Abstract
Polymorphisms in the cytochrome P450 1B1 (CYP1B1) and glutathione S-transferase (GST) drug metabolic enzymes, which are responsible for metabolic activation/detoxification of estrogen and environmental carcinogens, were analyzed for their association with breast cancer risk in 541 cases and 635 controls from a North Carolina population. Each polymorphism, altering the catalytic function of their respective enzymes, was analyzed in Caucasian and African-American women. As reported in previous studies, individual polymorphisms did not significantly impact breast cancer risk in either Caucasian or African-American women. However, African-American women exhibited a trend towards a protective effect when they had at least one CYP1B1 119S allele (OR=0.53; 95% CI=0.20–1.40) and increased risk for those women harboring at least one CYP1B1 432V allele (OR=5.52; 95% CI=0.50–61.37). Stratified analyses demonstrated significant interactions in younger (age ≤60) Caucasian women with the CYP1B1 119SS genotype (OR=3.09; 95% CI=1.22–7.84) and younger African-American women with the GSTT1 null genotype (OR=4.07; 95% CI=1.12–14.80). A notable trend was also found in Caucasian women with a history of smoking and at least one valine allele at GSTP1 114 (OR=2.12; 95% CI=1.02–4.41). In Caucasian women, the combined GSTP1 105IV/VV and CYP1B1 119AA genotypes resulted in a near 2-fold increase in risk (OR=1.96; 95% CI=1.04–3.72) and the three way combination of GSTP1 105IV/VV, CYP1B1 119AS/SS and GSTT1 null genotypes resulted in an almost 4-fold increase in risk (OR=3.97; 95% CI=1.27–12.40). These results suggest the importance of estrogen/carcinogen metabolic enzymes in the etiology of breast cancer, especially in women before the age of 60, as well as preventative measures such as smoking cessation.
Keywords: breast neoplasms, cytochrome P450, glutathione S-transferase
Introduction
Breast cancer is the second leading cause of cancer-related deaths among American women, with ~40,500 deaths expected this year alone (1). As less than half of breast cancer cases can be accounted for by known risk factors (2), there is a great need for continuing research on the etiological basis of breast carcinogenesis.
The potential involvement of environmental carcinogens in breast carcinogenesis is supported by in vitro and in vivo animal studies (3,4). Typically, the parent compound itself is inert although it can produce reactive metabolites that bind covalently to DNA following phase I enzyme metabolism. Breast tissue has been shown to be capable of this metabolic activation (5,6). Environmental contaminants are not the only source of reactive metabolites and the importance of endogenous estrogen exposure in lifetime breast cancer risk is well established. Estrogen can have an initiating as well as a promoting effect in breast carcinogenesis, through its DNA damaging capabilities as well as its ability to stimulate proliferative activity in mammary epithelial cells (7). Circulating estrogens are not the only source of exposure as breast tissue is capable of estrogen synthesis (8,9), creating a region of localized exposure (10).
The metabolism of environmental carcinogens and estrogen is carried out in part by the phase I and phase II enzymes. The phase I enzyme, cytochrome P450 1B1 (CYP1B1), is highly expressed in breast tissue (11) and plays an important role in estrogen as well as polycyclic aromatic hydrocarbon (PAH) and arylamine metabolism (12,13). Its predominant 4-hydroxylation of estrogen (6) results in the production of the ultimate carcinogenic metabolite, the depurinating quinone, as well as reactive oxygen species as reviewed by Cavalieri et al (14). It has recently been shown in vitro that CYP1B1 is the enzyme involved in the metabolism of estradiol to guanine and adenine adduct forming compounds, supporting its involvement in mammary carcinogenesis (15). Following oxidative metabolism and the activation by phase I enzymes, the phase II enzymes mediate detoxification of the reactive phase I metabolites. Glutathione S-transferases (GSTs) - such as GSTM1, GSTT1 and GSTP1-conjugate electrophilic compounds with glutathione rendering them less carcinogenic. Genetic polymorphisms, which can alter the activation and detoxification capabilities of these enzymes, could potentially influence the individual risk for breast cancer, as they may increase the amount of activated metabolites.
A single nucleotide polymorphism (SNP) in the CYP1B1 gene, which results in an amino acid substitution of L432V in the heme binding region, has been reported to alter the catalytic activity of the enzyme, producing a higher ratio of 4-OH to 2-OH catecholestrogen metabolites (16,17). However, other studies have not confirmed these findings (18). The A119S SNP does not appear to have as great a singular impact on the enzyme kinetics related to catecholestrogen metabolism (19). Haplotype analyses of the 119S and 432L allele, however, demonstrated a 1.2–1.5-fold increase in the activation of some PAHs compared to three other haplotypes (17), although the codon 48 and 119 linkage was not considered.
Polymorphisms in the phase II enzymes GSTM1, GSTT1, and GSTP1 also have known functional implications. The polymorphisms in GSTM1 and GSTT1 consist of deletions of the genes, resulting in a loss of expression and therefore loss of function (20). Individuals with the null alleles have a decreased capacity to metabolize the enzyme's respective substrates. The GSTP1 gene has two SNPs, I105V and A114V, which can affect enzyme activity depending on the substrate (20).
As variations in the ability to metabolize estrogen and carcinogens may be part of breast cancer etiology, the association of these polymorphisms with breast cancer risk has been the focus of multiple studies. Associations with breast cancer have been found in some studies (21,22), although these findings have generally not been confirmed by other laboratories (23–25). In some studies, analyzing individual genotypes did not demonstrate an association with risk for breast cancer, but including combinations of genotypes (26,27) or genotypes and menopausal status (28) demonstrated an increase in risk for breast cancer. Subpopulation analyses, such as in post-menopausal women, demonstrated an association with the GSTM1 polymorphism and breast cancer (27,28). A pooled meta-analysis found a slight increase in risk associated with the CYP1B1 432VV genotype (29).
Gene/environmental interaction studies have produced notable results. Studies focusing on CYP1B1 SNPs showed that the 432V allele was associated with breast cancer when smoking, body mass index (BMI), or hormone replacement therapy use were included in French and Finnish (30,31), Turkish (32) and Swedish (33) populations, respectively. Interactions with the phase II enzymes and smoking (22,34), alcohol consumption (27,35) and family history (36) have also been noted in these studies, though have not been confirmed in other laboratories (24,25,37). In light of these findings, this study utilized a case-control study design to determine if metabolic enzyme polymorphisms identified in Caucasian and African-American women interact with age, family history, smoking and each other to influence breast cancer risk.
