Abstract
The green fluorescent protein (GFP) is the most commonly used reporter protein for monitoring gene expression and protein localization in a variety of living and fixed cells, including not only prokaryotes, but also eukaryotes, e.g., yeasts, mammals, plants and fish. In general, it is thought that GFP is nontoxic to cells, although there are some reports on the side effect of GFP. Further, details of the molecular mechanism concerning the side effect of GFP remain unclear. Here we show that Ku80, but not XRCC4, plays an important role in the mechanism of the resistance to cytotoxicity induced by enhanced GFP (EGFP). EGFP inhibited both cell proliferation and colony formation, and induced cell death in Ku80-deficient hamster cells, i.e., xrs-6 cells. In addition, Ku80 attenuated EGFP-induced cytotoxicity in the xrs-6 cells. No EGFP-induced cytotoxicity was observed in the NHEJ core protein XRCC4-deficient hamster cells, i.e., XR-1 cells. Furthermore, EGFP markedly enhanced X-ray-induced cytotoxicity in the xrs-6 cells. These results suggest that Ku80 plays a key role in the novel NHEJ-independent defense mechanism against EGFP-induced cytotoxicity. Caution should be taken in considering of the potential influence by the stress response mechanism, namely, the Ku80-dependent elimination mechanism of EGFP-induced cytotoxicity, being activated, even when using EGFP-expressing cells in which Ku80 functions normally.
Keywords: Cell death, Cytotoxicity, GFP, Ku, XRCC4
Abbreviations: DNA-PKcs, DNA-dependent protein kinase catalytic subunit; DSBs, DNA double-strand breaks; ECFP, enhanced cyan fluorescent protein; EGFP, enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein; GFP, green fluorescent protein; NHEJ, nonhomologous end-joining
Highlights
▸ Enhanced GFP (EGFP) inhibits proliferation and induces cell death in Ku80-deficient cells. ▸ EGFP markedly enhances X-ray-induced cytotoxicity in Ku80-deficient cells. ▸ Ku80 attenuates EGFP-induced cytotoxicity in Ku80-deficient cells. ▸ No EGFP-induced cytotoxicity is observed in XRCC4-deficient cells. ▸ Ku80 plays a role in the NHEJ-independent defense mechanism against the cytotoxicity.
1. Introduction
The green fluorescent protein (GFP) from jellyfish Aequorea victoria is the most commonly used reporter protein for monitoring gene expression, protein localization, and protein–protein interactions in a variety of living and fixed cells, including not only prokaryotes, e.g., bacteria, but also eukaryotes, e.g., yeasts, plants, flies, fish and mammals including humans [1]. GFP was discovered by Shimomura et al. [2]. Since them, crucial breakthroughs have been made with the cloning of the GFP gene by Prasher et al. and the demonstrations by Chalfie et al. and Inouye and Tsuji that the expression of the GFP gene in other organisms creates fluorescence [1,3–5]. After the now commonly used enhanced GFP (EGFP) and its variants have been reported, the GFP technology has revolutionized developments in biology, and the recent explosion in the diversity of available fluorescent proteins promises a wide variety of new tools for biological imaging [6–9].
In general, GFP is considered nontoxic to cells in vitro and in vivo. In fact, EGFP expression is nontoxic in EGFP transgenic mice [10]. On the basis of this, GFP transgenic animals and their cells are also widely used in various fields of life sciences including basic biology and biomedical research. However, there are some reports on the side effect of GFP in in vitro and in vivo studies, although details of the molecular mechanism of such an effect remain unclear [11–15]. It is clear that GFP can induce apoptosis but how it triggers it remains unclear [11]. A report on GFP-induced cell death indicates that exciting GFP intensity for extended periods may generate free radicals that are genotoxic to cells. Recently, it has been reported that, in rat adult hepatic stem cells, no stable cell lines expressing EGFP have been established because these cells undergo cell death [16].
