Abstract
Gloeobacter violaceus PCC 7421 is considered, by molecular phylogenetic analyses, to be an early-branching cyanobacterium within the cyanobacterial clade. G. violaceus is the only known oxygenic photosynthetic organism that lacks thylakoid membranes. There is only one report on the development of a transformation system for G. violaceus [H. Guo, X. Xu, Prog. Nat. Sci. 14 (2004) 31–35] and further studies using the system have not been reported. In the present study, we succeeded in introducing an expression vector (pKUT1121) derived from a broad-host-range plasmid, RSF1010, into G. violaceus by conjugation. The frequency of transformation of our system is significantly higher than that described in the previous report. In addition, luciferase heterologously expressed in G. violaceus functioned as a reporter. The established system will promote the molecular genetic studies on G. violaceus.
Keywords: Cyanobacteria, Luciferase, RSF1010, Transformation, Gloeobacter violaceus PCC 7421
Abbreviations: CBB, Coomassie Brilliant Blue; PCR, polymerase chain reaction; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; Sm, streptomycin
Highlights
▸ We developed a transformation system for Gloeobacter violaceus PCC 7421. ▸ We succeeded in producing G. violaceus harboring a broad-host-range plasmid. ▸ Luciferase was introduced into G. violaceus as a reporter gene. ▸ In the resultant transformant, luciferase was functionally expressed.
1. Introduction
Cyanobacteria are considered to be the first oxygenic photosynthetic organisms that emerged about 2.7 billion years ago. Most of the genes that are responsible for photosynthesis are widely conserved from cyanobacteria to eukaryotic photosynthetic organisms, this conservation is a convincing evidence of the endosymbiotic acquirement of eukaryotic chloroplast from a cyanobacterium. This high conservation has prevented us from understanding of the evolution of photosynthetic mechanisms from the primordial one. Therefore, cyanobacteria diverged from early stage of the cyanobacterial evolution may be helpful in studying the evolution of photosynthetic mechanisms, because such cyanobacteria are expected to retain a part of primordial properties that had been lost during the evolution of other major cyanobacteria. However, “primordial cyanobacteria” that retain a part of primordial properties rarely exist nowadays.
Gloeobacter violaceus PCC 7421 (hereafter referred to as G. violaceus) is a unicellular cyanobacterium, and is considered to be an early-branching cyanobacterium within the cyanobacterial clade, by the molecular phylogenetic analyses [1–4]. Almost all oxygenic photosynthetic organisms form the internal membranes called thylakoid membranes, which are the site for the light reaction of photosynthesis. G. violaceus is the only known oxygenic photosynthetic organism that lacks the thylakoid membranes [5]. This unique property has been found only in this organism. Accordingly, both the photosystems and the respiratory chain in G. violaceus are localized at the cytoplasmic membrane. This indicates that photosynthetic activity per cell in G. violaceus is much lower than those in other cyanobacteria and eukaryotic photosynthetic organisms. For these unique characteristics, the complete genome of G. violaceus was sequenced in 2003 [6]. The genome sequence revealed that a part of the genes that are responsible for photosynthesis was not found in G. violaceus, whereas those genes are highly conserved among other oxygenic photosynthetic organisms [6]. Therefore, in recent years, protein complexes that are responsible for photosynthesis (e.g. photosystem I and phycobilisome) in G. violaceus were biochemically analyzed based on the genome information [7–11]. These recent results partly solved unique features previously reported [12–14]. Recently, it was reported that both the photosynthetic and respiratory complexes were concentrated at the respective domains, which may have specialized functions, in the cytoplasmic membrane of G. violaceus [15]. Moreover, the comparison of state transitions between G. violaceus and Synechocystis sp. PCC 6803 showed the commonalities and differences [16]. G. violaceus exhibited state transitions and non-photochemical fluorescence quenching like Synechocystis sp. PCC 6803 [16]. In G. violaceus, the structure of phycobilisome was quite different from other cyanobacterial phycobilisomes [12]. Nevertheless, orange carotenoid protein that binds to phycobilisome was also correlated with blue-light-induced heat dissipation in G. violaceus, like Synechocystis sp. PCC 6803 [16]. These results suggest that G. violaceus is an ideal organism for investigating the evolution of photosynthetic system by comparison of other cyanobacteria.
