Abstract
Human angiogenin (ANG) is an angiogenic molecule and a ribonucleolytic enzyme with significant amino acid sequence identity to pancreatic RNase A, plays a critical role in the establishment and growth of tumours. An association between ANG and cancer has been observed in more than 25 clinical studies to date. In addition, ANG has now been shown to be implicated in Amyotrophic Lateral Sclerosis (ALS) and Parkinson's Disease (PD). Structural and biochemical studies so far have showed several distinguishing features of ANG molecule compared to RNase A and provided details of the putative cell binding site, active site, nuclear translocation sequence and the roles of residues in binding and cleaving RNA. A key finding elucidated from the structural study on ANG is the presence of a ‘blocked’ C-terminus (part of the active site apparatus) compared with RNase A. Here we report the crystal structure of ANG with an ‘engineered-loop’ from eosinophil derived neurotoxin (a homologue of ANG) which has resulted with local perturbations (conformational flexibility) at the cell binding site and at the C-terminus of the molecule. This experimental observation will now provide a new avenue to design compounds (potent inhibitors) through a structure guided drug design route.
Keywords: Angiogenin, Eosinophil derived neurotoxin, Ribonuclease A, Crystal structure, Protein engineering, Conformational flexibility
Abbreviations: RNase A, - ribonuclease A; ANG, - Human angiogenin; EDN -, eosinophil derived neurotoxin; AEH -, angiogenin–eosinophil derived neurotoxin hybrid; RI -, ribonuclease inhibitor
Graphical abstract
Highlights
▸ A hybrid form (AEH) of human angiogenin (ANG) was created. ▸ The RI binding loop of ANG was substituted with the RI binding loop of EDN. ▸ Significant conformational changes were observed in the structure of AEH. ▸ Changes include the C-terminal segment and a putative cell binding domain of ANG. ▸ Binding of a chloride ion at the active site was observed
1. Introduction
Angiogenin (ANG), also known as ribonuclease 5 (RNase 5), is an angiogenic molecule that induces neovascularization [1]. It is a monomeric protein of 14.1 kDa that belongs to the pancreatic ribonuclease A (RNase A; EC 3.1.27.5) superfamily [2]. ANG has similar catalytic properties to RNase A – it cleaves preferentially on the 3′ side of pyrimidine and follows a transphosphorylation/hydrolysis mechanism [3,4]. The enzymatic activity of ANG, albeit several orders of magnitude lower than that of RNase A towards conventional RNase substrates [4], is essential to the angiogenic process [3,5].
The known biological properties of ANG encompass induction of angiogenesis to stimulating cell proliferation and promoting cell survival. Under growth conditions, ANG is translocated to nucleus (where it accumulates in nucleolus) and stimulates ribosomal RNA (rRNA) transcription, thus facilitating cell growth and proliferation [6]. Recently it has been established that under stress conditions, ANG accumulates in cytoplasmic compartments and modulates the production of tiRNA, a novel class of small RNA that is derived from tRNA and is induced by stress. ANG-mediated tiRNA reprogram protein translation, save anabolic energy, and promote cell survival [7–10].
Based on biochemical and structural data it has been established that ANG has three distinct functional sites – (a) a putative binding site for an unknown receptor, (b) a nuclear localization sequence (NLS), and (c) a catalytic site. The crystal structure of ANG has revealed the RNase A fold and the conservation of the catalytic triad – His13, Lys40 and His114 [11]. The loop region from Lys60 to Asn68 forms the receptor-binding site that interacts with a cell surface receptor [12]. Upon binding to the cell surface receptor, ANG is internalized and translocated to the nucleus [13]. The nuclear translocation process is mediated by an NLS located between amino acid residues Met30 and Gly34 [14]. The ribonucleolytic activity of ANG [5] is believed to function in stimulating rRNA transcription after ANG is localized in the nucleus. It is now firmly established that all three sites are essential for ANG to have angiogenic and growth stimulating activities.
Apart from the its vascular function, ANG has also been firmly associated with several neurodegenerative conditions. Studies by Greenway et al. [15] on a large cohort of Amyotrophic Lateral Sclerosis (ALS) patients (both familial and sporadic) showed for the first time that ANG is implicated in ALS. Since then several other studies worldwide have also identified ANG mutations in ALS patients. More recently a large multi-site study reported the association of mutations in ANG with ALS as well as Parkinson's disease (PD) [16,17]. Since the early reports on ANG association with ALS, it has been established that ANG also possesses neurotrophic and neuroprotective functions [18–20]. A recent detailed study has provided for the first time the structural and molecular insights into the mechanism of action of human ANG-ALS variants in neurons [21].
The role of ANG as a tumour angiogenic factor has been well documented. ANG has been shown to be upregulated in a variety of human cancers [22]. Elevated serum level and/or enhanced tissue expression of ANG have been noticed in all types of solid and blood cancers so far examined. A major mechanism by which ANG induces angiogenesis is related to its activity in stimulating ribosomal RNA (rRNA) transcription [22], a common downstream event of tumour angiogenesis [23]. In addition, ANG inhibitors have been shown to inhibit not only ANG-induced angiogenesis but also those induced by other angiogenic factors such as VEGF, FGF, and EGF [24]. ANG inhibitors would therefore have a profound effect in inhibiting tumour angiogenesis. Indeed, ANG inhibitors such as its siRNA [25], antisense [26], monoclonal antibodies [27], soluble binding proteins [28], enzymatic inhibitors [29], and nuclear translocation blockers [24,30–32] have all been shown to inhibit tumour angiogenesis and cancer progression in various animal models.