Recent studies using GFP-technology laser microbeam irradiation to induce DNA damage in the nucleus of living cells have shed light on the order of recruitment of DNA repair-core or related factors to sites of DNA damage as well as the kinetics of the repair process [17–21]. Nonhomologous DNA-end-joining (NHEJ) repair, which is responsible for repairing a major fraction of DNA double-strand breaks (DSBs) in somatic cells of multicellular eukaryotes, is considered to begin with the binding of Ku, i.e., a heterodimer of Ku70 and Ku80 [17,22,23]. The NHEJ repair requires Ku70, Ku80, a DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4, DNA ligase IV, Artemis, and XLF (also called Cernunnos) [17]. The mechanisms underlying the control of the subcellular localization of XRCC4 are dependent on Ku70 and Ku80 and might play a key role in the regulation of the physiological function of XRCC4 in both hamster and mouse lung cells [18,20]. The deletion or inactivation of any of these NHEJ core factors induces marked sensitivity to IR and anticancer drugs, e.g., bleomycin, as well as genomic instability and defects in V(D)J recombination [17]. However, it remains unclear whether GFP is toxic to cells, which is a result of the deletion or inactivation of any of these NHEJ core factors.
In this study, we examined whether EGFP is toxic to cells, which is a result of the deficiency in the NHEJ core factor Ku80.
2. Materials and methods
2.1. Cell lines, cultures, reagents and transfections
A Chinese hamster ovary cell line CHO-K1 (Riken Cell Bank), Ku80-deficient CHO-K1 mutant cell line (xrs-6), and XRCC4-deficient CHO-K1 mutant cell line (XR-1) were cultured as described in previous studies [24–26]. The xrs-6 cell line stably expressing EGFP-Ku80 or EGFP alone was described previously [27]. The XR-1 cell line stably expressing EGFP-XRCC4 or EGFP alone was described previously [24]. The transient transfections of pEGFP-Ku80, pEYFP (enhanced yellow fluorescent protein), or pEGFP was performed in cells using FuGene6 (Roche Diagnostics K.K., Indianapolis, IN), as described previously, and the cells were cultured for 2 days and then monitored under an FV300 confocal laser scanning microscope (Olympus, Tokyo, Japan), as previously described [20,21]. DNA of the fixed cells was stained with DAPI fluorescent dye.
2.2. Transient transfection assay of effect of EGFP on cell proliferation
The cell proliferation assay was performed according to the method of Mo and Dynan, as described previously [28]. In short, the cells were grown on a 35-mm dish the night before transfection. EGFP-Ku80, EGFP or EYFP vectors were introduced using FuGene6, as described above. The next day, the cells were replated on a glass slide. After 1 day, an index was used to determine whether the cells that show fluorescence underwent cell division. Fifty fluorescent clusters were counted in each experiment, and the procedure was repeated three times.
2.3. X-irradiation
Cells were exposed to X-rays at room temperature, as described previously [21,24]. X-rays were generated at 200 kVp/20 mA and filtered through 0.5-mm Cu and Al filters, as described previously [21,24].
2.4. Colony formation assay
Colony formation assay was performed as described previously [24].
2.5. Western blot analysis
Western blot analysis was performed as described previously [20,21,29]. The following antibodies were used: a rabbit anti-GFP polyclonal antibody (FL) (Santa Cruz Biotechnology, Santa Cruz, CA), a goat anti-Ku70 polyclonal antibody (C-19) (Santa Cruz Biotechnology), a goat anti-Ku80 polyclonal antibody (M-20) (Santa Cruz Biotechnology), a rabbit anti-H2AX polyclonal antibody (ab11175) (abcom, Cambridge, UK), a mouse anti-γ-H2AX monoclonal antibody (JBW301) (Upstate Biotechnology Inc., Charlottesville, VA), a rabbit anti-cleaved caspase-7 (Asp198) polyclonal antibody (9491) (Cell Signaling Technology Inc., Danvers, MA), or a mouse anti-β-actin monoclonal antibody (Sigma, St. Louis, MO, USA). The binding to corresponding proteins was visualized using an Advance Western blotting detection system (GE Healthcare Bio-Sci. Corp.), in accordance with the manufacturer's instructions.