Molecular genetics, such as the production and analysis of mutants, is a preferable method to analyze the function of individual genes in G. violaceus. Unfortunately, molecular genetic analysis cannot be applied to G. violaceus because of the lack of a highly-reproducible transformation system for this organism. Only one report on the development of a transformation system for G. violaceus has been published to date [17]. However, there is no subsequent paper that describes the functional expression of the foreign genes in G. violaceus using the system. In the present study, we re-examined the transformation system reported previously, and developed a highly-reproducible transformation system for G. violaceus. We succeeded in introducing an expression vector derived from a broad-host-range plasmid into G. violaceus by conjugal gene transfer. Using this system, we introduced a luciferase gene into G. violaceus, and the resultant transformant exhibited significant luciferase activity.
2. Materials and methods
2.1. Culture of G. violaceus
G. violaceus was grown photoautotrophically in BG11 medium [18] under the continuous white light (10 μmol photons m−2 s−1) at 25 °C, and air was supplied via an air filter (Millex-FG, Millipore, Massachusetts, USA). For transformants, 10 μg ml−1 streptomycin (Sm) was added to the medium. BG11 agar medium containing 1 mM TES–NaOH (pH 8.2) was used for solid culture.
2.2. Construction of plasmids and transformation of G. violaceus
We used a plasmid vector pKUT1121 [19], which was constructed from a broad-host-range plasmid RSF1010 [20], to establish a transformation system for G. violaceus. The coding region of firefly luciferase gene (luc) was amplified by polymerase chain reaction (PCR) using pGL3-Basic vector (Promega, Wisconsin, USA) as a template. The PCR product containing luc gene with additional restriction sites for NdeI and XhoI at the 5′- and 3′-ends, respectively, was amplified using the following primers: 5′-GGGCATATGGAAGACGCCAAAAACAT-3′, 5′-GCGGAAAGATCGCCGTGTAACTCGAGAAA-3′. After the PCR product was subcloned into pZErO-2 (Invitrogen, California, USA), the sequence of cloned luc gene was confirmed by sequencing. The luc gene was excised from the plasmid by NdeI and XhoI treatment, and subcloned into pKUT1121 to yield pKUT-luc.
Transformation was performed by diparental mating basically according to the method of Elhai and Wolk [21]. First, a conjugative helper plasmid, pRK2013 [22], was introduced into Escherichia coli XL1-Blue MRF′ (Agilent Technologies, California, USA). Subsequently, the expression vector (pKUT1121 or pKUT-luc) was introduced into XL1-Blue MRF′ (pRK2013). Equal amounts of resultant transformant cells and G. violaceus cells were mixed, and then aliquots of the mixture were spotted onto nitrocellulose membrane on a BG11 agar medium. Following a 48 h incubation under the light of 5 μmol photons m−2 s-1, the membrane was transferred onto BG11 agar medium containing 5 μg ml−1 Sm. Streptomycin-resistant colonies appeared after several months, and each colony was finally cultured in BG11 liquid medium containing 10 μg ml−1 Sm. Total DNA was prepared from G. violaceus cells using hexadecyl-trimethyl-ammonium bromide [23]. The presence of marker gene in the total DNA was checked by PCR.
