Key Points
Application of capturing/sequencing, copy number, and RNA analysis technologies ensures comprehensive molecular diagnosis of Fanconi anemia.
Abstract
Current methods for detecting mutations in Fanconi anemia (FA)–suspected patients are inefficient and often miss mutations. We have applied recent advances in DNA sequencing and genomic capture to the diagnosis of FA. Specifically, we used custom molecular inversion probes or TruSeq-enrichment oligos to capture and sequence FA and related genes, including introns, from 27 samples from the International Fanconi Anemia Registry at The Rockefeller University. DNA sequencing was complemented with custom array comparative genomic hybridization (aCGH) and RNA sequencing (RNA-seq) analysis. aCGH identified deletions/duplications in 4 different FA genes. RNA-seq analysis revealed lack of allele specific expression associated with a deletion and splicing defects caused by missense, synonymous, and deep-in-intron variants. The combination of TruSeq-targeted capture, aCGH, and RNA-seq enabled us to identify the complementation group and biallelic germline mutations in all 27 families: FANCA (7), FANCB (3), FANCC (3), FANCD1 (1), FANCD2 (3), FANCF (2), FANCG (2), FANCI (1), FANCJ (2), and FANCL (3). FANCC mutations are often the cause of FA in patients of Ashkenazi Jewish (AJ) ancestry, and we identified 2 novel FANCC mutations in 2 patients of AJ ancestry. We describe here a strategy for efficient molecular diagnosis of FA.
Introduction
Fanconi anemia (FA) is a rare recessive disorder characterized by debilitating congenital abnormalities, life-threatening bone marrow failure, and a predisposition to myeloid, head and neck squamous cell carcinoma and other malignancies.1 Because of the extensive underlying genetic heterogeneity, which is caused by a plethora of mutations in at least 15 genes involved in DNA repair and maintenance of DNA stability, understanding the underpinnings of FA has been challenging.2,3 However, a molecular understanding is critical for the diagnosis and clinical management of FA patients. Malignancies are often the first manifestation of FA, and conventional treatment can lead to devastating toxicities.4 Severe phenotypic consequences are associated with certain defective FA genes and, to an extent, specific mutations.5 Furthermore, the majority of FA patients develop bone marrow dysfunction, which may require hematopoietic stem cell transplantation. Screening family members as prospective bone marrow donors necessitates the forehand knowledge of both mutations segregating in the family. Therefore, finding both the defective gene and the disease-causing mutations for each patient is critical to appropriate, efficient, and timely care.
In addition to the large number of genes, the heterogeneous nature of mutations, including large deletions, makes the molecular diagnosis of FA a daunting task. The conventional screening process is a sequential, multistep approach in which the specific defective gene is discovered by implementing genetic complementation studies and then sequencing the exons of that gene for mutations.6 Cell lines from some patients are insensitive to diepoxybutane or mitomycin C treatment due to lymphocyte mosaicism and thus are not even amenable to complementation testing.7 In such cases, it is necessary to obtain cultured skin fibroblasts, an invasive and time-consuming procedure. The fact that some mutations may be intronic or regulatory makes the completion of many studies difficult. Particular challenges are associated with FANCD2, which has 2 known pseudogenes.8 In addition, although larger deletions contribute to a substantial proportion of the disease-causing mutations in FA,9 there has not been a reliable and comprehensive strategy that is sensitive enough to identify all of these deletions and their boundaries. Efficient, novel high-throughput strategies are therefore needed for the diagnosis of FA patients and complete discovery of the underlying mutations.
Multiple methodologies have been developed to capture a specific genomic region from patient DNA,10-13 and advances are being made in massively parallel sequencing technologies.14,15 We captured and sequenced the entire length of the ∼2-Mb genomic region representing all the FA genes as well as a series of related genes. We also screened this large region for deletions and duplications using high-resolution array comparative genomic hybridization (aCGH). Finally, we evaluated multiple capturing, enrichment, and massively parallel sequencing approaches for identification of the disease-causing mutations in all FA genes, including FANCD2.
We report here the molecular genetic analysis of 27 FA patients with no previously identified mutations. In all 27 patients, both the defective gene and the underlying mutations were identified. aCGH was critical in identifying deletions in FANCA, FANCC, and FANCD2 and 1 duplication in FANCB. By demonstrating the effect of the mutation on RNA splicing, RNA sequence analysis not only revealed exon skipping associated with some synonymous missense and nonsense mutations, but also identified 3 pathogenic mutations residing deep within introns. The genes and mutations we identified represented nearly all FA groups, demonstrating the generalizability of the approach.
Materials and methods
Study subjects
Genomic DNA samples and fibroblast and Epstein-Barr virus–immortalized lymphoblastoid cell lines (LCL) were obtained from individuals diagnosed with FA and registered in the International Fanconi Anemia Registry (IFAR), which requires informed written consent in accordance with the Declaration of Helsinki. These studies were approved by the Institutional Review Board of The Rockefeller University. The Office of Human Subjects Research at the National Institutes of Health and the Institutional Review Board of the National Human Genome Research Institute approved the reception of de-identified cell lines and DNA samples from The Rockefeller University and analysis of the underlying molecular variants.
DNA and RNA extraction and reverse-transcription polymerase chain reaction
DNA was isolated from blood and cell lines using the Puregene kit and DNeasy blood and tissue DNA extraction kit (Qiagen), respectively, and subjected to phenol/chloroform extraction and ethanol precipitation. Fibroblast and LCL cell lines were grown in Dulbecco’s modified Eagle’s medium (with 15% fetal bovine serum) and RPMI 1640 (with 20% fetal bovine serum) media, respectively. Both media were supplemented with 1% penicillin-streptomycin, 1% Fungizone, and 1% Glutamax-1. Total RNA was extracted from cell lines using the RNeasy Mini kit and treated with RNase-free DNase (Qiagen). Complementary DNA synthesis was carried out using the SuperScript First-Strand Synthesis System (Invitrogen) with oligo-dT primers.
MIP design, capture, and sequence
A total of 5136 100mer molecular inversion probes (MIP) were designed to capture the entire genomic region plus 1-kb flanking regions for the FA and related genes. Details of the design, capture, enrichment, library preparation, and sequencing were as described previously.10,16 Sequencing of the enriched libraries was performed on an Illumina GA-II platform in a single-end, 36-base configuration. Sequence reads were aligned to NCBI build 36 (hg18) of the human genome using ELAND (Illumina). Reads that could not be aligned by ELAND were then aligned to hg18 using cross-match (http://www.phrap.org).