3. Results
3.1. Role of Ku80 in EGFP-induced cytotoxicity
To examine whether EGFP and its variant protein EYFP are toxic to xrs-6 cells (derived from CHO-K1 cell line), which are deficient in the NHEJ core factor, Ku80, we transfected expression plasmids into xrs-6 cells. We found that the transfection with EGFP expression vector (pEGFP) yielded expressed cells less efficiently than the transfection with the analogous EYFP expression vector (pEYFP) (P < 0.05) (Fig. 1(A)). About 1.13% of the cells are EGFP positive, whereas about 5.13% of cells were EYFP positive. Some of the EGFP-expressing cells exhibited the apoptosis-like features, namely, nuclear condensation, round shape, and being detached, suggesting that EGFP is toxic to xrs-6 cells (Fig. 1(A), arrows). Expectedly, EYFP or EGFP was localized throughout the xrs-6 and CHO-K1 cells (Fig. 1(A) and (C)). Next, we examined the EGFP or EYFP expression levels in the transiently transfected xrs-6 or CHO-K1 cells by Western blot analysis. Consistent with previous reports [22,25,26], Ku80 was expressed in the CHO-K1 cells, but not in the xrs-6 cells (Fig. 1(B)). In addition, the Ku70 expression level was markedly reduced in the xrs-6 cells. Expectedly, β-actin was expressed in both cell lines. The expression level of EYFP was higher than that of EGFP in both cells, suggesting that there are more positive cells in the case of pEYFP transfection than in the case of pEGFP transfection (Fig. 1(B)). In addition, the expression levels of the two fluorescent proteins were higher in the CHO-K1 cells than in the xrs-6 cells, suggesting that EGFP and its variant protein EYFP are toxic to xrs-6 cells than to CHO-K1 cells. To confirm this and determine whether Ku80 protects xrs-6 cells against such cytotoxicity, pEGFP or pEGFP-Ku80 was individually transfected into the cells. After two days, more than 20% of the CHO-K1 cells with pEGFP or pEGFP-Ku80 and of the xrs-6 cells with pEGFP-Ku80 divided, whereas only less than 10% of the xrs-6 cells with pEGFP did (Fig. 1(D)). As shown in Fig. 1(E), there were many EGFP-expressing xrs-6 cells that exhibited the apoptosis-like features compared with the EGFP-Ku80-expressing xrs-6 cells. To examine whether EGFP triggered cell death is through apoptosis in xrs-6 cells, we investigated the expression of cleaved caspase-7. Western blotting analyses showed that the cleaved form of caspase-7 was expressed in the xrs-6 cells after transfection with pEGFP (Supplementary Fig. 1). Together, these results suggest that Ku80 inhibits EGFP-induced cell death in xrs-6 cells.
Fig. 1.