Materials and methods
Study populations
The population in this study is similar to one previously described (38). Breast cancer cases were initially recruited at the Breast Care Center at the North Carolina Baptist Hospital with additional cases recruited at the Moses H. Cone Memorial Hospital and the University of North Carolina (UNC) at Chapel Hill, Lineberger Comprehensive Cancer Center from ongoing studies conducted from November 1998 to May 2004. Cancer-free controls were recruited from women undergoing routine mammography screening at the Breast Screening and Diagnostic Center of the Comprehensive Cancer Center at Wake Forest University Baptist Medical Center. Controls were frequency-matched to cases by age (±5 years) and race. Controls were excluded if they had any previous history of cancer, chronic inflammatory diseases and abnormal current mammogram results. Eligibility requirements for cases and controls were designated as at least 18 years of age, English speaking, capable of comprehending informed consent and no history of other cancers, including skin cancer. Each participant was given a detailed description of the study protocol and signed her informed consent as approved by each of the institution's Institutional Review Boards. Following consent, a 20 ml blood sample was collected and each woman was given a self-administered questionnaire regarding demographic information, established breast cancer risk factors, medical and family history. The case-control status was confirmed by medical records and pathology reports.
Genotype analysis
DNA was isolated from each blood sample using the QIAamp® DNA Blood mini kit (Qiagen, Valencia, CA), as described in the manufacturer's protocol, for use in molecular genotyping of the metabolic enzymes. Two hundred microliters of blood was eluted in a final volume of 200 µl that, according to the manufacturer, yields 15–60 ng/µl of DNA. DNA from UNC breast cancer patient samples were obtained from lymphoblastoid cell lines as described previously (39). Paraffin-embedded tissue (PET) samples from breast cancer patients were used for DNA isolation in the event there was insufficient DNA isolated from the blood samples. In these cases, tissue from the block was de-paraffinized and digested in a buffer as described previously (40).
Each genotype was analyzed following polymerase chain reaction (PCR). The PCR reaction mixtures consisted of 2.5 mM MgCl2, 0.2 or 0.25 (GSTM1/T1 PET) mM dNTP mix (Promega, Madison, WI), 0.2 µM of respective primers, 1–2 units of either AmpliTaq Polymerase Gold (Applied Biosystems, Foster City, CA) or Taq DNA polymerase (Eppendorf, Westbury, NY) along with their manufacturer's supplied buffer mix and 2 µl of template DNA. Primer sequences have been described previously (40).
A modification of a previously used multiplex PCR was used for genotyping GSTM1 and GSTT1 (41,42) by analyzing for the presence of the GSTT1 (480 bp), GSTM1 (231 bp) and the PCR control GSTM4 (158 bp) bands. This procedure was modified for use with paraffin-embedded tissue and some blood samples, which utilized different primers for GSTT1 (111 bp product) and used the β-globin (268 bp) gene as the control for amplification (43,44).
The GSTP1 I105V SNP was initially analyzed using PCR/restriction fragment length polymorphism. The PCR product was digested in a 30 µl reaction consisting of 20 U of BsmB1 and supplied buffer (NE Biolabs, Beverly, MA) and analyzed for the wild-type 166 bp fragment or variant 94 and 72 bp fragments.
PCR followed by single-strand conformation polymorphism (SSCP) analysis was initially used for the genotyping of the A119S and L432V alleles of CYP1B1 (primers previously described) (21) as well as A114V of GSTP1. For each SSCP analysis, the PCR products were denatured and analyzed using the GenePhor electrophoresis unit (Amersham Biosciences, Piscataway, NJ) followed by staining with Plus One DNA Silver Staining kit (Amersham Biosciences). A119S PCR products were separated on a GeneGel SSCP gel rehydrated in GeneGel Clean 15/24 kit rehydration buffer maintained at 12°C. The L432V PCR products were separated on a precast GeneGel Clean 15/24 gel rehydrated in a 15/24 kit rehydration buffer maintained at 10°C. The A114V PCR products were separated on a precast GeneGel Clean 15/24 gel rehydrated in a Buffer B rehydration buffer maintained at 13°C. The variant and wild-type alleles were recognized by specific banding patterns on the gel. Initial products were sequenced by the DNA Sequencing Core of the Wake Forest University Biomolecular Resource Facility to confirm that the banding pattern corresponded to the correct polymorphism. Further into the study, SNP analyses were completed using MassARRAY® (Sequenom®, San Diego, CA) with the assistance of the Wake Forest University School of Medicine, Center for Human Genomics as well as by sequencing by the Biomolecular Resource Facility of Wake Forest University or MWG-Biotech Inc. (High Point, NC).
Statistical analysis
P-values for demographic information were associated with the Student's t- or Chi-square test. Genotype distributions were determined using the Chi-square or Fisher's exact test. Odds ratios were obtained by using logistic regression models with and without modifying covariates as indicated in each table. This was repeated for interaction models as well.
Gene-gene interactions were analyzed using the MARS-logit model as described previously (Lin et al: Proc Amer Assoc Cancer Res 2006 [abstract nr 1205]). Every frequency reported was from a complete data set for each of the genotypes as well as each of the covariates (age, family history, age at menarche, BMI, smoking history and age at first live birth) without any missing values. Therefore, 349 cases and 363 controls met the criteria and were included in the final model. Analyses were completed using the Statistical Analysis System (SAS Institute, Cary, NC) and MARS 2.0 (Salford Systems, San Diego, CA) for the MARS-logit analysis.
Results
This study included 541 cases and 635 controls. The demographic characteristics of this population are described in Table I. The case and control populations were similar for most criteria. Age, age at menarche, parity, BMI and smoking status showed no significant difference between the cases and controls. There was a significant difference in the age of first live birth as well as family history (FH) between cases and controls (P=0.019 and 0.007, respectively). Notably, cases were younger than controls at age at first live birth. These two factors are believed to contribute to overall breast cancer risk (45). As there was a significant difference in the distribution of race between the cases and controls (P=0.026), further analyses were completed analyzing the Caucasian and African-American populations independently. Among the demographic characteristics presented in Table I, age, family history, age at menarche, age at first live birth, BMI and smoking status were used as covariates in the adjusted odds ratios in Tables II–V. The CYP1B1 and GSTP1 genotypes were in Hardy-Weinberg equilibrium for the two control populations. As heterozygosity was not analyzed for GSTM1 and GSTT1, Hardy-Weinberg equilibrium could not be determined.
Table I.
Demographic characteristics of the study population.
