Abstract
Inhibition of Escherichia coli DNA replication by guanosine tetraphosphate (ppGpp) is demonstrated in vitro. This finding is compatible with impairment of the DnaG primase activity by this nucleotide. However, in agreement to previous reports, we were not able to detect a rapid inhibition of DNA synthesis in E. coli cells under the stringent control conditions, when intracellular ppGpp levels increase dramatically. We suggest that the process of ppGpp-mediated inhibition of DnaG activity may be masked in E. coli cells, which could provide a rationale for explanation of differences between ppGpp effects on DNA replication in E. coli and Bacillus subtilis.
Keywords: DNA replication, Guanosine tetraphosphate (ppGpp), In vitro DNA synthesis, Stringent response
Abbreviations: ppGpp, guanosine tetraphosphate; pppGpp, guanosine pentaphosphate.
Highlights
▸ ppGpp, but not DksA, inhibits Escherichia coli DNA replication in vitro. ▸ ppGpp, but not DksA, inhibits E. coli DnaG primase activity in vitro. ▸ Elongation of E. coli DNA replication is not halted during the stringent response. ▸ Effects of ppGpp on DnaG primase activity may be masked in E. coli cells.
Introduction
Guanosine tetraphosphate (ppGpp) is a specific nucleotide playing the role of a signal molecule involved in a global bacterial regulatory response to stress conditions, called the stringent response [1]. Although initially linked solely to amino acid starvation, the stringent response is now recognized as a process connected to various nutritional and environmental stresses [1,2]. For a long time, ppGpp had been considered as a signal molecule occurring exclusively in bacterial cells. However, recent analyses indicated occurrence of homologues of genes coding for enzymes of ppGpp metabolism in various organisms, from bacteria, through protists and plants, to animals, including Homo sapiens [3], implicating possible regulatory roles of ppGpp (in bacteria) or putative related nucleotides (in eukaryotes) in organisms from various domains of life. In addition to the stress response, ppGpp was reported to be one of the main regulators of the growth rate control in a model Gram-negative bacterium Escherichia coli [4]. Furthermore, recent studies on a model Gram-positive bacterium Bacillus subtilis led to the proposal that ppGpp is required to maintain physiological GTP levels even in the absence of starvation [5]. These recently published reports strongly suggest a global regulatory role for ppGpp, which is not restricted to conditions of nutrient limitation (when levels of this nucleotide are highly elevated).
In bacteria, shortly after the onset of starvation conditions, ppGpp is produced in large amounts [1]. In E. coli, this nucleotide directly interacts with RNA polymerase and modulates significantly its transcriptional properties. Therefore, dramatic changes in transcription of many genes are observed during the stringent response, and they are considered the primary effects of this cellular response, despite the fact that considerable changes in regulation of various cellular processes occur in starved cells [1,6,7]. The RNA polymerase-associated protein, DksA, was shown to be indispensable for the stringent response, and its role was suggested to enhance in vivo and in vitro effects of ppGpp, thus, DksA was proposed to be a co-factor of this regulation [8,9].
One of crucial processes which are severely affected under conditions of the stringent response is DNA replication. Specific inhibition of DNA synthesis was first described for chromosomes of B. subtilis and E. coli [10], but subsequent studies indicated that such a phenomenon occurs also in various other replicons (for reviews see Refs. [11,12]). Interestingly, for E. coli chromosome, the (p)ppGpp-mediated inhibition of replication was postulated to occur only at the initiation stage (Ref. [11] and references therein), whereas in B. subtilis, an arrest of the chromosomal replication forks was reported [10,13], strongly suggesting that ppGpp may impair DNA replication elongation. Subsequent studies demonstrated also a ppGpp-dependent cell cycle arrest at the stage of E. coli chromosome segregation [14], but no considerable inhibition of replication elongation could be detected in this bacterium.
A new light on the mechanism of ppGpp-mediated inhibition of DNA replication was shed by finding that B. subtilis primase activity is impaired by direct binding of this nulecotide [15]. These results suggested the molecular mechanism of negative regulation of replication elongation based on ineffective synthesis of primers. One could speculate that this might be potentially a reason for differences between effects of ppGpp on DNA replication in B. subtilis and E. coli. However, results of subsequent experiments, obtained by our group [16] and corroborated recently by others [17], led to the conclusion that E. coli DnaG primase is also directly inhibited by ppGpp; this inhibition occurs most probably due to direct obstruction of the primase active site by ppGpp [17]. Therefore, the question appeared whether ppGpp-mediated negative regulation of DNA replication elongation may also occur in E. coli. To address this question, we have studied effects of ppGpp on E. coli DNA replication in vitro in comparison to effects of the stringent response on DNA synthesis in vivo.
