Abstract
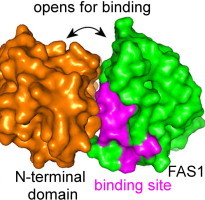
Fasciclin I (FAS1) domains have important roles in cell adhesion, which are not understood despite many structural and functional studies. Examples of FAS1 domain proteins include TGFBIp (βig-h3) and periostin, which function in angiogenesis and development of cornea and bone, and are also highly expressed in cancer tissues. Here we report the structure of a single-domain bacterial fasciclin I protein, Fdp, in the free-living photosynthetic bacterium Rhodobacter sphaeroides, and show that it confers cell adhesion properties in vivo. A binding site is identified which includes the most highly conserved region and is adjacent to the N-terminus. By mapping this onto eukaryotic homologues, which all contain tandem FAS1 domains, it is concluded that the interaction site is normally buried in the dimer interface. This explains why corneal dystrophy mutations are concentrated in the C-terminal domain of TGFBIp and suggests new therapeutic approaches.
Keywords: NMR structure, Cell adhesion, Biofilm, Autoinhibition, Domain interface
Abbreviations: FAS, fasciclin; Fdp, Fasciclin I Domain Protein
Graphical abstract
Highlights
▸ We report the NMR structure of a single-domain fasciclin I protein. ▸ We show that the protein enhances bacterial cell adhesion. ▸ A new alignment suggests that the binding site is in the H1 and H2 regions. ▸ We propose that in eukaryotic homologues, the binding site is buried in an interface.
1. Introduction
Members of the fasciclin I family of proteins (FAS1) occur in a wide range of vertebrates, invertebrates and microorganisms. A bioinformatics study concluded that the domain fold is ancient, traceable back to the Last Universal Common Ancestor [1], implying a likely common function across all phyla. They are generally cell-surface and membrane-anchored proteins involved in homophilic cell adhesion or symbiotic processes. One of the earliest and best studied examples is Drosophila FAS1, which is expressed during embryonic development, and guides axons from axon-generating neural cells to other target neurons or muscle cells [2–4]. FAS1 domains do not span the membrane, but are attached to the membrane via a lipid link that is developmentally regulated, resulting in variable levels of soluble and membrane-anchored proteins during embryogenesis [5,6]. Examples in mammals include transforming growth factor-β-induced gene product (TGFBIp, formerly known as βig-h3) [7], periostin [8–10], also known as osteoblast-specific factor 2 (OSF-2) [11], and stabilins 1 and 2, also known as scavenger receptor FEEL-1 and -2 proteins [12]. Mutations in TGFBIp are linked to corneal dystrophies, while periostin is required for development of tooth, bone and heart [13]. Many of these mammalian proteins are found expressed at high levels by tumour cells, presumably because of their roles in cell adhesion and angiogenesis, and they have been proposed both as tumour markers and therapeutic targets [13–15]. Several have been shown to bind to integrin cell surface receptors [8,10,16,17] including periostin which is suggested to be a ligand for αvβ5 integrin [16]. Knock-out mutations seldom exhibit discernible phenotypes. However, when combined with mutations in other linked signal transduction loci, distinct phenotypes can be observed, as shown by accompanying mutations in the abl tyrosine kinase in Drosophila, which results in defective axon tracts [2]. Amongst plants, fasciclin I-like domains occur widely as a major subgroup of the cell surface arabinogalactan proteins required for plant growth and development [18,19], and as the Arabidopsis thaliana SOS5 protein required for normal cell expansion [20,21]. Microbial fasciclin I proteins include the antigenic MPB70 protein secreted by Mycobacterium bovis, identical to M. tuberculosis MPT70 [22], and proteins important for symbiotic relationships of cyanobacteria [23] and in cnidarian–algal associations [24]. MPB70 is homologous to OSF-2, and adhesion of MPB70 to bone in neonates has been implicated in osteitis following BCG vaccination [25]. In symbiotic rhizobia such as Sinorhizobium meliloti, the fasciclin I protein Nex18 is required for normal nodule formation with leguminous plant partners [26].
