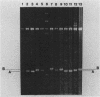
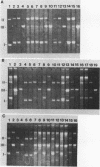
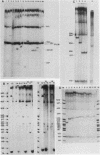
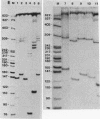
Abstract
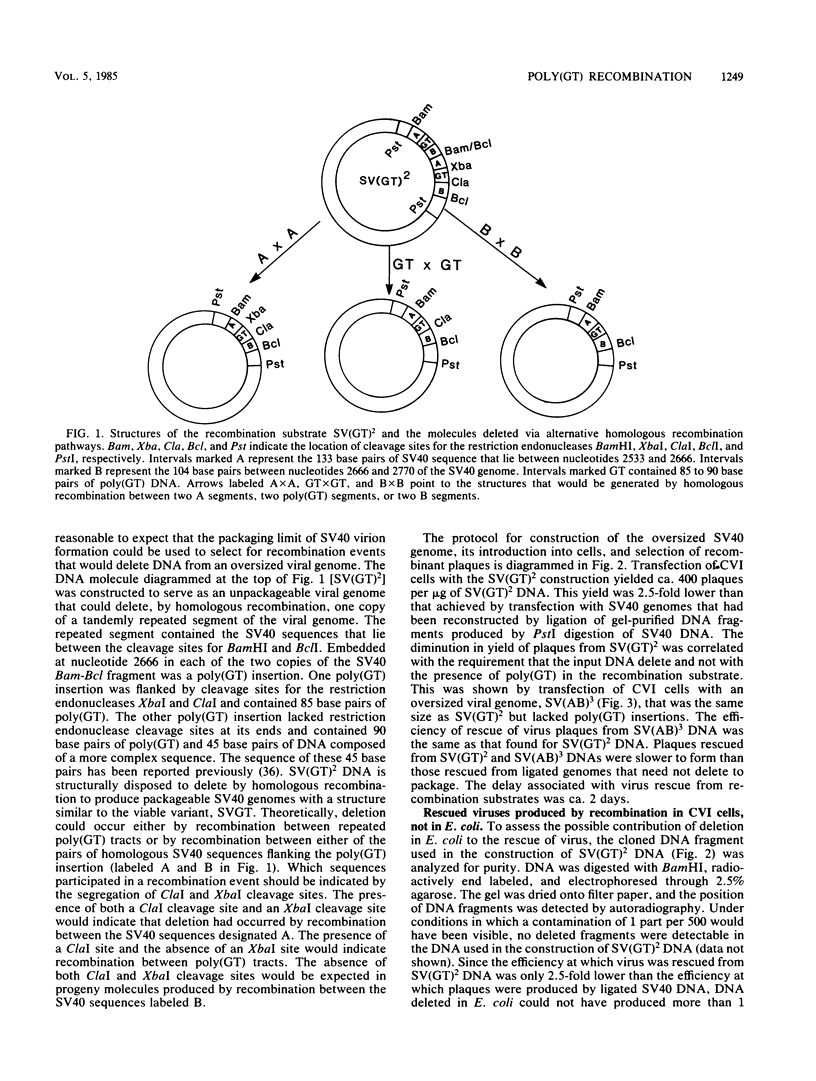
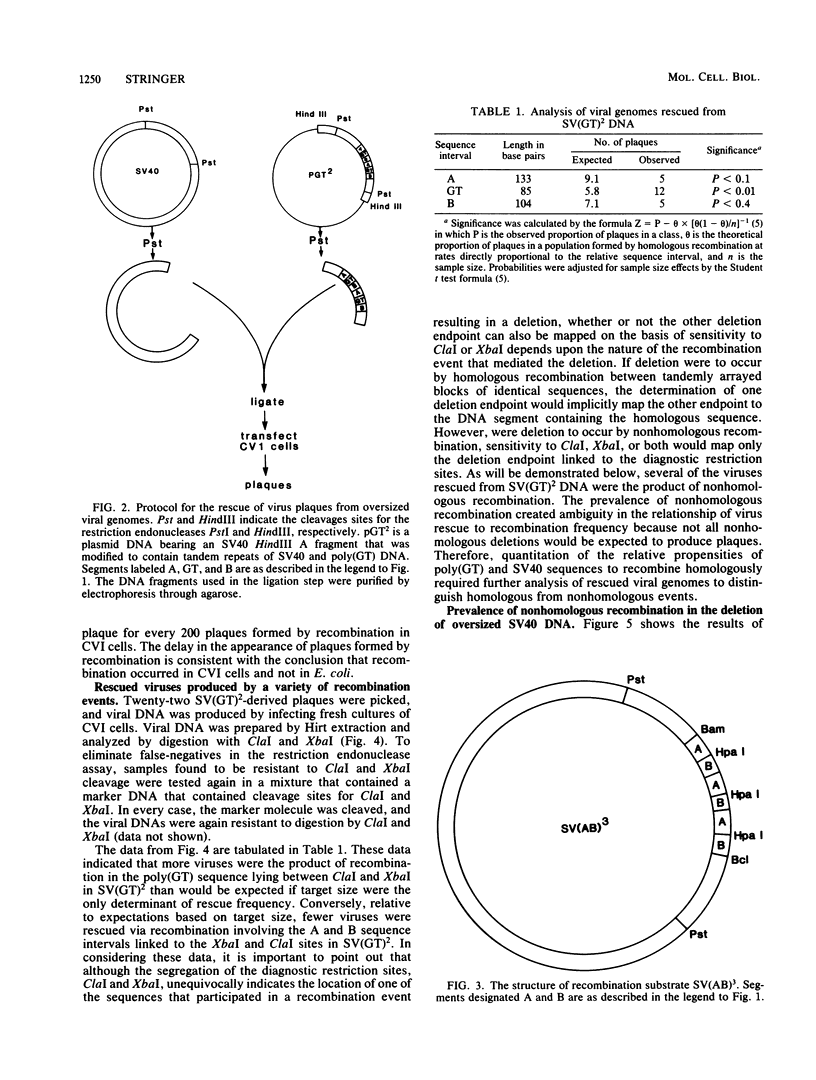
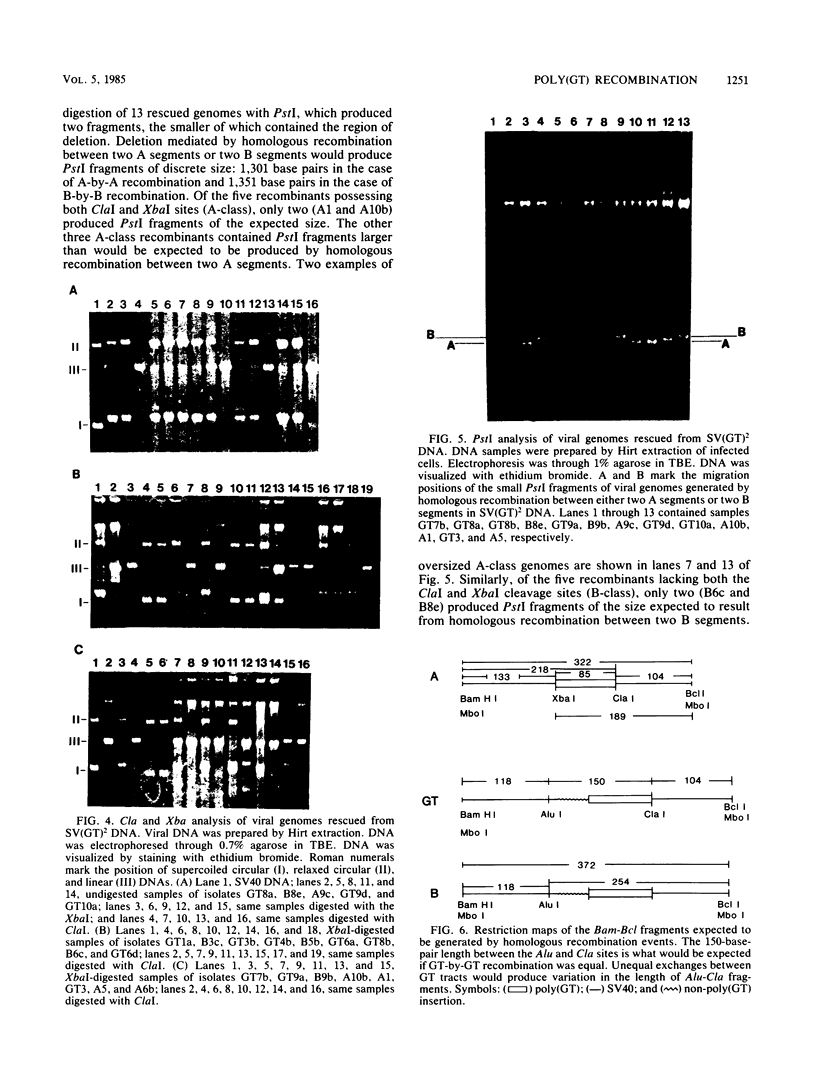
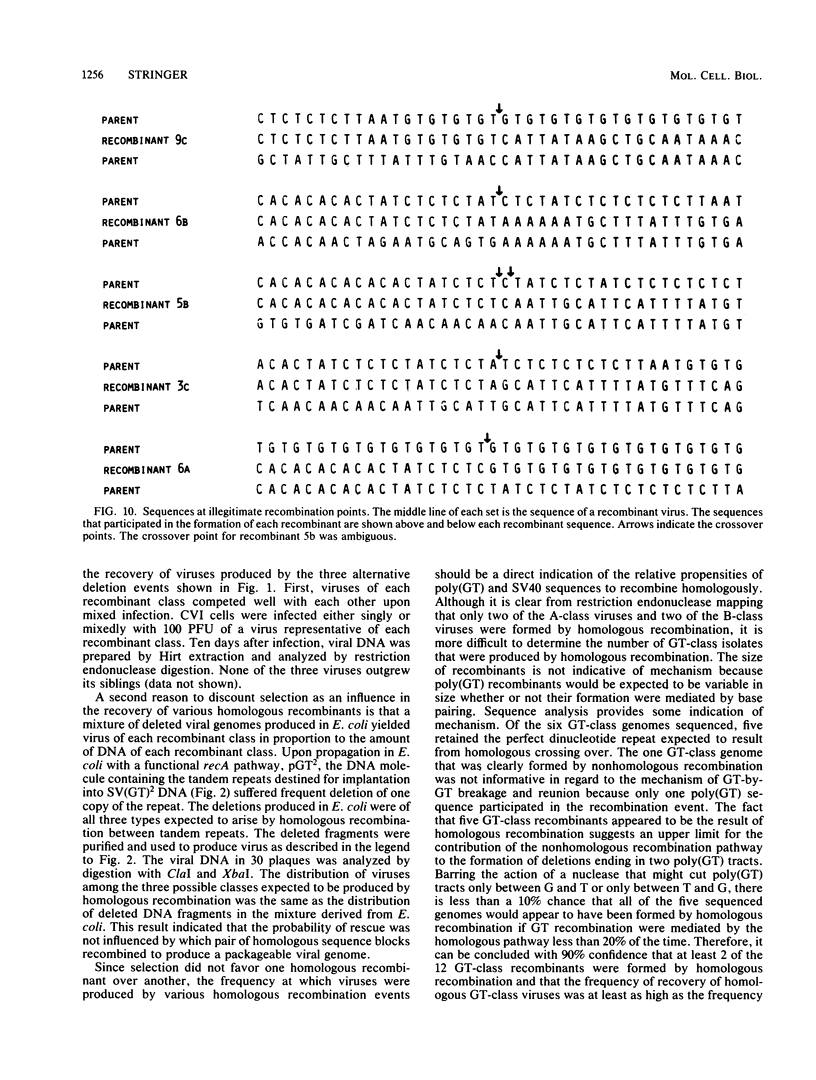
CVI cells were transfected with oversized simian virus 40 (SV40) genomes that could be reduced to packageable size by alternative homologous recombination pathways involving either two polydeoxyguanylic-thymidylic acid X polydeoxycytidylic-adenylic acid (poly[d(GT).d(CA)]; abbreviated hereafter as poly(GT)] tracts or two tracts of homologous SV40 sequence. Plaque-forming viruses rescued by this procedure were found to contain genomes formed by homologous and nonhomologous recombination events. Half of the viable viral DNA molecules recovered were the result of recombination between two tracts of poly(GT). Approximately 20% of the rescued viral genomes were produced by homologous recombination between tracts of SV40 DNA. Nonhomologous recombination involving SV40 sequences was also a major pathway of deletion, producing ca. 30% of the viral plaques. Tracts of poly(GT) generated by recombination were variable in length, suggesting that recombination between poly(GT) tracts was usually unequal. On a per-nucleotide basis, poly(GT) recombination occurred eight times more frequently than did recombination between homologous SV40 DNA. This eightfold difference is the maximum recombinatory enhancement attributable to poly(GT) sequences. Although DNA sequence analysis showed that tracts of poly(GT) generated by