2.3. SDS–PAGE and Western blotting
Total protein of G. violaceus cells was prepared by the following procedure. G. violaceus was resuspended with a buffer (20 mM MES–NaOH (pH 6.5), 1 mM MgCl2, 0.5 mM CaCl2, 1 mM NaCl, 0.6 M betaine). The suspended cells were disrupted by repeated agitation with glass beads (ϕ = 0.1 mm) at 4 °C. After the debris was removed by centrifugation (2000×g, 5 min, 4 °C), Triton X-100 was added to the supernatant at the final concentration of 1% to solubilize the membrane. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed according to Laemmli [24] using 12% (w/v) of polyacrylamide gel. Total proteins (10 μg) were loaded on each lane of a gel, and the gel was stained with Coomassie Brilliant Blue (CBB) after electrophoresis. For Western blotting, separated proteins in the gel were electroblotted onto a polyvinylidene difluoride membrane (Hybond-P, GE Healthcare, New Jersey, USA). Western blotting and chemiluminescence detection were performed according to the manufacturer's instructions. Anti-luciferase antibody (Luciferase (251-550), Santa Cruz Biotechnology, California, USA) was used as a primary antibody. After the treatment with secondary antibody (Anti-Rabbit IgG, Jackson Immuno Research Europe, Suffolk, UK), luciferase was detected by chemiluminescence (ECL Plus Western Blotting Detection System, GE Healthcare) with a luminescent image analyzer (LAS-3000 UV mini, Fujifilm, Tokyo Japan).
2.4. Luciferase assay
The concentration of G. violaceus cells was adjusted to 1.0 × 107 cells ml−1 with BG11 medium. After the cells were adapted to darkness for 5 min, background luminescence was measured with a luminometer (GloMax™ 20/20n Luminometer, Promega). Then, luciferin (Beetle Luciferin, Promega) was added to the cells at the final concentration of 100 μM, and the luminescence derived from luciferase reaction was measured.
3. Results
3.1. Antibiotic susceptibility of G. violaceus
First, we tried to culture G. violaceus at 28 °C under the light of 20 μmol photons m−2 s−1 according to Guo and Xu [17], however, cells were not able to survive. Therefore, we applied our routine culture conditions to further study. We examined the antibiotic susceptibility of G. violaceus for the use of antibiotic resistance genes as marker genes of transformant. We tested gentamicin, hygromycin, spectinomycin and zeocin in addition to antibiotics used in Guo and Xu [17]. Table 1 summarizes the result of antibiotic susceptibility test of wild type G. violaceus. Three antibiotics showed same susceptibility as described in Guo and Xu [17], however, the others showed different susceptibility (for details, see Section 4). Three of nine antibiotics, erythromycin, Sm and spectinomycin exhibited antibiotic activity against G. violaceus within the range of 1–50 μg ml−1. For these three antibiotics, we also checked the antibiotic activity against G. violaceus on the agar medium. G. violaceus cells adjusted to the concentration of 1.0 × 103 to 1.0 × 109 cells ml−1 were spotted onto nitrocellulose membrane on BG11 agar medium including each antibiotic. As a result, Sm was the most effective for killing cells at lower concentration (5 μg ml−1). Therefore, we chose Sm resistance gene (aadA) as a marker gene for the screening of transformant.
Table 1.
Antibiotic susceptibility of G. violaceus.
| Antibiotics | Concentration (μg ml−1) |
|||
|---|---|---|---|---|
| 1 | 5 | 15 | 50 | |
| Ap | R | R | R | R |
| Cm | R | R | R | S |
| Em | S | S | S | S |
| Gm | R | R | R | S |
| Hyg | R | R | R | R |
| Km | R | R | R | R |
| Sm | R | S | S | S |
| Sp | R | S | S | S |
| Zeo | R | R | R | R |
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin; Hyg, hygromycin; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Zeo, zeocin.
R, resistant; S, sensitive.
3.2. Development of transformation system for G. violaceus
Because G. violaceus was sensitive to Sm (Table 1), we tried to introduce a broad-host-range plasmid derived expression vector, pKUT1121 [19] that possesses Sm resistance gene cassette, by conjugal gene transfer. After the treatment of exconjugants with Sm, Sm resistant colonies appeared (Fig. 1A) . In contrast, no colony was formed in the spot of negative control (Fig. 1B ). The frequency of transformation of G. violaceus was approximately 1.2 × 10−4 per recipient cell for pKUT1121. Total DNA prepared from the Sm resistant strain and wild type were used as template of PCR (Fig. 2) to confirm successful introduction of the plasmid. As a marker gene, aadA was amplified by PCR, and a PCR product that exhibited the similar migration to that of positive control (Fig. 2A, lane 2) was observed in the Sm resistant strain by electrophoresis (Fig. 2A, lane 3). In contrast, no amplification was found in wild type (Fig. 2A, lane 1). Furthermore, we transformed two strains of E. coli (XL1-Blue MRF′ and DH5α) with the total DNA. For each E. coli strain, a lot of Sm resistant colonies appeared after the transformation with the total DNA prepared from Sm resistance strain. On the contrary, no colony formed after the transformation with the total DNA from wild type. Then, plasmids prepared from the E. coli transformants were digested with restriction enzymes. In agarose gel electrophoresis, the restriction patterns of the prepared plasmids were identical to that of the original pKUT1121 (data not shown). These results demonstrated that the Sm resistant strain harbors pKUT1121 as a plasmid. Therefore, we concluded that the transformation system for G. violaceus was established. We named the G. violaceus transformant pKUT1121 strain, and used this strain as a control for further experiments.