FAS1 domains in animals almost always occur in pairs: Drosophila FAS1 has two tandem pairs, as do TGFBIp and periostin, while the stabilins have seven tandem copies [27]. The best characterized system is TGFBIp, where a large number of mutations have been identified that lead to corneal dystrophies [28,29]. Over half of these derive from only two sites, one in FAS1 domain 1 (FAS1-1) and one in domain 4 (FAS1-4). However, almost all the other mutations are found in FAS1-4, the exception being one in the interface between FAS1-3 and FAS1-4.
Despite their low overall sequence conservation, fasciclin I domains are easily identifiable due to the presence of two conserved sequence motifs called H1 and H2. Several FAS1 structures have been reported, namely the crystal structure of a FAS1 domain pair from Drosophila [30], NMR and crystal structures of the FAS1-4 domain from TGFBIp [31] (Yoneyama et al., unpublished), and the single-domain MPB70 [32]. No clear binding site or mode of action has emerged [27,30], although a conserved Asp-Ile sequence was shown to be important [8]. In view of the growing clinical importance of FAS1 domains, a greater understanding of the function of these domains is urgently required.
Here we report on the identification of a new member of the fasciclin I family, Fdp (Fasciclin I Domain Protein), a simple single-domain protein found in the photosynthetic bacterium Rhodobacter sphaeroides, which is confirmed as a member of this protein family by determination of its structure. Our study defines a possible role for Fdp in adhesion properties of whole cells, which may be of significance for the bacterium in its natural environment. We identify a probable binding site on Fdp. On comparison to animal FAS1, we conclude that the physiological binding site of FAS1 is buried in a domain interface, and discuss therapeutic implications.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Escherichia coli strains were cultured aerobically in LB. Where appropriate, media were supplemented with 50 μg ml−1 ampicillin and/or 50 μg ml−1 kanamycin, or 500 μg ml−1 carbenicillin. Plasmid transfer into R. sphaeroides was by conjugative transfer from E. coli S17-1 [33]. R. sphaeroides NCIB 8253 was cultured at 34 °C in M22+ medium [33]; fdp mutants were cultured in M22+ containing 20 μg ml−1 kanamycin. Complementation plasmid pRKfdp was constructed by inserting a 1.1 kb BamHI fragment possessing the intact fdp gene into replicative pRK415 [34], and verified by sequencing.
2.2. Expression of recombinant fdp
Regions 57 to 470 (relative to ATG, where A is position 1) of fdp were amplified by PCR using primers 5′-TCAGCCATATGGAAACCGGAGACATCGTGGA-3′ (NdeI site underlined) and 5′-GCTAGGATCCGCATCAGGCGCCCGGCATCAGCAC-3′ (BamHI site underlined), using pSUP202fdp-13 as template. The 413-bp fragment was isolated and cloned into SmaI-digested pBluescript-SK to give pBlFDPtr. The presence of inserts with correct sequence was verified by restriction digest analysis and sequencing. The fdp fragment of BamHI NdeI-digested pBlFDPtr was cloned into pET14b (Novagen). The final expression construct, pETfdptr, expresses a Fdp protein with an N-terminal MGSS(H)6SSGLVPRGSHM sequence followed by Fdp starting at E19. Fdp was expressed and purified as described [35] and verified by N-terminal sequencing, electrospray mass spectrometry and Western blotting.
2.3. NMR studies
The fdp gene was cloned into a pET14b vector and expressed in E. coli BL21[DE3]. Labelled protein was produced by growth and IPTG induction in M9 minimal medium containing 13C and 15N. Cells were disrupted by sonication and the protein was purified using Ni-NTA chromatography (Qiagen). NMR experiments were recorded on Bruker DRX-500, 600 and 800 spectrometers at 298 K, using 1–2 mM protein in 50 mM sodium phosphate pH 7.0, 0.03% NaN3, in H2O containing 10% D2O. Processing and analysis of the spectra was carried out using Felix (Felix NMR Inc., San Diego, CA). Molecules were viewed with Pymol (DeLano Scientific, California; http://www.pymol.org). NOEs were assigned manually as much as possible, with a starting set of 1506 unambiguously assigned NOEs. The structure was calculated in CNS 1.1 [36] using a final set of 1788 distance restraints obtained from NOESY spectra (approximately 13 restraints per residue), and 148 angle restraints from TALOS [37]. Hydrogen bond restraints were added at a later stage in the structure calculation, after the secondary structure was already clearly established, to avoid biasing the calculation. Analysis of the structures calculated using the final set of restraints showed that 50 out of 100 structures calculated had closely similar energies and structures. Thirty of these were refined in ARIA 1.2 using explicit water refinement [38], which resulted in slightly worse restraint violations and a greater difference from ideal values, but a better Ramachandran distribution.
