Abstract
Accumulated evidence suggests that aberrant regulation of δ-catenin leads to pathological consequences such as mental retardation and cognitive dysfunction. This study revealed that 14-3-3ɛ/ζ stabilizes δ-catenin, with different binding regions involved in the interaction. Furthermore, the specific inhibition of the interaction of 14-3-3 with δ-catenin reduced levels of δ-catenin and significantly impaired the capacity of δ-catenin to induce dendritic branching in both NIH3T3 fibroblasts and primary hippocampal neurons. However, the S1094A δ-catenin mutant, which cannot interact with 14-3-3ζ, still retained the capability of inducing dendrogenesis. Taken together, these results elucidate the underlying events that regulate the stability of δ-catenin and δ-catenin-induced dendrogenesis.
Keywords: δ-Catenin, 14-3-3, Dendrogenesis, Neuron, Protein stability
Highlights
▸ Aberrant regulation of δ-catenin in neurons leads to several pathological consequences. 14-3-3ɛ/ζ Interacts with δ-catenin, increasing its stability. ▸ This interaction significantly affects the induction of dendritic branches in NIH 3T3 fibroblasts. ▸ A similar effect was seen in primary hippocampal neurons.
1. Introduction
δ-Catenin was first identified through yeast two-hybrid screening as an interacting molecule with Presenilin-1 (PS-1), the most prominently mutated gene in familial Alzheimer's disease (FAD) patients [1,2]. δ-Catenin is exclusively expressed in neurons, and δ-catenin deficient mice showed severe learning deficits and abnormal synaptic plasticity, suggesting a special role of δ-catenin at the synapse [3]. The interacting proteins of δ-catenin identified so far include PS-1, Cadherins [4], Kaiso [5], Cortactin [6], Sphingosin kinase [7], S-SCAM [8], Erbin [9], Densin-180 [10], ABP, GRIP [11], 14-3-3 [12], and p190RhoGEF [13], implicating that δ-catenin may play diverse roles in cells in addition to its role at the synapse. Interestingly, recent reports showed that δ-catenin is overexpressed in several human tumors [14,15]. In contrast, the hemizygous loss of the chromosomal 5p15.2 region, where the human δ-catenin gene is located, can result in severe mental retardation associated with Cri du Chat syndrome [16], suggesting that the expression of δ-catenin should be tightly regulated in terms of time and location. Even though we have previously demonstrated that PS-1 inhibits δ-catenin-induced cellular branching and promotes δ-catenin processing and turnover [17], few studies have been undertaken to identify the factors that regulate δ-catenin stability in cells.
Although previous studies suggested that 14-3-3 could be a new partner of δ-catenin [12,13], the functional significance of the interaction between δ-catenin and 14-3-3 is not completely understood. 14-3-3 Proteins are a highly conserved family of phospho-serine/threonine binding proteins with molecular weights in the range of 28–33 kD. They are composed of at least seven mammalian isoforms (β, ɛ, δ, ν, σ, τ, ζ) and regulate multiple signaling pathways involved in controlling the cell cycle, cell growth, differentiation, survival, apoptosis, migration and spreading [18–21]. More than 200 binding partners of 14-3-3 have been identified to date. For example, Bad and Forkhead proteins, which are important regulators in cell survival, can be phosphorylated by Akt and lead to the binding of 14-3-3 [22,23]. Most 14-3-3 proteins bind to the phospho-serine/threonine residue on a partner protein when it is phosphorylated by various kinases including protein kinase A, protein kinase C and Akt [21,24,25]. In this study, we found that 14-3-3ɛ/ζ can interact with δ-catenin, which as a result, significantly affects its stability. Transfection with sc138, a specific small peptide inhibitor of 14-3-3 in its substrate interaction, significantly reduced the induction of dendritic branches by δ-catenin in both NIH 3T3 fibroblasts and primary hippocampal neurons. The S1094A mutant of δ-catenin which cannot interact with 14-3-3ζ, however, is capable of inducing dendrogenesis. This suggests that either changes in the level of δ-catenin by another 14-3-3 isoform, i.e. 14-3-3ɛ, interact with δ-catenin or 14-3-3-mediated downstream signals play a key role in the regulation of the dendritic branching of NIH 3T3 fibroblasts and primary hippocampal neurons. Overall, these results suggest that the interaction between δ-catenin and 14-3-3 can act as a key modulator in regulating the stability of δ-catenin, δ-catenin-induced dendrogenesis and spine formation.