WES design, capture, and sequence
The TruSeq whole-exome sequencing (WES) kit (Illumina) was used to capture the 62 million bases of exomic sequences. The captured DNA was sequenced using Illumina HiSeq2000 as paired-end, 100-base reads, achieving sufficient coverage (∼40 million read pairs per sample) to call high-quality genotypes (see below) for at least 85% of targeted bases. Reads were mapped using ELAND. When at least 1 read in a pair mapped to a unique location in the genome, that read and its pair are then subjected to a more accurate gapped alignment to the 100-kb region surrounding the location with cross-match.
TruSeq-targeted design, capture, and sequence
The Illumina DesignStudio was used to design 4935 TruSeq custom enrichment oligos (95mer probes) targeting a total of 1 802 323 bp. Custom capture of targeted regions was performed on 24 indexed libraries constructed from 1 µg genomic DNA using Illumina’s TruSeq DNA Sample Prep Kit version 2. The capture was performed using Illumina’s TruSeq enrichment protocol. Libraries were pooled for sequencing. Sequences were collected and aligned to reference genome as described for whole-exome sequencing (WES).
Analysis of sequence from MIP, WES, and TruSeq capture
The alignments stored in BAM format were used for genotype determinations, including single-nucleotide and deletion/insertion variants, using the most probable genotype algorithm.16 Genotypes were considered high quality if the most probable genotype score was ≥10 and the score divided by the coverage was ≥0.5. VarSifter (http://research.nhgri.nih.gov/software/VarSifter/), a versatile software that can display and allow for sifting through sequence variants by both inclusive and exclusive criteria, was used for evaluation of sequence data.17 We chose to view unique exonic deleterious (nonsynonymous, indel, splice) variants by excluding those present in dbSNP in all the 15 FA genes. If we did not find 2 variants within a sample in an FA gene, the search was then extended to include synonymous changes and variants in the introns while excluding those present in dbSNP. The search was extended for variants in all of the 15 FA and 22 other genes that we performed capture and sequencing on. The program Integrative Genomics Viewer (IGV) (www.broadinstitute.org/igv/) was also used for visual inspection of the genomic variants.18
RNA-Seq
Indexed RNA sequencing (RNA-seq) libraries were constructed from 1 µg total RNA using a TruSeq RNA Sample Prep Kit version 2 (Illumina). The number of amplification cycles was set to 8 to avoid overamplification. Each library was sequenced in paired-end mode using 1 lane of Illumina HiSeq2000 flowcell, generating 2 × 100 bp reads. Raw-read data from the RNA-seq libraries were mapped to the human genome (hg18) using TopHat version 2.0.0. The TopHat output BAM file and its corresponding index file were loaded onto the UCSC Genome Browser (http://genome.ucsc.edu/) or IGV for visual evaluation of sequence alignments.
TopHat BAM files containing aligned reads were converted to BED format using the bamToBed script from the BedTools package,19 using the -split option, in addition to default parameters. The resulting BED file was then converted to WIG format using a custom C script and finally to BigWig format using the wigToBigWig (http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/wigToBigWig) utility from the UCSC Genome Browser toolkit.
Sanger sequencing
Polymerase chain reaction products were treated with USB ExoSAP-IT kit, and sequencing reactions were carried out using ABI Bigdye Terminator v3.1 Cycle Sequencing kit (ABI) and run on ABI3730XL sequencer.
aCGH
A custom CGH 12 × 135 K array was designed using NimbleDesign (NimbleGen) that consisted of 135 000 50mer probes (in triplicates). DNA from patients and reference DNA (human male DNA from Promega) were labeled with different fluorochromes, mixed, and hybridized to the 12 × 135 000 array. We used NimbleGen Service for CGH, and thus the manufacture, hybridization, scanning, and preliminary analysis was performed at their processing facility in Iceland. The data analysis was performed using NimbleScan and the intensity variations were visualized and displayed using SignalMap; both software programs were developed by NimbleGen.
Predicting pathogenicity of amino acid substitutions
Three programs—SIFT (http://sift-dna.org), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), and PANTHER (http://www.pantherdb.org/tips/tips_csnpScores.jsp)—were used for the analysis. PANTHER program calculates the subSPEC (substitution position-specific evolutionary conservation) scores 0 (neutral) to −10 (most likely to be deleterious), and Pdeleterious (probability that a given variant will cause a deleterious effect on protein function). A subSPEC cutoff of −3 corresponds to a 50% or higher probability that a score is deleterious. A score of −3 is equivalent to Pdeleterious 0.5.
Results
Targeted MIP capture, massively parallel sequencing, and aCGH for FA gene mutations
In order to fully interrogate all FA genes and a set of associated DNA repair genes, we targeted the entire length (intronic and exonic) of the FA genes along with 11 additional genes (total of 1 361 577 bp; supplemental Table 1) for sequencing and designed probes to capture 5136 regions of ∼200 bp across the targeted genomic region using the MIP strategy.10,16
We chose an initial set of 19 FA patients with no previously identified mutations, FA1 to FA19, for MIP capture, and we ensured a broad representation of FA genes by including 7 non-FANCA (excluded by sequencing for FANCA); 6 non-FANCA, C, or G (excluded by complementation for FANCA, FANCC, and FANCG groups); and 3 with prior assignment to FANCB, FANCG, and FANCL groups. Another set of 8 FA patients was analyzed using a different capture methodology that is described below. Ancestry and any prior knowledge of exclusion from a FA group for each patient are listed in supplemental Table 2.
Single-end, 36-base reads were generated for a library of the MIP-captured DNA and were aligned to the human reference genome (hg18). The genotype coverage (ie, percent bases covered by high-quality genotype) for the 19 samples was 74% to 89% of the targeted region and was even higher, at 89.68% to 95.63%, for the exonic regions (supplemental Table 3). Read depth was ∼200-fold. Sequence variants were confirmed by polymerase chain reaction amplification and Sanger sequencing of proband DNA as well as DNA from family members, if available (supplemental Figure 1). Assembled sequences revealed lack of coverage with high-quality genotypes for the 59-bp exon 12 in FANCL and the 290-bp exon 10 in FANCG, and samples found initially to have only 1 mutation in these 2 genes required Sanger sequencing of respective exons in search of the missing second mutation. Although RNA analysis was required to reveal the pathogenic nature of certain variants (see below), MIP capture and sequencing identified an FA gene carrying 1 or both inactivating variants in all 19 families except for FA11 and FA18 (Table 1).
Table 1.