2.4. Cell adherence assays
R. sphaeroides (∼9.6 × 107 cells) were suspended at 34 °C in 10 ml M22+, 10 mM glucose, and 200 μl aliquots introduced into 96-well microtitre plates fitted with 96-peg lids (68.1 mm2 submersed area). Adherent cells attached to pegs were counted after 5 days. Pegs were then removed and submersed in sterile distilled water to remove loosely-bound cells and transferred to 200 μl ¼-strength Ringer's diluent. Attached cells were removed into diluent using a sonicating water bath for 5 min. Suspensions of adherent cells were then spread-plated onto LBA agar for enumeration. Plates were incubated at 34 °C for 5 days. Additional assays were performed based on the crystal violet assay [39].
2.5. Isolation of the fdp gene and construction of insertionally-inactivated fdp mutants
The fdp gene is located on chromosome 1 (locus RSP1409; http://genome.ornl.gov), between two oppositely transcribed genes (one homologous to the endopeptidase Clp ATP-binding chain B of Mesorhizobium loti, and the other homologous to molybdopterin binding domains of oxidoreductase enzymes). Therefore, fdp is not co-transcribed with any flanking genes and forms a single-gene operon, and inactivation of fdp is not expected to exert any polarity effects on flanking genes. Sequencing and restriction mapping of the fdp region in the NCIB 8253 strain confirmed that the arrangement is identical to that of the 2.4.1 sequenced strain. An fdp fragment possessing the entire gene was amplified by PCR using primers 5′-ATGCATCGCCTCGTCGATCCGCAGC-3′ and 5′- CCGGGCTATGTGGGCTACGATGAG-3′. PCR was performed with 5% DMSO. The 1.9 kb product was purified and digested with BamHI. The 1.0 kb BamHI fdp fragment was then purified and labelled with digoxygenin using random priming. To isolate fdp-harbouring clones from a R. sphaeroides genomic DNA library by Southern hybridization, the labelled PCR product was used to screen a R. sphaeroides NCIB 8253 genomic library [40]. Hybridization was performed overnight at 65 °C. Membranes were washed in 0.2 × SSC, and detection of fdp-containing clones was by chemiluminescence. One positive clone (pSUP202fdp-13) possessed fdp approximately centrally on a 4.0 kb HindIII fragment. This fragment was isolated and ligated into HindIII-digested pUC19 to give pUCfdpH4-8 which has a unique SgrAI site which cleaves at base position 84 in the fdp gene. The 0.9 kb XmaI-ended Tn5 kanamycin resistance cassette of pUX-Km [41] was isolated and ligated into SgrAI-digested pUCfdpH4-8. The orientation of the kanamycin cassette in the clones was checked by restriction digestion, and also by PCR using primers 5′-GTTGTTGTAGTTCGAGATCTCCTCG-3′ (in the fdp promoter region), and 5′-TTGGTGGTCGAATGGGCAGGTAGCC-3′ (in the kanamycin resistance gene). The correct construct was called pUCfdp4-KM. The 4.9 kb HindIII fdp::kan fragment was isolated and cloned into HindIII-digested pSUP202. The resulting plasmid pSUPfdpKM was checked by restriction analysis and introduced into R. sphaeroides NCIB 8253. Kanamycin-resistant transconjugants were screened by Southern hybridization to check for loss of the suicide plasmid, and insertion of the kanamycin resistance cassette at the correct chromosomal position. Additional confirmation was obtained by PCR using the primers above with mutant genomic DNA as template.