| Characteristics | Categories | Cases (n=541) n(%) |
Controls (n=635) n(%) |
P-valuea |
|---|---|---|---|---|
| Age (years) | Mean ± S.D. | 58.03±12.95 | 59.10±1.56 | 0.140 |
| ≤50 | 158 (29.2) | 169 (26.6) | 0.211 | |
| 51–60 | 147 (27.2) | 154 (24.3) | ||
| 61–70 | 121 (22.4) | 173 (27.2) | ||
| >70 | 115 (21.3) | 139 (21.9) | ||
| Race | African-American | 63 (11.8) | 103 (16.4) | 0.026 |
| Caucasian | 469 (88.2) | 524 (83.6) | ||
| Others | 9 | 8 | ||
| Family history | None | 411 (76.5) | 524 (82.9) | 0.007 |
| Mother and/or Sister | 126 (23.5) | 108 (17.1) | ||
| Missing | 4 | 3 | ||
| Age at menarche | ≤12 | 240 (45.4) | 286 (45.6) | 0.129 |
| 12.5–14 | 210 (39.8) | 272 (43.4) | ||
| ≥14.5 | 78 (14.8) | 69 (11.0) | ||
| Missing | 13 | 8 | ||
| Parity | Nulliparous | 75 (14.0) | 88 (13.9) | 0.954 |
| ≥1 | 460 (86.0) | 545 (86.1) | ||
| Missing | 6 | 2 | ||
| Age at first live birth | Nulliparous | 75 (14.0) | 88 (14.0) | 0.019 |
| ≤24 | 308 (57.6) | 310 (49.3) | ||
| 24.5–29 | 102 (19.1) | 150 (23.9) | ||
| ≥29.5 | 50 (9.4) | 81 (12.9) | ||
| Missing | 6 | 6 | ||
| Body mass index (kg/m2) | Mean ± S.D. | 27.79±6.15 | 27.29±5.79 | 0.155 |
| Smoking status | Never | 319 (59.6) | 361 (57.1) | 0.180 |
| Former | 143 (26.7) | 198 (31.3) | ||
| Current | 73 (13.6) | 73 (11.6) | ||
| Missing | 6 | 3 |
P-value associated with either the Student's t- or Chi-square test, others and missing values are not included in the tests.
Table II.
Drug-metabolism genotypes in Caucasian breast cancer cases and controls.
| Genes | Genotype | Controls | Cases |
|---|---|---|---|
| GSTM1 | Wild-type | 198 | 185 |
| Null | 268 | 206 | |
| OR (95% CI)a | 0.82 (0.60–1.11) | ||
| P-valueb | 0.157 | ||
| GSTT1 | Wild-type | 384 | 322 |
| Null | 82 | 69 | |
| OR (95% CI)a | 1.08 (0.72–1.62) | ||
| P-valueb | 0.985 | ||
| GSTP1 | II | 179 | 160 |
| I105V | IV | 179 | 183 |
| VV | 35 | 42 | |
| IV vs. II OR (95% CI)a | 1.22 (0.87–1.71) | ||
| VV vs. II OR (95% CI)a | 1.46 (0.84–2.54) | ||
| IV/VV vs. II OR (95% CI)a | 1.26 (0.91–1.74) | ||
| VV vs. II/IV OR (95% CI)a | 1.31 (0.77–2.22) | ||
| P-valueb | 0.435 | ||
| GSTP1 | AA | 337 | 328 |
| A114V | AV | 47 | 56 |
| VV | 1 | 2 | |
| AV vs. AA OR (95% CI)a | 1.37 (0.87–2.18) | ||
| VV vs. AA OR (95% CI)a | 2.17 (0.19–25.10) | ||
| AV/VV vs. AA OR (95% CI)a | 1.39 (0.88–2.19) | ||
| VV vs. AA/AV OR (95% CI)a | 2.08 (0.18–24.13) | ||
| P-valueb | 0.527 | ||
| CYP1B1 | AA | 201 | 199 |
| A119S | AS | 173 | 148 |
| SS | 32 | 32 | |
| AS vs. AA OR (95% CI)a | 0.83 (0.60–1.16) | ||
| SS vs. AA OR (95% CI)a | 1.09 (0.60–1.99) | ||
| AS/SS vs. AA OR (95% CI)a | 0.87 (0.64–1.19) | ||
| SS vs. AA/AS OR (95% CI)a | 1.18 (0.66–2.12) | ||
| P-valueb | 0.598 | ||
| CYP1B1 | LL | 149 | 121 |
| L432V | LV | 188 | 184 |
| VV | 75 | 76 | |
| LV vs. LL OR (95% CI)a | 1.24 (0.87–1.76) | ||
| VV vs. LL OR (95% CI)a | 1.29 (0.83–2.01) | ||
| LV/VV vs. LL OR (95% CI)a | 1.25 (0.90–1.74) | ||
| VV vs. LL/LV OR (95% CI)a | 1.14 (0.77–1.69) | ||
| P-valueb | 0.418 |
Odds Ratio (OR) adjusted for age, family history, smoking history, age at menarche, age at first live birth and body mass index using logistic regression.
Chi-square or Fisher's exact test for genotype distributions.
Table V.
Association between drug-metabolism genotypes and breast cancer risk for African-Americans, by age, family history and smoking history.
| Age | Family history | Smoking history | ||||
|---|---|---|---|---|---|---|
| Genotype | ≤60 Control/Case |
>60 Control/Case |
No Control/Case |
Yes Control/Case |
No Control/Case |
Yes Control/Case |
| GSTM1 | ||||||
| Low risk (null) | 18/13 | 10/6 | 25/14 | 3/4 | 15/9 | 13/10 |
| High risk (wild-type) | 35/29 | 24/8 | 54/31 | 4/6 | 33/19 | 25/18 |
| OR (95% CI)a | 1.04 (0.37–2.92) | 0.10 (0.01–1.46) | 0.63 (0.25–1.59) | 3.37 (0.48–63.49) | 0.60 (0.16–2.20) | 0.40 (0.10–1.70) |
| LRTb P-value | 0.19 | 0.388 | 0.51 | |||
| GSTT1 | ||||||
| Low risk (wild-type) | 46/29 | 25/12 | 64/31 | 6/9 | 42/21 | 28/20 |
| High risk (null) | 7/13 | 9/2 | 15/14 | 1/1 | 6/7 | 10/8 |
| OR (95% CI)a | 4.07 (1.12–14.80) | 2.75 (0.21–35.99) | 2.79 (0.95–8.20) | - | 4.30 (0.84–22.04) | 2.42 (0.60–9.73) |
| LRT P-value | 0.26 | 0.49 | ||||
| GSTP1 | ||||||
| I105V | ||||||
| Low risk (II/IV) | 35/33 | 29/10 | 59/33 | 4/9 | 35/23 | 28/20 |
| High risk (VV) | 10/9 | 3/4 | 11/11 | 2/2 | 9/5 | 4/8 |
| OR (95% CI)a | 1.12 (0.30–4.15) | 6.39 (0.47–87.51) | 2.36 (0.72–7.80) | - | 1.12 (0.21–6.07) | 8.23 (0.77–88.30) |
| LRT P-value | 0.35 | 0.08 | ||||
| GSTP1 | ||||||
| A114V | ||||||
| Low risk (AA) | 39/36 | 31/13 | 66/39 | 4/9 | 41/25 | 29/24 |
| High risk (AV/VV) | 3/5 | 1/0 | 2/3 | 2/2 | 2/2 | 2/3 |
| OR (95% CI)a | 2.10 (0.38–11.71) | - | 2.95 (0.42–20.62) | - | 1.65 (0.20–13.74) | 1.26 (0.12–13.11) |
| LRT P-value | 0.94 | |||||
| CYP1B1 | ||||||
| A119S | ||||||
| Low risk (AA/AS) | 32/31 | 26/9 | 54/33 | 3/6 | 33/24 | 24/16 |
| High risk (SS) | 13/10 | 6/5 | 16/10 | 3/5 | 11/4 | 8/11 |
| OR (95% CI)a | 0.52 (0.17–1.59) | 2.52 (0.32–19.96) | 0.87 (0.31–2.48) | - | 0.28 (0.04–1.83) | 2.48 (0.68–8.95) |
| LRT P-value | 0.20 | 0.02 | ||||
| CYP1B1 | ||||||
| L432V | ||||||
| Low risk (LL/LV) | 22/17 | 12/7 | 31/20 | 3/3 | 19/12 | 15/12 |
| High risk (VV) | 23/23 | 20/5 | 39/21 | 3/7 | 25/15 | 17/13 |
| OR (95% CI)a | 1.50 (0.54–4.20) | 0.49 (0.07–3.52) | 0.87 (0.35–2.13) | 0.81 (0.01–112.60) | 1.07 (0.35–3.28) | 0.69 (0.19–2.50) |
| LRT P-value | 0.24 | 1.000 | 0.68 | |||
For age stratification, OR adjusted for family history (FH), age at menarche, age at first live birth, smoking history and body mass index using logistic regression. For family history stratification, OR adjusted for age, age at menarche, age at first live birth, smoking history and body mass index. For smoking history stratification, OR adjusted for age, FH, age at menarche, age at first live birth and body mass index.