Results
Until now, effects of ppGpp on E. coli DNA replication were tested in vivo, using stringent (wild-type) and relaxed (not able to produce ppGpp in amino acid-starved cells) strains [10,18,19]. It was speculated that ppGpp may influence oriC-initiated replication initiation indirectly, through changes in efficiency of transcription from promoters whose functions are important in either expression of genes coding for replication proteins or in transcriptional activation of the origin [18,20], similarly to the mechanism actually described for plasmids derived from bacteriophage λ [21]. Nevertheless, since results of those experiments strongly suggested that E. coli chromosome replication is inhibited during the stringent response at the stage of initiation rather than elongation, we have tested effects of ppGpp on E. coli DNA replication in vitro.
We have employed a semi-purified in vitro replication system, in which a cellular fraction containing all proteins necessary for the replication process (called Fraction II) is used [22]. We found a marked inhibition of DNA synthesis in vitro in the presence of ppGpp (Fig. 1). These results are compatible with the ppGpp-mediated inhibition of DnaG primase activity, reported previously [16] and confirmed in this work (Fig. 2).
Fig. 1.
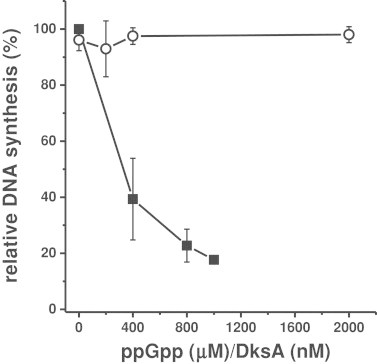
Effects of ppGpp and DksA on in vitro DNA replication. The reactions were performed in the presence of the Fraction II from wild type bacteria and increasing ppGpp concentrations (closed squares), and the Fraction II isolated from the dksA mutant and increasing concentrations of DksA (open circles). The value obtained in experiments with [3H]thymidine incorporation without additional factors was set as 100%. This value corresponds to 68 pmol of synthesized DNA. Mean values from three experiments with error bars representing SD are shown.
Fig. 2.
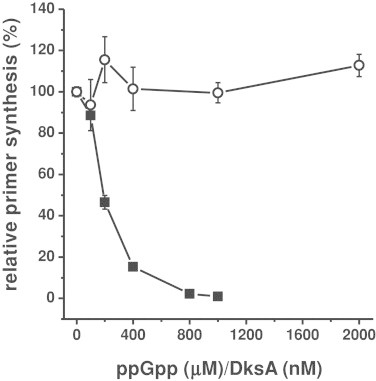
Effects of ppGpp and DksA on the DnaG primase activity. Primer synthesis was performed by DnaG in the presence of either ppGpp (closed squares) or DksA (open circles). The synthesis with no additional factors was set as 100%. Mean values from three independent experiments with error bars representing SD are shown.
Since DksA is considered as a co-factor of the stringent response, we asked whether it can affect DNA replication. The addition of purified DksA protein to the in vitro replication assay showed no effect of this protein (Fig. 1). Contrary to ppGpp, this protein also did not inhibit E. coli DnaG primase activity in vitro (Fig. 2).
In the light of the results of in vitro experiments, we have investigated kinetics of DNA replication in E. coli cells under conditions of amino acid starvation. Agents resulting in inhibition of DNA replication elongation cause a quick impairment in incorporation of labeled precursors, which is exemplified by the effects of the presence of DNA-intercalating antibiotic, mitomycin C (Fig. 3A). On the other hand, if only replication initiation is affected, minor effects on DNA synthesis can be observed shortly after induction of the inhibiting conditions. In fact, such a phenomenon was observed in amino acid-starved stringent strain (Fig. 3A), which massively produced ppGpp under these conditions (Fig. 3C), and whose growth was rapidly inhibited upon the starvation onset (Fig. 3B). A lack of both ppGpp synthetases (RelA and SpoT proteins) in the ppGpp-null strain leads to inability of ppGpp production (Fig. 3B) but did not influence DNA synthesis in starved and unstarved E. coli cells (Fig. 4). These results corroborate previously reported data [18] suggesting that ppGpp may influence E. coli chromosome replication in vivo only at the initiation stage.
Fig. 3.