3. Results
3.1. Structure of fdp
The fdp gene is designated ORF RSP1409 on chromosome 1 in the R. sphaeroides 2.4.1 database (http://genome.ornl.gov/microbial/rsph). The protein is predicted to possess 155 residues (excluding initiating fMet), with residues 1–18 (RKTLLALSLGLLAAPAFA) constituting a signal peptide for translocation across the inner membrane. This results in a mature 137-residue protein, possessing the N-terminal sequence ETGDIVETATGA. Here, we number the protein as in the full-length sequence, so that the first residue is residue 19. By PSI-BLAST, the closest sequence similarity (60% identical; 74% similar) is to S. meliloti Nex18. Fdp is also related (39% identity; 55% similarity) to M. bovis MPB70 major secreted protein and Drosophila FAS1-4 (29% identity) (Fig. 1a). The sequence similarities are striking, since FAS1 domains generally exhibit low overall sequence conservation (<20%) [30]. The two regions of high conservation recognized for the FAS1 superfamily (H1 and H2) are also strongly conserved in Fdp. It is a single-domain protein, and is not co-transcribed with any other gene.
Fig. 1.
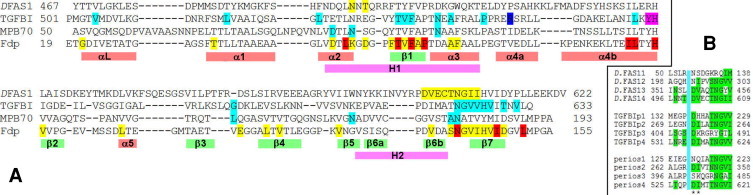
Sequence alignments of FAS1 domains. (A) Alignments based on structural similarity. Sequences are those of the four FAS1 domains with three-dimensional structures: The Drosophila FAS1-3/4 pair (DFAS1: PDB 1o70, domain 4); the fourth FAS1 domain of human TGFBIp (TGFBI: PDB 2vxp); the M. bovis secreted protein MPB70 (MPB70: PDB 1nyo); and Fdp (Fdp: PDB 1w7d). The NMR structure of the fourth FAS1 domain of human TGFBIp (PDB 1x3b) is very similar to the crystal structure and was not used as an independent structure. Colour code: in DFAS1, yellow denotes residues described here as being interacting residues. In TGFBIp, blue denotes R555, one of the two major sites giving rise to corneal dystrophy. Other disease-causing sites are indicated in cyan. The sequence YH, suggested as a possible binding site [16], is shown in magenta. In MPB70, cyan indicates suggested interaction sites [32]. Highly conserved and completely conserved residues are indicated on Fdp in yellow and red respectively. Locations of regular secondary structure, and the conserved regions H1 and H2, are indicated below the sequences. (B) Domains 1 through 4 from Drosophila FAS1, TGFBIp and periostin, each of which contains four tandem FAS1 domains. The alignments encompass the two regions (separated by a blue box) discussed here as being binding sites. Comparisons are more reliable in the second sequence, which is longer and better conserved. Conserved residues are highlighted in green; the important DI/V sequence is marked by asterisks. Domains 2 and 4 are more highly conserved. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fdp was expressed in E. coli with an N-terminal His6 tag for purification, as residues 19–155 of the full-length protein, which corresponds to the mature protein after cleavage of the N-terminal signal sequence. It constituted 12% of total soluble E. coli proteins and typical yields were 7.5 mg per litre of culture.
The NMR spectrum was sharp and well resolved and was assigned using standard triple resonance experiments on double labelled protein [42]. NMR spectra (particularly 15N relaxation experiments, not shown) indicate that the protein behaves as a monomer in solution, even at NMR concentrations. The structure was calculated using simulated annealing based on distance and angle restraints, and is shown in Fig. 2, with structural statistics in Table 1. It is an α + β structure, consisting of a wedge-shaped β-sandwich of approximately 30 Å diameter made up of two β-sheets, with six α-helices covering one face of the wedge. The structure is similar to those of other FAS1 domains whose structures have been determined: backbone RMSDs to TGFBIp, FAS1-4 and MBP70 are 2.4, 2.4 and 2.2 Å respectively (Fig. 2C and D). The structure of Fdp does not contain the helix α5 present in FAS1-4 (Fig. 2C), and has therefore a clearer split between the α-domain and the β-domain than does FAS1-4.