We have identified different types of inhibitors of ANG based on our structural, mutagenesis and biophysical studies. These will be used to probe the mechanistic details of ANG. In particular, our aim is to study the nucleotide inhibitors that target the enzymatic active site of ANG and also monoclonal antibodies against ANG. The angiogenic activity of ANG is critically dependent on its enzymatic activity and inhibitors that bind to its enzymatic active site also inhibit its angiogenic activity. Our results from ANG inhibitors have demonstrated proof of principle, but design of structure-based small molecule nucleotide inhibitors so far proved to be elusive due to the blockage of the active site predominantly by Gln117 and the remaining C-terminal segment of the ANG molecule [11,33–37]. Mutation of Gln117 to Gly/Ala has shown to increase the ribonucleolytic activity of ANG [38]. However, the structures of ANG in complex with – (a) a leucine rich, natural protein inhibitor (50 kDa) of ANG (the ribonuclease inhibitor – RI; [39]; PDB ID: 1A4Y) and (b) an antibody fragment 26-2F (shown to inhibit ANG) ([40]; PDB id: 1H0D) have shown significant conformational changes at the cell-binding region of ANG. This region is close to the active site of ANG and thus it was predicated that induction of similar conformational change would allow the C-terminal β-strand (Fig. 1A) to undergo small movement to accommodate the substrate/inhibitor without significant alteration in the catalytic property of the molecule.
Fig. 1.
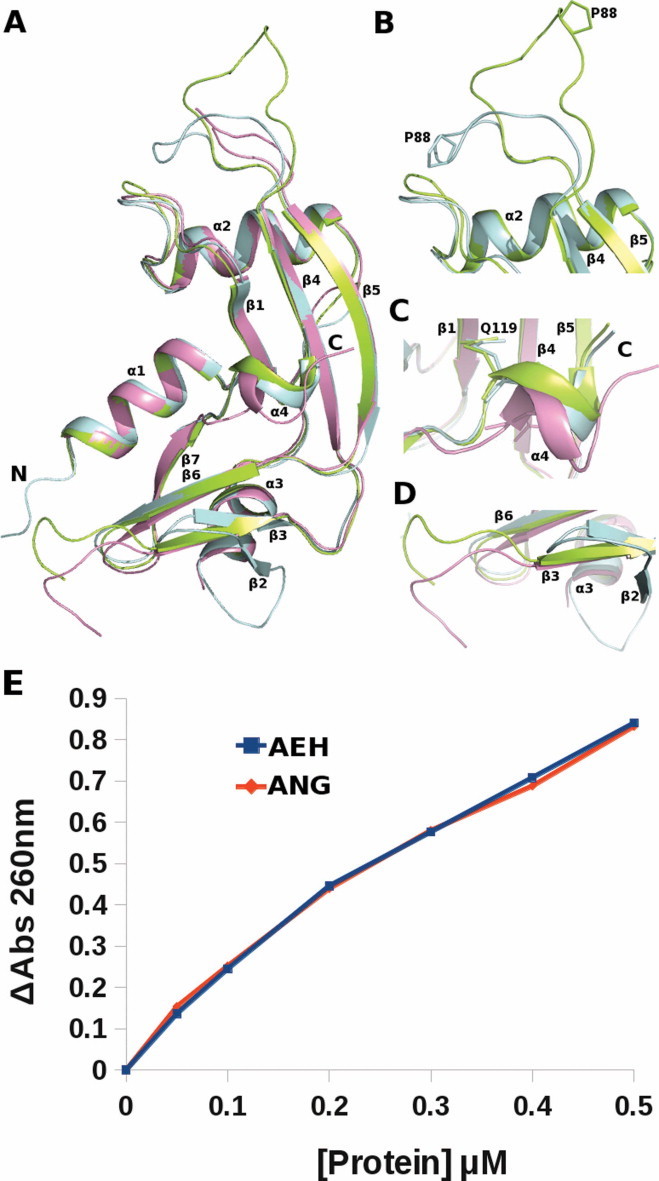
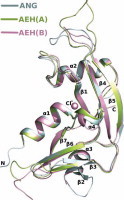
General features and flexible regions of the AEH molecule (A–D). (A) Superposition of AEH [limon (molecule A) and pink (molecule B)] on ANG (pale cyan). Structural and conformational changes as observed in AEH (limon) vs. ANG (pale cyan); (B) Loop1: ribonuclease inhibitor (RI) binding region of ANG. Molecule B not shown as the loop region is disordered; (C) Conformational changes in C-terminal region showing the displacement of Q119 (Q117 of ANG) away from the active site; (D) Loop2: putative cell binding site of ANG; and (E) Ribonucleolytic activity of AEH vs ANG. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In this study we have substituted 84HGGSPWPP91 (a loop that interacts with RI) of ANG [39] with 86TTPSPQNISN95 of eosinophil-derived neurotoxin (EDN, also shown to interact with RI) [41] and constructed an ANG–EDN hybrid (AEH) protein molecule of molecular weight ∼14.5 kDa. EDN is also a member of RNase A superfamily (known as RNase 2), possesses weak ribonucleolytic activity, antiviral property [42–45] and neurotoxicity [46,47]. Based on our previous crystal structure analysis of ANG–RI [39] and EDN–RI [41] complexes it was realistic to assume that AEH molecule would also retain RI binding property. The experimental data presented in this report shows that the design of AEH molecule proved to be useful to study the relationship between the RI binding loop and the structural changes in the cell-binding region. In addition, the high resolution crystal structure of the AEH molecule reported here shows that the ‘engineered loop’ causes conformational changes that would perturb the network of interactions from the nuclear localization sequence to cell binding region extending to the active site (through inclusion of the C-terminal segment of the molecule) without altering the enzymatic activity. This information may prove useful for the design of ‘ANG–nucleotide inhibitor complexes’. Similar studies have been reported previously amongst RNase A family members and also other proteins [48–52].