2. Material and methods
2.1. Plasmids and antibodies
The construction of δ-catenin full-length (FL-) in pEGFP-C1 has been previously described [26]. The S1094A mutant was made using the QuickChange site-directed mutagenesis kit (Stratagene) with wild-type δ-catenin as a template. sc138 and sc174 constructs were kindly provided by Dr. Haian Fu [27,28]. The antibodies were obtained as follows: anti-δ-catenin (Upstate biotechnology); anti-GFP (Clontech, BD Biosciences); and anti-β-tubulin (Sigma). HA epitope was detected using media from 12CA5 hybridoma.
2.2. Cell culture, transfection and CHX treatment
Mouse embryonic fibroblast (MEF) and NIH 3T3 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C with 5% CO2. The cells were transfected using the calcium phosphate transfection method or with the Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. For the protein stability assays, the cells were transfected, and then were treated with cycloheximide (40 μg/ml, Sigma) 12 h after transfection to inhibit de novoprotein synthesis. The cells were then harvested at different time points as indicated.
Cultured hippocampal neurons were prepared from embryonic day 18 (E-18) fetal Sprague–Dawley rats and were plated on poly-d-lysine coated five 18 mm glass coverslips at a density of 200,000 cells/60 mm dish. The cultures were grown in Neurobasal medium (Gibco) supplemented with 2% B-27, and 0.5 mM l-glutamine. The neurons were transfected at 16 DIV using the calcium-phosphate mediated method.
2.3. Immunoblotting
Immunoprecipitation/immunoblotting was performed as previously described [29].
2.4. Image acquisition and data analysis
The cells were fixed in 4% paraformaldehyde/4% sucrose/PBS for 15 min, washed 2 × 5 min, permeabilized for 5 min in 0.25% Triton X-100/PBS. The images were obtained using an Olympus IX71 microscope (Olympus) with 40× N.A. 1.0 or 60× N.A. 1.4 oil lens using a CoolSNAP-Hq CCD camera (Roper Scientific) driven by MetaMorph imaging software (Universal Imaging Co.). Light from a mercury lamp was shuttered using a VMM1 Uniblitz shutter (Vincent Associates). The number of dendritic branches was analyzed with a sphere, 100 μm in radius, centered at the soma. The number of intersections between the dendritic branches and a sphere was averaged. Statistical analysis was performed using ANOVA and Tukey's HSD post-hoc test. The analysis and quantification of the data were performed using MetaMorph software and SigmaPlot 8.0 (Systat Software). Expression of each construct was confirmed by retrospective immunostaining using specific antibodies, and only immunopositive neurons were included in the analysis. Data analysis was performed in a blinded manner. For sc138 inhibition experiments, primary dendrite length was measured using the REGION MEASUREMENTS and TRACE object functions. The number of primary dendrites and branch points were determined using the REGION MEASUREMENTS and COUNT functions. The data was compared using one-way ANOVA with Tukey's HSD post-hoc test. The data is presented as mean ± SE.