Likelihood ratio test.
The impact of each of the genotypes alone on breast cancer risk is described in Tables II and III for Caucasians and African-Americans, respectively. There were no statistically significant interactions between any of the genotypes and breast cancer in either Caucasians or African-Americans. There was an indication of an increased risk for Caucasian women homozygous for the valine allele at the GSTP1 114 locus (OR=2.17; 95% CI=0.19–25.10) compared to 114AA, though this was not statistically significant (P=0.53). This polymorphism has not been analyzed in most studies; however, in one previous study, alleles containing the 114V polymorphism were shown to be protective against breast cancer (46). African-American women who had at least one 119S allele in CYP1B1 exhibited a protective effect compared to the 119AA alleles (OR=0.53; 95% CI=0.20–1.40), while African-American women who had at least one valine allele at codon 432 appeared to exhibit an increased breast cancer risk (OR=5.52; 95% CI=0.50–61.37), though neither of these associations reached statistical significance.
Table III.
Drug-metabolism genotypes in African-American breast cancer cases and controls.
| Genes | Genotype | Controls | Cases |
|---|---|---|---|
| GSTM1 | Wild-type | 59 | 37 |
| Null | 28 | 19 | |
| OR (95% CI)a | 1.35 (0.57–3.15) | ||
| P-valueb | 0.828 | ||
| GSTT1 | Wild-type | 71 | 41 |
| Null | 16 | 15 | |
| OR (95% CI)a | 2.37 (0.86–6.55) | ||
| P-valueb | 0.234 | ||
| GSTP1 | II | 25 | 14 |
| I105V | IV | 39 | 29 |
| VV | 13 | 13 | |
| IV vs. II OR (95% CI)a | 1.01 (0.39–2.67) | ||
| VV vs. II OR (95% CI)a | 1.74 (0.48–6.26) | ||
| IV/VV vs. II OR (95% CI)a | 1.14 (0.45–2.90) | ||
| VV vs. II/IV OR (95% CI)a | 1.72 (0.57–5.18) | ||
| P-valueb | 0.525 | ||
| GSTP1 | AA | 70 | 49 |
| A114V | AV | 4 | 5 |
| VV | - | - | |
| AV vs. AA OR (95% CI)a | 1.48 (0.33–6.65) | ||
| VV vs. AA OR (95% CI)a | - | ||
| AV/VV vs. AA OR (95% CI)a | 1.48 (0.33–6.65) | ||
| VV vs. AA/AV OR (95% CI)a | - | ||
| P-valueb | 0.492 | ||
| CYP1B1 | AA | 17 | 19 |
| A119S | AS | 41 | 21 |
| SS | 19 | 15 | |
| AS vs. AA OR (95% CI)a | 0.51 (0.18–1.45) | ||
| SS vs. AA OR (95% CI)a | 0.57 (0.18–1.79) | ||
| AS/SS vs. AA OR (95% CI)a | 0.53 (0.20–1.40) | ||
| SS vs. AA/AS OR (95% CI)a | 0.88 (0.35–2.22) | ||
| P-valueb | 0.177 | ||
| CYP1B1 | LL | 6 | 1 |
| L432V | LV | 28 | 23 |
| VV | 43 | 28 | |
| LV vs. LL OR (95% CI)a | 6.96 (0.58–83.48) | ||
| VV vs. LL OR (95% CI)a | 4.88 (0.43–55.87) | ||
| LV/VV vs. LL OR (95% CI)a | 5.52 (0.50–61.37) | ||
| VV vs. LL/LV OR (95% CI)a | 0.87 (0.38–1.99) | ||
| P-valueb | 0.316 |
Odds ratio (OR) adjusted for age, family history, smoking history, age at menarche, age at first live birth and body mass index using logistic regression.
Chi-square or Fisher's exact test for genotype distributions.
Stratified analyses were used to determine if there were interactions between each of the genotypes and age, family history and smoking history in the Caucasian and African-American populations. The results are summarized in Tables IV and V. In the Caucasian population, women with a history of smoking and the GSTP1 114V SNP had an approximate 2-fold increase in breast cancer risk (OR=2.12; 95% CI=1.02–4.41), while in non-smoking women there was no association. The women diagnosed on or before the age of 60 with the CYP1B1 119SS alleles had a 3-fold increase in risk (OR=3.09; 95%CI=1.22–7.84), while these alleles were protective in the older population. None of the polymorphisms showed significant interactions with family history of breast cancer. In African-Americans, the null GSTT1 genotype showed a significantly higher risk for breast cancer among younger women (≤60) (OR=4.07, 95% CI=1.12–14.80). Due to a limited sample size, this finding could not be replicated in the >60 age group. Also among African-Americans, the difference in ORs between never and ever smokers with the GSTP1 105VV genotype approached significance (P=0.08) (OR=1.12, 95% CI=0.21–6.07 vs. OR=8.23 95% CI=0.77–88.30).
Table IV.