Fig. 1.
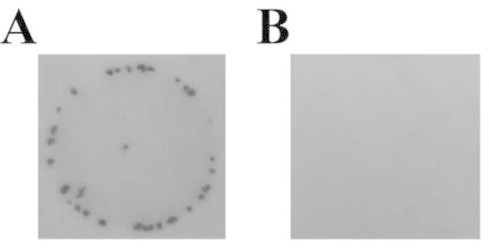
Sm resistant colonies formed by conjugation. (A) Conjugation of G. violaceus and XL1-Blue MRF′ (pRK2013, pKUT1121). (B) Wild type G. violaceus as a negative control.
Fig. 2.
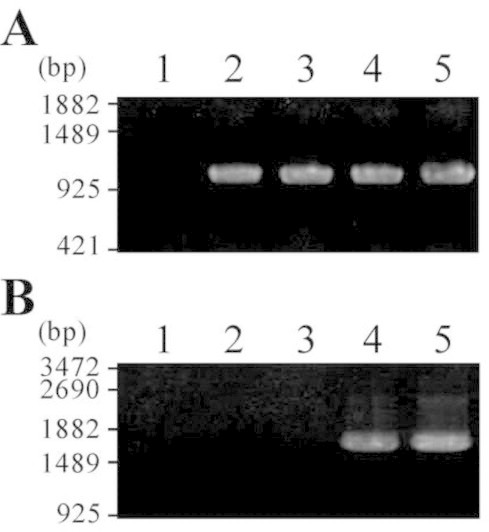
Confirmation of the production of G. violaceus transformants. Coding regions of aadA (A) and luc (B) were amplified by PCR. Templates for PCR are as follows: lane 1, total DNA prepared from wild type G. violaceus; lane 2, pKUT1121 prepared from E. coli; lane 3, total DNA prepared from pKUT1121 strain; lane 4, pKUT-luc prepared from E. coli; lane 5, total DNA prepared from pKUT-luc strain.
3.3. Introduction of a luc gene into G. violaceus as a reporter gene
Because we succeeded in developing the transformation system for G. violaceus by conjugation, we evaluated the use of a luc gene as a reporter gene in G. violaceus cells. The luc gene was subcloned into pKUT1121, and the resultant vector (pKUT1121-luc) was introduced into G. violaceus. By conjugation, Sm resistant colonies were obtained at the frequency of transformation of approximately 3.6 × 10−5 per recipient cell. Total DNA was prepared from this Sm resistant strain (pKUT1121-luc strain), and used as a template of PCR. Two marker genes, aadA and luc were successfully amplified by PCR from total DNA of pKUT1121-luc strain (Fig. 2, lane 5), whereas luc gene was not amplified from the total DNA of both wild type and pKUT1121 strain (Fig. 2B, lanes 1 and 3). Transformation of E. coli with the total DNA prepared from pKUT1121-luc strain confirmed that pKUT1121-luc was maintained as a plasmid in pKUT1121-luc strain.