3. Results
3.1. The interaction between δ-catenin and 14-3-3ɛ or 14-3-3ζ increases its stability
The published literature showed that 14-3-3ζ bound directly to Ser-1094 in mouse δ-catenin [12]. In order to confirm the binding between δ-catenin and 14-3-3, a full-length δ-catenin construct tagged with GFP together with either 14-3-3ɛ or 14-3-3ζ tagged with HA was co-transfected into mouse embryonic fibroblast (MEF) cells and examined by immunoprecipitation assay. As shown in Fig. 1A, δ-catenin interacted well with both 14-3-3ɛ and 14-3-3ζ. However, as we have previously demonstrated, the binding domain in δ-catenin with 14-3-3 can be different depending on 14-3-3 isoforms (supplemental data of [13]). The levels of δ-catenin were significantly reduced when the interaction between 14-3-3 and δ-catenin was interrupted using sc138, a small peptide inhibitor of 14-3-3 binding. In contrast, treatment with sc174, a mutant inhibitor, did not show such a decrease (Fig. 1B). The effects of increased amounts of 14-3-3 on the levels of δ-catenin were examined in order to demonstrate that the interaction between δ-catenin and 14-3-3 indeed increases its stability. The results shown in Fig. 1C and D show that 14-3-3ɛ/ζ increased the levels of δ-catenin. Treatment with cycloheximide, a specific protein synthesis inhibitor, showed that 14-3-3 in fact increased the stability of δ-catenin (Fig. 1E). The half-life of δ-catenin in NIH 3T3 fibroblasts was extended from ∼8 to ∼12 h by co-transfecting 14-3-3ɛ/ζ.
Fig. 1.
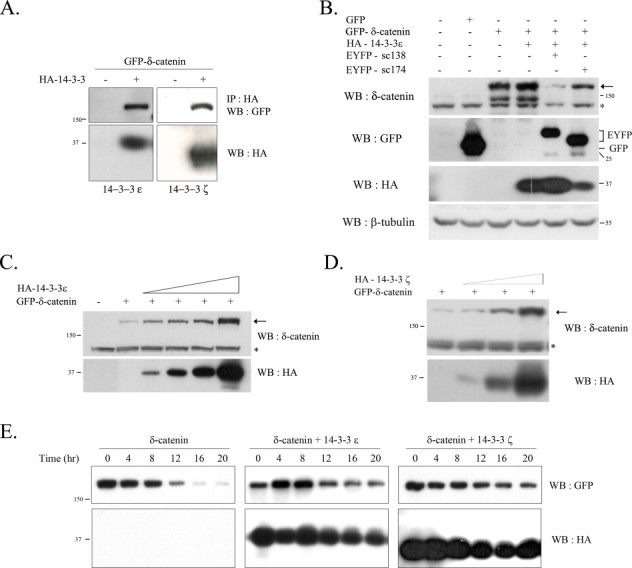
The interaction between δ-catenin and 14-3-3ɛ/ζ increases stability of δ-catenin. (A) The wt MEF cells were transfected with GFP-tagged δ-catenin with HA-14-3-3ɛ (left panel) or HA-14-3-3ζ (right panel), respectively. To examine the binding of δ-catenin and 14-3-3ɛ/ζ, immunoprecipitation was performed with the anti-HA antibody, and blotted with the anti-GFP antibody. (B) The wt MEF cells were co-transfected with GFP-δ-catenin or with both GFP-δ-catenin and HA-14-3-3 as indicated. Some cells were cotransfected with sc138, a specific inhibitor for 14-3-3 substrate binding, or sc174, a non-functional mutant. The expression of GFP-δ-catenin and YFP-sc138/sc174, and HA-14-3-3 was analyzed using the anti-δ-catenin, anti-GFP, and HA antibody, respectively. The bottom panel indicates the loading control (β-tubulin). (C and D) The effects of different dose of 14-3-3ɛ or 14-3-3ζ on the level of δ-catenin. The wt MEF cells were transfected with GFP-δ-catenin together with different doses of HA-14-3-3ɛ (C) or HA-14-3-3ζ (D). In contrast to others, one-tenth (0.1 µg) amount of δ-catenin expression vector was transfected, and for 14-3-3s, 0.1, 0.3, 05, 1.0 µg (for 14-3-3ɛ) and 0.1, 0.5, 1.0 µg (for 14-3-3ζ) were transfected. The level of δ-catenin was examined using the anti-δ-catenin antibody (upper panel; the arrows indicate δ-catenin bands and asterisks indicate non-specific bands), and the expression of 14-3-3ɛ/ζ was analyzed using the HA antibody (bottom panel). (E) The wt MEF cells were transfected with GFP-δ-catenin alone or together with HA-14-3-3ɛ/ζ, and then treated with cycloheximide for different durations after transfection, as indicated. The expression of δ-catenin and 14-3-3ɛ/ζ were analyzed using the anti-GFP antibody (upper panel) and HA antibody (bottom panel), respectively.
