and tumour areas were delimited by a pathologist and macrodissected with a surgical blade to ensure at least 80% tumour content. DNA extraction was carried according to standard protocol including overnight proteinase K digestion and using QIAamp DNA mini kit (Qiagen, Westburg, Leusden, The Netherlands) according to manufacturer's instructions. Labelling of test and reference DNA was performed with the Enzo Genomic DNA labeling kit (Enzo Life Sciences, Farmingdale, NY, USA) as described previously. In 45 out of 72 hybridisations (62%), patient-matched normal DNA was used as a reference (in 26 from conserved normal tissue and in 19 from the patient's peripheral blood). In the remaining 27 hybridizations (38%), a pool of normal DNA from healthy females was used as a reference (http://www.kreatech.com/products/megapool-reference-dna.html).
Hybridisations, scanning and image acquisition
Hybridisations were performed on slides of four arrays, each containing 180 880 in situ synthesised 60-mer oligonucleotides (4 × 180 K, Agilent Technologies, Palo Alto, CA, USA) representing 169 793 unique chromosomal locations evenly distributed across the genome (space∼17 kb), and 4548 additional unique oligonucleotides, located at 238 of the Cancer Census genes (http://www.sanger.ac.uk/genetics/CGP/Census/). Oligonucleotide positions were defined according to the NCBI36/hg18 assembly (March 2006). Hybridisation, scanning and feature extraction were carried out as previously described (Buffart et al, 2008). The aCGH data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO accession number GSE41253 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41253).
Preprocessing and processing
Data normalisation, segmentation and calling were performed in R (v.2.8.2 and 2.13; http://www.r-project.org), using median normalisation and Wave Smoothing and CGH-call packages (van de Wiel and Zhang, 2007). For visualisation and downstream processing, data were analysed in Nexus Copy Number v5.1 (BioDiscovery, Inc., El Segundo, CA, USA). The WECCA (Weighted Clustering of Called aCGH Data) R package (Van Wieringen et al, 2008) was used for unsupervised hierarchical clustering (total linkage and overall similarity algorithms).
Degree of genomic instability: number and length of alterations
To determine the degree of genomic instability in each subgroup of tumours (BRCA1/2/X and sporadic), total number of alterations and of a particular type (homozygous deletions (HDs)/losses/gains/amplifications) were calculated per sample. Total size of altered genome and size accounted for by gains and losses was calculated by adding up the lengths of individual segments. Next, average number of changes and average size of altered genome were calculated for sporadic tumours and for each group of familial tumours. To determine the relative contribution of each type of change (losses or gains), the ratio of the average number of losses to the average number of gains within each tumour subtype was computed. Similarly, the ratio of the average length of the lost genome to the average length of gained was calculated.
Common and potentially specific regions of copy number changes
To visualise the general pattern of chromosomal changes, frequency plots and a list of recurrent minimal common regions (MCRs) of alterations were generated for each tumour subtype (BRCA1/2/X and sporadic) using Nexus Copy Number v5.1 (BioDiscovery, Inc.). Potentially group-specific alterations were defined using Fisher's Exact Test (FET) (P-value<0.05) implemented in Nexus and chi-square test within CGHtest (R package) with correction for multiple testing (FDR<0.02).
The list of potentially group-specific regions was further refined using data from the TCGA ovarian study (TCGA, 2011) as described in Supplementary Materials.
Immunohistochemical analysis
To validate our hybridisation and analytical approaches, we selected three high-amplitude events (HDs at the CDKN2A and RB1 loci and amplification at the CCNE1 locus) to determine the consistency between the assigned DNA copy number status and the expression levels of the target proteins. Details are explained in Supplementary Materials and shown in Supplementary Table S2.
Statistical analyses
Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Comparison of continuous variables (number and size of alterations between different tumour groups and clusters) was done using Student's two-tailed t-test (for variables of normal distribution) or the Mann–Whitney test (non-parametric distributions). For categorical data (FIGO stage, BRCA1/2 mutation status, etc.), chi-square or Fisher's Exact Test were applied, depending on the size of the compared groups.
Results
Tumour characteristics
In our series of 57 familial tumours, 4 were classified as borderline lesions and 53 corresponded to carcinomas. The borderline tumours were analysed separately (data not shown) and excluded from this study. All of them belonged to the BRCAX group. Most tumours from BRCA1/2 mutation carriers were serous, high grade and high FIGO stage. In contrast, BRCAX tumours were more heterogeneous and presented a wider range of histological subtypes and stages. As expected, hereditary patients were diagnosed at a significantly younger age than sporadic ones (51 vs 62 years, P=0.001). Patient and tumour characteristics are summarised in Supplementary Table 1.
Number and length of copy number alteration across tumour subtypes
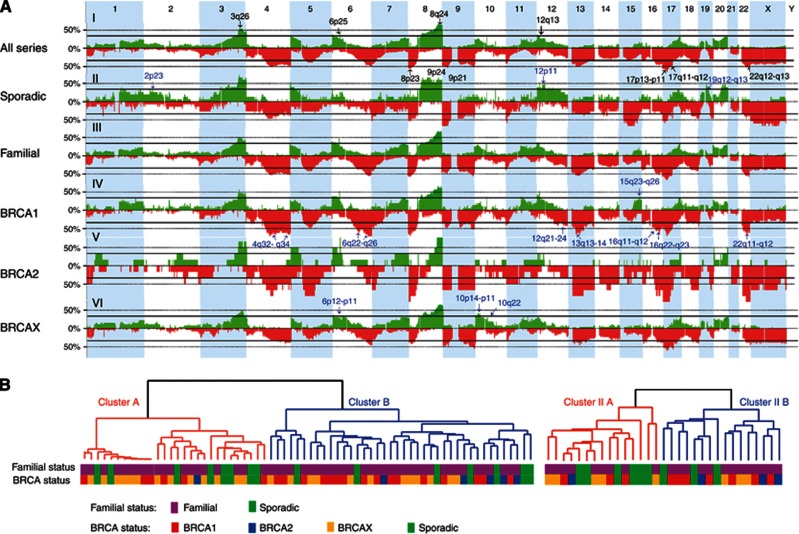
Overall, the pattern of copy number alterations was not substantially different between familial (all subtypes) and sporadic tumours (Figure 1A). Likewise, there were no significant differences between familial and sporadic tumours regarding the average total number of alterations and the average total length of genome altered per tumour (considering all carcinomas and the subgroup of type II neoplasms, the latter defined as described in Materials and methods) (Supplementary Table S3). Both familial and sporadic tumours were characterised by a high level of genomic instability, which in the subgroup of type II carcinomas was exemplified by an average of >60 aberrations per tumour that involved >1 Mb of the genome (Supplementary Table S3). Despite this general similarity, a separate analysis of gains and losses and stratification of familial tumours according to their BRCA1/2 mutation status revealed some differences.
Figure 1.