3.4. Activity of luciferase expressed in the pKUT1121-luc strain
We examined whether the luc gene was functionally expressed in the pKUT1121-luc strain, by Western blotting and luciferase assay. Total proteins prepared from cells were analyzed by SDS–PAGE and following Western blotting. Although no specific band was found among wild type, pKUT1121 strain and pKUT1121-luc strain by CBB staining of the gel (Fig. 3A) , luciferase was immunochemically detected only in the pKUT1121-luc strain (Fig. 3B ). For wild type and transformants, we measured luciferase activity in vivo (Table 2). The background luminescence were measured after the dark adaptation of the cells, and the values were quite low in all samples. After the addition of luciferin to the cells, approximately 1000 times higher luminescence than background was observed in pKUT1121-luc strain, whereas virtually no difference was found in wild type and pKUT1121 strain (Table 2).
Fig. 3.
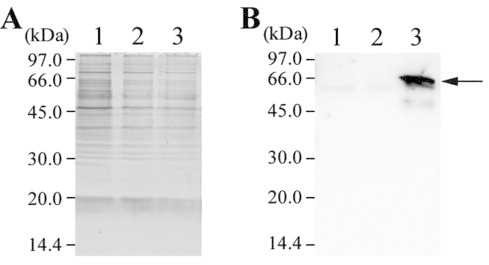
Detection of luciferase by Western blotting. Total proteins were used for SDS–PAGE and Western blotting. Lane 1, wild type G. violaceus; lane 2, pKUT1121 strain; lane 3, pKUT1121-luc strain. (A) The gel image of CBB staining after SDS–PAGE. (B) The result of Western blotting using anti-luciferase antibody. The arrow indicates the position of luciferase in the membrane.
Table 2.
Luciferase activity of wild type and G. violaceus transformants.
| Samples | Background (RLU)* | +Luciferin (RLU)* |
|---|---|---|
| Wild type | 118 ± 16 | 106 ± 32 |
| pKUT1121 strain | 83 ± 5 | 87 ± 9 |
| pKUT-luc strain | 87 ± 11 | 110437 ± 10431 |
The values represent the averages and standard deviations of triplicate measurements.
RLU, relative luminescence units.
4. Discussion
Development of the transformation system for G. violaceus was already reported by Guo and Xu [17]. They demonstrated the introduction of a RSF1010 derived plasmid into G. violaceus by conjugation. However, we could not reproduce even their culture condition (28–30 °C, 10–20 μmol photons m−2 s−1). Because G. violaceus was reported to be a cyanobacterium that can grow only under the dim light [5], we presumed that G. violaceus used by Guo and Xu [17] was adapted to slightly stronger light condition during long cultivation. Since the higher-light adapted G. violaceus was not obtained in our experiment, we examined proper conditions suitable for the transformation of G. violaceus using our routine culture. Sensitivity of G. violaceus to erythromycin and Sm (Table 1) was similar to that described in Guo and Xu [17]. However, sensitivity to ampicillin and chloramphenicol was different from the former result, which demonstrated that ampicillin was effective at the concentration of 15 μg ml−1 and chloramphenicol was ineffective up to 100 μg ml−1 [17]. We newly evaluated the four antibiotics (gentamicin, hygromycin, spectinomycin and zeocin) for G. violaceus. Among them, only spectinomycin was effective.
We used expression vectors derived from a broad-host-range plasmid RSF1010, because G. violaceus have no endogenous plasmid [6]. Guo and Xu [17] reported that no transformant was obtained by the transformation of E. coli DH5α with DNA prepared from plasmid-introduced G. violaceus whereas transformants appeared with high efficiency in the case of a methylation-restriction mutant, DH10B as a host strain. They concluded that the difference in the transformation efficiency reflect the presence of DNA methylation in G. violaceus, which caused the restriction by E. coli. On the other hand, our results using DH5α and a methylation-restriction mutant, XL1-Blue MRF′ demonstrated that there is no difference in the transformation efficiency of two strains. Guo and Xu [17] used a vector, pKT210 [25], which is different from our vector, pKUT1121 that was recently constructed [19]. Both vectors are derived from a broad-host-range plasmid RSF1010, however, pKUT1121 is 3.3 kb smaller than pKT210. Therefore, the transformation efficiency of pKUT1121 might be higher than that of pKT210 for E. coli. However, we assumed that the difference is not related to DNA methylation in G. violaceus because DH10B was transformed with high efficiency using pKT210 [17]. Hence, this inconsistency also indicated that the properties of G. violaceus reported in 2004 was quite different from our strain.