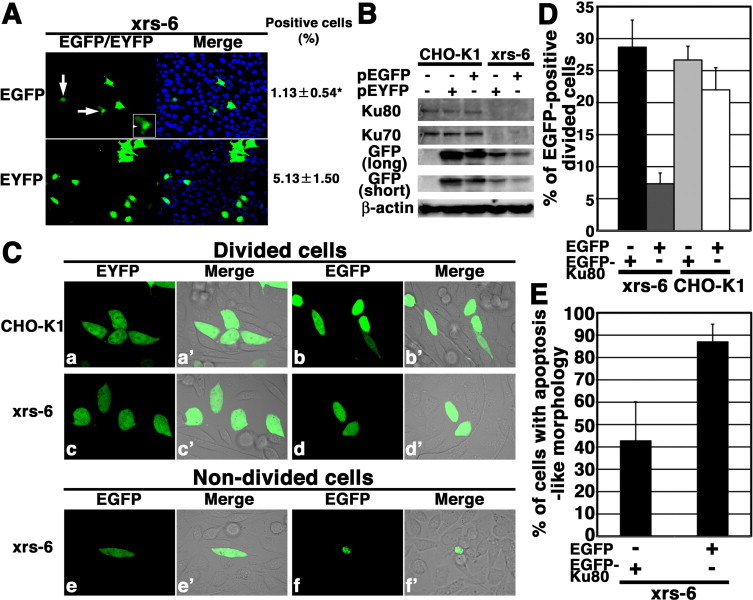
EGFP inhibits cell proliferation and induces cell death in DNA repair protein Ku80-deficient hamster cells, xrs-6. (A) Xrs-6 cells were transfected with pEGFP (upper panel) or pEYFP (lower panel). Two days following transfection, the cells were fixed and stained with DAPI. Three representative fields (more than 300 cells, each) were scored for each transfection and data represent the mean ± standard deviations. A significant difference is represented by * (P < 0.05, t test). Arrows indicate the cells showing an abnormal morphology. White panel: an enlarged image of the typical cell. (B) Comparison of EGFP or EYFP expression levels in xrs-6 and CHO-K1 cells 2 days after transfection. Total cell lysates from each cell line following transfection were analyzed by Western blot analysis using an anti-Ku70, anti-Ku80, anti-GFP or anti-β-actin antibody. Both EGFP and EYFP were detected by the anti-GFP antibody used. Short, short exposure; long, long exposure. (C) Imaging of living EYFP- (a, a’, c, c’) or EGFP-transfected (b, b’, d, d’, e, e’, f, f’) xrs-6 (c–f, c’–f’) or CHO-K1 (a, a’, b, b’) cells. Typical images of cells that underwent division (a–d’), nondividing cells with normal nucleus (e, e’) or nondividing cells with a condensed nucleus (f, f’) are shown. (D) The graph shows the percentage of EGFP-positive divided xrs-6 cells 2 days after transfection with pEGFP or pEGFP-Ku80. (E) The graph shows the percentage of cells with apoptosis-like morphology in the non-divided xrs-6 cells 2 days after transfection with pEGFP or pEGFP-Ku80. Error bars represent SD.
3.2. Role of NHEJ repair in EGFP-induced cytotoxicity
Next, we examined other side effects of EGFP in stable EGFP-expressing cells. First, the colony formation abilities of the CHO-K1 and xrs-6 cells were analyzed by colony formation assay. The colony formation ability of the CHO-K1 cells was slightly higher than that of the xrs-6 cells (data not shown). Previously, we established stable cell lines expressing functional EGFP-Ku80 or EGFP [27]. To confirm the expression of EGFP or EGFP-Ku80 by Western blot analysis, whole-cell extracts prepared from the CHO-K1 cells, xrs-6 cells, and xrs-6 transformants containing pEGFP (EGFP/xrs-6) or pEGFP-Ku80 (EGFP-Ku80/xrs-6) were used. In the extract of the EGFP-Ku80/xrs-6 cells stably transformed with pEGFP-Ku80, an EGFP-Ku80 signal was detected by Western blot analysis with an anti-Ku80 and anti-GFP polyclonal antibody. We also detected Ku70 in the extract prepared from the EGFP-Ku80/xrs-6 cells, but not from the EGFP/xrs-6 cells (Fig. 2(A)), and confirmed that in the stable transformants, exogenous Ku80 tagged with EGFP, as well as endogenous native Ku80, also stabilizes Ku70. Using those cell lines, we determined by colony formation assay whether a stable EGFP expression affects the colony formation ability of the xrs-6 cells. As shown in Fig. 2(B) and (C), the EGFP-Ku80/xrs-6 cells had the