(A) Frequency plots of copy number gains (in green) and losses (in red) defined in all carcinomas and subgroups. The proportion of tumours with gained/lost regions is plotted on the y axis versus genomic location on the x axis. Common recurrently altered regions across all four subgroups or present in >55% of the whole series are marked with black arrows on the general plot (for all series), while group-specific regions identified for sporadic, BRCA1 and BRCAX tumours are identified on the corresponding plots with blue arrows. Simplified chromosomal locations are given next to the arrows (using the same colour code). (B) Dendrograms derived from unsupervised hierarchical clustering based on copy number alterations of epithelial ovarian carcinomas (n=68) (left panel) and a subgroup of type II carcinomas (n=31) (right panel) with colour labels defining the familial or sporadic condition of the tumour as well as the BRCA gene mutation status of the sample.
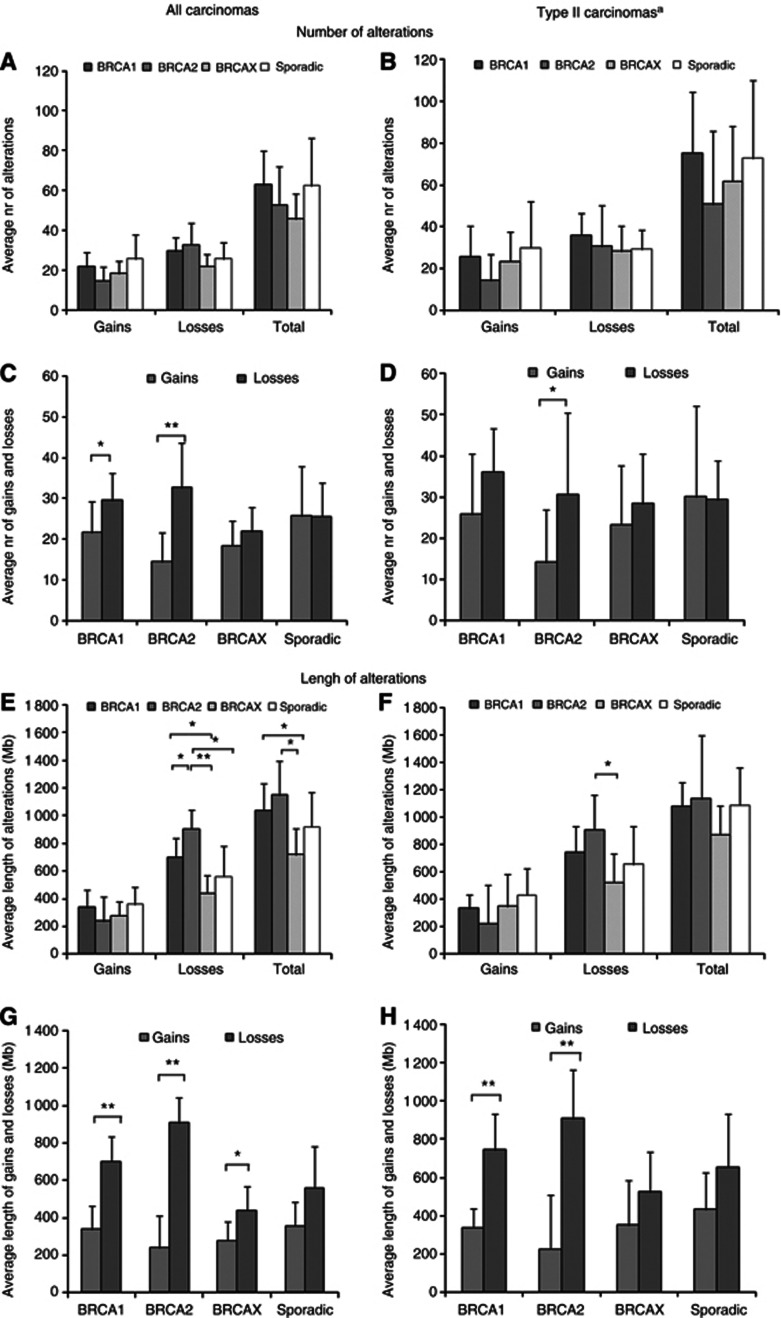
BRCA1 and BRCA2 tumours presented a greater average number of losses and HDs than sporadic or BRCAX tumours, while sporadic cases presented the highest average number of gains and amplifications of all tumour subtypes (Figure 2A; Supplementary Table S3). A similar pattern was observed when only high-grade tumours were considered (Figure 2B; Supplementary Table S3). Comparisons of gains and losses within each tumour subtype revealed that sporadic tumours presented a similar average number of both events (25.7 vs 25.6, respectively). However, in familial tumours the average number of losses was 1.4 times greater than the average number of gains, with differences mostly attributed to BRCA1 (29.6 losses vs 21.7 gains, P=0.02) and BRCA2 tumours (32.7 losses vs 14.5 gains, P=0.009) (Figure 2C; Supplementary Table S3). This was also observed in the subgroup of type II tumours, with significant and borderline significant differences between numbers of gains and losses in BRCA2 and BRCA1 tumours, respectively (Figure 2D; Supplementary Table S3).
Figure 2.
Average number (A–D) and length (E–H) of copy number alterations in different groups of ovarian carcinomas (A, C, E, D) and a subgroup of type II carcinomas (B, D, F, H). Significant differences in number and length of alterations between (A, B, E, F) and within (C, D, G, H) tumour groups are indicated with *(P<0.05) or **(P<0.01). Error bars represent 95% Confidence Intervals. aSubgroup of type II carcinomas as defined in Materials and methods.
In agreement with the analysis of the number of alterations, we found that BRCA1 and BRCA2 tumours presented a significantly higher average length of genome altered due to losses than sporadic or BRCAX tumours (Figure 2E; Supplementary Table S3). This pattern was partially maintained in the subgroup of type II carcinomas, with significant differences between tumours from BRCA2 and BRCAX patients (Figure 2F; Supplementary Table S3). Interestingly, in all tumour subtypes, including the sporadic group, which showed similar average numbers of both alterations, more genetic material was lost than gained (Figures 2G and H; Supplementary Table S3). In BRCA1 and BRCA2 tumours, the average length of lost genome was 2.1 and 3.8-fold greater than the length of gained material, respectively. In sporadic and BRCAX tumours, differences were less marked (1.6-fold in both) (Figure 2G; Supplementary Table S3). Differences between length of genome gained and lost per tumour were statistically significant in all familial tumours (BRCA1, BRCA2 and BRCAX), while only a trend was observed in sporadic cases (P=0.09). In the subgroup of type II tumours, only differences in BRCA1/2 carriers remained significant (P<0.001) (Figure 2H; Supplementary Table S3).
Common and group-specific copy number alterations
A summary of common gains and losses identified as recurrent in at least three of the analysed tumour subtypes (BRCA1, BRCA2, BRCAX and sporadic) is shown in Table 1. Regions recurrently gained in the four tumour subtypes were 6p25.3, 8q24.2-q24.3 and 12p13.33-p13.32, and regions exhibiting the highest frequencies were 3q26.2 and 8q24.2-q24.3 (Figure 1A–I; Table 1). These regions and others defined as recurrently gained included from 1 up to 45 genes and spanned well-known or potential oncogenes such as MECOM, PIK3CA, FOXQ1, MYC, CCND2 and CANT1. Regions recurrently lost in all four subgroups were defined at 9p24.3, 9p21.3, 17q11.2-q12, 22q12.3, 22q13.1 and 22q13.31-q13.33. Alterations of the highest incidence were found at 8p23.3-p23.1 and 17p13.3-p11.2 (Figure 1A–I; Table 1). Many of these deleted regions and the others qualifying as recurrent across tumour subtypes encompassed tumour suppressors previously linked to ovarian carcinogenesis (e.g., MCPH1, CDKN2A, CDKN2B and NF1). However, other regions pointed to less well-characterised suppressors not previously associated with ovarian cancer (e.g., FANCC, TSC1, CREBBP, CDH11 and EDA2R).