2. Materials and methods
2.1. Cloning, expression and purification of AEH
The ANG–EDN hybrid protein (AEH) was prepared using recombinant DNA technology where the loop region (84HGGSPWPP91) of ANG was substituted with the corresponding segment (loop region) (86TTPSPQNISN95) of EDN. The pET22b(+) Escherichia coli expression plasmid was provided by Dr. Robert Shapiro, Harvard Medical School (USA). The protein expression trials were carried out in BL21-DE3 Codon plus (RIPL) cells in the culture medium terrific broth. The required AEH protein was purified from inclusion bodies using the protocol described by Holloway et al. [53]. Purity of the protein was analysed on an SDS–PAGE.
2.2. Mass spectrometry
Purified protein mass of AEH was experimentally confirmed (in addition to amino acid sequence verification) using electrospray ionization mass spectrometry (ESMS) service at University of Bath. Protein solutions were prepared for mass spectrometry by dilution of a desalted standard stock solution of protein in water (1 mg/mL, 70 μM) in a 50:50 ratio with HPLC-grade acetonitrile containing 0.1% formic acid. ESMS was performed in positive ion mode on a QStar XL System (Applied Biosystems), fitted with a NanoMate (Advion Biosciences) automated nanospray source. A delivery pressure of 0.3 psi of nitrogen gas and a spray voltage of 1.6 kV were applied to the sample to generate the nanospray plume. Transformed mass spectra were calculated by use of the Bayesian protein reconstruct algorithm provided as part of the MDS Sciex Analyst 1.1 software package.
2.3. RNase activity assay
The activity of AEH protein toward yeast tRNA was determined by measuring the formation of acid-soluble fragments as described by Shapiro et al. [4]. Assay mixtures contained 2 mg/mL yeast tRNA (Sigma), 0.1 mg/mL bovine serum albumin (BSA), and 0.05, 0.1, 0.2, 0.3, 0.4, or 0.5 μM test protein in 33 mM Na-Hepes and 33 mM NaCl, pH 7.0. After 2 h of incubation at 37 °C, reactions were terminated by the addition of 2.3 volumes of ice-cold 3.4% perchloric acid. The mixture was then centrifuged at 13,000 rpm for 10 min at 4 °C. The absorbance of the supernatant was measured at 260 nm.
2.4. X-ray crystallography
X-ray diffraction data were recorded (at 100 K) from a single crystal of AEH protein grown in 1.4–2.5 M NaCl at 16 °C on a Mar CCD 165 at station PX 10.1, Synchrotron Radiation Source, Daresbury, UK. No cryoprotectant was used. The protein was crystallized using hanging-drop vapour diffusion method at 1:1 drop ratio of protein (10 mg/ml in water) to mother liquor. The data were processed and scaled in primitive orthorhombic P212121 space group (2 molecules/asymmetric unit) using HKL2000 software suite [54] to a 1.76 Å resolution. The data were complete to 91.9% overall with a merging R-value of 0.082 (Table 1). Initial phases for AEH molecule were obtained by molecular replacement method using the human ANG structure (PDB ID: 1ANG; [11]) as the starting model. The software used for molecular replacement trials were MOLREP [55] and PHASER [56]. The resultant model of AEH protein was taken through a rigid body refinement (after deletion of the substituted loop region (84HGGSPWPP91)). The Rcryst and Rfree values after the first round of rigid body refinement were 0.491 and 0.505 respectively. Further model building and restrained refinement were carried out using COOT [57] and PHENIX [58] respectively. During this process all the side chain atoms were placed in the electron density map, the substituted loop region was modelled and fitted into the electron density using Fourier difference density map. Finally the water molecules and chloride ions (from the crystallization medium) were modelled into the structure. The structure was refined to a final Rcryst and Rfree value of 0.239 and 0.285 respectively. There are a total of 83 water molecules and 4 chloride ions in the final structure. A few regions of the AEH structure exhibit a high degree of conformational flexibility (see below). The structure was validated using MolProbity validation tool [59,60]. There were no residues in the disallowed region of the Ramachandran plot. Crystallographic data statistics are summarized in Table 1. All figures were drawn with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA) and rendered with POV-ray.
Table 1.
X-ray data collection and refinement statistics.
| Space group | Orthorhombic – P212121 |
| Cell dimensions | a = 38.4 Å; b = 54.7 Å; c = 94.7 Å |
| Resolution range (Å) | 50–1.76 |
| Rsymma (outer shell) | 0.082 (0.299) |
| I/σI (outer shell) | 14.5 (2.9) |
| Completeness (outer shell) % | 91.9 (65.8) |
| Total number of reflections | 230,027 |
| Number of unique reflections | 20,481 |
| Redundancy (outer shell) | 4.8 (2.5) |
| Wilson B-factor (Å2) | 29.0 |
| Rcrystb/Rfreec | 0.239/0.285 |
| Average B-factor (Å2) | |
| Overall | 34.7 |
| Protein (chain A, B) | 33.6, 35.5 |
| Chloride ion | 36.9 |
| Solvent | 37.1 |
| RMS deviation | |
| Bond length (Å) | 0.007 |
| Bond angle (°) | 1.012 |
| Ramachandran plot statistics | |
| Favoured (%) | 97.71 |
| Additional favoured (%) | 2.29 |
| PDB ID | 4B36 |
Rsymm = Σh Σi |I(h) − Ii(h)|/Σh ΣiIi(h), where Ii(h) and I(h) are the ith and the mean measurements of the intensity of reflection h, respectively.