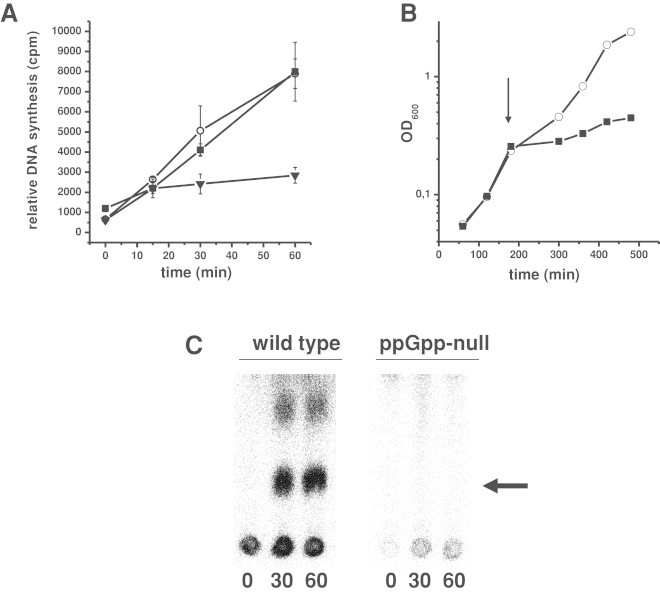
Effects of amino acid starvation and mitomycin C on in vivo DNA synthesis (A), bacterial growth (B), and ppGpp production (C) in E. coli. Panel A: [3H]thymidine-labelled DNA (quantified in cpm) was synthesized with no addition (open circles), with 1 mg/ml l-valine (closed squares) or with 1 mg/ml mitomycin C (closed triangles). The results are mean values from three independent experiments with SD indicated. Panel B: growth of E. coli cell culture was monitored with no addition (open circles) and with 1 mg/ml l-valine (closed squares) added at the time indicated by arrow (note that this time corresponds to time = 0 at panel A). Results of a representative experiment are shown. Panel C: the thin layer chromatography showing ppGpp accumulation in the wild type and ppGpp-deficient strains after the addition of 1 mg/ml l-valine. Arrow indicates the spot corresponding to ppGpp.
Fig. 4.
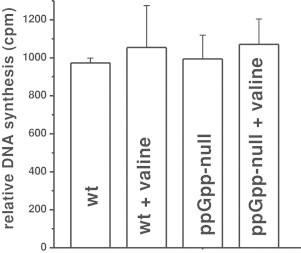
The effect of ppGpp accumulation on DNA synthesis in vivo. [3H]thymidine incorporation was measured in wild type and ppGpp-null strains in the absence or in the presence of 1 mg/ml l-valine. The results show the DNA synthesis 30 min after l-valine addition. Mean values from three independent experiments with error bars representing SD are shown.
Discussion
While the stringent response is a global response of vast majority of bacteria to nutritional stresses, it appears that specific regulatory mechanisms, mediated by ppGpp, the alarmone of this response, may differ between various species [1]. One example is DNA replication, the crucial cellular process, which is inhibited under stringent response conditions in both E. coli and B. subtilis, while its different stages are affected in these bacteria. It is generally accepted that replication initiation is specifically inhibited in E. coli, while the elongation process is affected in B. subtilis [13,18,23].
The discovery that B. subtilis primase is inhibited by ppGpp [15] implied that this may be a major mechanism for replication elongation impairment, which could also distinguish the regulatory processes occurring in B. subtilis and E. coli. However, E. coli primase was subsequently shown to be also inhibited by ppGpp [16,17].
In this report, we have demonstrated for the first time that ppGpp can directly inhibit E. coli DNA replication in vitro. Although Fraction II, used in our experimental system, contains some RNA polymerase molecules, indirect effects of ppGpp on in vitro DNA replication, through modulating transcription efficiency, are unlikely since neither gene expression nor transcriptional activation of oriC are required under these conditions [24]. Therefore, we suggest that the observed impairment of DNA synthesis in the presence of ppGpp, as shown in Fig. 1, may be caused by depression of the DnaG primase activity (Fig. 2). In this light, it is intriguing why replication elongation is apparently unaffected in E. coli cells during the stringent response (Fig. 3). One possibility is that in vivo there is/are factor(s) masking effects of ppGpp on DnaG primase by preventing its binding to this protein. Such factor(s) would be absent in our in vitro assays for measurements of kinetics of primase activity and DNA synthesis. Another hypothesis which may explain the mechanism of masking the ppGpp potential to impair DnaG activity is that the primase is not efficiently inhibited in E. coli cells due to competition for binding of ppGpp to this protein and to RNA polymerase. There are many (2000–3000) RNA polymerase holoenzymes in bacterial cells [25], which could outcompete DnaG primase molecules (about 50 molecules per cell) for binding of ppGpp. This might cause a lack of inhibition of primer synthesis and unrestricted DNA replication elongation even at high levels of this nucleotide. In this light it is worth mentioning that B. subtilis RNA polymerase does not bind ppGpp, and transcription inhibition in starved cells of this bacterium is based rather on changes in nucleotide pools [26,27]. Therefore, one might speculate that in B. subtilis, ppGpp is responsible for blocking the primase activity, while in E. coli, ppGpp fails to inhibit DnaG sufficiently strongly to stop replication elongation when it is involved mostly in interactions with RNA polymerase.