Table 1. Recurrent minimal common regions (MCRs) of gains and losses shared across tumour subtypes.
| Cytoband | Region | Size (bp) | Frequencya (%) | No. of genes | Genes of interestb | Other genes in the regionc |
|---|---|---|---|---|---|---|
|
Gains | ||||||
| 3q26.1 | 161 858 336–163 987 311 | 2 128 975 | 37 | 6 | ARL14 PPM1L B3GALNT1 NMD3 C3orf57 OTOL1 | |
| 3q26.2 | 170 336 781–170 346 939 | 10 158 | 56 | 1 | MECOM | |
| 3q26.32 | 180 277 629–180 352 072 | 74 443 | 54 | 1 | PIK3CA | |
| 5p14.3-p14.2 | 19 297 228–23 809 228 | 4 512 000 | 28 | 5 | CDH18 CDH12 PRDM9 | CDH18 GUSBP1 PMCHL1 CDH12 PRDM9 |
| 6p25.3 | 1 189 126–1 285 230 | 96 104 | 38 | 1 | FOXQ1 | |
| 6p25.1-p24.3 | 5 776 997–8 041 186 | 2 264 189 | 29 | 16 | NRN1 TXNDC5 | |
| 6p24.1 | 13 408 897–13 486 913 | 78 016 | 32 | 2 | TBC1D7 | GFOD1 |
| 7q32.2-q32.3 | 129 620 363–130 651 373 | 1 031 010 | 28 | 18 | ||
| 7q33-q34 | 134 477 470–142 682 704 | 8 205 234 | 30 | 80 | CREB3L2 KIAA1549 BRAF | |
| 7q35-q36.1 | 144 378 243–147 685 879 | 3 307 636 | 28 | 3 | CNTNAP2 MIR548F4 MIR548T | |
| 8q24.21 | 128 807 376–129 291 694 | 484 318 | 68 | 2 | MYC | PVT1 |
| 8q24.3 | 141 522 457–143 860 659 | 2 338 202 | 65 | 21 | TRAPPC9 PSCA | |
| 10q11.23 | 51 473 718–51 664 989 | 191 271 | 31 | 3 | FAM21A FAM21B ASAH2 | |
| 12p13.33-p13.32 | 54 933–5 102 331 | 5 047 398 | 37 | 45 | KDM5A CCND2 | |
| 17q25.1-q25.3 | 70 225 044–72 286 125 | 2 061 081 | 30 | 26 | FOXJ1 | |
| 17q25.3 | 74 387 898–78 774 742 | 4 386 844 | 26 | 93 | CANT1 ASPSCR1 | |
| 20p13 |
0–3 018 638 |
3 018 638 |
41 |
61 |
ANGPT4 |
|
|
Losses | ||||||
| 4q24 | 103 855 981–104 152 060 | 296 079 | 51 | 4 | MANBA UBE2D3 CISD2 NHEDC1 | |
| 4q28.3-q31.21 | 136 561 996–142 584 570 | 6 022 574 | 46 | 19 | ||
| 4q34.3-q35.1 | 182 303 764–182 934 067 | 630 303 | 49 | 1 | NCRNA00290 | |
| 6q26 | 161 740 093–161 828 041 | 87 948 | 44 | 1 | PARK2 | |
| 8p23.3-p23.1 | 1 190 650–6 260 759 | 5 070 109 | 56 | 9 | MCPH1 | |
| 8p21.2-p21.1 | 26 260 243–28 304 348 | 2 044 105 | 47 | 21 | ||
| 9p24.3 | 0–1 631 746 | 1 631 746 | 51 | 11 | ||
| 9p24.1 | 8 056 737–8 434 111 | 377 374 | 50 | 1 | PTPRD | |
| 9p23-p22.3 | 12 386 932–14 108 018 | 1 721 086 | 51 | 5 | NFIB | TYRP1 C9orf150 MPDZ FLJ41200 |
| 9p22.3 | 14 622 555–16 229 616 | 1 607 061 | 35 | 8 | ||
| 9p22.3-p21.3 | 16 479 078–20 933 035 | 4 453 957 | 51 | 17 | MLLT3 | |
| 9p21.3 | 21 969 065–22 051 061 | 81 996 | 54 | 3 | CDKN2A CDKN2B | CDKN2BAS |
| 9q21.33 | 88 865 092–89 129 765 | 264 673 | 44 | 2 | LOC494127 C9orf170 | |
| 9q22.32 | 96 898 992–97 116 235 | 217 243 | 43 | 1 | FANCC | |
| 9q22.33-q31.1 | 100 644 626–101 678 237 | 1 033 611 | 43 | 3 | NR4A3 | GALNT12 COL15A1 TGFBR1 ALG2 SEC61B |
| 9q33.1 | 117 953 363–118 381 663 | 428 300 | 46 | 3 | PAPPA LOC100128505 ASTN2 | |
| 9q33.2 | 123 060 025–123 208 621 | 148 596 | 46 | 2 | GSN STOM | |
| 9q33.3 | 128 975 869–129 006 542 | 30 673 | 35 | 1 | RALGPS1 | |
| 9q34.13-q34.2 | 133 483 430–135 001 392 | 1 517 962 | 51 | 17 | TSC1 | |
| 9q34.2 | 135 339 532–135 424 778 | 85 246 | 53 | 2 | TMEM8C ADAMTSL2 | |
| 11p15.5-p15.4 | 826 091–2 876 898 | 2 050 807 | 41 | 44 | ||
| 13q12.13 | 25 719 218–25 817 850 | 98 632 | 44 | 1 | CDK8 | |
| 16p13.3 | 3 759 661–3 782 779 | 23 118 | 35 | 1 | CREBBP | |
| 16q21 | 62 541 693–64 974 691 | 2 432 998 | 40 | 3 | CDH11 | LOC283867 CDH5 |
| 16q22.3-q23.1 | 73 046 913–74 862 172 | 1 815 259 | 40 | 20 | ||
| 17p13.3 | 1 431 769–1 639 791 | 208 022 | 57 | 11 | ||
| 17p11.2 | 19 082 703–20 046 930 | 964 227 | 63 | 15 | ||
| 17q11.2-q12 | 26 420 214–29 077 825 | 2 657 611 | 54 | 4 | NF1 | TMEM98 SPACA3 ACCN1 |
| 18q21.2 | 48 644 312–51 715 504 | 3 071 192 | 40 | 9 | ||
| 18q21.32-q21.33 | 55 721 386–57 214 344 | 1 492 958 | 40 | 2 | PMAIP1 MC4R | |
| 18q23 | 75 912 794–76 117 153 | 204 359 | 44 | 3 | ADNP2 LOC100130522 PARD6G | |
| 22q12.3 | 30 817 999–32 856 500 | 2 038 501 | 43 | 12 | ||
| 22q12.3 | 32 856 500–34 737 030 | 1 880 530 | 53 | 10 | ||
| 22q12.3 | 34 737 030–35 049 039 | 312 009 | 43 | 6 | MYH9 | RBFOX2 APOL3 APOL4 APOL2 APOL1 |
| 22q13.1 | 37 668 125–37 724 138 | 56 013 | 53 | 2 | APOBEC3A APOBEC3B | |
| 22q13.1 | 43 530 191–46 673 931 | 3 143 740 | 53 | 32 | ||
| 22q13.31-q13.33 | 46 673 931–49 472,215 | 2 798 284 | 54 | 40 | ||
| Xq11.1-q12 | 62 673 834–65 512 219 | 2 838 385 | 35 | 14 | ||
| Xq12 | 65 512 219–66 014 702 | 502 483 | 46 | 1 | EDA2R | |
| Xq12-q13.1 | 66 014 702–68 066 344 | 2 051 642 | 35 | 5 | AR | OPHN1 YIPF6 STARD8 EFNB1 |
| Xq27.3 | 144 141 243–146 657 619 | 2 516 376 | 41 | 21 | ||
Gained and lost regions listed as shared across tumour subtypes if present among the top 60 most frequently altered regions (minimum frequency=25%) in at least three tumour subtypes (BRCA1, B1; BRCA2, B2; BRCAX, BX; Sporadic, S). In bold regions found to be recurrent in all four tumour groups.