Rcryst = Σh |Fo − Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h, respectively.
Rfree is equal to Rcryst for a randomly selected 5.0% subset of reflections not used in the refinement.
3. Results and discussion
3.1. General features
All ANG structures determined so far [11,33–37] have a highly conserved RNase A fold – a characteristic feature of proteins belonging to this family (Fig. 1A). Briefly, the RNase A fold (a kidney-shaped molecule) consist of α and β secondary structure elements forming the core of the molecule connected by loop structures. In ANG molecule there are three α-helices, seven β-strands and a 310 helix (at the C-terminus) (Fig. 1A). The core of the molecule is stabilized by three disulphide bridges. The catalytic site [formed by residues His13 (α1), Lys40 (loop connecting α2 and β1) and His114 (β7)] is located on the concave structure of the molecule. The NLS residues lie on helix α2, RI binding residues in the loop connecting β4 and β5, and receptor binding site includes the loop connecting helix α3 and strand β2 and β2. The AEH structure retains these features in both molecules of the asymmetric unit but displays significant conformational flexibility in the loop regions of the molecule (Fig. 1B–D, see below).
Comparison of native ANG structure [11] with the AEH structure (present study) result with a root mean square deviation of 1.34 and 1.71 Å (Cα atoms) for molecule A and B (for the two copies in the hybrid molecule in the asymmetric unit) respectively. The active site of AEH molecule comprises the catalytic triad residues His13, Lys40 and His116 (His13, Lys40 and His114 in ANG) which form links with residues Gln12, Asn43, 117Leu-Asp-Gln119 (115Leu-Asp-Gln117 in ANG) either through direct hydrogen bonding or through a water-mediated network. A detailed analyses of the catalytic site of AEH against a few members of the RNase A superfamily determined at atomic resolution – the prototype molecule RNase A (PDB: 2E3W, 1.05 Å resolution [61]), EDN (PDB: 1GQV, 0.98 Å resolution [62]), and ANG (PDB: 4AOH, 1.04 Å resolution [21]) showed the presence of a highly conserved solvent network involving four water molecules (labelled as 1–4 in Fig. 2A–E). It is likely that these solvent molecules might play an important role in substrate recognition and/or binding. For example, in an ANG-ALS variant Gln12Leu structure [33], the enzyme lost both its ribonucleolytic and angiogenic properties (required for the action of ANG), where the mutation from Gln to Leu perturbed the network of interactions (including the conserved solvent molecules) involving the catalytic residue His13.
Fig. 2.
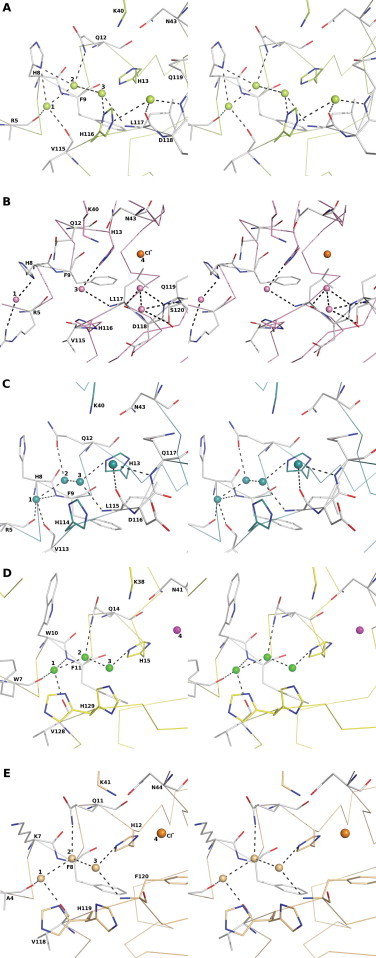
Stereo representation of conserved solvent network in (A) AEH – molecule A; (B) AEH – molecule B; (C) ANG; (D) EDN (solvent molecule equivalent to 4 or chloride ions are coloured in magenta) and (E) RNase A. Conserved water molecules are labelled as 1–4. Chloride ions are coloured in orange. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Conformational flexibility
3.2.1 RI-binding loop
A large movement in the engineered loop region (86TTPSPQNISN95) was observed where residue Pro88 undergoes a major shift of ∼90° in comparison to its position in the native ANG structure (Figs. 1B, 3). In the ANG structure this region (also called as the RI-binding loop) is involved in binding to RI as observed in the crystal structure of ANG–RI complex [39] through several main- and side-chain interactions. In both the molecules of AEH, the RI-binding loop adopts similar conformation, but significantly different from that observed in the ANG–RI complex or in the native ANG structure (Figs. 1B, 3). Hence we predict that this change would perturb RI binding even though AEH molecule seems to possess similar ribonucleolytic activity as native ANG (Fig. 1E). The Ki value of RI for ANG and EDN are 7.1 × 10−16 M [63,64] and 6.7 × 10−15 M [41] respectively. The structural superposition of native EDN (PDB: 1GQV [62]), EDN-RI (PDB: 2BEX [41]), native ANG (PDB: 4AOH [21]), ANG–RI (PDB: 1A4Y [39]) against AEH molecules show that the conformational changes observed are unique to AEH molecules. However, the nuclear translocation sequence 31RRRGL35 in the AEH molecule does not undergo any conformational change and retains similar conformation as in native ANG.