Fig. 2.
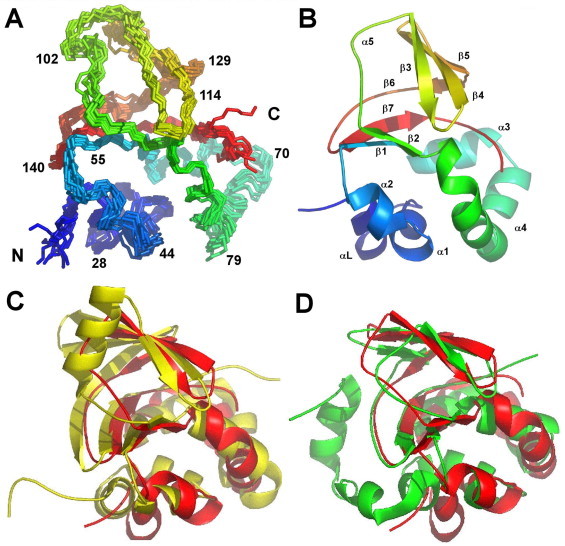
The solution structure of Fdp. (A) 10 overlaid structures, shown as a rainbow view, from blue at the N-terminus to red at the C-terminus. Only backbone atoms (Cα, C′, N) are shown. (B) Best structure as a cartoon, same colour scheme and orientation. The α-helix and β-sheet numbering is indicated. Labelling of helices and sheets follows that in [30]. This means that the first helix is αL rather than α1, α4 has a large bend in the middle, α5 is a helical turn rather than a full helix, and β6 is a short strand followed by a longer extended strand. (C) Best fit superposition to Drosophila FAS1-4 (Fdp red, FAS1 yellow). (D) Best fit superposition to M. tuberculosis MPB70 (Fdp red, MPB70 green). Superpositions and RMSD values in the text were based on the most highly conserved regions of secondary structure, corresponding to residues 45–50, 55–65, 82–94, 102–104, 114–122, 126–128 and 135–151 from Fdp. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Structural statistics for Fdp structure determination.
| Unrefined ensemblea | Refined bestb | |
|---|---|---|
| Restraint violations | ||
| NOE violations > 0.2 Å | 0 | 1 |
| Dihedral violations > 5° | 0 | 0 |
| RMSD from experimental restraints | ||
| Distance restraints (Å)c | 0.12 ± 0.0009 | 0.039 |
| Dihedral restraints (°)d | 0.14 ± 0.02 | 0.75 |
| Coordinate precision (Å)e | ||
| Backbone | 0.87 ± 0.17 | NA |
| All heavy atoms | 1.29 ± 0.55 | NA |
| Ramachandran analysisf | ||
| Most favoured region (%) | 79.1 | 86.4 |
| Additionally allowed region (%) | 19.1 | 11.8 |
| Generously allowed region (%) | 0.9 | 0.9 |
| Disallowed region (%) | 0.9 | 0.9 |
| Energy (kcal/mol)g | ||
| Overall | 165.4 ± 12.3 | −4126.8 |
| Bond | 7.4 ± 0.7 | 33.8 |
| Angle | 74.3 ± 4.0 | 178.6 |
| Dihedral | 9.8 ± 1.1 | 619.6 |
| VdW | 54.4 ± 6.4 | −369.4 |
| Electrostatics | NA | −4863.9 |
| NOE | 19.4 ± 2.9 | 124.0 |
| Dihedral (TALOS) | 0.2 ± 0.1 | 5.1 |
| Difference from ideal values | ||
| Bonds (Å) | 0.012 ± 0.001 | 0.0042 |
| Angles (°) | 0.37 ± 0.01 | 0.57 |
| Impropers (°) | 0.26 ± 0.02 | 2.12 |
Ensemble of 30 lowest energy structures picked from 100 in CNS 1.1.
Best structure after energy refinement in water using ARIA 1.2.
1788 Restraints, consisting of 619 intra-residue, 494 sequential, 304 medium-range (2 ≤ i − j ≤ 4), 317 long-range (i − j > 4) and 27 pairs of hydrogen bond restraints.