Global frequency of the alteration in whole tumour set; frequencies greater or equal to 55% highlighted in bold.
Genes of interest selected from Cancer Census (in bold) or based on their function and previously published data.
Rest of the genes in the defined region listed if less than seven.
Despite the similar profile of copy number changes exhibited by hereditary and sporadic tumours, supervised analysis unveiled some regions potentially associated with particular tumour subtypes (FDR<0.2) (Figure 1A; Supplementary Table S4). Among the alterations more recurrently found in BRCA1 tumours compared with sporadic cases were losses at 4q32.3-q34.1, 6q22.33-q26 or 12q21.2-q23.2 (Figure 1A and IV) while gains at 6p12-p11, 10p14-p11 and 10q22 were found to be more frequent in BRCAX tumours (compared with BRCA1 cases; Figure 1A and IV). Gains at 2p23.3, 12p11.22-p11.1 and 19q12-q13.11 were identified more recurrently in sporadic cases (Figure 1A and II), with the latter containing the CCNE1 gene that was also amplified in these tumours.
High level amplifications and HDs
The high resolution of our platform allowed us to identify focal high-amplitude copy number changes. Fifty-nine narrow amplifications (median length of 739 Kb spanning on average 11 genes) were identified in at least two cases (Supplementary Table S5, part A). For instance amplification at 8q22.1, found only in BRCA1 tumours, presented a MCR that spanned just one gene, the cancer-associated LAPTM4B. The most frequent regions of amplification with their distributions across the groups of tumours are shown in Table 2. In addition, 57 focal HDs (median length of 465 Kbp spanning 6 genes on average) were identified in at least 2 samples (Table 2; Supplementary Table S5, part B). The two most common HDs (9% each) present in all tumour groups were found at 17q11.2 and 13q14.2; each deletion encompassed only one gene (NF1 and RB1, respectively). Other frequent HDs common for sporadic and familial tumours were defined at 8p23.2-p23.1, spanning the early DNA damage-response gene MCPH1, and at the fragile site on chromosome 3 (3p14.2) containing the known tumour suppressor FHIT.
Table 2. Distribution of most frequent amplifications and homozygous deletions across tumour subtypes.
| |
|
|
|
No. of samples with alteration |
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytoband | Region | Size (bp) | Frequencya % | All | B1 | B2 | BX | S | No. of genes | Genes of interestb | Other genes in the regionc |
|
Amplifications | |||||||||||
| 3q26.2 |
170 112 026–170 346 939 |
234 913 |
5.9 |
4 |
1 |
|
|
3 |
1 |
MECOM |
MECOM |
| 3q26.32-q26.33 |
179 526 769–181 046 005 |
1 519 236 |
5.9 |
4 |
|
|
1 |
3 |
12 |
PIK3CA |
|
| 7q36.3 |
155 939 892–158 642 803 |
2 702 911 |
5.9 |
4 |
2 |
|
2 |
|
16 |
|
|
|
8q21.2-q21.3 |
85 047 239–87 355 110 |
2 307 871 |
5.9 |
4 |
2 |
|
1 |
1 |
15 |
|
|
|
8q21.3 |
92 323 386–92 443 508 |
120 122 |
7.4 |
5 |
3 |
|
1 |
1 |
1 |
|
SLC26A7 |
| 8q22.1 |
94918806–95 029 680 |
110874 |
5.9 |
4 |
3 |
|
1 |
|
1 |
|
PDP1 |
|
8q22.1 |
96 742 090–97 658 770 |
91 668 |
7.4 |
5 |
3 |
|
1 |
1 |
6 |
|
LOC100500773 GDF6 UQCRB |
| |
|
|
|
|
|
|
|
|
|
|
MTERFD1 PTDSS1 SDC2 |
| 8q22.1 |
98 814 783–98 930 489 |
115 706 |
8.8 |
6 |
6 |
|
|
|
1 |
LAPTM4B |
LAPTM4B |
|
8q22.3 |
101 926 982–102 101 553 |
174 571 |
11.8 |
8 |
5 |
|
1 |
2 |
1 |
YWHAZ |
YWHAZ |
| 8q23.1 |
107 121 208–108 087 190 |
965 982 |
10.3 |
7 |
4 |
|
3 |
|
2 |
|
OXR1 ABRA |
| 8q23.1-q23.2 |
110 438 342–111 177 707 |
739 365 |
11.8 |
8 |
4 |
|
4 |
|
4 |
|
PKHD1L1 EBAG9 SYBU KCNV1 |
| 8q24.11 |
118 543 986–118 626 272 |
82 286 |
14.7 |
10 |
3 |
1 |
6 |
|
1 |
|
MED30 |
| 8q24.13 |
122 701 184–123 202 812 |
501 628 |
13.2 |
9 |
3 |
1 |
5 |
|
2 |
|
HAS2 HAS2AS |
| 8q24.21 |
128 537 094–128 997 964 |
460 870 |
19.1 |
13 |
5 |
2 |
6 |
|
2 |
MYC |
MYC PVT1 |
| 8q24.23-q24.3 |
139 929 002–142 315 091 |
2 386 089 |
22.1 |
15 |
7 |
2 |
6 |
|
8 |
TRAPPC9 |
COL22A1 KCNK9 TRAPPC9 CHRAC1 EIF2C2 PTK2 DENND3 SLC45A4 |
|
11q13.1 |
64 958 966–65 036 226 |
77 260 |
11.8 |
8 |
3 |
|
2 |
3 |
2 |
MALAT1 |
MIR612 MALAT1 |
| 17q12-q21.1 |
35 083 091–35 415 740 |
332 649 |
5.9 |
4 |
|
|
2 |
2 |
11 |
ERBB2 |
|
| 19p13.2 |
8 939 055–10 745 326 |
1 806 271 |
5.9 |
4 |
|
|
2 |
2 |
33 |
MUC16 |
|
| 20q13.2 |
51 530 308–52 731 992 |
1 201 684 |
5.9 |
4 |
|
|
2 |
2 |
7 |
|
TSHZ2 ZNF217 SUMO1P1 BCAS1 CYP24A1 PFDN4 DOK5 |
|
20q13.31 |
54 436 225–55 409 396 |
973 171 |
5.9 |
4 |
1 |
|
2 |
1 |
12 |
|
|
|
Homozygous deletions | |||||||||||