Fig. 3.
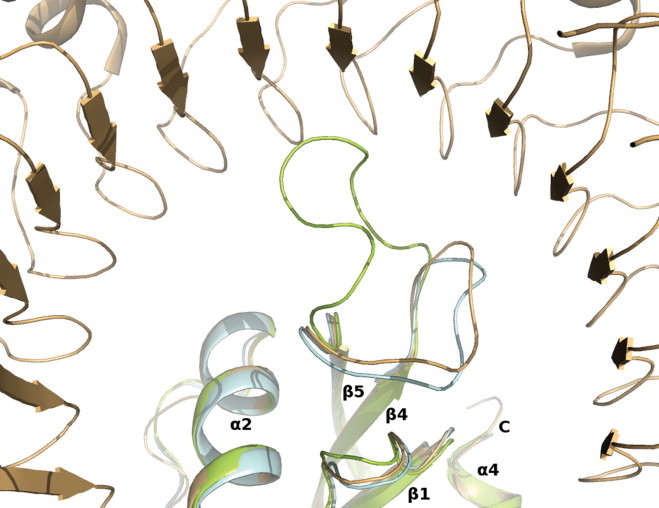
Cartoon representation of AEH molecule docked to ribonuclease inhibitor (RI – PDB id: 1A4Y). In pale cyan is native ANG, light brown is ANG bound to RI (PDB ID: 1A4Y) and in limon is AEH. There are no large conformational changes observed between native ANG and ANG bound to RI. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.2 Cell receptor binding loop
The second structural change was observed at the secondary structural element β2 strand of ANG, which transformed to a loop region (Gly62–His65) in the AEH structure (Fig. 1A, D), and the loop connecting the strand β2 and α3 could not be modelled due to lack of visible electron density. This region is a part of the putative cell binding segment of ANG [12,65,66]. This segment has shown to undergo significant conformational change by adopting an extended conformation based on the structural details observed from the crystal structures of ANG–RI [39] and ANG–Fab antibody [40] complex reported previously from our laboratory.
3.2.3 C-terminal region
An unexpected conformational flexibility was observed at the C-terminal segment of AEH structure starting from Leu117 (Leu115 based on ANG numbering). This structural movement at the active site displaces the Gln119 of AEH (Gln117 of ANG) by ∼90° inwards (away from the active site) (Figs. 1C, 4). In the most predominantly observed state (as observed in the native structure of ANG [11]), the side chain atoms OE1 and NE2 of Gln117/Gln119 of ANG/AEH respectively are involved in direct hydrogen bonding interactions with active site residue His13 and Thr44, thus blocking substrate binding and the availability of His13 and Thr44 for catalysis. As observed in one of the molecule of AEH structure, the displacement of Gln119 (Gln117 of ANG) opens the active site and enhances the binding of the substrate. The side chain atoms OE1 and NE2 of Gln119 (Gln117 of ANG) of AEH interact with Arg103, Arg104 and Val106 through hydrogen bonding interactions. A chloride ion (due to high concentration of NaCl used in crystallization medium) was bound in the place of Gln119 following the displacement of Gln119 (Fig. 4). We tested the AEH protein for its catalytic activity in the presence of 0.1–2.5 M NaCl concentration and did not observe any alteration in the RNase activity and hence conclude that the observed chloride ion at this site has no direct role in catalysis. Comparison of electrostatic potential between ANG and the AEH (in both molecules) (Fig. 5) clearly shows the apparent change in potential charge at the active site due to the conformational change at the C-terminal region – resulting in larger basic surface area for the nucleotide substrate to bind. Thus for the first time the AEH structure has provided a glimpse of conformational flexibilities at the functional regions of the parent molecule.
Fig. 4.
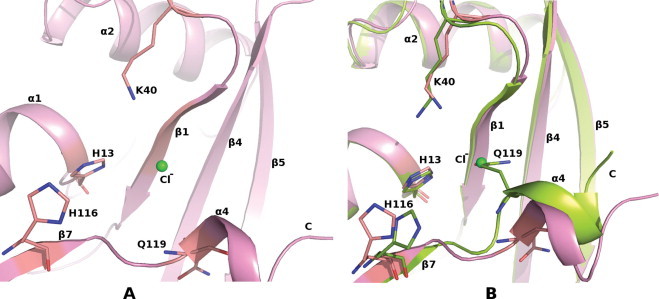
The C-terminal pocket of AEH molecule. (A) Chloride ion bound at the active site of AEH and (B) Superposition of both the molecules of AEH in the asymmetric unit. One of the molecule of AEH (limon) shows the position of Q119 overlapping with the chloride ion from the second molecule of AEH (pink). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
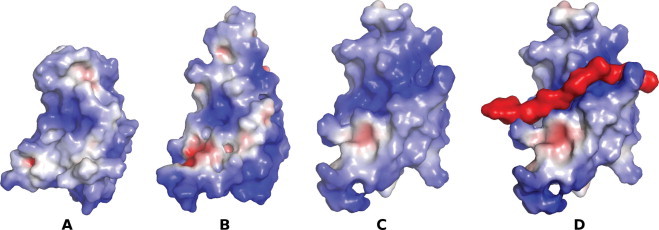
Surface potential diagram – (A) for ANG and (B, C) for AEH molecule. In (B) the surface charge near the C-terminal region (active site) is similar to ANG, but in (C) the C-terminal region undergoes a conformational change and forms a long groove where the substrate possibly binds. Differences are due to the changes observed in the conformations of the loop regions 1 and 2 for the AEH molecule. (D) An oligomer of DNA backbone (coloured in red) (10-mer, with only phosphate and ribose moieties) docked on to the AEH molecule B. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusions
We have compared the solution structure of ANG (determined by nuclear magnetic resonance) (PDB: 1AWZ [67]) with the high resolution structure of ANG in the crystalline state (determined by X-ray crystallography, PDB: 4AOH [21]) and found that the loop regions retain similar conformation in both structures. Thus, substitution of ANG loop region (84HGGSPWPP91) by EDN amino acid residues (86TTPSPQNISN95) in the AEH structure seem to have resulted in conformational changes at the functional regions of the molecule (that are observed for the first time in the ANG structure determined so far). The hybrid molecule AEH retains the full complement of ribonucleolytic activity and do not exhibit any perturbation at the catalytic centre (involving the catalytic triad). However, significant movement of the cell-binding segment of AEH molecule was evident. In addition, the C-terminus of AEH molecule (in one copy of the asymmetric unit) adopts a non-obstructive conformation (hitherto unseen) in the hybrid molecule. These observations will have implications for the future design of ANG nucleotide based inhibitors.