148 (74 φ and 74 ψ) obtained from TALOS.
After alignment of backbone atoms of residues 22–154.
Calculated using Procheck-NMR [54].
Calculated using CNS 1.1 and ARIA 1.2.
3.2. Fdp is involved in adherence properties of whole cells
Three independent insertionally inactivated fdp knockout mutants were constructed in R. sphaeroides and compared with wild type in adherence assays. Growth rates of mutant and wild type strains were similar under aerobic, semi-aerobic and anaerobic (photosynthetic) conditions, and there were no significant differences in levels of photosynthetic complexes as revealed by spectrophotometric analyses of dark/semi-aerobically cultured cells (data not shown). The assay measured the ability of stationary phase cells to clump together and thereby adhere to pegs in 96-well plates. Cell adherence was significantly reduced in the fdp mutants compared with the wild type strain (Fig. 3), from 8.8 × 103 cells mm−2 in wild type to 0.87 ± 0.15 × 102 cells mm−2 in the three mutants, confirming a clear role for Fdp in ability to adhere to external surfaces (Fig. 3A). This effect was confirmed to be specific for the fdp mutants and not attributable to the presence of the kanamycin resistance cassette present in these mutants by conducting experiments with other unrelated mutants containing this cassette, in which levels of adherent cells were comparable to wild type (data not shown).
Fig. 3.
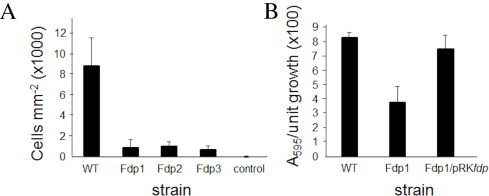
Adherence of R. sphaeroides NCIB 8253 strains. (A) Viable cells growing in biofilm attached to 68.1 mm2 pegs following 5 days aerobic growth at 34 °C. WT, wild type; Fdp1, Fdp2 and Fdp3, mutants possessing an insertionally-inactivated fdp gene; control, uninoculated medium (incorporating extremely low cell numbers that arise upon sonication). Standard deviation derived from nine replicate samples. (B) Biofilm formation during complementation studies of Fdp1 using pRKfdp determined by crystal violet staining, and expressed as a percentage of total culture growth in wells. Culturing was after 5 days at 34 °C. Fdp1/pRKfdp is a complementation by Fdp1 harbouring fdp inserted on pRK415. Standard deviation derived from nine replicate samples. Similar results were obtained for the other mutants.
An alternative adherence assay used crystal violet to measure adherence to the well [39]. In this assay, adherence was only reduced 2.5-fold (Fig. 3B). However, when the mutants were transformed with the complementation vector pRKfdp, almost full complementation (91%) by the fdp gene was achieved.
We have thus shown that Fdp in R. sphaeroides has a similar function to that in other members of the FAS1 family, namely cell adhesion. In bacteria, cell adhesion plays many important roles, particularly in the formation of biofilms, which is an important feature of many colonizing bacteria [43]. R. sphaeroides is however not pathogenic and lives in aquatic environments. It can grow chemoheterotrophically in the dark or light, photosynthetically in anaerobic environments or by anaerobic respiration in the dark [44]. A regulatable ability to aggregate would give it much greater control over its location. The ability to adjust its depth in the water column in response to environmental signals is thus likely to be crucial to its ability to move to suitable locations. In this context, it is significant that the expression of Fdp is regulated by redox status, being downregulated by the Prr oxygen-dependent regulatory system (Phillips-Jones et al., unpublished observations).
Our results do not provide any information on the nature of the binding partner of Fdp, except that Fdp shows no indications of dimerizing, even at NMR concentrations, implying that homomeric interactions are unlikely. There are also no other identified fasciclin domains in the R. sphaeroides genome, further ruling out homomeric interactions. In eukaryotic homologs, the ligand is a cell-surface integrin glycoprotein, which is the most likely type of binding partner.
3.3. Database
The atomic coordinates for FDP have been deposited with the Protein Data Bank; PDB: 1w7e (ensemble) and PDB: 1w7d (minimized best structure).