| 1p36.32 |
4 132 552–4 616 506 |
483 954 |
4.4 |
3 |
3 |
|
|
|
2 |
AJAP1 |
LOC284661 AJAP1 |
|
3p14.2 |
60 530 653–60 943 377 |
412 724 |
4.4 |
3 |
|
1 |
1 |
1 |
1 |
FHIT |
FHIT |
|
5q15 |
95 249 110–95 428 565 |
179 455 |
4.4 |
3 |
1 |
|
1 |
1 |
1 |
|
ELL2 |
| 8p23.3 |
0–206 224 |
206 224 |
7.4 |
5 |
4 |
1 |
|
|
3 |
|
OR4F21 RPL23AP53 ZNF596 |
|
8p23.2-p23.1 |
6 177 456–6 455 595 |
278 139 |
7.4 |
5 |
3 |
1 |
|
1 |
2 |
MCPH1 |
MCPH1 ANGPT2 |
| 8p22-p21.3 |
17 000 420–19 725 139 |
2 724 719 |
5.9 |
4 |
3 |
|
|
1 |
17 |
PCM1 MTUS1 |
|
| 8p21.3-p21.2 |
22 096 663–26 106 924 |
4 010 261 |
7.4 |
5 |
4 |
|
|
1 |
37 |
DOCK5 CDCA2 EBF2 |
|
| 8p21.1-p12 |
28 636 935–33 247 321 |
4 610 386 |
4.4 |
3 |
2 |
|
|
1 |
21 |
WRN DUSP4 HMBOX1 NRG1 |
|
|
13q14.2 |
47 841 552–47 847 231 |
5679 |
8.8 |
6 |
2 |
|
3 |
1 |
1 |
RB1 |
RB1 |
| 16q23.3 |
80 791 319–82 209 820 |
1 418 501 |
4.4 |
3 |
3 |
|
|
|
2 |
CDH13 |
CDH13 MIR3182 |
|
17q11.2 |
26 516 613–26 548 438 |
31 825 |
8.8 |
6 |
1 |
1 |
2 |
2 |
1 |
NF1 |
NF1 |
|
19q13.43 |
61 855 409–62 293 274 |
437 865 |
4.4 |
3 |
1 |
|
1 |
1 |
6 |
PEG3 ZIM2 |
LOC147670 ZNF835 ZIM2 |
| |
|
|
|
|
|
|
|
|
|
|
PEG3AS PEG3 MIMT1 |
| 22q13.31-q13.33 |
46 034 061–48 551 789 |
2 517 728 |
4.4 |
3 |
2 |
|
1 |
|
4 |
|
FLJ46257 MIR3201 FAM19A5 C22orf34 |
|
Xp22.11-p21.3 |
24 744 413–25 039 949 |
295 536 |
4.4 |
3 |
1 |
|
1 |
1 |
2 |
|
POLA1 ARX |
|
Xp21.1 |
31 763 941–33 579 700 |
1 815 759 |
5.9 |
4 |
1 |
|
1 |
2 |
1 |
|
DMD |
|
Xp11.4 |
38 519 008–39 470 641 |
951 633 |
4.4 |
3 |
1 |
|
1 |
1 |
1 |
|
MID1IP1 |
|
Xp11.3 |
43 295 963–43 705 583 |
409 620 |
5.9 |
4 |
1 |
|
2 |
1 |
3 |
|
MAOA MAOB NDP |
| Xp11.3 |
44 198 148–44 786 777 |
588 629 |
4.4 |
3 |
1 |
|
2 |
|
3 |
KDM6A |
FUNDC1 DUSP21 KDM6A |
| Xp11.3-p11.23 |
46 905 791–48 631 317 |
1 725 526 |
4.4 |
3 |
2 |
|
1 |
|
45 |
SSX4 WAS GATA1 |
|
| Xp11.23-p11.22 | 49 634 473–49 949 828 | 315 355 | 4.4 | 3 | 1 | 1 | 1 | 10 | |||
Amplifications and homozygous deletions with frequencies greater than 5% (⩾4 samples) and 4% (⩾3 samples), respectively. Cytobands in bold indicate regions found in sporadic and at least two of the familial subtypes. BRCA1, B1; BRCA2, B2; BRCAX, BX; Sporadic, S.
Global frequency of the alteration in whole tumour set; frequencies >5% are highlighted in bold.
Genes of interest selected from Cancer Census (in bold) or based on their function and previously published data.
Rest of genes in the defined region (a maximum of six genes per region is shown).
Immunohistochemical validation of aCGH results
To validate our aCGH results, we assessed the correlation between the assigned DNA copy number and the immunohistochemical expression of three genes targeted by high-amplitude events: CDKN2A and RB1 located at homozygously deleted regions, and CCNE1 that was found amplified. Immunohistochemical analysis showed complete lack or much lower expression of CDKN2A and RB1 in tumours with HD at these loci compared with the mean value of samples with a flat profile at 9p21.3 and 13q14.2, respectively (Supplementary Figure S1A and B). Tumours exhibiting CCNE1 amplification presented much higher expression compared with the mean value of tumours with normal DNA copy number at this locus (Supplementary Figure S1C).
Unsupervised analysis
In addition to systematic comparison of copy number alterations across different tumour subtypes, we also carried out unsupervised analysis of the aCGH data to unveil possible associations between particular patterns of genomic changes and the sporadic or familial status of tumours (or hereditary subtype, BRCA1/2/X). Unsupervised hierarchical clustering stratified ovarian carcinomas (n=68), based on their copy number changes, into two main clusters (A and B) (Figure 1B, left panel). Significant differences were not found between tumours from both clusters (or from smaller subgroups) either according to their general familial or sporadic condition or according to their specific BRCA mutation status. In contrast, clustering was associated with genomic instability level, FIGO stage and histological subtype. The cluster with more genomically instable tumours (cluster B) was significantly enriched in high FIGO stage (P=0.03) and serous type carcinomas (vs all other subtypes, P=0.001). Unsupervised hierarchical clustering of the subgroup of type II tumours (n=31) also rendered two clusters (II-A and II-B) without significant enrichment in tumours from particular BRCA subgroups (Figure 1B, right panel).