highest density, and the largest colony among the three cell lines. Interestingly, the EGFP/xrs-6 cells was had a lower density and a smaller colony than the xrs-6 cells. These results suggest that a stable expression of EGFP affects the colony formation ability of xrs-6 cells. NHEJ repair is responsible for repairing a major fraction of DSBs and requires the NHEJ core factor Ku80. Meanwhile, cells that sustain DSBs can undergo cell death and cell growth arrest. To evaluate whether EGFP expression induces the accumulation of spontaneous DSBs, we performed Western blot analysis with the antibody to γ-H2AX, which is a golden standard marker of DSBs. Expectedly, as shown in Fig. 2(D), a low expression level of γ-H2AX in the EGFP-Ku80/xrs-6 cells was observed in contrast to those in the EGFP/xrs-6 cells and xrs-6 cells. Surprisingly, the stable EGFP expression did not increase the expression level of γ-H2AX in the EGFP/xrs-6 cells, and the expression levels were comparable to those in the xrs-6 cells. Expectedly, β-actin and H2AX were expressed in all three cell lines. Next, we tested whether a stable EGFP expression affects the colony formation ability of XR-1 cells, which are derived from the Chinese hamster ovary and are deficient in the NHEJ core factor XRCC4. As shown in Fig. 3, the colony formation rate of the XR-1 cells was comparable to those of the EGFP-XRCC4/XR-1 cells and two EGFP/XR-1 cells. Taken together, these results suggest that XRCC4-deficient cells are insensitive to EGFP-induced cytotoxicty.
Fig. 2.
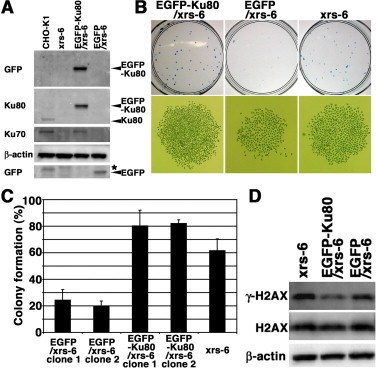
Characterization of cytotoxicity in stable EGFP-Ku80 and EGFP transformants. (A) Cells stably expressing the constructs denoted on the top, xrs-6, and CHO-K1 were lysed and analyzed by Western blotting using an anti-GFP, anti-Ku70, anti-Ku80, or anti-β-actin antibody. *, nonspecific band. (B) Representive images of dishes used in colony formation assay. (C) The graph shows the percentage of colonies formed in the xrs-6, two EGFP-Ku80/xrs-6 and two EGFP/xrs-6 cells. Error bars represent SD. (D) No excessive accumulation of γ-H2AX in EGFP/xrs6 cells. Total cell lysates from each cell line were analyzed by Western blotting using an anti-γ-H2AX, anti-H2AX, or anti-β-actin antibody.
Fig. 3.
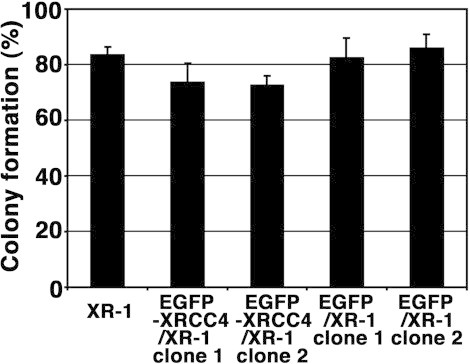
Nondetection of EGFP cytotoxicity in XRCC4-deficient cells, i.e., XR-1 cells. The graph shows the percentage of colonies formed in the XR-1, two EGFP-XRCC4/XR-1 and two EGFP/XR-1 cells. Error bars represent SD.
3.3. EGFP enhances X-ray-induced cytotoxicity in Ku80-deficient cells
To determine whether EGFP expression affects the sensitivity of xrs-6 cells to X-rays, we examined the radiosensitivity of EGFP/xrs-6 cells and xrs-6 cells by colony formation assay. As shown in Fig. 4, the EGFP/xrs-6 cells were highly sensitive to X-irradiation compared with the xrs-6 cells, suggesting that EGFP expression enhanced the cytotoxicity of xrs-6 cells to X-rays.