Association between drug-metabolism genotypes and breast cancer risk for Caucasians, by age, family history and smoking history.
| Age | Family history | Smoking history | ||||
|---|---|---|---|---|---|---|
| Genotype | ≤60 Control/Case |
>60 Control/Case |
No Control/Case |
Yes Control/Case |
No Control/Case |
Yes Control/Case |
| GSTM1 | ||||||
| Low risk (null) | 124/108 | 144/98 | 215/162 | 52/44 | 162/125 | 104/78 |
| High risk (wild-type) | 99/101 | 99/84 | 159/138 | 39/45 | 107/107 | 91/76 |
| OR (95% CI)a | 1.26 (0.81–1.98) | 1.24 (0.81–1.89) | 1.24 (0.88–1.75) | 1.24 (0.63–2.45) | 1.21 (0.81–1.79) | 1.26 (0.77–2.04) |
| LRTb P-value | 1.00 | 0.92 | 0.96 | |||
| GSTT1 | ||||||
| Low risk (wild-type) | 182/169 | 202/153 | 309/246 | 74/74 | 222/190 | 161/127 |
| High risk (null) | 41/40 | 41/29 | 65/54 | 17/15 | 47/42 | 34/27 |
| OR (95% CI)a | 1.07 (0.60–1.92) | 1.15 (0.64–2.07) | 1.02 (0.65–1.62) | 1.34 (0.52–3.43) | 1.08 (0.64–1.83) | 1.10 (0.58–2.10) |
| LRT P-value | 0.93 | 0.65 | 0.92 | |||
| GSTP1 | ||||||
| I105V | ||||||
| Low risk (II/IV) | 171/180 | 187/163 | 292/263 | 65/78 | 205/204 | 152/15 |
| High risk (VV) | 14/24 | 18/18 | 24/32 | 11/10 | 21/24 | 14/17 |
| OR (95% CI)a | 1.50 (0.68–3.30) | 1.06 (0.51–2.20) | 1.41(0.76–2.61) | 1.10 (0.39–3.08) | 1.07 (0.55–2.09) | 1.99 (0.82–4.82) |
| LRT P-value | 0.50 | 0.70 | 0.33 | |||
| GSTP1 | ||||||
| A114V | ||||||
| Low risk (AA) | 163/183 | 174/145 | 272/248 | 64/78 | 192/200 | 144/123 |
| High risk (AV/VV) | 22/26 | 26/32 | 39/47 | 9/11 | 31/30 | 17/28 |
| OR (95% CI)a | 1.22 (0.60–2.50) | 1.67 (0.91–3.07) | 1.40 (0.85–2.33) | 1.57 (0.52–4.71) | 1.06 (0.59–1.91) | 2.12 (1.02–4.41) |
| LRT P-value | 0.58 | 0.98 | 0.14 | |||
| CYP1B1 | ||||||
| A119S | ||||||
| Low risk (AA/AS) | 183/181 | 191/166 | 298/268 | 76/77 | 216/202 | 157/140 |
| High risk (SS) | 10/22 | 22/10 | 27/22 | 4/10 | 16/22 | 16/10 |
| OR (95% CI)a | 3.09 (1.22–7.84) | 0.55 (0.23–1.31) | 1.06 (0.55–2.06) | 1.67 (0.46–6.08) | 1.64 (0.75–3.56) | 0.76 (0.30–1.92) |
| LRT P-value | 0.005 | 0.55 | 0.24 | |||
| CYP1B1 | ||||||
| L432V | ||||||
| Low risk (LL) | 69/70 | 80/51 | 124/92 | 24/28 | 89/70 | 59/49 |
| High risk (LV/VV) | 128/132 | 135/128 | 207/201 | 56/58 | 145/154 | 118/103 |
| OR (95% CI)a | 1.07 (0.66–1.75) | 1.36 (0.86–2.15) | 1.37 (0.94–1.98) | 0.89 (0.42–1.91) | 1.46 (0.95–2.24) | 0.99 (0.58–1.68) |
| LRT P-value | 0.45 | 0.32 | 0.23 | |||
For age stratification, OR adjusted for family history (FH), age at menarche, age at first live birth, smoking history and body mass index using logistic regression. For family history stratification, OR adjusted for age, age at menarche, age at first live birth, smoking history and body mass index. For smoking history stratification, OR adjusted for age, FH, age at menarche, age at first live birth and body mass index.
Likelihood ratio test.
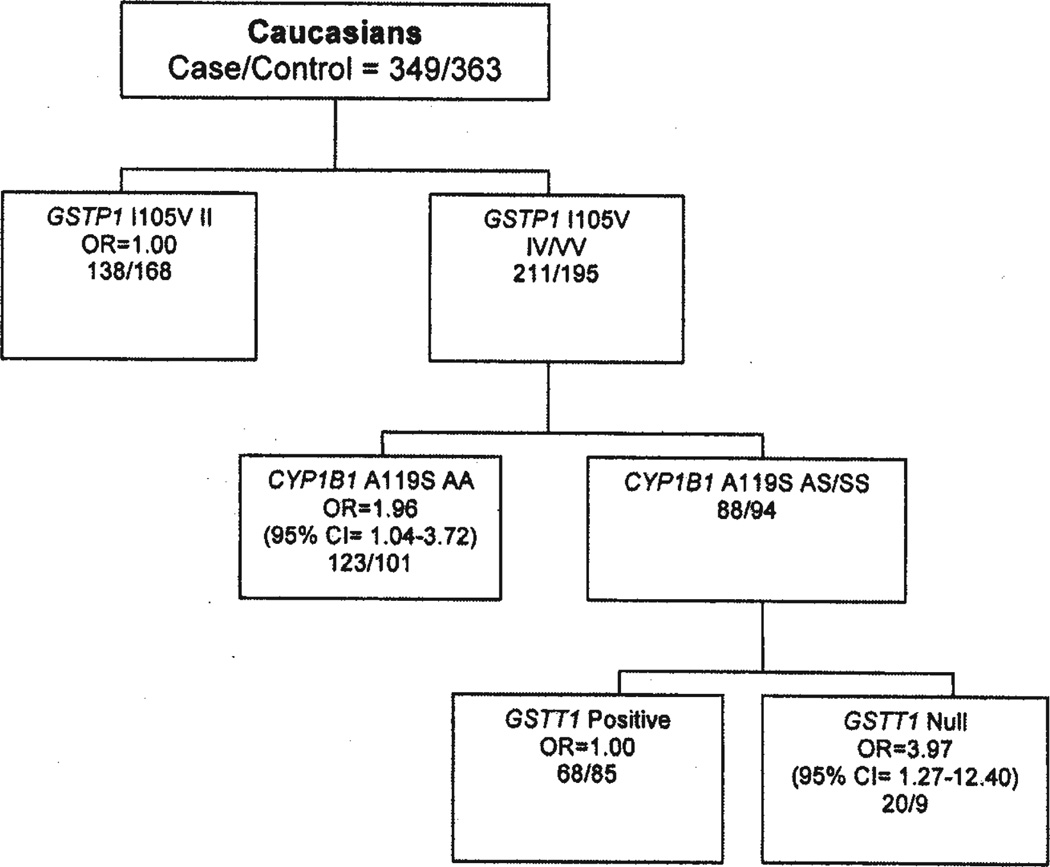
Potential gene-gene interactions were also explored to identify high-risk subpopulations using the MARS-logit model as described previously (Lin HY et al: Proc Amer Assoc Cancer Res 2006 [abstract nr 1205]). Fig. 1 shows the best predictive risk model for Caucasians. In Caucasians, breast cancer risk was significantly higher in two groups of subjects: i) A two-way interaction between GSTP1 105IV/VV and CYP1B1 119AA genotypes (OR=1.96, 95%CI=1.04–3.72); ii) A three-way interaction between GSTP1 105IV/VV, CYP1B1 119AS/SS and GSTT1 null genotypes (OR=3.97, 95% CI=1.27–12.40).