Discussion
Few studies have addressed the characterisation of DNA copy number changes arising in hereditary ovarian tumours, especially in comparison with more extensively studied familial breast tumours (Hedenfalk et al, 2003; Gronwald et al, 2005; Jonsson et al, 2005; Mangia et al, 2008; Melchor et al, 2008; Joosse et al, 2009; Stefansson et al, 2009; Waddell et al, 2010; Didraga et al, 2011; Focken et al, 2011). The few existing studies have rendered contradictory results, either supporting that mutations in BRCA1/2 affect the particular chromosomal alterations occurring throughout cancer progression (Ramus et al, 2003, 2007), or reporting very few copy number changes specifically associated to tumours harbouring such mutations (Israeli et al, 2003; TCGA, 2011). Given these antecedents, the recently confirmed relevance of copy number changes as drivers of ovarian oncogenesis (TCGA, 2011) and the growing clinical implications of the BRCA1/2 mutation status, we aimed to determine how hereditary and sporadic ovarian tumours relate to genomic instability and to define common and/or distinct events occurring in the genesis and evolution of these neoplasms. Different from most prior studies, we analysed tumours not only from carriers of BRCA1/2 mutations, but also from non-BRCA1/2 hereditary patients (BRCAX tumours) as these have been particularly poorly characterised. Also, we used a high-resolution aCGH platform and separately analysed gains and losses (in contrast to other studies correlating BRCA1/2 impairment only with global instability).
Our findings indicate lack of substantial differences in the pattern of DNA copy number changes displayed by sporadic carcinomas and the different subtypes of familial tumours. This similarity was illustrated by the existence of shared regions found to be recurrently altered in each individual group of tumours. These common events point to the involvement of genes fundamental for ovarian carcinogenesis, selected throughout the evolution of the tumours and providing advantage to any cancer cell, independently of the existence of germinal mutations in the BRCA1/2 genes. As possible candidates, we found known players previously related to EOC such as PIK3CA, MECOM, MYC, NF1 and RB, but also less characterised genes, whose gains of function (CDH12, FOXQ1, TXNDC5, CCND2, FOXJ1) and/or abrogation (FANCC, TSC1, CREBBP, CDH11, EDA2R) might be crucial for ovarian cancer development and/or progression. Exploration of the therapeutic opportunities provided by these targets, to which a majority of tumours are likely to be addicted, is an attractive possibility. For instance, it has been suggested that modulation of cellular activities of the forkhead transcription factor FOXQ1 may have an application in cancer therapy since its inhibition blocks epithelial to mesenchymal transition and results in cancer cell sensitisation to a variety of chemotherapeutic agents (Qiao et al, 2011). Therapeutic approaches based on repression of cyclin D gene have also been investigated (Tiedemann et al, 2008; Dong et al, 2010) and may be applicable to EOCs presenting aberrant CCND2 expression due to DNA-copy number gains. Likewise, m-TORC1-directed therapies may be more effective in cancer patients whose tumours present TSC1 (tuberous sclerosis complex 1) genomic losses as it has been proposed for patients whose tumour harbour TSC1 somatic mutations (Iyer et al, 2012).
Also exemplifying the absence of marked differences in the profile of genomic changes of carriers and non-carriers of BRCA1/2 mutations, unsupervised hierarchical clustering did not stratify tumours according to their familial or sporadic condition, nor did it according to their BRCA1/2 mutation status. This is different from what has been reported on familial BRCA1 and BRCA2 breast tumours, which show association with particular molecular subtypes (defined with expression arrays) and specific patterns of copy number changes (Jonsson et al, 2005; Bergamaschi et al, 2006; Melchor et al, 2008; Stefansson et al, 2009). Lack of segregation of ovarian tumours from carriers and non-carriers of BRCA1/2 germline mutations based on their genomic instability pattern would support a model according to which HR repair loss (through inactivation of the BRCA1/2 genes or other members of the pathway) would not only be a frequent event in high-grade serous ovarian tumours, as recently demonstrated (TCGA, 2011), but also an event occurring in the initial phases of tumour growth.
Interestingly, despite this general picture of similarity between sporadic and hereditary tumours, differences concerning the overall degree of genomic instability were revealed when gains and losses were analysed separately. Copy number losses were particularly abundant in BRCA1 and BRCA2 tumours, both in global terms (comparison of losses made across tumour subtypes) and relative to the number of gains (comparison within each tumour subtype). Although greater contribution of losses than gains was observed in all tumour subtypes, the extent of this phenomenon was more prominent in carriers of BRCA1 and BRCA2 mutations. Some prior studies, including the most comprehensive one conducted by the TCGA Research network in high-grade serous ovarian carcinomas, reported no differences in the global degree of instability between tumours with BRCA1/2 inactivating events and those with functional BRCA1/2 genes (TCGA, 2011; Ramus et al, 2003). However, no distinction was made between gains and losses, and only comparison of total changes was conducted. Earlier studies already suggested the relevance of loss of heterozygosity (LOH) in ovarian tumours from BRCA1 and BRCA2 mutation carriers (Walsh et al, 2008; Leunen et al, 2009; Wang et al, 2012), but included very few familial cases (Walsh et al, 2008; Leunen et al, 2009) or used low-resolution platforms (Leunen et al, 2009). Our results derived from analysis made across tumour types, within each tumour subgroup and particularly when taking into account only high-grade tumours highlight the relevance of genomic loss in BRCA1 and BRCA2 tumours, a phenomenon that would not merely reflect differences related to the higher grade or more prevalent serous histotype of hereditary tumours. Interestingly, correlation between extent of LOH and therapy sensitivity has been recently reported in high-grade serous ovarian tumours (Wang et al, 2012).
Our findings suggest that in the oncogenesis of ovarian tumours, and in particular of hereditary BRCA1 and BRCA2 tumours, loss of function of tumour suppressor genes might be under greater selection pressure than gain of function of proto-oncogenes, at least through DNA copy number-related mechanisms. However, lack of clear segregation of BRCA1 and BRCA2 tumours in the unsupervised analysis would indicate that most of the genomic loss in carriers does not involve a defined group of critical regions (or specific suppressor genes) recurrently selected during evolution of these particular tumours. Alternatively, greater involvement of loss events in ovarian tumours might be related to impairment of HR function, with grosser effects in BRCA1 and BRCA2 tumours due to the central role played by both genes within the pathway.
Although a consistent distinct pattern of copy number changes does not seem to characterise BRCA1 and BRCA2 hereditary ovarian tumours, we were able to define several individual regions potentially associated with BRCA1 tumours (mainly losses) and a few other aberrations (gains and amplifications) potentially associated with sporadic tumours. For instance, losses defined at 4q32.1-q35.2, 13q13.3-q14.3, 17q11.1-q11.2, 17q12, 17q21.32-q21.33, 17q24.3-q25.1 and 22q13.31 found to be related more specifically to BRCA1 cases in our series, were also previously reported to be associated with this tumour group (Zweemer et al, 2001; Ramus et al, 2003; Domanska et al, 2010). The ovarian TCGA study reported only two regions being significantly enriched in one tumour subtype, amplifications at 19p13.13 and 19q12 associated with sporadic tumours (vs BRCA-altered tumours). Interestingly, gains and amplifications at 19q12 encompassing CCNE1 were also defined as potentially associated with sporadic tumours in our analysis. This finding reinforces the proposed role for CCNE1 and of other proteins implicated in cell-cycle progression as important contributors to ovarian carcinogenesis in tumours with intact BRCA function (Bowtell, 2010; TCGA, 2011; Berns and Bowtell, 2012)
In summary, we have performed a comprehensive analysis of the DNA-copy number changes that occur in hereditary EOCs. We have found that overall hereditary and sporadic EOCs exhibit a similar pattern of DNA copy number alterations. However, greater contribution of losses was revealed to be a hallmark of BRCA1 and BRCA2 mutation carriers. Importantly, this feature may help to identify BRCA-related patients who have been shown to respond to PARP inhibitors and to present better prognosis when treated with standard regimes.