We have previously demonstrated that Akt phosphorylates Thr-454 residue on δ-catenin [13]. As 14-3-3 is also one of the targets of Akt, the two mutant 14-3-3ζ constructs, S58A (Akt target site Ser-58 residue is point mutated to Ala) and S58E (Akt target site Ser-58 residue is point mutated to Glu), were used to investigate whether the phosphorylation of 14-3-3 by Akt is essential for its effect on the increased stability of δ-catenin. As shown in Fig. 2, both forms of the mutant 14-3-3ζ, S58A (no phosphorylation but forms dimers) and S58E (phosphorylation-mimic form but no dimers), increased the levels of δ-catenin. This suggests that the phosphorylation and dimerization of 14-3-3 regulated by certain kinase like Akt may not have a significant role in effecting on the increased stability of δ-catenin. However, it is worth notion that the monomeric form of 14-3-3ζ may have a favorable effect on the stabilization of δ-catenin than the dimeric form as an increment of δ-catenin level was higher in some extent in S58E mutant than S58A mutant.
Fig. 2.
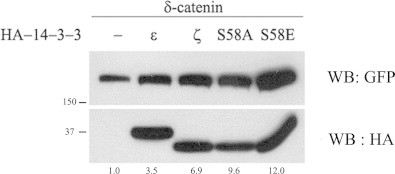
The phosphorylation state at the Ser-58 residue of 14-3-3 affects the level of δ-catenin. The wt MEF cells were transfected with GFP-δ-catenin or with GFP-δ-catenin together with different forms of HA-14-3-3 (epsilon or zeta wt or HA-14-3-3ζ mutants S58A or S58E). The expression of δ-catenin and 14-3-3 protein were detected using the anti-GFP antibody (upper panel) and anti-HA antibody (bottom panel), respectively. The numbers below the lane indicate relative intensity of the δ-catenin bands which is normalized by the level of each 14-3-3 band from two independent experiments.
3.2. The interaction between δ-catenin and 14-3-3 is important for the dendritic branches
As 14-3-3 interaction increased the levels of δ-catenin, we hypothesized that inhibition of 14-3-3 interaction affects the δ-catenin-induced dendritic branching processes. As expected, treatment with okadaic acid, a specific Ser/Thr phosphatase inhibitor, and co-transfection with 14-3-3ζ enhanced δ-catenin-induced dendritic branching. In contrast, transfection with sc138, a specific inhibitor of the 14-3-3 interaction with its substrate, significantly impaired the capacity of full length δ-catenin to induce branching in both NIH 3T3 fibroblasts (Fig. 3A) and primary hippocampal neurons (Fig. 3B). In comparison, treatment with okadaic acid and transfection with 14-3-3 or sc138 did not induce any noticeable morphological changes in GFP-transfected NIH 3T3 fibroblasts (sc138, top panel in Fig. 3A; okadaic acid and 14-3-3, data not shown). In primary hippocampal neurons, there was only a marginal decrease in the number of primary dendrites, and no difference in the primary dendrite length. However, the number of branch points induced by δ-catenin was significantly reduced when the cells were co-transfected with sc138 (22.6 ± 2.1/100 μm for δ-catenin; 3.66 ± 0.6/100 μm for δ-catenin + sc138, p < 0.05). This suggests that the interaction between δ-catenin and 14-3-3 has great importance in inducing dendritic branches, possibly through its enhanced stability and/or recruitment of specific membrane compartments.
Fig. 3.