recombination retained the alternating G-T repeat motif throughout their length, the contribution of the nonhomologous pathway to poly(GT) recombination cannot be ruled out, and the relative proclivity of a given length of d(GT).d(CA) sequence to undergo homologous recombination is probably less than eight times greater than that of an SV40 sequence of the same length.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brockman W. W., Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: characterization of cloned complementing variants. Virology. 1975 Jul;66(1):36–52. doi: 10.1016/0042-6822(75)90177-4. [DOI] [PubMed] [Google Scholar]
- Brockman W. W., Lee T. N., Nathans D. The evolution of new species of viral DNA during serial passage of simian virus 40 at high multiplicity. Virology. 1973 Aug;54(2):384–397. doi: 10.1016/0042-6822(73)90151-7. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. G., Lefranc M. P., Rabbitts T. H. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984 Mar;36(3):681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Radding C. M. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983 Sep;34(2):647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T., Seidman M., Stollar B. D. Characterization of genomic poly(dT-dG).poly(dC-dA) sequences: structure, organization, and conformation. Mol Cell Biol. 1984 Dec;4(12):2610–2621. doi: 10.1128/mcb.4.12.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions. Nature. 1983 Apr 14;302(5909):632–634. doi: 10.1038/302632a0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Höchtl J., Zachau H. G. A novel type of aberrant recombination in immunoglobulin genes and its implications for V-J joining mechanism. Nature. 