4. Discussion
4.1. Location of the protein interaction site
Here we report the structure of a new member of the FAS1 family, which unusually has only a single FAS1 domain. This new member is predicted to possess a signal peptide at the N-terminus, and the program PSORTb v3.0 [45] predicts a very high probability that it is attached to the cytoplasmic membrane, presumably via a C-terminal covalent anchor, consistent with its role in cell adhesion. Attempts to raise antibodies specific enough to identify the location of Fdp have proven unsuccessful.
The ubiquity of this domain across phyla suggests that it may represent an evolutionarily ancient cell adhesion domain, most likely functioning by binding to cell-surface proteins [1,18]. We therefore looked for residues that are conserved across a wide range of species, and are likely to be functionally important. To this end, we have prepared a new sequence alignment that is based on structural similarity rather than simply sequence similarity, using the existing structures as guides. Our structure of Fdp is important in guiding the alignment, because of the low level of sequence similarity between existing sequences, and the presence of insertions and deletions. The alignment is shown in Fig. 1, and identifies a number of highly conserved residues, in particular the H1 and H2 regions previously identified (Fig. 1a). These regions are adjacent in the structure, and form a large surface patch, followed by two β-strands that form the protein core and emerge on the opposite surface (Fig. 4A). Thus much of the conserved sequence appears to be essential because of its role in maintaining the structure, leaving the most likely binding site as the contiguous surface patch comprising residues 136–144 from H2 plus K50 and D52 from H1 (Fig. 4B).
Fig. 4.
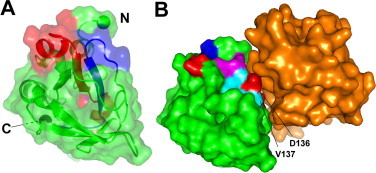
The binding surface on Fdp. (A) The surface of Fdp in partially transparent view. Conserved regions H1 and H2 are shown in red and blue respectively. They form a large patch at the top of the structure, run through the protein as two parallel β strands, and emerge on the opposite face. The N and C termini are indicated by spheres. (B) Suggested binding surface of Fdp; residues 50, 52 and 136–144, all other residues being green. Acidic residues are shown in red, basic in blue, hydrophobic in cyan, and hydrophilic in magenta. The key binding residues D136 and V137 are indicated. The orientation is the same as in (A). The orange surface is domain 3 of Drosophila FAS1, from the crystal structure of domains 3 and 4 [30], oriented so that domain 4 aligns with Fdp. For clarity, domain 4 is not shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
There have previously been several attempts to identify binding sites on FAS1 domains. An analysis of M. tuberculosis MPB70, based on highly conserved residues and disease-inducing mutations, identified the same region as being important, together with other residues on the opposite face of the protein that were suggested to form a second interaction site [32]. The best-supported site is the two residues DI or DV (136–137 in Fdp), at the start of H2. These residues have been shown to be important in cell adhesion of TGFBIp via integrin α3β1, since mutations in these positions showed loss of function, and synthetic pentapeptides containing this sequence blocked cell adhesion [8,17,46].
Other binding sites have also been proposed. In particular, residues Tyr71-His72 were suggested to form an alternative binding site specific for αvβ5 integrin [16]. However, His72 is largely buried in both Fdp and FAS1, implying that it is unlikely to be involved in protein recognition [47]. In addition, several hydrophobic residues flanking Tyr71-His72 were identified as important for interaction with αvβ5 integrin [16]. In Fdp these are generally either absent or buried, again making it unlikely that this site is important for Fdp. We conclude that the most likely binding site for FAS1 domains is residues 136–144 plus 50 and 52 (Fig. 1a).
It is of course possible that the remarkable conservation of the H1 and H2 regions, from bacteria to plants and humans, is unrelated to function, and that our imputation of a binding region here is incorrect. Against this we would argue that the cell adherence function of the fasciclin I domain is strongly conserved, and that as far as is known, the ligand type is also conserved [1]; that Fdp has a 29% sequence identity with Drosophila FAS1-4, this being a high enough similarity to make similarity of function very likely [48]; that the conserved residues identified here are surface-exposed and have no obvious structural role; and that most studies to date on a range of FAS domains have agreed in highlighting this region as the most likely binding site.