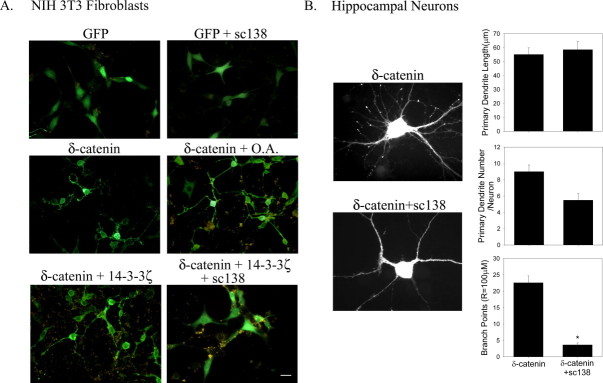
Effects of sc138, a specific peptide inhibitor of the 14-3-3 interaction with its substrate, on the dendrite-like process formation in NIH 3T3 fibroblasts and on dendrogenesis in primary hippocampal neurons. (A) The NIH 3T3 fibroblast cells transfected with GFP-δ-catenin, and were further treated with okadaic acid, a specific Ser/Thr phosphatase inhibitor, or co-transfected with 14-3-3ζ alone or 14-3-3ζ and sc138, a specific inhibitor peptide of 14-3-3 interaction with its substrate. At 24 h post-transfection, the cells were fixed, and a fluorescent image was taken. (B) The hippocampal neurons obtained from embryonic day 18 (E18) were plated onto poly-l-lysine coated coverslips at a density of 60,000 neurons/coverslip. At DIV 16, the neurons were transfected with the full-length RFP-tagged δ-catenin and/or an YFP tagged sc138. The fixed cells were immunostained with anti-δ-catenin Ab and visualized using a Ziess Axiovert S100 microscope. The images were captured, stored, and analyzed using MetaMorph software (Universal Imaging). The data were compared using one-way ANOVA with a Tukey's HSD post-hoc test. The data are presented as mean ± SEM (*p < 0.05).
To examine whether 14-3-3 itself is crucial for δ-catenin-induced dendrogenesis, we generated a δ-catenin S1094A mutant which is reported not to interact with 14-3-3ζ [12] and examined if δ-catenin-induced dendrogenesis can be observed with this mutant. As previously reported, the binding of the δ-catenin S1094A mutant with 14-3-3ζ showed a dramatic reduction compared with that of wild type δ-catenin while the binding of mutant with 14-3-3ɛ remained intact (Fig. 4A). In agreement with our results shown in Fig. 1, only 14-3-3ɛ which can bind to δ-catenin S1094A stabilized the S1094A mutant of δ-catenin suggesting that the regulation of δ-catenin stability by 14-3-3 occurred through their interaction (Fig. 4B). Interestingly, however, this mutant was able to induce dendrogenesis (Fig. 4C) implicating that 14-3-3ζ interaction plays a certain role in regulating δ-catenin stability rather than in the process of dendrogenesis itself. Taken together, these results suggest that interaction of δ-catenin with 14-3-3ɛ/ζ stabilized δ-catenin and stabilized δ-catenin, in turn, can affect dendrite-like process formation.
Fig. 4.