The structural flexibility observed in the AEH molecule gives us some insight about the network of interactions involving the NLS and the cell-binding sites through the active site and C-terminal segments. These changes appear to be a sequential event during catalysis. The perturbation of the C-terminus moves the Gln119 residue away from its obstructive state and Arg121 points outwards – leading to an enhanced surface accessibility and gain in net positive charge. In order to assess the feasibility of substrate binding, we have docked an oligomer of DNA backbone (containing only the ribose and the phosphate moiety) to this cavity (Fig. 5D). The length of the DNA backbone used was a 10-mer. From the modelling studies we predict that Arg5, His8, Gln12, Lys40, Asp41, Lys82, His116 (equivalent to H114 of ANG) and Arg124 make hydrogen bonding interactions with the phosphate and ribose molecules.
Database
The atomic coordinates and structure factors for AEH (with code 4B36) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Acknowledgements
This work was supported by the Wellcome Trust (UK) through a programme grant (number 083191) and an equipment grant (number 088464) to K.R.A. We thank Dr. Robert Shapiro for providing the plasmid carrying the engineered segment of human angiogenin; Dr. Matthew Baker, Jodie Gibson and Claire Padgett for their contribution to structural and protein biochemistry part of the project.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Fett J.W., Strydom D.J., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma-cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 2.Strydom D.J., Fett J.W., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F. Amino acid sequence of human tumor derived angiogenin. Biochemistry. 1985;24:5486–5494. doi: 10.1021/bi00341a031. [DOI] [PubMed] [Google Scholar]
- 3.Harper J.W., Vallee B.L. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bind generated by regional mutagenesis. Biochemistry. 1989;28:1875–1884. doi: 10.1021/bi00430a067. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro R., Riordan J.F., Vallee B.L. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry. 1986;25:3527–3532. doi: 10.1021/bi00360a008. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro R., Vallee B.L. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin alanine derivatives inhibit angiogenin induced angiogenesis. Biochemistry. 1989;28:7401–7408. doi: 10.1021/bi00444a038. [DOI] [PubMed] [Google Scholar]
- 6.Li S., Hu G.F. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell Physiol. 2012;227:2822–2826. doi: 10.1002/jcp.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 11.Acharya K.R., Shapiro R., Allen S.C., Riordan J.F., Vallee B.L. Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc. Natl. Acad. Sci. USA. 1994;91:2915–2919. doi: 10.1073/pnas.91.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallahan T.W., Shapiro R., Vallee B.L. Dual site model for the organogenic activity of angiogenin. Proc. Natl. Acad. Sci. USA. 1991;88:2222–2226. doi: 10.1073/pnas.88.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroianu J., Riordan J.F. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc. Natl. Acad. Sci. USA. 1994;91:1677–1681. doi: 10.1073/pnas.91.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moroianu J., Riordan J.F. Identification of the nucleolar targeting signal of human angiogenin. Biochem. Biophys. Res. Commun. 1994;203:1765–1772. doi: 10.1006/bbrc.1994.2391. [DOI] [PubMed] [Google Scholar]
- 15.Greenway M.J., Andersen P.M., Russ C., Ennis S., Cashman S., Donaghy C. Loss-of-function ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 16.Rayaprolu S., Soto-Ortolaza A., Rademakers R., Uitti R.J., Wszolek Z.K., Ross O.A. Angiogenin variation and Parkinson disease. Ann. Neurol. 2012;71:725–727. doi: 10.1002/ana.23586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Es M.A., Schelhaas H.J., van Vught P.W., Ticozzi N., Andersen P.M., Groen E.J. Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann. Neurol. 2011;70:964–973. doi: 10.1002/ana.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian V., Crabtree B., Acharya K.R. Human angiogenin is a neuroprotective factor and amyotrophic lateral sclerosis associated angiogenin variants affect neurite extension/pathfinding and survival of motor neurons. Hum. Mol. Genet. 2008;17:130–149. doi: 10.1093/hmg/ddm290. [DOI] [PubMed] [Google Scholar]
- 19.Kieran D., Sebastia J., Greenway M.J., King M.A., Connaughton D., Concannon C.G. Control of motoneuron survival by angiogenin. J. Neurosci. 2008;28:14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian V., Feng Y. A new role for angiogenin in neurite growth and pathfinding-implications for amyotrophic lateral sclerosis. Hum. Mol. Genet. 2007;16:1445–1453. doi: 10.1093/hmg/ddm095. [DOI] [PubMed] [Google Scholar]