Figure 1.
A MARS-logit model of breast cancer risk in Caucasians.
Discussion
Early in vitro and in vivo animal studies suggest that breast carcinogenesis may result from exposure to environmental agents (3,4). The cellular components of breast tissue make it an ideal location for storage of known mammary genotoxins, as the abundance of fatty tissue in the breast can contribute to the storage and concentration of lipophilic compounds such as PAHs (47,48). This is supported by the frequent presence of aromatic DNA adducts in the normal adjacent tissues of breast cancer cases than in a cancer free control population (49). A major source of PAH exposure for those using cigarettes can be found in tobacco smoke (50). The effects of smoking on breast cancer risk are controversial due to the potential for anti-estrogenic effects (51). Based on results published during or before 2002, it was concluded that smoking most likely did not decrease risk, though whether it increases risk is still unclear (52). The GSTP1 enzyme is known to metabolize carcinogenic compounds such as those found in cigarette smoke. Variations in the ability to metabolize these carcinogens may impact the effects of cigarette smoking on breast cancer. The alteration of enzyme activity as a result of the codon 105 SNP most likely is related to the structural effects at the H-site (53,54). The 114 SNP, however, is not in the active site, though may have a minor effect on enzyme activity (53,55).
As in several previous studies, no significant associations were found when examining individual genotypes and breast cancer risk in either Caucasian or African-American women. In the two populations of women examined in this study, the effect of the GSTP1 SNPs appeared to be modulated by smoking history. The Caucasian women exhibited a near 2-fold increase in risk in women that smoked and had the GSTP1 105VV alleles, though the difference was not significant (P=0.33). A 2-fold increase in risk was also observed in Caucasian women smokers with the 114V allele (OR=2.12; 95% CI=1.02–4.41). In African-American women, an 8-fold increase in risk was observed in women with a history of smoking and the GSTP1 105VV genotype, although not significant. Several studies have analyzed the effect of the GSTP1 I105V SNP and smoking on breast cancer risk, though these investigations have produced conflicting results (27,28,36). A pooled analysis of Caucasian women found no association with any of the GSTP1 I105V genotypes alone or when combined with smoking (25).
The CYP1B1 enzyme is one of the predominant cytochrome P450 isozymes responsible for the metabolism of estrogen and PAHs in breast tissue (6,11,13). In this study, the CYP1B1 119SS genotype risk was significantly higher in younger Caucasian women (P=0.005), with a 3-fold increase in risk associated with this population of women (≤60 years old). In younger African-American women, risk was significantly higher in those with the GSTT1 null alleles. Breast cancer in young women, particularly those younger than 35 years, is associated with a poorer prognosis (56,57). The differences in tumor histopathology and patient prognosis may be related to the poor differentiation of tumors, their higher grade and the presence of mutations in tumor suppressor genes such as p53 (58,59). Recent studies from our laboratory, utilizing a case-case study design, have demonstrated that combinations of specific genotypes in phase I and phase II enzymes or interactions between specific genotypes and smoking predisposed women to mutations in the p53 gene (40). The impact of combinations of polymorphisms on risk of disease was explored in the Caucasian population. Alone the GSTP1 105IV/VV genotype did not have much impact on breast cancer risk, although in women with these alleles along with the CYP1B1 119AA genotype an almost 2-fold increase in risk is observed. Breast cancer risk was increased nearly 4-fold in women with the GSTP1 105IV/VV, CYP1B1 119AS/SS and GSTT1 null genotypes. These results suggest that women with specific combinations of SNPs, particularly in combination with environmental or endogenous exposures, may be predisposed to an earlier onset of the disease. Future studies with larger patient samples will be needed in order to confirm and extend these findings.
The biological plausibility of the association of these polymorphisms, due to functional effects on metabolism, and breast cancer risk supports the findings observed in this study. The number of women included in this study was of sufficient size to analyze the interaction of metabolic genotypes with potential risk factors in the Caucasian and African-American women. One limitation of this study, however, was the small number of African-American women recruited, which prevented analyses of the interactions between the genotypes. This study focused on individual polymorphisms within genes containing multiple SNPs. Future studies should include haplotype analyses, which would take into account the effects of the SNPs when present on the same allele.
The results of this study provide further data demonstrating that individual genotypes may not impact breast cancer risk, as no significant associations were noted in either the Caucasian or African-American populations. There were interesting trends none-the-less with the CYP1B1 SNPs in the African- American population that were not seen with the Caucasian women. The differences between races were not surprising as it has been well established that breast cancer incidence and prognosis differs between African-American and Caucasian women (60). A recent study demonstrated that African- American women more frequently exhibit the basal-like subtype of breast cancer, which is associated with a poorer prognosis (61).
In contrast, combining genotypes with each other as well as other plausible breast cancer risk factors did provide notable findings. We noted that the less common GSTP1 A114V SNP genotypes exhibited a trend towards an increased breast cancer risk in Caucasian women who smoked. The significant interactions between the CYP1B1 A119S SNP and age of diagnosis may give some insight into the etiology of breast cancer in women diagnosed before the age of 60. Additional studies would be required to determine the mechanism behind this association. As there are not many studies that focus on either of these polymorphisms, further studies would be required in order to conclude that there is truly an association with breast cancer risk. The interactions among the phase I and phase II enzyme polymorphisms support the concept that genetic alterations in the ability to activate and detoxify potential breast carcinogens can influence cancer risk. This information is vital in understanding the etiology of this disease, which can assist in the generation of effective breast cancer prevention strategies.