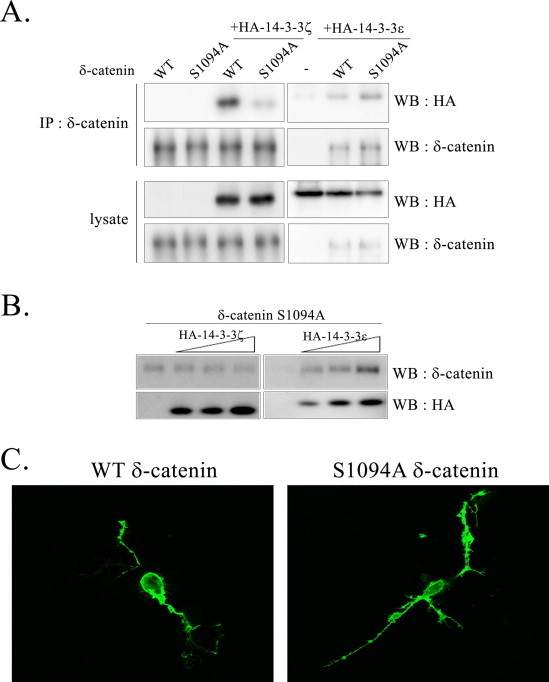
δ-Catenin-induced dendrogenesis of 14-3-3ζ binding null mutant. (A) The wt MEF cells were transfected with indicated plasmids, and immunoprecipitation was performed with the anti-δ-catenin antibody, and blotted with indicated antibody. (B) The wt MEF cells were transfected with GFP-δ-catenin S1094A mutant together with different doses of HA-14-3-3ɛ or HA-14-3-3ζ. In contrast to others, one-tenth (0.1 µg) amount of δ-catenin expression vector was transfected, and for 14-3-3s, 0.1, 0.5, 1.0 µg were transfected. (C) The NIH 3T3 fibroblast cells were transfected either with the wt or S1094 mutant of δ-catenin. At 24 h post-transfection, the cells were fixed, and an image was taken with a confocal microscope.
4. Discussion
In this report, we revealed three findings: (1) both 14-3-3ɛ and 14-3-3ζ interact with δ-catenin; (2) the interaction of 14-3-3ɛ/ζ with δ-catenin increases its stability; and (3) stabilization of δ-catenin by 14-3-3 significantly affects the induction of dendritic branches in both NIH 3T3 fibroblasts and primary hippocampal neurons. As transfection of sc138, a specific inhibitor of 14-3-3 in its substrate interaction, significantly reduced the induction of dendritic branches by δ-catenin in both NIH 3T3 fibroblasts and primary hippocampal neurons, stabilization of δ-catenin by 14-3-3 seems to be essential in δ-catenin-induced dendrogenesis. In fact, the δ-catenin S1094A mutant which lacks the ability to interact with 14-3-3ζ is still capable of inducing dendritic branching in NIH 3T3 fibroblasts (Fig. 4C), suggesting that stabilized δ-catenin by other 14-3-3 isoforms including 14-3-3ɛ may be necessary for δ-catenin-induced dendrogenesis, if not, another 14-3-3-interacting protein may be critical for the dendrogenesis. As we have previously demonstrated, the δ-catenin T454A mutant, a defective form in binding to p190RhoGEF, still interacted with both 14-3-3 isoforms, ɛ and ζ, but did not induce any noticeable morphological changes, suggesting that p190RhoGEF binds directly to the domains containing Thr-454 in δ-catenin in a Akt-phosphorylation-dependent manner. Also, this shows that the association of δ-catenin with p190RhoGEF is essential for the δ-catenin-induced dendrite-like process formation in fibroblasts. Therefore, the interaction of 14-3-3 with δ-catenin is necessary but not sufficient to induce morphologic changes in fibroblasts and neuronal cells. However, the possibility that 14-3-3ɛ has favorable effects on the association between p190RhoGEF and δ-catenin either as a scaffolder for the interaction or by promoting the formation of heterodimers with other 14-3-3 isoforms should be investigated in future researches.
At this time, we do not know how the interaction of 14-3-3ɛ/ζ with δ-catenin increases its stability. Complex formation of 14-3-3ɛ/ζ with δ-catenin may recruit δ-catenin into a specific cellular compartment, which reduces the degradation of δ-catenin. Alternatively, conformational changes of δ-catenin induced by 14-3-3 interaction may cause steric hindrance from its degradation machinery. It should be noted that similar to δ-catenin, the 14-3-3 family is also involved in neuronal development and regulates synaptic plasticity and neurite outgrowth by associating with its various binding partners [30–34]. For example, Drosophilas with single allelic mutations in Leonardo (14-3-3ζ) are poor learners [31], and neurite outgrowth of hippocampal neurons are stimulated by 14-3-3ζ through promoting L1 phosphorylation [32]. Mice deficient in 14-3-3ɛ show defects in brain development and neuronal migration [33], and NR1-knock down mice which has reduced synapse number demonstrated reduction of 14-3-3ɛ together with DISC1 (Disrupted in Schizophrenia-1) [34]. As there are countless 14-3-3 binding partners in cells, it is very speculative to mention that δ-catenin, a newly identified 14-3-3ɛ/ζ binding partner, is a key player in 14-3-3ɛ/ζ-mediated brain development, learning process, and neuronal migration. However, as δ-catenin is abundantly expressed in the brain and has been implicated in cognitive function, future researches are required to answer how the interaction of 14-3-3 with δ-catenin affects its cellular localization, association with other binding partners, and dendrogenesis.