Genes and mutations identified in 27 FA families by next-generation sequencing technologies and aCGH
| Mutation 1 (maternal) | Mutation 2 (paternal) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Gene | Exon-intron | Nucleotide (hg18) | cDNA | Protein | Comments | Exon-intron | Nucleotide (hg18) | cDNA | Protein | Comments |
| MIP-targeted capture | |||||||||||
| FA1 | FANCA | 16 | chr16:88376916 | c.1566G>A | p.K522K | Novel, skips exon 16 | 16-17 | del chr16:88374749-88377749† | — | — | — |
| FA2 | FANCB | 9 | chrX:14772684 | c.2027T>C | p.L676P | De novo, novel, subsequently complemented by FANCB | — | — | — | — | — |
| FA3 | FANCB | 9 | chrX:14772652 | c.2059G>T | p.E687* | Novel | — | — | — | — | — |
| FA4 | FANCC | 2 | chr9:97051386-97051387 | c.8_9delAA | p.Q3Rfs*7 | Novel | 2 | chr9:97051386-97051387 | c.8_9delAA | p.Q3Rfs*7 | Novel |
| FA5 | FANCD1 | 11 | chr13:31809756 | c.3264dupT | p.P1088Pfs*15 | — | 19i | chr13:31842697 | c.8487+3A>G | — | — |
| FA6 | FANCF | 1 | chr11:22603448- 22603453 | c.480_485delCT | p.L162Dfs*102 | — | 1 | chr11:22603448-22603453 | c.480_485delCT | p.L162Dfs*102 | — |
| FA7 | FANCG | 10 | chr9:35065742 | c.1153C>A | p.P385T | De novo, novel | 13 | chr9:35064381 | c.1747G>T | p.E583* | Novel, skips exon 13 |
| FA8 | FANCJ | 17 | chr17:57148196 | c.2390A>G | p.K797R | Novel | 17 | chr17:57148194 | c.2392C>T | p.R798* | — |
| FA9 | FANCA | 20i | chr16:88369725 | c.1827-1G>A | — | — | 36 | chr16:88338972-88338977 | c.3520_3522delTGG | p.W1174del | — |
| FA10 | FANCC | 1‡ | del chr9:97116249-97124749† | — | — | Novel, RNA not expressed | 5i | chr9:96974136 | c.456+4A>T | — | — |
| FA11 | FANCB | 2 | dup chrX:14788000-14797000† | — | — | Novel | — | — | — | — | — |
| FA12 | FANCG | 10 | chr9:35065737-35065742 | c.1158dupC | p.S387Lfs*8 | Novel | 8 | chr9:35066497 | c.1008dupA | p.P337Tfs*40 | — |
| FA13 | FANCL | 5i | chr2:58286898† | c.375-2033C>G | — | Novel, skips exons 4, 6, and 7 | 12 | chr2:58242172-58242174 | c.1007_1009delTAT | p.I336-C337delinsS | — |
| FA14 | FANCI | 13 | chr15:87621097 | c.1264G>A | p.G422R | — | 16i | chr15:87626212† | c.1583+142C>T | — | Novel, inserts part of intron 16 |
| FA15 | FANCJ | 7 | chr17:57240777 | c.751C>T | p.R251C | — | 9 | chr17:57231397 | c.1186C>G | p.H396D | — |
| FA16 | FANCF | 1 | chr11:22603448-22603453 | c.480_485delCT | p.L162Dfs*102 | — | 1 | chr11:22603448-22603453 | c.480_485delCT | p.L162Dfs*102 | — |
| FA17 | FANCL | 13 | chr2:58240747† | c.1092G>A | p.K364K | Novel, skips exon13 | 13 | chr2:58240747 | c.1092G>A | p.K364K | Novel, skips exon13 |
| MIP-targeted capture and WES | |||||||||||
| FA18 | FANCD2 | 15i | chr3:10063413 | c.1278+6T>C§ | — | Novel | 18 | del chr3:10066949-10071649† | — | — | — |
| FA19 | FANCD2 | 16 | chr3:10064601† | c.1279G>T | p.V427F | Novel, skips exon16 | 7i | chr3:10053024 | c.491+1G>A§ | — | Novel |
| TruSeq-targeted capture | |||||||||||
| FA20 | FANCA | 39 | chr16:88333953 | c.3884T>G | p.L1295* | — | 37-43|| | del chr16:88172538-88337600† | — | — | — |
| FA21 | FANCD2 | 12i | chr3:10060167 | c.990-1G>A | — | — | 38 | chr3:10108889 | c.3802T>G | p.W1268G | Novel |
| FA22 | FANCA | 14 | chr16:88385367 | c.1304G>T | p.R435L | Novel | 20i | chr16:88369725 | c.1827-1G>A | — | |
| FA23 | FANCA | 33i | chr16:88342567 | c.3348+1G>A | — | — | 36 | chr16:88338972-88338978 | c.3520_3522delTGG | p.W1174del | — |
| FA24 | FANCA | 13 | chr16:88385943-88385946 | c.1115_1118delTTGG | p.V372Afs*42 | — | 15 | chr16:88378855 | c.1378C>T | p.R460* | Novel |
| FA25 | FANCC | 2 | chr9:97051328-97051330 | c.67delG | p.D23Ifs*23 | — | 7 | chr9:96952159 | c.553C>T | p.R185* | — |
| FA26 | FANCL | 5i | chr2:58286898† | c.375-2033C>G | — | Novel, multiple splicing aberrations¶ | 11 | chr2:58243534-58243540 | c.871_874delGATT | p.D291Ffs*49 | Novel |
| FA27 | FANCA | 20i | chr16:88369725 | c.1827-1G>A | — | — | 1-5# | del chr16:88403999-88437249† | — | — | — |
All genomic coordinates refer to Build 36.1 (hg18). The mutation nomenclature is in accordance with the Human Genome Variation Society (http://www.hgvs.org/mutnomen/standards.html#aalist). Paternal DNA is unavailable for FA16, FA23, and FA25, and maternal DNA is unavailable for FA9. Both parental DNAs are unavailable for FA18, FA20, FA21, FA26, and FA27, and for these families, the assignment of the maternal and paternal mutations is arbitrary. FA2, FA3, and FA11 are male individuals belonging to the X-linked FANCB group and will have only 1 mutation. Pathogenicity prediction for p.R435L by SIFT (not tolerated), PolyPhen2 (probably damaging), and PANTHER (subPSEC, −5.18; Pdeleterious, 0.89). Pathogenicity prediction for p.K797R by SIFT (not tolerated), PolyPhen2 (probably damaging), and PANTHER (subPSEC, −3.85; Pdeleterious, 0.70). Pathogenicity prediction for p.W1268G by SIFT (not tolerated), PolyPhen2 (probably damaging), and PANTHER (subPSEC, −4.36; Pdeleterious, 0.80). Pathogenicity prediction for p.L676P by SIFT (not tolerated), PolyPhen2 (probably damaging), and PANTHER (not available). Pathogenicity prediction for p.P385T by SIFT (tolerated), PolyPhen2 (probably damaging), and PANTHER (subPSEC, −3.08087; Pdeleterious, 0.52).
cDNA, complementary DNA.