In the transformation of bacteria, restriction-modification system in the cell affects the frequency of transformation. In the case of Anabaena sp. PCC 7120, type II DNA restriction-modification system is major barrier against transformation [21]. This problem was overcome by the coexpression of DNA methylases that were associated with type II restriction enzymes in the E. coli [21]. Guo and Xu [17] reported that the frequency of transformation of G. violaceus raised from 4.63 × 10−6 to 1.67 × 10−5 by the coexpression of three DNA methylases that were effective for the transformation of Anabaena [26]. In our result, the frequency of transformation of pKUT1121 strain was 1.2 × 10−4 without coexpression of Anabaena DNA methylases. The frequency of transformation of our system is significantly higher than that described in the previous report. Moreover, no genes for type II restriction-modification system was found in the genome of G. violaceus [6]. These results indicate that the barrier of the restriction system for transformation is not high in G. violaceus unlike Anabaena.
The luciferase assay revealed that there is no significant background activity in G. violaceus (Table 2). Therefore, the luc gene can be available for promoter assay using G. violaceus as a host. Although we could not succeed in accumulating luciferase at a high level (Fig. 3), this low-level expression will be enough for the metabolic engineering of G. violaceus. The genes for enzymes that are responsible for chlorophyll, carotenoid and lipid biosynthesis are candidates for alteration, because those molecules affect photosynthetic activity.
In the present study, we established the highly-reproducible transformation system on G. violaceus, and demonstrated the introduction and functional expression of the luc gene in G. violaceus. Using our system, other molecular genetic techniques such as transposon tagging and gene targeting will be developed in the future, and the analyses of G. violaceus will progress by the novel techniques.
Acknowledgments
This work was supported by Grant-in-Aid for Creative Research from Japan Society for the Promotion of Science (No. 17GS0314 to M.M.), and by Grant-in-Aid from Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 22370017 to M.M. and T.T., and No. 24658080 to T.T.).
References
- 1.Nelissen B., Van de Peer Y., Wilmotte A., De Wachter R. An early origin of plastids within the cyanobacterial divergence is suggested by evolutionary trees based on complete 16S rRNA sequences. Mol. Biol. Evol. 1995;12:1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- 2.Swingley W.D., Blankenship R.E., Raymond J. Integrating Markov clustering and molecular phylogenetics to reconstruct the cyanobacterial species tree from conserved protein families. Mol. Biol. Evol. 2008;25:643–654. doi: 10.1093/molbev/msn034. [DOI] [PubMed] [Google Scholar]
- 3.Falcón L.I., Magallón S., Castillo A. Dating the cyanobacterial ancestor of the chloroplast. ISME J. 2010;4:777–783. doi: 10.1038/ismej.2010.2. [DOI] [PubMed] [Google Scholar]
- 4.Gupta R.S., Mathews D.W. Signature proteins for the major clades of cyanobacteria. BMC Evol. Biol. 2010;10:24. doi: 10.1186/1471-2148-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rippka R., Waterbury J., Cohen-Bazire G. A cyanobacterium which lacks thylakoids. Arch. Microbiol. 1974;100:419–436. [Google Scholar]
- 6.Nakamura Y., Kaneko T., Sato S., Mimuro M., Miyashita H., Tsuchiya T., Sasamoto S., Watanabe A., Kawashima K., Kishida Y., Kiyokawa C., Kohara M., Matsumoto M., Matsuno A., Nakazaki N., Shimpo S., Takeuchi C., Yamada M., Tabata S. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 7.Inoue H., Tsuchiya T., Satoh S., Miyashita H., Kaneko T., Tabata S., Tanaka A., Mimuro M. Unique constitution of photosystem I with a novel subunit in the cyanobacterium Gloeobacter violaceus PCC 7421. FEBS Lett. 2004;578:275–279. doi: 10.1016/j.febslet.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Koyama K., Tsuchiya T., Akimoto S., Yokono M., Miyashita H., Mimuro M. New linker proteins in phycobilisomes isolated from the cyanobacterium Gloeobacter violaceus PCC 7421. FEBS Lett. 2006;580:3457–3461. doi: 10.1016/j.febslet.2006.04.098. [DOI] [PubMed] [Google Scholar]