Fig. 4.
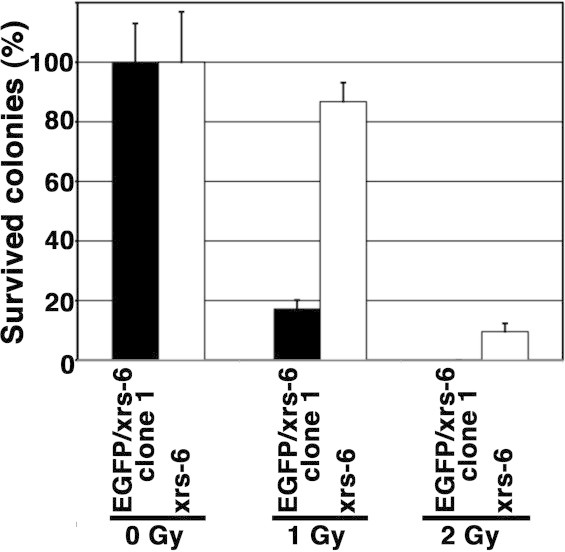
Enhancement of X-ray-induced cytotoxicity by EGFP in xrs-6 cells. The graph shows the percentage of colonies formed in the xrs-6 and EGFP/xrs-6 cells after X-irradiation (1 Gy or 2 Gy). Error bars represent SD.
4. Discussion
EGFP and its variants are effective reporter proteins for analyzing protein behavior and provide good temporal and spatial resolutions in living cells. In this study, we examined whether EGFP is toxic to cells deficient in the NHEJ core factor Ku80. In general, it is considered that EGFP is nontoxic to cells in vitro and in vivo [10]. Surprisingly, our results showed that EGFP inhibits both cell proliferation and colony formation, and induces cell death in Ku80-deficient hamster cells, i.e., xrs-6 cells. In addition, our data showed that Ku80 attenuates EGFP-induced cytotoxicity in xrs-6 cells. On the other hand, no EGFP-induced cytotoxicity was observed in the XR-1 cells, which are deficient in the NHEJ core protein XRCC4.
There is a link between GFP expression and apoptosis induction in several cell lines, i.e., NIH3T3 from mouse fibroblasts, BHK-21 from a baby hamster kidney, and HepG2 and Huh-7 from human hepatoma cancer [11], although its molecular mechanism remains unclear. In this study, we found that some EGFP expressing cells derived from the hamster ovary undergo cell death. Although not well known, generally, GFP or its variants have other side effects in vitro. Zhang et al. reported that GFP selectively induces the HSP70-mediated upregulation of the COX-2 expression in endothelial cells, but not in other cell types such as cancer cell lines [30]. In addition, Baens et al. reported that EGFP and EGFP fusion proteins inhibit polyubiquitination, a posttranslational modification that controls a wide variety of cellular processes, like the activation of kinase signaling or protein degradation by the proteasome [15]. On the other hand, it had reported that EGFP expression is nontoxic in EGFP-transgenic mice [10], supporting the idea that EGFP is nontoxic to cells in vivo. However, the transgenic expression of GFP could cause dilated cardiomyopathy in transgenic mice [12]. Furthermore, Agbulut et al. showed that EGFP expression impairs actin–myosin interactions by binding to the actin binding site of myosin, thereby causing excitation–contraction uncoupling and an impaired contractile function of muscle cells [13,14]. Taken together, previous findings and ours suggest that the expression of GFP or its variant affects cell fate and molecular processes such as post-translational modification and protein–protein interaction in a cell or tissue-type-dependent manner, although GFP or its variant is conveniently used to visualize organelles, organs, cell populations, protein localizations and subcellular processes in vitro and in vivo.