Indicates STOP condons in Protein columns.
Finding these pathogenic variants required aCGH and/or RNA-seq.
Plus 5 kb upstream.
From WES data.
Plus 160 kb downstream.
ins c.375-2034_2066, c.375-2300_2360, and skipping of exons 4, 6, and 7.
Plus 25 kb upstream.
In order to identify large deletions and duplications, we employed an aCGH strategy with probes designed for the entire length of all FA genes and related genes and up to 200 kb on either side of each gene (supplemental Table 4). The aCGH revealed a deletion in FANCC (FA10), FANCD2 (FA18), and FANCA (FA1) and a duplication in FANCB (FA11) (Figure 1). RNA analysis revealed deleterious consequences associated with a genomic deletion of a noncoding FANCC exon in FA10; a synonymous FANCA variant (c.1566G>A, p.K522K) in FA1, a homozygous synonymous FANCL variant (c.1092G>A, p.K364K) in FA17, and variants deep in introns in FANCL in FA13 (c.375-2033C>G) and FANCI in FA14 (c.1583+142C>T) (see below). The mutations and their pathological consequences are listed in Table 1. The mutations, as expected based on family selection, represented multiple FA groups: FANCA (2), FANCB (3), FANCC (2), FANCD1 (1), FANCD2 (2), FANCF (2), FANCG (2), FANCI (1), FANCJ (2), and FANCL (2). Using MIP capture and sequencing and aCGH and RNA analysis methods, an FA gene with 1 mutation was identified for all of the 19 families tested, and 2 mutations were found for 17 of the 19 families.
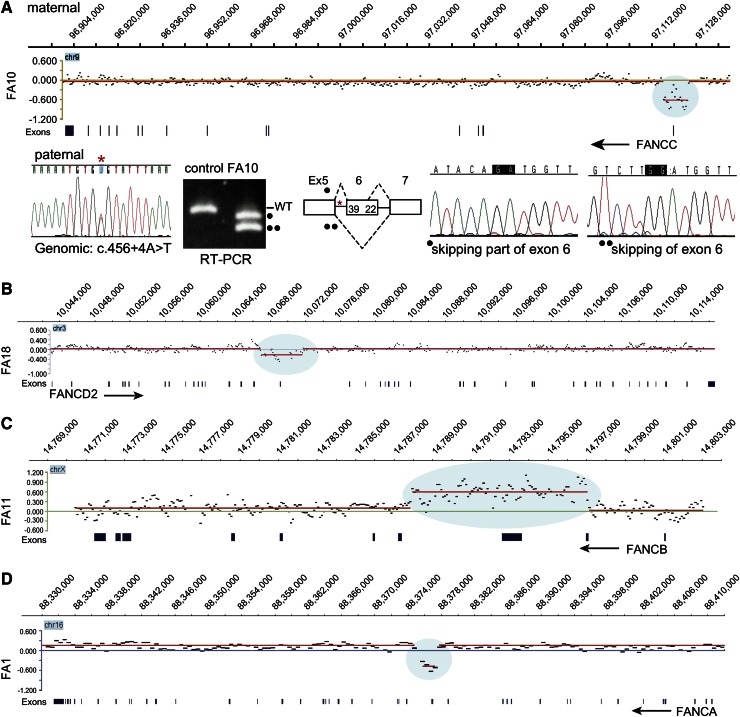
Figure 1.
aCGH identifies deletions in FANCA, FANCC, and FANCD2 and duplication in FANCB. (A) Deletion in FANCC. The CGH data for the FANCC gene region in FA10 DNA are displayed in the top panel, genomic coordinates are above, and exons are below. The display was generated using SignalMap (Nimblegen). The y-axis shows the intensity ratio (log value) between the test sample and the reference DNA. A “0” represents 2 copies and thus no change in copy number, but the region of decreased ratio (blue shading) indicates a deletion. The red line connects the individual data points (black dots displaying a 500-bp moving average) with a similar intensity ratio. The arrow indicating FANCC points in the direction of transcription of the gene, right to left. This deletion removes exon 1 and 5 kb upstream. The CGH data were generated from the maternal DNA. The Sanger sequencing trace shows the paternal mutation (*) in the genomic DNA. RT-PCR using FA10 RNA for the region of paternal mutation is shown along with that of a control RNA. The normal-size product present in the control lane is absent in the FA10 RNA lane. The 2 lower-size bands in FA10 lane (indicated by • and ••) represent the alternatively spliced products caused by the paternal mutation in intron 5 skipping either the entire or a portion (39 bases) of exon 6. The maternal allele that carries the deletion is not expressed. (B) Deletion in FANCD2. The CGH data for the FANCD2 gene region in FA18 DNA are displayed, along with genomic coordinates above, and the exons below. The FANCD2 gene transcription is from left to right (arrow). The 4.7-kb deletion (blue shading) highlighted by the reduced intensity ratio indicates the loss of exon 18. (C) Duplication in FANCB. The CGH data displayed for the FANCB region in FA11 are shown. The reference DNA is from a male, and thus the intensity ratio for FANCB on the X chromosome is still “0”. The arrow (right to left) points to the direction of transcription of FANCB. The blue shading indicates an increased ratio and represents a duplication that includes exons 2 and 3. (D) Deletion in FANCA. The CGH data for the FANCA region in FA1 are displayed. The intensity ratios, shaded blue, indicate that the deletion in FA1 removes exons 16 to 17. The arrow (right to left) points in the direction of transcription of FANCA.
A wide spectrum of FA gene mutations includes large deletions and duplications in FANCA, FANCB, FANCC, and FANCD2
aCGH identified deletions in FANCA, FANCC, and FANCD2, and a duplication in FANCB (Figure 1). The FANCA deletion in FA1 removed exons 16 to 17 (Figure 1D). The duplication in FANCB included exons 2 and 3 (FA11), and the deletion in FANCD2 included exon 18 (FA18) (Figure 1 B-C). Interestingly, the 8.5-kb maternal deletion in a FANCC patient (FA10) did not remove any coding region but eliminated noncoding exon 1 together with a 5-kb upstream region. Reverse-transcription polymerase chain reaction (RT-PCR) analysis revealed that the only transcripts present were those that were alternatively spliced as a result of the paternal mutation c.456+4A>T. The allele with the 8.5-kb maternal deletion is not expressed at all, as the deletion appears to have eliminated the required promoter element for FANCC transcription (Figure 1A).