- 9.Sicora C.I., Brown C.M., Cheregi O., Vass I., Campbell D.A. The psbA gene family responds differentially to light and UVB stress in Gloeobacter violaceus PCC 7421, a deeply divergent cyanobacterium. Biochim. Biophys. Acta. 2008;1777:130–139. doi: 10.1016/j.bbabio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Dreher C., Hielscher R., Prodöhl A., Hellwig P., Schneider D. Characterization of two cytochrome b6 proteins from the cyanobacterium Gloeobacter violaceus PCC 7421. J. Bioenerg. Biomembr. 2010;42:517–526. doi: 10.1007/s10863-010-9279-6. [DOI] [PubMed] [Google Scholar]
- 11.Mendoza-Hernández G., Pérez-Gómez B., Krogmann D.W., Gutiérrez-Cirlos E.B., Gómez-Lojero C. Interactions of linker proteins with the phycobiliproteins in the phycobilisome substructures of Gloeobacter violaceus. Photosynth. Res. 2010;106:247–261. doi: 10.1007/s11120-010-9601-5. [DOI] [PubMed] [Google Scholar]
- 12.Guglielmi G., Cohen-Bazire G., Bryant D.A. The structure of Gloeobacter violaceus and its phycobilisomes. Arch. Microbiol. 1981;129:181–189. [Google Scholar]
- 13.Koenig F., Schmidt M. Gloeobacter violaceus – investigation of an unusual photosynthetic apparatus. Absence of the long wavelength emission of photosystem I in 77 K fluorescence spectra. Physiol. Plant. 1995;94:621–628. [Google Scholar]
- 14.Mangels D., Kruip J., Berry S., Rögner M., Boekema E.J., Koenig F. Photosystem I from the unusual cyanobacterium Gloeobacter violaceus. Photosynth. Res. 2002;72:307–319. doi: 10.1023/A:1019822316789. [DOI] [PubMed] [Google Scholar]
- 15.Rexroth S., Mullineaux C.W., Ellinger D., Sendtko E., Rögner M., Koenig F. The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell. 2011;23:2379–2390. doi: 10.1105/tpc.111.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernát G., Schreiber U., Sendtko E., Stadnichuk I.N., Rexroth S., Rögner M., Koenig F. Unique properties vs. common themes: the atypical cyanobacterium Gloeobacter violaceus PCC 7421 is capable of state transitions and blue-light-induced fluorescence quenching. Plant Cell Physiol. 2012;53:528–542. doi: 10.1093/pcp/pcs009. [DOI] [PubMed] [Google Scholar]
- 17.Guo H., Xu X. Broad host range plasmid-based gene transfer system in the cyanobacterium Gloeobacter violaceus which lacks thylakoids. Prog. Nat. Sci. 2004;14:31–35. [Google Scholar]
- 18.Allen M.M. Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 1968;4:1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya T., Mizoguchi T., Akimoto S., Tomo T., Tamiaki H., Mimuro M. Metabolic engineering of the Chl d-dominated cyanobacterium Acaryochloris marina: production of a novel Chl species by the introduction of the chlorophyllide a oxygenase gene. Plant Cell Physiol. 2012;53:518–527. doi: 10.1093/pcp/pcs007. [DOI] [PubMed] [Google Scholar]
- 20.Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 21.Elhai J., Wolk C.P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 22.Figurski D.H., Helinski D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., editors. Current Protocols in Molecular Biology. John Wiley & Sons; Hoboken, NJ: 1997. pp. 2.4.1–2.4.5. [Google Scholar]
- 24.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Bagdasarian M., Lurz R., Rückert B., Franklin F.C.H., Bagdasarian M.M., Frey J., Timmis K.N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 26.Elhai J., Vepritskiy A., Muro-Pastor A.M., Flores E., Wolk C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