Acknowledgements
This study was supported by the Korea Healthcare Technology R&D project (#A101474), Ministry for Health, Welfare & Family Affairs, and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0022501; 2009-0065189), Republic of Korea (K.K.), and in part by Sunchon National University Research Fund in 2012 and research grant funded by Sunchon Research Center for Natural Medicines to H.K.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Hangun Kim, Email: hangunkim@sunchon.ac.kr.
Kwonseop Kim, Email: koskim@chonnam.ac.kr.
References
- 1.Zhou J., Liyanage U., Medina M., Ho C., Simmons A.D., Lovett M., Kosik K.S. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 2.Tanahashi H., Tabira T. Isolation of human delta-catenin and its binding specificity with presenilin 1. Neuroreport. 1999;10:563–568. doi: 10.1097/00001756-199902250-00022. [DOI] [PubMed] [Google Scholar]
- 3.Israely I., Costa R.M., Xie C.W., Silva A.J., Kosik K.S., Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 4.Lu Q., Paredes M., Medina M., Zhou J., Cavallo R., Peifer M., Orecchio L., Kosik K.S. Delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 1999;144:519–532. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodova M., Kelly K.F., VanSaun M., Daniel J.M., Werle M.J. Regulation of the rapsyn promoter by kaiso and delta-catenin. Mol. Cell. Biol. 2004;24:7188–7196. doi: 10.1128/MCB.24.16.7188-7196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez M.C., Ochiishi T., Majewski M., Kosik K.S. Dual regulation of neuronal morphogenesis by a delta-catenin–cortactin complex and Rho. J. Cell Biol. 2003;162:99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita T., Okada T., Hayashi S., Jahangeer S., Miwa N., Nakamura S. Delta-catenin/NPRAP (neural plakophilin-related armadillo repeat protein) interacts with and activates sphingosine kinase 1. Biochem. J. 2004;382:717–723. doi: 10.1042/BJ20040141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ide N., Hata Y., Deguchi M., Hirao K., Yao I., Takai Y. Interaction of S-SCAM with neural plakophilin-related Armadillo-repeat protein/delta-catenin. Biochem. Biophys. Res. Commun. 1999;256:456–461. doi: 10.1006/bbrc.1999.0364. [DOI] [PubMed] [Google Scholar]
- 9.Laura R.P. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 10.Izawa I., Nishizawa M., Ohtakara K., Inagaki M. Densin-180 interacts with delta-catenin/neural plakophilin-related armadillo repeat protein at synapses. J. Biol. Chem. 2002;277:5345–5350. doi: 10.1074/jbc.M110052200. [DOI] [PubMed] [Google Scholar]
- 11.Silverman J.B., Restituito S., Lu W., Lee-Edwards L., Khatri L., Ziff E.B. Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 2007;27:8505–8516. doi: 10.1523/JNEUROSCI.1395-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackie S., Aitken A. Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J. 2005;272:4202–4210. doi: 10.1111/j.1742-4658.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim H. Delta-catenin-induced dendritic morphogenesis. An essential role of p190RhoGEF interaction through Akt1-mediated phosphorylation. J. Biol. Chem. 2008;283:977–987. doi: 10.1074/jbc.M707158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burger M.J., Tebay M.A., Keith P.A., Samaratunga H.M., Clements J., Lavin M.F., Gardiner R.A. Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancer. Int. J. Cancer. 2002;100:228–237. doi: 10.1002/ijc.10468. [DOI] [PubMed] [Google Scholar]