WES for FANCD2 mutations
Subsequent to the MIP capture and sequencing and aCGH, we had 2 families with only 1 mutation each in the FANCD2 gene: FA19 had a missense mutation (p.V427F) and FA18 had a 4.7-kb deletion that eliminated exon 18. Therefore, in both cases the second pathogenic mutation was unknown. The presence of 2 pseudogenes, homologous to parts of the FANCD2 gene, prevented the adequate design of unique probes for capturing and aligning the sequences to the reference genome. MIP capture is a polymerase-based strategy that requires designing unique probes capable of recognizing and annealing to ∼20 bp at each end of an ∼200-bp target region. Although MIP probes could be designed for all but 2 FANCD2 exons, it was apparent that after the capture, sequence, and alignment steps, sequence coverage was not adequate for some regions in FANCD2 in FA19 and FA18 (Figure 2A). In addition, the single-end shorter sequence lengths (36-base reads) would have affected unambiguous alignment to the authentic FANCD2 gene. We therefore turned to an alternative strategy.
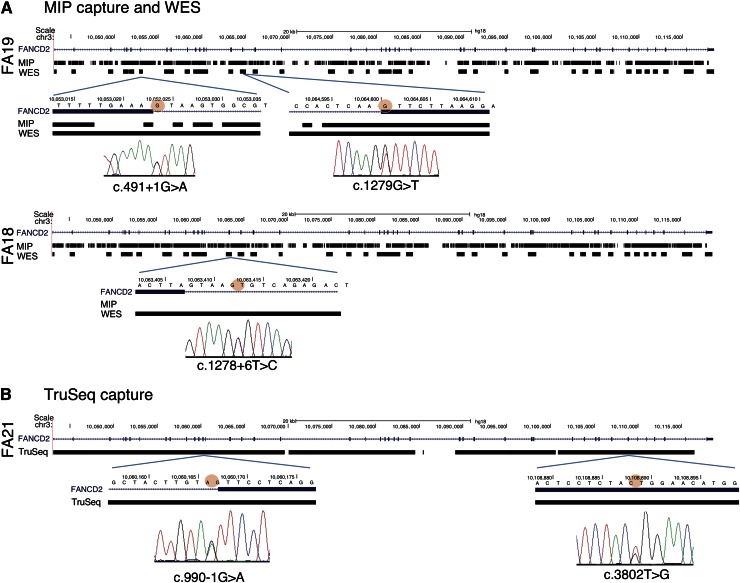
Figure 2.
Evaluation of MIP, WES, and TruSeq capturing-sequencing technologies for FANCD2 mutations. (A) FANCD2 mutations identified by MIP and WES capture methods. The genotype coverage generated by the MIP and the WES methods is aligned with the FANCD2 gene track from the UCSC browser (hg18) for patients FA19 (upper panel) and FA18 (lower panel). The regions of high quality genotype coverage are indicated by solid rectangles; gaps indicate no coverage. The regions harboring the mutations are expanded below, and the circle with red shading points to the base with a mutation in the respective patient DNA. Sanger sequencing traces showing the mutations are shown below. MIP capture sequence includes the c.1279G>T location, but does not include the other mutation (c.491+G>A) for the FA19 DNA or that for the mutation in FA18. Coverage generated using the WES method, however, is nearly complete, albeit only exonic (plus immediate flanking) regions. (B) FANCD2 mutations in FA21 by TruSeq capture sequencing. The UCSC browser track for the FANCD2 gene is aligned with the genotype coverage by the TruSeq method for FA21. Sequences are recovered for nearly the entire gene. The regions harboring the mutations are expanded below (mutant base marked with red shading), along with the Sanger sequencing traces that indicate the mutations.
We took advantage of advances in capture and sequencing technologies and used a WES approach on FA19 and FA18, which allowed hybridization-based capturing with long oligos (95mer) and paired-end sequencing with longer read lengths (100 bases). WES genotype coverage and sequence depth were 89.2% and 91.3% and 55- and 85-fold for FA19 and FA18, respectively. WES generated comprehensive data on all FANCD2 exons as well as exons from all other FA genes (supplemental Figure 2), which allowed us to find the missing second mutations. FA18 harbored c.1278+6T>C in intron 15 and FA19 had c.491+1 G>A in intron 7. Both mutations affected splice donor signals and thus accounted for the missing second FANCD2 mutations (Figure 2A; Table 1).
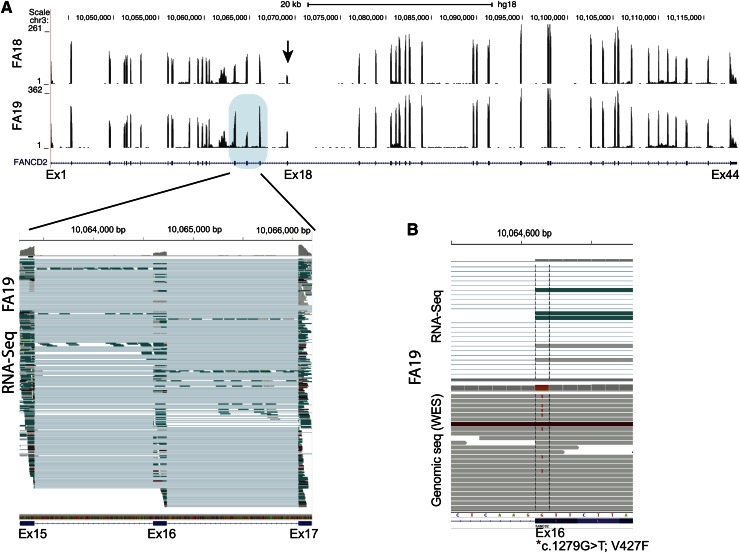
We also performed sequencing using RNA isolated from FA18 and FA19 LCL cell lines. We noted that FANCD2 sequence reads for exon 18 in FA18 were decreased, reflecting a genomic deletion that encompassed the exon (Figure 3A). Surprisingly, a similar reduction in RNA-seq reads was observed for exon 16 in FA19, and it appeared to be the consequence of p.V427F, the missense mutation caused by the G>T variant in the first nucleotide of the exon. About half of the RNA sequence reads extending from exon 15 to exon 17 lacked sequences from, and thus skipping of, exon 16 (Figure 3A lower left panel). Evaluation of both RNA-seq and DNA-sequencing data together (Figure 3B top and bottom panels, respectively) demonstrate that while the T (mutant) allele is present in DNA along with the G (wild-type) allele, it is absent in RNA-seq reads, suggesting that skipping of the p.V427F-bearing allele in the FANCD2 transcript occurred during RNA splicing.
Figure 3.