It was reported that colonies of transiently EGFP-expressing cells can not be propagated as stable cell strains, whereas both transiently EYFP- and enhanced cyan fluorescent protein (ECFP)-expressing cell colonies have 100% cloning efficiencies in rat hepatic adult stem cell studies in vitro [16]. In this study, we observed that the cytotoxicity of EYFP was lower than that of EGFP in not only the Ku80-wild-type cells (CHO-K1), but also the Ku80-deficient cells (xrs-6). These findings strongly suggest that the selection of the fluorescence protein to be used is important to prevent the effect of cell-type-specific side effect of some fluorescence proteins, e.g., EGFP.
The NHEJ repair pathway, which is responsible for repairing a major fraction of DSBs, requires Ku80 and XRCC4 [17]. In this study, our results showed that Ku80, but not XRCC4, inhibits EGFP-induced cell death in hamster ovary cells. Meanwhile, the stable EGFP expression did not cause DSB accumulation, although DSBs can induce cell death and cell growth arrest. These results strongly suggest that the EGFP-induced cell death pathway is protected by the novel Ku80-dependent and XRCC4-independent defense mechanism in hamster ovary cells. On the basis of these findings, we consider that EGFP-expressing cells are always exposed to stress, which arises with EGFP expression; in such cells, the unknown mechanism that eliminates EGFP-induced cytotoxicity is always functioning. In addition, our results indicate that EGFP expression markedly enhances X-ray-induced cytotoxicity in xrs-6 cells. Therefore, caution should be taken in considering the potential effect of the stress response mechanism, namely Ku80-dependent elimination mechanism of EGFP-induced cytotoxicity, being activated, when using cells in which Ku80 functions normally with or without exogenous stress.
In conclusion, our data demonstrated that Ku80, but not XRCC4, plays an important role in the mechanism of the resistance to EGFP-induced cytotoxicity. These findings and further study of the molecular mechanism underlying EGFP-induced cytotoxicity in Ku80-deficient cells will be useful for the study not only of novel Ku80 functions and the molecular mechanism of cellular response to EGFP-induced stress, but also of the development of next-generation fluorescence proteins.
Acknowledgments
We thank Dr. P. Jeggo for providing xrs-6 cells and Dr. T.D. Stamato for providing the XR-1 cells. We also thank Mr. S. Ikenaga for technical assistance with the immunoblotting. This work was supported in part by Grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary material
Expression of apoptosis-related proteins in xrs-6 cells 2 days after transfection with pEGFP. Total cell lysates from cells following transfection were analyzed by Western blot analysis using an anti-cleaved-caspase-7 or anti-β-actin antibody. β-actin was used as the loading control.
References
- 1.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Shimomura O., Johnson F.H., Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 3.Prasher D.C., Eckenrode V.K., Ward W.W., Prendergast F.G., Cormier M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 4.Chalfie M., Tu Y., Euskirchen G., Ward W.W., Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Inouye S., Tsuji F.I. Aequorea green fluorescent protein. Expression of the gene and fluorescence characteristics of the recombinant protein. FEBS Lett. 1994;341:277–280. doi: 10.1016/0014-5793(94)80472-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G., Gurtu V., Kain S.R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- 7.Cormack B.P., Valdivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 8.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 9.Chudakov D.M., Matz M.V., Lukyanov S., Lukyanov K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 10.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 11.Liu H.S., Jan M.S., Chou C.K., Chen P.H., Ke N.J. Is green fluorescent protein toxic to the living cells. Biochem. Biophys. Res. Commun. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 12.Huang W.Y., Aramburu J., Douglas P.S., Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 2000;6:482–483. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 13.Agbulut O., Coirault C., Niederländer N., Huet A., Vicart P., Hagège A., Puceat M., Menasché P. GFP expression in muscle cells impairs actin–myosin interactions: implications for cell therapy. Nat. Methods. 2006;3:331 doi: 10.1038/nmeth0506-331. [DOI] [PubMed] [Google Scholar]