Although one might suppose that differences between effects of ppGpp on DNA replication in B. subtilis and E. coli cell could arise from higher sensitivity to this alarmone of DnaG primase from the former bacterium relative to the latter one, previously published results strongly suggest that it is not the case. Namely, 50% inhibition of in vitro primer formation was observed at 0.5 mM ppGpp for B. subtilis DnaG primase [15], and at 0.2 mM ppGpp for the E. coli enzyme [16].
The obvious question is whether ppGpp concentrations used in in vitro experiments are relevant to in vivo conditions. This problem has been addressed in previous studies on ppGpp-mediated inhibition of DnaG activity [16]. Since direct measurement of ppGpp concentrations in cells is technically challenging, this parameter has been calculated on the basis of estimation of ppGpp amount per dry cell weight or moles of this compound per optical density (OD) of bacterial culture. Namely, in exponentially growing E. coli cultures, the levels of ppGpp were reported to be 55–76 nmol/g of dry cell weight or 3–40 pmol/OD [28,29]. Taking these values, and considering that bacterial cells contain about 30% dry mass, and OD600 = 1 of an E. coli culture corresponds roughly to 109 cells per ml, one may calculate a possible ppGpp concentration in the cytoplasm of exponentially growing bacteria to be about a few μM. Since after induction of the stringent response the ppGpp level increases about 20–100 times (Ref. [1] and Fig. 3C in this report), concentrations of this nucleotide in cells may be in ranges of those which affected primer synthesis (Fig. 2) and DNA replication (Fig. 1) in vitro.
Methods
Bacterial strains and plasmids
E. coli MG1655 strain and its relA spoT (ppGpp-null) derivative [30] were used. The C600 strain [31] was also employed, and the DksA-deficient derivative of this strain was constructed by P1 transduction from the dksA strain [32]. Plasmid pBSoriC was employed as a template for in vitro replication [33].
Proteins and nucleotides
Cellular fraction of replication proteins (Fraction II) was purified as described previously [22]. The method of purification of DnaG primase has already been described [34]. DksA was purified according to the previously reported procedure [35]. Nucleotides and [3H]thymidine were purchased from Fermentas Bioscience and Hartmann Analytic, respectively. ppGpp was purified as described previously [36].
In vitro DNA replication
The in vitro DNA replication assay was performed essentially as described [22], but plasmid pBSoriC was used as a template.
DnaG primase activity
The primase activity was assessed as described previously [16].
Measurement of DNA synthesis in vivo
The assay was performed essentially as described previously [37] with modifications concerning growth conditions. Bacteria were cultivated in the MM minimal medium [38] supplemented with 1 μg/ml non-labeled thymidine and 5 μCi/ml [3H]thymidine. The isoleucine starvation was induced by addition of l-valine to the final concentration of 1 mg/ml.