FANCD2 expression analysis from RNA-seq data for FA18 and FA19 LCL cell lines. (A) Wiggle plot displaying RNA-seq read coverage along with the UCSC FANCD2 gene track (shown below). Data from FA18 and FA19 are shown on top and bottom, respectively. The number of sequence reads (range) is indicated on the y-axis and is reflected by the height of the peak for each exon. The decreased number of sequence reads for exon18 (arrow) is apparent for FA18. This reflects the 4.7-kb genomic deletion that removes this exon. Multiple individual RNA sequence reads from FA19 spanning exons 15 to 17 (blue shade) are displayed in the lower panel (generated using the IGV program). Each horizontal line is an independent sequence. The thicker rectangle at each exon shows the mapped RNA sequences, while the thin line connects the gaps (introns) and connects the sequences from a single read. It is apparent that several sequence reads that include both exon 15 and 17 do not include the sequence for exon 16 (thin line), which is evidence of exon skipping. At the top of each exon, the gray color reflects the number of sequences at single-base resolution. Reduction in the reads for exon 16 compared with the 2 flanking exons is readily apparent. (B) Display of a cross section of RNA-seq (top) and genomic (bottom) sequence read alignments for FA19 in the region spanning the first nucleotide of exon 16 (*) that carries a missense mutation (c.1279G>T; p.V427F). Some of the genomic sequence reads show the heterozygous mutant T allele while RNA-Seq shows no reads with the mutant allele, indicating that the allele carrying the exon 16 mutation is skipped during messenger RNA splicing.
A comprehensive screening strategy for mutations in all FA genes: TruSeq-targeted capture of both introns and exons, followed by sequencing
The capturing and sequencing strategy employed for WES was successful in uncovering the missing mutations in FANCD2 and would allow for identification of mutations present in FA exons (supplemental Figure 2). However, coverage is limited to exons, and critical mutations exist outside the exon boundary. Therefore, we employed a custom liquid hybridization strategy similar to WES (TruSeq) to capture the entire length of all 15 FA genes and 22 related genes (Table 2).
Table 2.
Targeted gene regions and design coverage for TruSeq capture
| Target gene | Alternate name(s) | Target region (hg18) | Design coverage (%) |
|---|---|---|---|
| CDKN2A | chr9:21957751-21984490 | 99 | |
| DKC1 | Dyskerin | chrX:153644344-153659154 | 97 |
| FAAP100 | C17orf70 | chr17:77117387-77129871 | 99 |
| FAAP24 | C19orf40 | chr19:38154988-38159800 | 99 |
| FANCA | chr16:88331460-88410566 | 99 | |
| FANCB | chrX:14771450-14801105 | 99 | |
| FANCC | chr9:96901157-97119812 | 97 | |
| FANCD1 | BRCA2 | chr13:31787617-31871809 | 97 |
| FANCD2 | chr3:10043113-10116344 | 92 | |
| FANCE | chr6:35528116-35542859 | 96 | |
| FANCF | chr11:22600655-22603963 | 100 | |
| FANCG | chr9:35063835-35070013 | 100 | |
| FANCI | chr15:87588198-87661366 | 96 | |
| FANCJ | BRIP1 | chr17:57114767-57295537 | 98 |
| FANCL | chr2:58239882-58322019 | 90 | |
| FANCM | chr14:44674886-44739843 | 99 | |
| FANCN | PALB2 | chr16:23521984-23560179 | 98 |
| FANCO | RAD51C | chr17:54124962-54166691 | 99 |
| FANCP | SLX4 (BTBD12) | chr16:3571184-3601586 | 94 |
| GAR1 | chr4:110956115-110965342 | 97 | |
| MRE11 | chr11:93790115-93866688 | 93 | |
| NBN | chr8:91014740-91066075 | 99 | |
| NHP2 | chr5:177509072-177513567 | 100 | |
| NOP10 | chr15:32421209-32422654 | 100 | |
| p53 | TP53 | chr17:7512445-7531588 | 98 |
| POT1 | chr7:124249676-124357273 | 94 | |
| RAD50 | chr5:131920529-132007494 | 97 | |
| RAD51AP1 | chr12:4518317-4539475 | 99 | |
| RAP1 | chr1:111963928-112057624 | 91 | |
| Rb | chr13:47775884-47954027 | 96 | |
| TCAB1 | WRAP53 | chr17:7532520-7547544 | 98 |
| TERC | chr3:170964979-170965655 | 100 | |
| TERF1 | chr8:74083651-74122541 | 96 | |
| TERF2 | chr16:67947034-67977374 | 99 | |
| TERT | chr5:1306287-1348162 | 94 | |
| TINF2 | TIN2 | chr14:23778691-23781720 | 100 |
| TPP1 | chr11:6590573-6597268 | 100 |
We tested 8 additional families, FA20 to FA27, with FA diagnoses but with no a priori knowledge of the complementation groups or mutations, with the single exception that FA21 and FA26 were known to be non-FANCA. The sequences provided excellent coverage (98.7%-98.9%) and depth (194-fold to 750-fold) over the targeted regions, including all FA genes (supplemental Figure 3). We identified the complementation group and both mutations in all 8 families. They belonged to FANCD2 (1), FANCL (1), FANCC (1), and FANCA (5) groups (Table 1). FANCD2 coverage was nearly complete, as illustrated by FA21, which was found to harbor 2 mutations (Figure 2B). With the exception of the 2 deleterious mutations in FA21, no other FANCD2 mutations were found in the 8 families, indicating a lack of confounding effect from pseudogene sequences. aCGH and RNA analysis were needed to fully characterize the families. Two FANCA patients carried large deletions: 1 removed exons 37 to 43 and additional 160 kb downstream of the gene (FA20), and the other removed exons 1 to 5 plus 25 kb upstream (FA27) (Table 1). We observed splicing defects due to an intronic mutation in a FANCL patient (FA26) (see below). With these data, we were now able to identify biallelic mutations in all of the 27 families tested (Table 1). The targeted sequencing (TruSeq) ensures very high coverage of all FA genes including FANCD2 and, together with aCGH, is a method of choice for comprehensive molecular diagnosis for families with no a priori knowledge other than a clinical diagnosis of FA.