Acknowledgments
We are very grateful to María del Carmen González-Neira and Victoria Fernández (from the CNIO Human Cancer Genetics group) and to the CNIO Tumor Bank Network, in particular to María Jesús Artiga, for exceptional coordination of tumour collection and clinical data retrieval. We also thank the CNIO Histology and Immunohistochemistry Core Unit, especially Lydia Sánchez for excellent assistance. Also, we would like to acknowledge Paul P Eijk, Daniëlle Israeli, François Rustenburg, Dirk Vanessen and Jolanda Reek (VU University Medical Center, Department of Pathology, Amsterdam, The Netherlands) for their technical and laboratory support. MJG is the recipient of a research contract from the Instituto de Salud Carlos III, Ministerio Español de Sanidad y Consumo (Miguel Servet Program, CP07/00113). MMK receives financial support from the Fundación La Caixa. This study was funded by the Fondo de Investigación Sanitaria (FIS), Instituto de Salud Carlos III (grants CP07/00113 and PS09/01094).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Alsop K, Fereday S, Meldrum C, Defazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C, Friedlander M, Fox S, Bowtell D, Mitchell G. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian ovarian cancer study group. J Clin Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- Berns EM, Bowtell DD. The changing view of high-grade serous ovarian cancer. Cancer Res. 2012;72:2701–2704. doi: 10.1158/0008-5472.CAN-11-3911. [DOI] [PubMed] [Google Scholar]
- Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, Lambrechts D, Despierre E, Barrowdale D, McGuffog L, Healey S, Easton DF, Sinilnikova O, Benitez J, Garcia MJ, Neuhausen S, Gail MH, Hartge P, Peock S, Frost D, Evans DG, Eeles R, Godwin AK, Daly MB, Kwong A, Ma ES, Lazaro C, Blanco I, Montagna M, D'Andrea E, Nicoletto MO, Johnatty SE, Kjaer SK, Jensen A, Hogdall E, Goode EL, Fridley BL, Loud JT, Greene MH, Mai PL, Chetrit A, Lubin F, Hirsh-Yechezkel G, Glendon G, Andrulis IL, Toland AE, Senter L, Gore ME, Gourley C, Michie CO, Song H, Tyrer J, Whittemore AS, McGuire V, Sieh W, Kristoffersson U, Olsson H, Borg A, Levine DA, Steele L, Beattie MS, Chan S, Nussbaum RL, Moysich KB, Gross J, Cass I, Walsh C, Li AJ, Leuchter R, Gordon O, Garcia-Closas M, Gayther SA, Chanock SJ, Antoniou AC, Pharoah PD. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- Buffart TE, Israeli D, Tijssen M, Vosse SJ, Mrsic A, Meijer GA, Ylstra B. Across array comparative genomic hybridization: a strategy to reduce reference channel hybridizations. Genes Chromosomes Cancer. 2008;47:994–1004. doi: 10.1002/gcc.20605. [DOI] [PubMed] [Google Scholar]
- Didraga MA, van Beers EH, Joosse SA, Brandwijk KI, Oldenburg RA, Wessels LF, Hogervorst FB, Ligtenberg MJ, Hoogerbrugge N, Verhoef S, Devilee P, Nederlof PM. A non-BRCA1/2 hereditary breast cancer sub-group defined by aCGH profiling of genetically related patients. Breast Cancer Res Treat. 2011;130:425–436. doi: 10.1007/s10549-011-1357-x. [DOI] [PubMed] [Google Scholar]
- Domanska K, Malander S, Staaf J, Karlsson A, Borg A, Jonsson G, Nilbert M. Genetic profiles distinguish different types of hereditary ovarian cancer. Oncol Rep. 2010;24:885–895. doi: 10.3892/or.2010.885. [DOI] [PubMed] [Google Scholar]
- Dong Q, Meng P, Wang T, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focken T, Steinemann D, Skawran B, Hofmann W, Ahrens P, Arnold N, Kroll P, Kreipe H, Schlegelberger B, Gadzicki D. Human BRCA1-associated breast cancer: no increase in numerical chromosomal instability compared to sporadic tumors. Cytogenet Genome Res. 2011;135:84–92. doi: 10.1159/000332005. [DOI] [PubMed] [Google Scholar]
- Gilks CB, Ionescu DN, Kalloger SE, Kobel M, Irving J, Clarke B, Santos J, Le N, Moravan V, Swenerton K. Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol. 2008;39:1239–1251. doi: 10.1016/j.humpath.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Gronwald J, Jauch A, Cybulski C, Schoell B, Bohm-Steuer B, Lener M, Grabowska E, Gorski B, Jakubowska A, Domagala W, Chosia M, Scott RJ, Lubinski J. Comparison of genomic abnormalities between BRCAX and sporadic breast cancers studied by comparative genomic hybridization. Int J Cancer. 2005;114:230–236. doi: 10.1002/ijc.20723. [DOI] [PubMed] [Google Scholar]
- Hedenfalk I, Ringner M, Ben-Dor A, Yakhini Z, Chen Y, Chebil G, Ach R, Loman N, Olsson H, Meltzer P, Borg A, Trent J. Molecular classification of familial non-BRCA1/BRCA2 breast cancer. Proc Natl Acad Sci USA. 2003;100:2532–2537. doi: 10.1073/pnas.0533805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli O, Gotlieb WH, Friedman E, Goldman B, Ben-Baruch G, Aviram-Goldring A, Rienstein S. Familial vs sporadic ovarian tumors: characteristic genomic alterations analyzed by CGH. Gynecol Oncol. 2003;90:629–636. doi: 10.1016/s0090-8258(03)00375-5. [DOI] [PubMed] [Google Scholar]
- Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, Viale A, Heguy A, Peng L, Chan TA, Bochner B, Bajorin DF, Berger MF, Taylor BS, Solit DB. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR, Greshock JD, Luts L, Olsson H, Rahman N, Stratton M, Ringner M, Borg A, Weber BL. Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res. 2005;65:7612–7621. doi: 10.1158/0008-5472.CAN-05-0570. [DOI] [PubMed] [Google Scholar]
- Joosse SA, van Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, Ligtenberg MJ, Wessels LF, Axwijk P, Verhoef S, Hogervorst FB, Nederlof PM. Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat. 2009;116:479–489. doi: 10.1007/s10549-008-0117-z. [DOI] [PubMed] [Google Scholar]
- Kalloger SE, Kobel M, Leung S, Mehl E, Gao D, Marcon KM, Chow C, Clarke BA, Huntsman DG, Gilks CB. Calculator for ovarian carcinoma subtype prediction. Mod Pathol. 2011;24:512–521. doi: 10.1038/modpathol.2010.215. [DOI] [PubMed] [Google Scholar]
- Kobel M, Kalloger SE, Carrick J, Huntsman D, Asad H, Oliva E, Ewanowich CA, Soslow RA, Gilks CB. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- Koul A, Malander S, Loman N, Pejovic T, Heim S, Willen R, Johannsson O, Olsson H, Ridderheim M, Borg AA. BRCA1 and BRCA2 mutations in ovarian cancer: Covariation with specific cytogenetic features. Int J Gynecol Cancer. 2000;10:289–295. doi: 10.1046/j.1525-1438.2000.010004289.x. [DOI] [PubMed] [Google Scholar]
- Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2011;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leunen K, Gevaert O, Daemen A, Vanspauwen V, Michils G, De Moor B, Moerman P, Vergote I, Legius E. Recurrent copy number alterations in BRCA1-mutated ovarian tumors alter biological pathways. Hum Mutat. 2009;30:1693–1702. doi: 10.1002/humu.21135. [DOI] [PubMed] [Google Scholar]
- Malpica A, Deavers MT, Lu K, Bodurka DC, Atkinson EN, Gershenson DM, Silva EG. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- Mangia A, Chiarappa P, Tommasi S, Chiriatti A, Petroni S, Schittulli F, Paradiso A. Genetic heterogeneity by comparative genomic hybridization in BRCAx breast cancers. Cancer Genet Cytogenet. 2008;182:75–83. doi: 10.1016/j.cancergencyto.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Melchor L, Honrado E, Garcia MJ, Alvarez S, Palacios J, Osorio A, Nathanson KL, Benitez J. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–3175. doi: 10.1038/sj.onc.1210975. [DOI] [PubMed] [Google Scholar]
- Milne RL, Osorio A, Cajal TR, Vega A, Llort G, de la Hoya M, Diez O, Alonso MC, Lazaro C, Blanco I, Sanchez-de-Abajo A, Caldes T, Blanco A, Grana B, Duran M, Velasco E, Chirivella I, Cardenosa EE, Tejada MI, Beristain E, Miramar MD, Calvo MT, Martinez E, Guillen C, Salazar R, San Roman C, Antoniou AC, Urioste M, Benitez J. The average cumulative risks of breast and ovarian cancer for carriers of mutations in BRCA1 and BRCA2 attending genetic counseling units in Spain. Clin Cancer Res. 2008;14:2861–2869. doi: 10.1158/1078-0432.CCR-07-4436. [DOI] [PubMed] [Google Scholar]
- Patael-Karasik Y, Daniely M, Gotlieb WH, Ben-Baruch G, Schiby J, Barakai G, Goldman B, Aviram A, Friedman E. Comparative genomic hybridization in inherited and sporadic ovarian tumors in Israel. Cancer Genet Cytogenet. 2000;121:26–32. doi: 10.1016/s0165-4608(00)00224-7. [DOI] [PubMed] [Google Scholar]
- Pennington KP, Swisher EM. Hereditary ovarian cancer: beyond the usual suspects. Gynecol Oncol. 2012;124:347–353. doi: 10.1016/j.ygyno.2011.12.415. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi SC, Yu Q. FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 2011;71:3076–3086. doi: 10.1158/0008-5472.CAN-10-2787. [DOI] [PubMed] [Google Scholar]
- Ramus SJ, Harrington PA, Pye C, DiCioccio RA, Cox MJ, Garlinghouse-Jones K, Oakley-Girvan I, Jacobs IJ, Hardy RM, Whittemore AS, Ponder BA, Piver MS, Pharoah PD, Gayther SA. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat. 2007;28:1207–1215. doi: 10.1002/humu.20599. [DOI] [PubMed] [Google Scholar]
- Ramus SJ, Pharoah PD, Harrington P, Pye C, Werness B, Bobrow L, Ayhan A, Wells D, Fishman A, Gore M, DiCioccio RA, Piver MS, Whittemore AS, Ponder BA, Gayther SA. BRCA1/2 mutation status influences somatic genetic progression in inherited and sporadic epithelial ovarian cancer cases. Cancer Res. 2003;63:417–423. [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Silverberg SG. Histopathologic grading of ovarian carcinoma: a review and proposal. Int J Gynecol Pathol. 2000;19:7–15. doi: 10.1097/00004347-200001000-00003. [DOI] [PubMed] [Google Scholar]
- Soslow RA, Han G, Park KJ, Garg K, Olvera N, Spriggs DR, Kauff ND, Levine DA. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol. 2012;25:625–636. doi: 10.1038/modpathol.2011.183. [DOI] [PubMed] [Google Scholar]
- Stefansson OA, Jonasson JG, Johannsson OT, Olafsdottir K, Steinarsdottir M, Valgeirsdottir S, Eyfjord JE. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res. 2009;11:R47. doi: 10.1186/bcr2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann RE, Mao X, Shi CX, Zhu YX, Palmer SE, Sebag M, Marler R, Chesi M, Fonseca R, Bergsagel PL, Schimmer AD, Stewart AK. Identification of kinetin riboside as a repressor of CCND1 and CCND2 with preclinical antimyeloma activity. J Clin Invest. 2008;118:1750–1764. doi: 10.1172/JCI34149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wiel DF, Zhang WL. Identification of pork quality parameters by proteomics. Meat Sci. 2007;77:46–54. doi: 10.1016/j.meatsci.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Van Wieringen WN, Van De Wiel MA, Ylstra B. Weighted clustering of called array CGH data. Biostatistics. 2008;9:484–500. doi: 10.1093/biostatistics/kxm048. [DOI] [PubMed] [Google Scholar]
- Waddell N, Arnold J, Cocciardi S, da Silva L, Marsh A, Riley J, Johnstone CN, Orloff M, Assie G, Eng C, Reid L, Keith P, Yan M, Fox S, Devilee P, Godwin AK, Hogervorst FB, Couch F, Grimmond S, Flanagan JM, Khanna K, Simpson PT, Lakhani SR, Chenevix-Trench G. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res Treat. 2010;123:661–677. doi: 10.1007/s10549-009-0653-1. [DOI] [PubMed] [Google Scholar]
- Walsh CS, Ogawa S, Scoles DR, Miller CW, Kawamata N, Narod SA, Koeffler HP, Karlan BY. Genome-wide loss of heterozygosity and uniparental disomy in BRCA1/2-associated ovarian carcinomas. Clin Cancer Res. 2008;14:7645–7651. doi: 10.1158/1078-0432.CCR-08-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZC, Birkbak NJ, Culhane A, Drapkin RI, Fatima A, Tian R, Schwede M, Alsop K, Daniels KE, Piao H, Liu J, Etemadmoghadam D, Miron A, Salvesen HB, Mitchell G, Defazio A, Quackenbush J, Berkowitz RS, Iglehart JD, Bowtell DD, Matulonis UA. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clin Cancer Res. 2012;18:5806–5815. doi: 10.1158/1078-0432.CCR-12-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . Histological Typing of Ovarian Tumours. Springer: Berlin; 2004. [Google Scholar]
- Zweemer RP, Ryan A, Snijders AM, Hermsen MA, Meijer GA, Beller U, Menko FH, Jacobs IJ, Baak JP, Verheijen RH, Kenemans P, van Diest PJ. Comparative genomic hybridization of microdissected familial ovarian carcinoma: two deleted regions on chromosome 15q not previously identified in sporadic ovarian carcinoma. Lab Invest. 2001;81:1363–1370. doi: 10.1038/labinvest.3780350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.