Acknowledgements
The authors would like to thank Dr Tasha Smith and Jenny Goa for their assistance with the DNA isolation, Ms. Elyse Jung of the DNA Sequencing Core for assistance in sequencing of the single nucleotide polymorphisms, Dr Lilly Zheng of the Center for Human Genomics for the MassARRAY analyses, Dr Hui-Yi Lin for assistance with statistical analyses and Dr Lynn Dressler and the Tissue Procurement and Analysis/Immunohistochemistry Core Facilities for managing the tissues from UNC. Our research was supported by grants RO1 CA81330 (to MSM), RO1 CA91221 (to JJH), Cancer Center Support Grant P30 CA12197, M01RR00046 (to LAC) and training grant T32 CA79448 (for BOV) from the National Cancer Institute, as well as grants from the Susan G. Komen Breast Cancer Foundation, the Breast Cancer Research Foundation and Friends for an earlier breast cancer test foundation.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Madigan MP, Ziegler RG, Benichou J, Byrne C, Hoover RN. Proportion of breast cancer cases in the United States explained by well-established risk factors. J Natl Cancer Inst. 1995;87:1681–1685. doi: 10.1093/jnci/87.22.1681. [DOI] [PubMed] [Google Scholar]
- 3.MacNicoll AD, Easty GC, Neville AM, Grover PL, Sims P. Metabolism and activation of carcinogenic polycyclic hydrocarbons by human mammary cells. Biochem Biophys Res Commun. 1980;95:1599–1606. doi: 10.1016/s0006-291x(80)80081-7. [DOI] [PubMed] [Google Scholar]
- 4.Sukumar S, McKenzie K, Chen Y. Animal models for breast cancer. Mutat Res. 1995;333:37–44. doi: 10.1016/0027-5107(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 5.Stampfer MR, Bartholomew JC, Smith HS, Bartley JC. Metabolism of benzo[a]pyrene by human mammary epithelial cells: toxicity and DNA adduct formation. Proc Natl Acad Sci USA. 1981;78:6251–6255. doi: 10.1073/pnas.78.10.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Yager JD. Endogenous estrogens as carcinogens through metabolic activation. J Natl Cancer Inst Monogr. 2000:67–73. doi: 10.1093/oxfordjournals.jncimonographs.a024245. [DOI] [PubMed] [Google Scholar]
- 8.Nimrod A, Ryan KJ. Aromatization of androgens by human abdominal and breast fat tissue. J Clin Endocrinol Metab. 1975;40:367–372. doi: 10.1210/jcem-40-3-367. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Nichols JE, Bulun SE, Mendelson CR, Simpson ER. Aromatase P450 gene expression in human adipose tissue. Role of a Jak/STAT pathway in regulation of the adipose-specific promoter. J Biol Chem. 1995;270:16449–16457. doi: 10.1074/jbc.270.27.16449. [DOI] [PubMed] [Google Scholar]
- 10.Jefcoate CR, Liehr JG, Santen RJ, et al. Tissue-specific synthesis and oxidative metabolism of estrogens. J Natl Cancer Inst Monogr. 2000:95–112. doi: 10.1093/oxfordjournals.jncimonographs.a024248. [DOI] [PubMed] [Google Scholar]
- 11.Hellmold H, Rylander T, Magnusson M, Reihner E, Warner M, Gustafsson JA. Characterization of cytochrome P450 enzymes in human breast tissue from reduction mammaplasties. J Clin Endocrinol Metab. 1998;83:886–895. doi: 10.1210/jcem.83.3.4647. [DOI] [PubMed] [Google Scholar]
- 12.Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents-DNA adducts and mutations. J Natl Cancer Inst Monogr. 2000:75–93. doi: 10.1093/oxfordjournals.jncimonographs.a024247. [DOI] [PubMed] [Google Scholar]
- 15.Belous AR, Hachey DL, Dawling S, Roodi N, Parl FF. Cytochrome P450 1B1-mediated estrogen metabolism results in estrogen-deoxyribonucleoside adduct formation. Cancer Res. 2007;67:812–817. doi: 10.1158/0008-5472.CAN-06-2133. [DOI] [PubMed] [Google Scholar]
- 16.Shimada T, Watanabe J, Inoue K, Guengerich FP, Gillam EM. Specificity of 17beta-oestradiol and benzo[a]pyrene oxidation by polymorphic human cytochrome P4501B1 variants substituted at residues 48, 119 and 432. Xenobiotica. 2001;31:163–176. doi: 10.1080/00498250110043490. [DOI] [PubMed] [Google Scholar]
- 17.Shimada T, Watanabe J, Kawajiri K, et al. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis. 1999;20:1607–1613. doi: 10.1093/carcin/20.8.1607. [DOI] [PubMed] [Google Scholar]
- 18.Sissung TM, Price DK, Sparreboom A, Figg WD. Pharmacogenetics and regulation of human cytochrome P450 1B1: implications in hormone-mediated tumor metabolism and a novel target for therapeutic intervention. Mol Cancer Res. 2006;4:135–150. doi: 10.1158/1541-7786.MCR-05-0101. [DOI] [PubMed] [Google Scholar]
- 19.McLellan RA, Oscarson M, Hidestrand M, et al. Characterization and functional analysis of two common human cytochrome P450 1B1 variants. Arch Biochem Biophys. 2000;378:175–181. doi: 10.1006/abbi.2000.1808. [DOI] [PubMed] [Google Scholar]
- 20.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology. 2000;61:154–166. doi: 10.1159/000028396. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe J, Shimada T, Gillam EM, et al. Association of CYP1B1 genetic polymorphism with incidence to breast and lung cancer. Pharmacogenetics. 2000;10:25–33. doi: 10.1097/00008571-200002000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Zheng T, Holford TR, Zahm SH, et al. Cigarette smoking, glutathione-s-transferase M1 and t1 genetic polymorphisms, and breast cancer risk (United States) Cancer Causes Control. 2002;13:637–645. doi: 10.1023/a:1019500109267. [DOI] [PubMed] [Google Scholar]
- 23.Bailey LR, Roodi N, Dupont WD, Parl FF. Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer. Cancer Res. 1998;58:5038–5041. [PubMed] [Google Scholar]
- 24.Bailey LR, Roodi N, Verrier CS, Yee CJ, Dupont WD, Parl FF. Breast cancer and CYPIA1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res. 1998;58:65–70. [PubMed] [Google Scholar]
- 25.Vogl FD, Taioli E, Maugard C, et al. Glutathione S-transferases M1, T1, and P1 and breast cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2004;13:1473–1479. [PubMed] [Google Scholar]
- 26.Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006;15:754–761. doi: 10.1016/j.breast.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Helzlsouer KJ, Selmin O, Huang HY, et al. Association between glutathione S-transferase M1, P1, and T1 genetic polymorphisms and development of breast cancer. J Natl Cancer Inst. 1998;90:512–518. doi: 10.1093/jnci/90.7.512. [DOI] [PubMed] [Google Scholar]
- 28.Mitrunen K, Jourenkova N, Kataja V, et al. Glutathione S-transferase M1, M3, P1, and T1 genetic polymorphisms and susceptibility to breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:229–236. [PubMed] [Google Scholar]
- 29.Paracchini V, Raimondi S, Gram IT, et al. Meta- and pooled analyses of the cytochrome P-450 1B1 Val432Leu polymorphism and breast cancer: a HuGE-GSEC review. Am J Epidemiol. 2007;165:115–125. doi: 10.1093/aje/kwj365. [DOI] [PubMed] [Google Scholar]