RNA analysis unveils pathogenicity of unsuspected variants in FANCL, FANCI, and FANCC
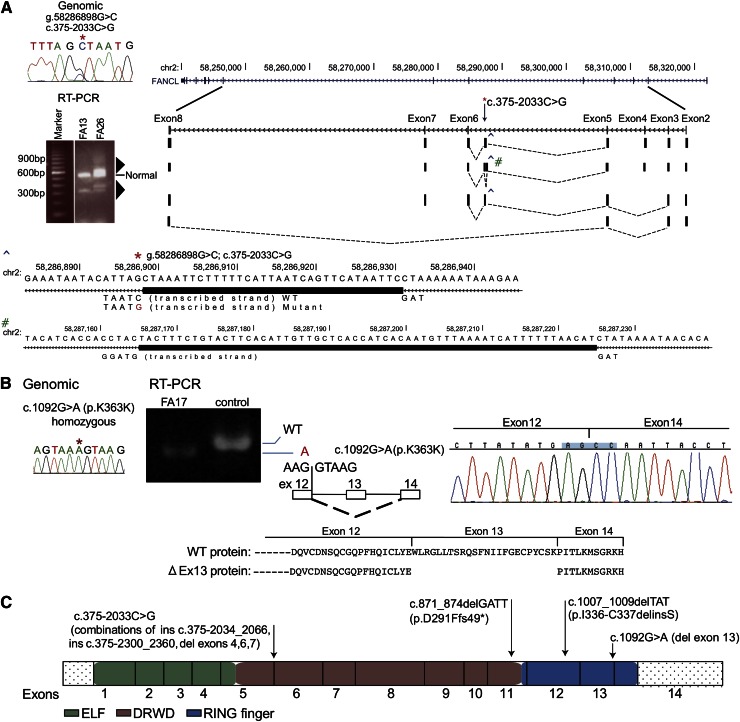
Among the 27 families in which we identified both the FA complementation group and underlying mutations, there were 3 FANCL families. This is a rare complementation group.20,21 A total of 5 of the 6 mutations in the 3 FANCL families (Figure 4) were novel, and intriguingly, 2 would not easily be recognized as pathogenic. The homozygous mutation in the last nucleotide of exon 13 in FA17, inherited in each case from a heterozygous carrier parent, did not alter the encoded amino acid (p.K364K), but RNA analysis revealed skipping of exon 13 in the messenger RNA, resulting in deletion of 72 nucleotides encoding 24 amino acids from a RING finger domain (Figure 4B). Two other FANCL families each carried a distinct mutation, c.1007_1009delTAT (FA13) and c.871_874delGATT (FA26), but their second mutation was initially obscure. However, both were eventually found to carry a variant within intron 5 (c.375-2033C>G), 2 kb away from the closest exon, which was exon 6. RT-PCR analysis of this region from FA13 and FA26 RNA using primers derived from the flanking exons 2 and 8 generated a product of expected size as well as multiple additional aberrant products that were both larger and smaller than expected (Figure 4A triangles). Cloning and sequencing of these RT-PCR products displayed 4 unique and alternatively spliced products. No other sequence variant was apparent in the vicinity of these 4 splicing events, and thus each was presumably caused by the same intronic variant, c.375-2033 C>G (Figure 4A, supplemental Figure 4A-D). The mutations in the 3 FANCL families are shown in Figure 4C.
Figure 4.
Biallelic FANCL gene mutations from 3 families and their effect on RNA splicing. (A) RT-PCR analysis of the FANCL region harboring the mutation (c.375-2033C>G) that is shared by FA26 and FA13 (*). The germline mutation is displayed using Sanger sequencing. RT-PCR from FA26 and FA13 using primers located in exons 2 and 8 shows additional multiple products (triangles) that are longer and shorter than the correctly sized product. RT-PCR products from FA26 RNA were cloned and individual colonies representing different size products were sequenced. The sequences and their representation of the alternate splice patterns between exons 2 and 8 are aligned with the UCSC browser for the FANCL gene. Four unique and alternatively spliced products were identified. A larger product represents a 33-bp insertion (ins c.375-2033_2066), and this was generated using the splice donor signal created by the variant (CTAAT>GTAAT), and a TAG acceptor, 34 bases away (^). This region is expanded below with the thick rectangle, showing the bases inserted by the mutation. A second alternative transcript includes the 33-bp insertion (^) and an additional 61-bp insertion (#) from intron 5 (ins c.375-2300_2360), resulting from cryptic splice signals (GTAAG and TAG) on either side of this insertion. The minus strand is transcribed for FANCL. The third variant includes the 33-nt insertion but exon 4 is skipped, and in the fourth variant exons 4, 6, and 7 are skipping. Supplemental Figure 4 provides additional detail. (B) Homozygous, synonymous FANCL mutation in FA17 results in exon skipping. The Sanger sequence trace displays a genomic mutation, c.1092G>A (p.K364K) (*). RT-PCR analysis for the mutation shows only a smaller-than-expected product, and no product of reduced size is observed in the control lane. Sequence traces for the RT-PCR product are displayed along with a diagram showing the skipping of exon 13. The wild-type protein, along with the predicted mutant protein resulting from in-frame removal of 24 amino acids, is shown. (C) Mutations in the FANCL gene. The mutations identified in this study are on top of the FANCL coding region, displayed as exons. The ELF, DRWD, and RING finger domains23 are color-coded.
We found a maternally inherited FANCI missense mutation for FA14. One of the two paternally inherited unique variants in the intronic regions, c.1583+142C>T in intron 16, was found to alter RNA splicing by using the splice donor created by the mutation and creating an aberrant transcript with the insertion of 140 nt from the adjacent intron (ins c.1583+1_140). (supplemental Figure 5). The intronic variants in both FANCL and FANCI could not have been discovered without a sequencing strategy that included introns, and their pathogenic consequences could not have been recognized without inclusion of RNA analysis.
Discussion
We employed MIP, WES, and TruSeq capture methods to pilot use of new technologies in the molecular diagnosis of FA. Though MIP works well for capturing the genomic regions of interest, the improvements in probe design and capture from WES and TruSeq methodologies and the sequencing improvements from the HiSeq platform improved the ability to determine genotypes in problematic regions. Thus, we were able to identify mutations in each individual across many FA genes including FANCD2, which presents challenges due to the presence of pseudogenes.
Targeted capture using TruSeq is our current method of choice and, unlike WES, it allows for sequencing the intronic regions as well. This is important, as demonstrated by the intronic mutations described for FANCL and FANCI genes in this study. We can further supplement understanding of molecular diagnosis with the addition of aCGH and RNA-seq technologies. The deletion of the noncoding exon in FANCC initially appeared inconsequential, but it includes 5-kb region upstream, eliminating the expression of the allele. This clearly illustrates the importance of the design and use of aCGH.
RNA sequence analysis was also critical in understanding the pathogenicity of several variants, particularly those in the intronic regions, as it can reveal aberrantly spliced products. Two unsuspecting variants affected splicing: one in FANCI intron 16 (FA14), 142 nt away from exon 16, and the other in FANCL intron 5 (FA13 and FA26), 2 kb from exon 6. Once applied to a larger scope of patients, the DNA-sequencing strategies optimized here will reveal a large number of likely pathogenic variants, and RNA-seq will provide a way to quickly evaluate accompanying changes in transcripts production.