- 21.Thiyagarajan N., Ferguson R., Subramanian V., Acharya K.R. Structural and molecular insights into the mechanism of action of human angiogenin-ALS variants in neurons. Nat. Commun. 2012;3:1121. doi: 10.1038/ncomms2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., Yu W., Kishikawa H., Hu G.F. Angiogenin prevents serum withdrawal-induced apoptosis of P19 embryonal carcinoma cells. FEBS J. 2010;277:3575–3587. doi: 10.1111/j.1742-4658.2010.07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishimoto K., Liu S., Tsuji T., Olson K.A., Hu G.F. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- 24.Hirukawa S., Olson K.A., Tsuji T., Hu G.F. Neamine inhibits xenografic human tumor growth and angiogenesis in athymic mice. Clin. Cancer Res. 2005;11:8745–8752. doi: 10.1158/1078-0432.CCR-05-1495. [DOI] [PubMed] [Google Scholar]
- 25.Ibaragi S., Yoshioka N., Kishikawa H., Hu J.K., Sadow P.M., Li M. Angiogenin-stimulated rRNA transcription is essential for initiation and survival of AKT-induced prostate intraepithelial neoplasia. Mol. Cancer Res. 2009;7:415–424. doi: 10.1158/1541-7786.MCR-08-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson K.A., Byers H.R., Key M.E., Fett J.W. Prevention of human prostate tumor metastasis in athymic mice by antisense targeting of human angiogenin. Clin. Cancer Res. 2001;7:3598–3605. [PubMed] [Google Scholar]
- 27.Olson K.A., Byers H.R., Key M.E., Fett J.W. Inhibition of prostate carcinoma establishment and metastatic growth in mice by an antiangiogenin monoclonal antibody. Int. J. Cancer. 2002;98:923–929. doi: 10.1002/ijc.10282. [DOI] [PubMed] [Google Scholar]
- 28.Olson K.A., Fett J.W., French T.C., Key M.E., Vallee B.L. Angiogenin antagonists prevent tumor growth in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:442–446. doi: 10.1073/pnas.92.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao R.Y., Jenkins J.L., Olson K.A., Key M.E., Fett J.W., Shapiro R. A small-molecule inhibitor of the ribonucleolytic activity of human angiogenin that possesses antitumor activity. Proc. Natl. Acad. Sci. USA. 2002;99:10066–10071. doi: 10.1073/pnas.152342999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibaragi S., Yoshioka N., Li S., Hu M.G., Hirukawa S., Sadow P.M. Neamine inhibits prostate cancer growth by suppressing angiogenin-mediated rRNA transcription. Clin. Cancer Res. 2009;15:1981–1988. doi: 10.1158/1078-0432.CCR-08-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshioka N., Wang L., Kishimoto K., Tsuji T., Hu G.F. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc. Natl. Acad. Sci. USA. 2006;103:14519–14524. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper J.W., Vallee B.L. Mutagenesis of aspartic acid-116 enhances the ribonucleolytic activity and angiogenic potency of angiogenin. Proc. Natl. Acad. Sci. USA. 1988;85:7139–7143. doi: 10.1073/pnas.85.19.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabtree B., Thiyagarajan N., Prior S.H., Wilson P., Iyer S., Ferns T. Characterization of human angiogenin variants implicated in amyotrophic lateral sclerosis. Biochemistry. 2007;46:11810–11818. doi: 10.1021/bi701333h. [DOI] [PubMed] [Google Scholar]
- 34.Holloway D.E., Chavali G.B., Hares M.C., Subramanian V., Acharya K.R. Structure of murine angiogenin: features of the substrate- and cell-binding regions and prospects for inhibitor-binding studies. Acta Crystallogr. D. 2005;61:1568–1578. doi: 10.1107/S0907444905029616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonidas D.D., Shapiro R., Subbarao G.V., Russo A., Acharya K.R. Crystallographic studies on the role of the C-terminal segment of human angiogenin in defining enzymatic potency. Biochemistry. 2002;41:2552–2562. doi: 10.1021/bi015768q. [DOI] [PubMed] [Google Scholar]
- 36.Leonidas D.D., Shapiro R., Allen S.C., Subbarao G.V., Veluraja K., Acharya K.R. Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth. J. Mol. Biol. 1999;285:1209–1233. doi: 10.1006/jmbi.1998.2378. [DOI] [PubMed] [Google Scholar]
- 37.Acharya K.R., Shapiro R., Riordan J.F., Vallee B.L. Crystal structure of bovine angiogenin at 1.5 Å resolution. Proc. Natl. Acad. Sci. USA. 1995;92:2949–2953. doi: 10.1073/pnas.92.7.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo N., Shapiro R., Acharya K.R., Riordan J.F., Vallee B.L. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc. Natl. Acad. Sci. USA. 1994;91:2920–2924. doi: 10.1073/pnas.91.8.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papageorgiou A.C., Shapiro R., Acharya K.R. Molecular recognition of human angiogenin by placental ribonuclease inhibitor – an X-ray crystallographic study at 2.0 Å resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavali G.B., Papageorgiou A.C., Olson K.A., Fett J.W., Hu G., Shapiro R. The crystal structure of human angiogenin in complex with an antitumor neutralizing antibody. Structure. 2003;11:875–885. doi: 10.1016/s0969-2126(03)00131-x. [DOI] [PubMed] [Google Scholar]