- 14.Agbulut O., Huet A., Niederländer N., Puceat M., Menasché P., Coirault C. Green fluorescent protein impairs actin–myosin interactions by binding to the actin-binding site of myosin. J. Biol. Chem. 2007;282:10465–10471. doi: 10.1074/jbc.M610418200. [DOI] [PubMed] [Google Scholar]
- 15.Baens M., Noels H., Broeckx V., Hagens S., Fevery S., Billiau A.D., Vankelecom H., Marynen P. The dark side of EGFP: defective polyubiquitination. PLoS One. 2006;20 doi: 10.1371/journal.pone.0000054. 1:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taghizadeh R.R., Sherley J.L. (2008) CFP and YFP, but Not GFP, provide stable fluorescent marking of rat hepatic adult stem cells. J. Biomed. Biotechnol. doi:10.1155/2008/453590 [DOI] [PMC free article] [PubMed]
- 17.Mahaney B.L., Meek K., Lees-Miller S.P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M., Houtsmuller A.B., Gent D.C. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike M., Koike A. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 2008;314:1061–1070. doi: 10.1016/j.yexcr.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Koike M., Yutoku Y., Koike A. Accumulation of Ku70 at DNA double-strand breaks in living epithelial cells. Exp. Cell Res. 2011;317:2429–2437. doi: 10.1016/j.yexcr.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Koike M., Yutoku Y., Koike A. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 2011;412:39–43. doi: 10.1016/j.bbrc.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 22.Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. 2002;43:223–236. doi: 10.1269/jrr.43.223. [DOI] [PubMed] [Google Scholar]
- 23.Hopfner K.P., Putnam C.D., Tainer J.A. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 2002;12:115–122. doi: 10.1016/s0959-440x(02)00297-x. [DOI] [PubMed] [Google Scholar]
- 24.Koike M., Yutoku Y., Koike A. Establishment of hamster cell lines with EGFP-tagged human XRCC4 and protection from low-dose X-ray radiation. J. Vet. Med. Sci. 2012;74:1269–1275. doi: 10.1292/jvms.12-0112. [DOI] [PubMed] [Google Scholar]
- 25.Singleton B.K., Priestley A., Steingrimsdottir H., Gell D., Blunt T., Jackson S.P., Lehmann A.R., Jeggo P.A. Molecular and biochemical characterization of xrs mutants defective in Ku80. Mol. Cell. Biol. 1997;17:1264–1273. doi: 10.1128/mcb.17.3.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike M., Shiomi T., Koike A. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 2001;276:11167–11173. doi: 10.1074/jbc.M010902200. [DOI] [PubMed] [Google Scholar]
- 27.Koike M., Koike A. Establishment and characterization of stable cell lines expressing human Ku80 tagged with enhanced green fluorescent protein. J. Radiat. Res. 2004;45:119–125. doi: 10.1269/jrr.45.119. [DOI] [PubMed] [Google Scholar]
- 28.Mo X., Dynan W.S. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell. Biol. 2002;22:8088–8099. doi: 10.1128/MCB.22.22.8088-8099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koike M., Awaji T., Kataoka M., Tsujimoto G., Kartasova T., Koike A., Shiomi T. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 1999;112:4031–4039. doi: 10.1242/jcs.112.22.4031. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F., Hackett N.R., Lam G., Cheng J., Pergolizzi R., Luo L., Shmelkov S.V., Edelberg J., Crystal R.G., Rafii S. Green fluorescent protein selectively induces HSP70-mediated up-regulation of COX-2 expression in endothelial cells. Blood. 2003;102:2115–2121. doi: 10.1182/blood-2003-01-0049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of apoptosis-related proteins in xrs-6 cells 2 days after transfection with pEGFP. Total cell lysates from cells following transfection were analyzed by Western blot analysis using an anti-cleaved-caspase-7 or anti-β-actin antibody. β-actin was used as the loading control.