The reduced number of RNA-sequence reads for exon 18 in FA18 revealed the consequences of a genomic deletion of exon 18. A similar reduction in sequence reads for exon 16 in FA19 confirmed the pathogenicity of p.V427F. It is interesting that both these observations from RNA-seq data for FANCD2 result in in-frame deletions: the former leads to c.1546_1656del111, p.G516_Q552del, and the latter to c.1279_1413del, p.V427_Q471del. In fact, these are not unlike a reported 459-bp deletion in a FANCD2 patient that eliminates a 132-bp exon 17, and is predicted to express protein with an in-frame loss of 44 amino acids, p.E472_K515del.8 In addition, of the 6 mutations in FANCD2 families we describe here, 1 missense, 3 splice, and 1 genomic deletion all appear to be affecting splicing of RNA. These observations are consistent with the earlier report regarding FANCD2 in suggesting that at least 1 of the mutations is typically milder, and a majority of the mutations affect splicing.8
Our sequence analysis of 27 FA families contributed 18 novel mutations to the repertoire of known variants (see Table 1). The prediction programs—SIFT, PolyPhen2, and PANTHER—find the novel missense variants are likely to be pathogenic (Table 1), but confirmation of their pathogenicity would require functional assays. FANCA mutations account for the disease in ∼65% of FA patients. However, this group of patients was not selected to represent an accurate statistical distribution of complementation groups in the FA patient population. By including a subset of non-FANCA patients, we demonstrate that the methods presented here are comprehensive and can find mutations in any FA gene. Of particular interest is that we add 3 new FANCL families to the small number reported thus far.20,21
Two of the patients in this study who were of Ashkenazi Jewish (AJ) ancestry had novel mutations in FANCC. While FA10 carried the common AJ founder mutation (c.456+4A>T; intron 5), the second mutation was a novel deletion in the noncoding region of the gene, which resulted in nonexpression of RNA. The second AJ subject, FA4, was homozygous for a novel mutation, c.8_9delAA, in FANCC.
We observed several instances of additional variants in a second FA gene, including a FANCD1 (c.951A>G; p.N317S) variant in a FANCC patient (FA4) and a FANCJ variant (c.3737C>T; p.P1246L) in a FANCL patient (FA26). These variants are not in the Fanconi Anemia Mutation Database (www.rockefeller.edu/fanconi/mutate), dbSNP, or the 1000Genome database, and thus are unique but not necessarily disease associated. Availability of the parental DNA for the former suggests that it is paternally inherited and not a de novo change.
Since our sequencing target included causative genes for other chromosomal instability syndromes, such as Bloom syndrome, it is not surprising that we observed a BLM variant in an FA patient. In the FANCI patient (FA14), we observed a maternally inherited BLM variant (c.1237 G>A; p.E413L). Based on an observation that FANCJ and BLM interact, crosstalk between the BLM and FA pathways has been proposed.22 However, recognition of any contribution from a BLM variant on the phenotype of a FANCI patient can only emerge when a substantial number of such instances are carefully evaluated.
Our efforts here are illustrative of how the application of evolving new technologies can help mutation detection in genetically heterogeneous diseases become more economical, affordable, and efficient. The necessity of finding both mutations in an FA patient, and the mutation status of siblings and validation in relatives, is an important part of the diagnostic profile of each FA patient, and its importance cannot be overestimated. We demonstrate here, for the first time, that it is possible to identify both complementation group and both mutations for a given FA patient in a quick and economical way.
Supplementary Material
Acknowledgments
The authors thank Pedro Cruz for help designing MIP primers and Julia Fekecs for help with figures. The authors are most grateful to the individuals and families who participated in this study.
This work was supported in part by a grant from the National Institutes of Health National Center for Research Resources (grant UL1RR024143) (A.D.A. and A.S.), by the Anderson Cancer Center at the Rockefeller University Burroughs Wellcome Fund Career Award for Medical Scientists (A.S.), and by a grant from the Fanconi Anemia Research Fund (S.C.C.). A.S. is a Rita Allen Foundation, Irma T. Hirschl, and Alexandrine and Alexander Sinsheimer Foundation scholar and is a recipient of a Doris Duke Clinical Scientist Development Award. E.A.O., S.C.C., J.K.T., D.C.K., A.K., F.X.D., E.F., and S.K.S. gratefully acknowledge the Intramural Program of the National Human Genome Research Institute, Bethesda, MD.
Footnotes
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.C.C. designed research, analyzed and interpreted data, and wrote the manuscript; F.L.P., E.S., and A.S. contributed vital reagents; J.K.T., D.C.K., A.K., F.X.D., E.F., S.K.S., and S.T. performed research and analyzed and interpreted results; A.D.A. contributed reagents and interpreted data; and E.A.O. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.K.T. is Department of Biomedical Informatics, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL.
Correspondence: Arleen D. Auerbach, Human Genetics and Hematology Program, The Rockefeller University, 1230 York Ave, New York, NY 10065; e-mail: auerbac@mail.rockefeller.edu; and Settara C. Chandrasekharappa, Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, 50 South Dr, Building 50, Room 5232, Bethesda, MD 20892; e-mail: chandra@mail.nih.gov.
References
- 1.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668(1-2):4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soulier J. Fanconi anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:492-497. [DOI] [PubMed]
- 3.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26(13):1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkeland AC, Auerbach AD, Sanborn E, et al. Postoperative clinical radiosensitivity in patients with fanconi anemia and head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2011;137(9):930–934. doi: 10.1001/archoto.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neveling K, Endt D, Hoehn H, Schindler D. Genotype-phenotype correlations in Fanconi anemia. Mutat Res. 2009;668(1-2):73–91. doi: 10.1016/j.mrfmmm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Chandra S, Levran O, Jurickova I, et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther. 2005;12(5):976–984. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Pinto FO, Leblanc T, Chamousset D, et al. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 2009;94(4):487–495. doi: 10.3324/haematol.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalb R, Neveling K, Hoehn H, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet. 2007;80(5):895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ameziane N, Errami A, Léveillé F, et al. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat. 2008;29(1):159–166. doi: 10.1002/humu.20625. [DOI] [PubMed] [Google Scholar]
- 10.Porreca GJ, Zhang K, Li JB, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4(11):931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 11.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4(11):907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 12.Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 13.Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 14.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bainbridge MN, Wang M, Burgess DL, et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11(6):R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teer JK, Bonnycastle LL, Chines PS, et al. NISC Comparative Sequencing Program. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20(10):1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28(4):599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35(2):165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 21.Ali AM, Kirby M, Jansen M, et al. Identification and characterization of mutations in FANCL gene: a second case of Fanconi anemia belonging to FA-L complementation group. Hum Mutat. 2009;30(7):E761–E770. doi: 10.1002/humu.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhasini AN, Brosh RM., Jr Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012;28(1):7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17(3):294–298. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.