- 41.Iyer S., Holloway D.E., Kumar K., Shapiro R., Acharya K.R. Molecular recognition of human eosinophil-derived neurotoxin (RNase 2) by placental ribonuclease inhibitor. J. Mol. Biol. 2005;347:637–655. doi: 10.1016/j.jmb.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 42.Domachowske J.B., Dyer K.D., Bonville C.A., Rosenberg H.F. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 1998;177:1458–1464. doi: 10.1086/515322. [DOI] [PubMed] [Google Scholar]
- 43.Domachowske J.B., Rosenberg H.F. Eosinophils inhibit retroviral transduction of human target cells by a ribonuclease-dependent mechanism. J. Leukoc. Biol. 1997;62:363–368. doi: 10.1002/jlb.62.3.363. [DOI] [PubMed] [Google Scholar]
- 44.Gleich G.J., Jacoby D.B., Fryer A.D. Eosinophil-associated inflammation in bronchial asthma: a connection to the nervous system. Int. Arch. Allergy Immunol. 1995;107:205–207. doi: 10.1159/000236978. [DOI] [PubMed] [Google Scholar]
- 45.Hamann K.J., Barker R.L., Loegering D.A., Gleich G.J. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987;73:523–529. [PubMed] [Google Scholar]
- 46.Sorrentino S., Glitz D.G., Hamann K.J., Loegering D.A., Checkel J.L., Gleich G.J. Eosinophil-derived neurotoxin and human liver ribonuclease. Identity of structure and linkage of neurotoxicity to nuclease activity. J. Biol. Chem. 1992;267:14859–14865. [PubMed] [Google Scholar]
- 47.Durack D.T., Ackerman S.J., Loegering D.A., Gleich G.J. Purification of human eosinophil-derived neurotoxin. Proc. Natl. Acad. Sci. USA. 1981;78:5165–5169. doi: 10.1073/pnas.78.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raines R.T., Toscano M.P., Nierengarten D.M., Ha J.H., Auerbach R. Replacing a surface loop endows ribonuclease A with angiogenic activity. J. Biol. Chem. 1995;270:17180–17184. doi: 10.1074/jbc.270.29.17180. [DOI] [PubMed] [Google Scholar]
- 49.Hedstrom L., Szilagyi L., Rutter W.J. Converting trypsin to chymotrypsin: the role of surface loops. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 50.Allemann R.K., Presnell S.R., Benner S.A. A hybrid of bovine pancreatic ribonuclease and human angiogenin: an external loop as a module controlling substrate specificity? Protein Eng. 1991;4:831–835. doi: 10.1093/protein/4.7.831. [DOI] [PubMed] [Google Scholar]
- 51.Hynes T.R., Kautz R.A., Goodman M.A., Gill J.F., Fox R.O. Transfer of a beta-turn structure to a new protein context. Nature. 1989;339:73–76. doi: 10.1038/339073a0. [DOI] [PubMed] [Google Scholar]
- 52.Harper J.W., Vallee B.L. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry. 1989;28:1875–1884. doi: 10.1021/bi00430a067. [DOI] [PubMed] [Google Scholar]
- 53.Holloway D.E., Hares M.C., Shapiro R., Subramanian V., Acharya K.R. High-level expression of three members of the murine angiogenin family in Escherichia coli and purification of the recombinant proteins. Protein Expression Purif. 2001;22:307–317. doi: 10.1006/prep.2001.1434. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 56.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen V.B., Arendall 3rd W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boerema D.J., Tereshko V.A., Kent S.B. Total synthesis by modern chemical ligation methods and high resolution (1.1 Å) X-ray structure of ribonuclease A. Biopolymers. 2008;90:278–286. doi: 10.1002/bip.20800. [DOI] [PubMed] [Google Scholar]
- 62.Swaminathan G.J., Holloway D.E., Veluraja K., Acharya K.R. Atomic resolution (0.98 Å) structure of eosinophil-derived neurotoxin. Biochemistry. 2002;41:3341–3352. doi: 10.1021/bi015911f. [DOI] [PubMed] [Google Scholar]
- 63.Lee F.S., Vallee B.L. Binding of placental ribonuclease inhibitor to the active site of angiogenin. Biochemistry. 1989;28:3556–3561. doi: 10.1021/bi00434a061. [DOI] [PubMed] [Google Scholar]
- 64.Lee F.S., Shapiro R., Vallee B.L. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 65.Shapiro R., Vallee B.L. Identification of functional arginines in human angiogenin by site-directed mutagenesis. Biochemistry. 1992;31:12477–12485. doi: 10.1021/bi00164a026. [DOI] [PubMed] [Google Scholar]
- 66.Hallahan T.W., Shapiro R., Strydom D.J., Vallee B.L. Importance of asparagine-61 and asparagine-109 to the angiogenic activity of human angiogenin. Biochemistry. 1992;31:8022–8029. doi: 10.1021/bi00149a036. [DOI] [PubMed] [Google Scholar]
- 67.Lequin O., Thuring H., Robin M., Lallemand J.Y. Three-dimensional solution structure of human angiogenin determined by 1H,15N-NMR spectroscopy–characterization of histidine protonation states and pKa values. Eur. J. Biochem. 1997;250:712–726. doi: 10.1111/j.1432-1033.1997.00712.x. [DOI] [PubMed] [Google Scholar]