- 15.Lu Q., Dobbs L.J., Gregory C.W., Lanford G.W., Revelo M.P., Shappell S., Chen Y.H. Increased expression of delta-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Hum. Pathol. 2005;36:1037–1048. doi: 10.1016/j.humpath.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Medina M., Marinescu R.C., Overhauser J., Kosik K.S. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–164. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- 17.Kim J.S. Presenilin-1 inhibits delta-catenin-induced cellular branching and promotes delta-catenin processing and turnover. Biochem. Biophys. Res. Commun. 2006;351:903–908. doi: 10.1016/j.bbrc.2006.10.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muslin A.J., Tanner J.W., Allen P.M., Shaw A.S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 19.Fu H., Subramanian R.R., Masters S.C. 14-3-3 Proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 20.Yaffe M.B., Elia A.E. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 2001;13:131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 21.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta S.R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M.E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 23.Burgering B.M., Medema R.H. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J. Leukoc. Biol. 2003;73:689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- 24.Powell D.W., Rane M.J., Chen Q., Singh S., McLeish K.R. Identification of 14-3-3zeta as a protein kinase B/Akt substrate. J. Biol. Chem. 2002;277:21639–21642. doi: 10.1074/jbc.M203167200. [DOI] [PubMed] [Google Scholar]
- 25.Kovacina K.S., Park G.Y., Bae S.S., Guzzetta A.W., Schaefer E., Birnbaum M.J., Roth R.A. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 26.Kim K., Sirota A., Chen Yh Y.H., Jones S.B., Dudek R., Lanford G.W., Thakore C., Lu Q. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp. Cell Res. 2002;275:171–184. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- 27.Masters S.C., Fu H. 14-3-3 Proteins mediate an essential anti-apoptotic signal. J. Biol. Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 28.Kim D. Regulation of Dyrk1A kinase activity by 14-3-3. Biochem. Biophys. Res. Commun. 2004;323:499–504. doi: 10.1016/j.bbrc.2004.08.102. [DOI] [PubMed] [Google Scholar]
- 29.Kim H., Ki H., Park H.S., Kim K. Presenilin-1 D257A and D385A mutants fail to cleave Notch in their endoproteolyzed forms, but only presenilin-1 D385A mutant can restore its gamma-secretase activity with the compensatory overexpression of normal C-terminal fragment. J. Biol. Chem. 2005;280:22462–22472. doi: 10.1074/jbc.M502769200. [DOI] [PubMed] [Google Scholar]
- 30.Kent C.B. 14-3-3 Proteins regulate protein kinase a activity to modulate growth cone turning responses. J. Neurosci. 2010;30:14059–14067. doi: 10.1523/JNEUROSCI.3883-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philip N., Acevedo S.F., Skoulakis E.M. Conditional rescue of olfactory learning and memory defects in mutants of the 14-3-3zeta gene leonardo. J. Neurosci. 2001;21:8417–8425. doi: 10.1523/JNEUROSCI.21-21-08417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramser E.M., Wolters G., Dityateva G., Dityatev A., Schachner M., Tilling T. The 14-3-3zeta protein binds to the cell adhesion molecule L1, promotes L1 phosphorylation by CKII and influences L1-dependent neurite outgrowth. PLoS One. 2010;5:e13462. doi: 10.1371/journal.pone.0013462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyo-oka K. 14-3-3Epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller–Dieker syndrome. Nat. Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 34.Ramsey A.J. Impaired NMDA receptor transmission alters striatal synapses and DISC1 protein in an age-dependent manner. Proc. Natl. Acad. Sci. USA. 2011;108:5795–5800. doi: 10.1073/pnas.1012621108. [DOI] [PMC free article] [PubMed] [Google Scholar]