Acknowledgments
We are grateful to Aleksandra Gołąb and Dr. Anna Szambowska for their help in Fraction II purification, and to Dr. Mike Cashel for discussion. This work was supported by National Science Center (Poland) (Project Grant No. 2011/02/A/NZ1/00009 to G.W.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Potrykus K., Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Szalewska-Palasz A., Johansson L.U., Bernardo L.M., Skärfstad E., Stec E., Brännström K., Shingler V. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J. Biol. Chem. 2007;22:18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson G.C., Tenson T., Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potrykus K., Murphy H., Philippe N., Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriel A., Bittner A.N., Kim S.H., Liu K., Tehranchi A.K., Zou W.Y., Rendon S., Chen R., Tu B.P., Wang J.D. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell. 2012;48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterji D., Fujita N., Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;15:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 7.Toulokhonov I.I., Shulgina I., Hernandez V.J. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta’-subunit. J. Biol. Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- 8.Paul B.J., Ross W., Gaal T., Gourse R.L. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 9.Paul B.J., Berkmen M.B., Gourse R.L. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine A., Vannier F., Dehbi M., Henckes G., Seror S.J. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J. Mol. Biol. 1991;219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- 11.Węgrzyn G. Replication of plasmids during bacterial response to amino acid starvation. Plasmid. 1999;41:1–16. doi: 10.1006/plas.1998.1377. [DOI] [PubMed] [Google Scholar]
- 12.Węgrzyn G., Węgrzyn A. Stress responses and replication of plasmids in bacterial cells. Microb. Cell Fact. 2002;1:2. doi: 10.1186/1475-2859-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine A., Autret S., Seror S.J. A checkpoint involving RTP, the replication terminator protein, arrests replication downstream of the origin during the stringent response in Bacillus subtilis. Mol. Microbiol. 1995;15:287–295. doi: 10.1111/j.1365-2958.1995.tb02243.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferullo D.J., Lovett S.T. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J.D., Sanders G.M., Grossman A.D. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciag M., Kochanowska M., Lyzeń R., Wegrzyn G., Szalewska-Pałasz A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Rymer R.U., Solorio F.A., Tehranchi A.K., Chu C., Corn J.E., Keck J.L., Wang J.D., Berger J.M. Binding mechanism of metal NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zyskind J.W., Smith D.W. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992;69:5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber G., Ron E.Z., Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr. Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- 20.Chiaramello A.E., Zyskind J.W. Coupling of DNA replication to growth rate in Escherichia coli: a possible role for guanosine tetraphosphate. J. Bacteriol. 1990;172:2013–2019. doi: 10.1128/jb.172.4.2013-2019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szalewska-Pałasz A., Wegrzyn A., Herman A., Wegrzyn G. The mechanism of the stringent control of lambda plasmid DNA replication. EMBO J. 1994;13:5779–5785. doi: 10.1002/j.1460-2075.1994.tb06916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narajczyk M., Barańska S., Szambowska A., Glinkowska M., Wegrzyn A., Wegrzyn G. Modulation of lambda plasmid and phage DNA replication by Escherichia coli SeqA protein. Microbiology. 2007;153:1653–1663. doi: 10.1099/mic.0.2006/005546-0. [DOI] [PubMed] [Google Scholar]
- 23.Levine A., Autret S., Seror S.J. A checkpoint involving RTP, the replication terminator protein, arrests replication downstream of the origin during the stringent response in Bacillus subtilis. Mol. Microbiol. 1995;15:287–295. doi: 10.1111/j.1365-2958.1995.tb02243.x. [DOI] [PubMed] [Google Scholar]
- 24.Crooke E. DNA synthesis initiated at oriC: in vitro replication reactions. Methods Enzymol. 1995;262:500–506. doi: 10.1016/0076-6879(95)62041-9. [DOI] [PubMed] [Google Scholar]
- 25.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F.C., editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. American Society for Microbiology Press; Washington, DC, USA: 1996. pp. 1553–1569. [Google Scholar]
- 26.Krasny L., Gourse R.L. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tojo S., Satomura T., Kumamoto K., Hirooka K., Fujita Y. Molecular mechanism underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J. Bacteriol. 2008;190:6143–6147. doi: 10.1128/JB.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neubauer P., Ahman M., Tornkvist M., Larsson G., Enfors S.O. Response of guanosine tetraphosphate to glucose fluctuations in fedbatch cultivations of Escherichia coli. J. Biotechnol. 1995;43:195–204. doi: 10.1016/0168-1656(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 29.Slominska M., Neubauer P., Wegrzyn G. Regulation of bacteriophage λ development by guanosine 5′-diphosphate-3′-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- 30.Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 31.Appleyard R.K. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang P.J., Craig E.A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J. Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker T.A., Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA–DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 34.Tougu K., Peng H., Marians K.J. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- 35.Aberg A., Shingler V., Balsalobre C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 2006;60:1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- 36.Cashel M. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal. Biochem. 1974;57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 37.Ulanowska K., Tkaczyk A., Konopa G., Wegrzyn G. Differential antibacterial activity of genistein arising from global inhibition of DNA, RNA and protein synthesis in some bacterial strains. Arch. Microbiol. 2006;184:271–278. doi: 10.1007/s00203-005-0063-7. [DOI] [PubMed] [Google Scholar]
- 38.Węgrzyn G., Neubauer P., Krueger S., Hecker M., Taylor K. Stringent control of replication of plasmids derived from coliphage λ. Mol. Gen. Genet. 1991;225:94–98. doi: 10.1007/BF00282646. [DOI] [PubMed] [Google Scholar]