1983 Mar 17;302(5905):260–263. doi: 10.1038/302260a0. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Davis M., Sinn E., Patten P., Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981 Dec;27(3 Pt 2):573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Angelides K. J., Holloman W. K. Left-handed DNA and the synaptic pairing reaction promoted by Ustilago rec1 protein. Cell. 1985 Jan;40(1):139–145. doi: 10.1016/0092-8674(85)90317-4. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Synapsis promoted by Ustilago rec1 protein. Cell. 1984 Mar;36(3):593–598. doi: 10.1016/0092-8674(84)90338-6. [DOI] [PubMed] [Google Scholar]
- Lai C. J. Mapping simian virus 40 mutants by marker rescue. Methods Enzymol. 1980;65(1):811–816. doi: 10.1016/s0076-6879(80)65075-7. [DOI] [PubMed] [Google Scholar]
- Liu C. P., Tucker P. W., Mushinski J. F., Blattner F. R. Mapping of heavy chain genes for mouse immunoglobulins M and D. Science. 1980 Sep 19;209(4463):1348–1353. doi: 10.1126/science.6774414. [DOI] [PubMed] [Google Scholar]
- Marvo S. L., King S. R., Jaskunas S. R. Role of short regions of homology in intermolecular illegitimate recombination events. Proc Natl Acad Sci U S A. 1983 May;80(9):2452–2456. doi: 10.1073/pnas.80.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Krystal M., Arnheim N. A member of a new repeated sequence family which is conserved throughout eucaryotic evolution is found between the human delta and beta globin genes. Nucleic Acids Res. 1981 Nov 25;9(22):5931–5947. doi: 10.1093/nar/9.22.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba P. K., Safaya S. K. Similarity of the nucleotide sequences of rat alpha-lactalbumin and chicken lysozyme genes. Nature. 