- 30.Saintot M, Malaveille C, Hautefeuille A, Gerber M. Interactions between genetic polymorphism of cytochrome P450-1B1, sulfotransferase 1A1, catechol-o-methyltransferase and tobacco exposure in breast cancer risk. Int J Cancer. 2003;107:652–657. doi: 10.1002/ijc.11432. [DOI] [PubMed] [Google Scholar]
- 31.Sillanpaa P, Heikinheimo L, Kataja V, et al. CYP1A1 and CYP1B1 genetic polymorphisms, smoking and breast cancer risk in a Finnish Caucasian population. Breast Cancer Res Treat. 2007;104:287–297. doi: 10.1007/s10549-006-9414-6. [DOI] [PubMed] [Google Scholar]
- 32.Kocabas NA, Sardas S, Cholerton S, Daly AK, Karakaya AE. Cytochrome P450 CYP1B1 and catechol O-methyltransferase (COMT) genetic polymorphisms and breast cancer susceptibility in a Turkish population. Arch Toxicol. 2002;76:643–649. doi: 10.1007/s00204-002-0387-x. [DOI] [PubMed] [Google Scholar]
- 33.Rylander-Rudqvist T, Wedren S, Granath F, et al. Cytochrome P450 1B1 gene polymorphisms and postmenopausal breast cancer risk. Carcinogenesis. 2003;24:1533–1539. doi: 10.1093/carcin/bgg114. [DOI] [PubMed] [Google Scholar]
- 34.van der Hel OL, Peeters PH, Hein DW, et al. NAT2 slow acetylation and GSTM1 null genotypes may increase postmenopausal breast cancer risk in long-term smoking women. Pharmacogenetics. 2003;13:399–407. doi: 10.1097/00008571-200307000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Zheng T, Holford TR, Zahm SH, et al. Glutathione S-transferase M1 and T1 genetic polymorphisms, alcohol consumption and breast cancer risk. Br J Cancer. 2003;88:58–62. doi: 10.1038/sj.bjc.6600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millikan R, Pittman G, Tse CK, Savitz DA, Newman B, Bell D. Glutathione S-transferases M1, T1, and P1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:567–573. [PubMed] [Google Scholar]
- 37.Egan KM, Cai Q, Shu XO, et al. Genetic polymorphisms in GSTM1, GSTP1, and GSTT1 and the risk for breast cancer: results from the Shanghai Breast Cancer Study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2004;13:197–204. doi: 10.1158/1055-9965.epi-03-0294. [DOI] [PubMed] [Google Scholar]
- 38.Smith TR, Levine EA, Perrier ND, et al. DNA-repair genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:1200–1204. [PubMed] [Google Scholar]
- 39.Kaufmann WK, Filatov L, Oglesbee SE, et al. Radiation clastogenesis and cell cycle checkpoint function as functional markers of breast cancer risk. Carcinogenesis. 2006;27:2519–2527. doi: 10.1093/carcin/bgl103. [DOI] [PubMed] [Google Scholar]
- 40.Van Emburgh BO, Hu JJ, Levine EA, et al. Polymorphisms in drug metabolism genes, smoking, and p53 mutations in breast cancer. Mol Carcinog. 2008;47:88–99. doi: 10.1002/mc.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirvonen A, Husgafvel-Pursiainen K, Anttila S, Vainio H. The GSTM1 null genotype as a potential risk modifier for squamous cell carcinoma of the lung. Carcinogenesis. 1993;14:1479–1481. doi: 10.1093/carcin/14.7.1479. [DOI] [PubMed] [Google Scholar]
- 42.Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300(Pt 1):271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl Cancer Inst. 1993;85:1159–1164. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 44.Li R, Boerwinkle E, Olshan AF, et al. Glutathione S-transferase genotype as a susceptibility factor in smoking-related coronary heart disease. Atherosclerosis. 2000;149:451–462. doi: 10.1016/s0021-9150(99)00483-9. [DOI] [PubMed] [Google Scholar]
- 45.American Cancer Society. Breast Cancer Facts & Figures 2005–2006. Atlanta: American Cancer Society, Inc.; 2006. [Google Scholar]
- 46.Maugard CM, Charrier J, Pitard A, et al. Genetic polymorphism at the glutathione S-transferase (GST) P1 locus is a breast cancer risk modifier. Int J Cancer. 2001;91:334–339. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1057>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 47.Grover PL, Martin FL. The initiation of breast and prostate cancer. Carcinogenesis. 2002;23:1095–1102. doi: 10.1093/carcin/23.7.1095. [DOI] [PubMed] [Google Scholar]
- 48.Obana H, Hori S, Kashimoto T, Kunita N. Polycyclic aromatic hydrocarbons in human fat and liver. Bull Environ Contam Toxicol. 1981;27:23–27. doi: 10.1007/BF01610981. [DOI] [PubMed] [Google Scholar]
- 49.Li D, Wang M, Dhingra K, Hittelman WN. Aromatic DNA adducts in adjacent tissues of breast cancer patients: clues to breast cancer etiology. Cancer Res. 1996;56:287–293. [PubMed] [Google Scholar]
- 50.Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 51.Baron JA, La Vecchia C, Levi F. The antiestrogenic effect of cigarette smoking in women. Am J Obstet Gynecol. 1990;162:502–514. doi: 10.1016/0002-9378(90)90420-c. [DOI] [PubMed] [Google Scholar]
- 52.Terry PD, Rohan TE. Cigarette smoking and the risk of breast cancer in women: a review of the literature. Cancer Epidemiol Biomarkers Prev. 2002;11:953–971. [PubMed] [Google Scholar]
- 53.Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem. 1997;272:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 54.Hu X, O'Donnell R, Srivastava SK, et al. Active site architecture of polymorphic forms of human glutathione S-transferase P1-1 accounts for their enantioselectivity and disparate activity in the glutathione conjugation of 7beta,8alpha-dihydroxy-9alpha, 10alpha-ox y-7,8,9,10-tetrahydrobenzo(a)pyrene. Biochem Biophys Res Commun. 1997;235:424–428. doi: 10.1006/bbrc.1997.6777. [DOI] [PubMed] [Google Scholar]
- 55.Hu X, Xia H, Srivastava SK, et al. Activity of four allelic forms of glutathione S-transferase hGSTP1-1 for diol epoxides of polycyclic aromatic hydrocarbons. Biochem Biophys Res Commun. 1997;238:397–402. doi: 10.1006/bbrc.1997.7311. [DOI] [PubMed] [Google Scholar]
- 56.El Saghir NS, Seoud M, Khalil MK, et al. Effects of young age at presentation on survival in breast cancer. BMC Cancer. 2006;6:194. doi: 10.1186/1471-2407-6-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Figueiredo JC, Ennis M, Knight JA, et al. Influence of young age at diagnosis and family history of breast or ovarian cancer on breast cancer outcomes in a population-based cohort study. Breast Cancer Res Treat. 2007;105:69–80. doi: 10.1007/s10549-006-9433-3. [DOI] [PubMed] [Google Scholar]
- 58.Pratap R, Shousha S. Breast carcinoma in women under the age of 50: Relationship between p53 immunostaining, tumour grade, and axillary lymph node status. Breast Cancer Res Treat. 1998;49:35–39. doi: 10.1023/a:1005993220824. [DOI] [PubMed] [Google Scholar]
- 59.Sidoni A, Cavaliere A, Bellezza G, Scheibel M, Bucciarelli E. Breast cancer in young women: clinicopathological features and biological specificity. Breast. 2003;12:247–250. doi: 10.1016/s0960-9776(03)00095-x. [DOI] [PubMed] [Google Scholar]
- 60.Ademuyiwa FO, Olopade OI. Racial differences in genetic factors associated with breast cancer. Cancer Metastasis Rev. 2003;22:47–53. doi: 10.1023/a:1022259901319. [DOI] [PubMed] [Google Scholar]
- 61.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]