1984 Mar 22;308(5957):377–380. doi: 10.1038/308377a0. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. H., Smithies O. Human globin psi B2 is not a globin-related sequence. Nucleic Acids Res. 1982 Dec 11;10(23):7809–7818. doi: 10.1093/nar/10.23.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Gauss P., Doherty D. H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982 Nov;31(1):25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Construction of a viable simian virus 40 variant that carries a poly[d(GT) . d(CA)] insertion. J Virol. 1985 Feb;53(2):698–701. doi: 10.1128/jvi.53.2.698-701.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., Parker R. C., Varmus H. E., Bishop J. M. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc Natl Acad Sci U S A. 1983 May;80(9):2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasicek T. J., McDevitt B. E., Freeman M. W., Fennick B. J., Hendy G. N., Potts J. T., Jr, Rich A., Kronenberg H. M. Nucleotide sequence of the human parathyroid hormone gene. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2127–2131. doi: 10.1073/pnas.80.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Defined oligomeric SV40 DNA: a sensitive probe of general recombination in somatic cells. Cell. 1980 Aug;21(1):141–148. doi: 10.1016/0092-8674(80)90121-x. [DOI] [PubMed] [Google Scholar]
- Winocour E., Keshet I. Indiscriminate recombination in simian virus 40-infected monkey cells. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4861–4865. doi: 10.1073/pnas.77.8.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]