4.2. Corneal dystrophy mutations affect structural integrity not binding
There have been detailed studies of mutations in TGFBIp, which lead to a range of corneal dystrophies, characterized by amyloid-like protein deposits in the eye. Over half of the cases studied are caused by two mutations, at R124 in FAS1-1 and R555 in FAS1-4. The equivalent position to R555 is not well conserved in Fdp (Fig. 1; in Fig. 2 it is residue 75, just above the text α4 in Fig. 2B). In the Drosophila FAS1 structure, the equivalent residue is in a turn, and it was concluded that it should also be exposed in TGFBIp, and consequently mutations here could affect interactions with other proteins [30]. It is however diametrically opposite to the interaction site suggested here, and in our structure corresponds to a partially buried valine. We therefore suggest that mutations of R555 may lead to restructuring of the loop, and thus perturbation to the adjacent H1/H2 strands. In support of this suggestion, we note that different mutations at R555 can have either stabilizing or destabilizing effects [49,50]. Almost all the other disease-causing mutations are at sites that are buried in Fdp, and are therefore likely to lead to instability and consequent amyloid formation, rather than loss of interactions, as also suggested by others [27,29,30,32].
4.3. The interaction site is at the dimer interface
The N-terminus of Fdp is immediately adjacent to the proposed binding site, while the C-terminus is on the opposite face of the protein (Fig. 4). Assuming that the membrane attachment site is in its usual location at the C-terminus, then the Fdp binding site is in the most exposed region of the protein, as expected.
There is an important difference for eukaryotic homologs. In these proteins, the FAS1 domains generally occur in pairs. Our most detailed understanding comes from the crystal structure of the FAS1-3/4 pair from Drosophila, in which there is a substantial domain interface of 1700 Å2 [30]. Mutational studies of the homologous TGFBIp, discussed above, implicate the C-terminal domain as being by far the most important for function. The importance of the C-terminal domain can also be seen by sequence comparisons of Drosophila FAS1, TGFBIp and periostin (Fig. 1b), which show that the binding site residues are much more highly conserved in domains 2 and 4 (i.e., the C-terminal domain from each pair) than in the other two domains [51]. Studies using recombinant proteins and antagonist peptides identified domains 2 and 4 as both being important [8]. The clear implication is that the binding site in these proteins is located mainly or entirely on domains 2 and 4, which means that the binding site is more than 50% obscured by the domain/domain interaction (Fig. 4B), implying that binding must involve a competition between intramolecular and intermolecular binding (Fig. 5). The key residues D136 and V137 are almost completely buried in the interface (Fig. 4B). Further support for this hypothesis comes from the observation that one of the two mutations in TGFBIp that is not within domain 4 (P501T) is in the interface between domains 3 and 4, which could potentially disrupt the domain reorientation. Small-angle X-ray scattering (SAXS) has suggested that TGFBIp has a ‘beads on a string’ structure, with the four domains roughly extended in solution: there is thus clearly some motional freedom between domains, allowing the C-terminal domain to open out and expose the binding surface when required [52]. Inspection of the Drosophila FAS1 structure shows that the interdomain loop is long enough to allow considerable flexibility.
Fig. 5.
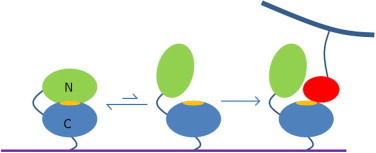
A model for the function of eukaryotic FAS1 domains. The binding site (orange) is located on the C-terminal domain of a pair of FAS1 domains, and is normally hidden by binding of the domain to its N-terminal partner. Access to the binding site requires dissociation of the N-terminal domain, and is therefore less favourable than it would be for the C-terminal domain on its own. Binding of a ligand (red) [or activation by other means such as binding of a third protein or post-translational modification] opens up the binding site on the C-terminal domain. The FAS1 pair and its ligand are shown as being anchored to cell walls. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.4. Implications of the binding site location
It is common to observe binding sites that are obscured by weak intramolecular binding. Such behaviour is often termed autoinhibition [53], and is used to regulate binding, such that the binding site is not available ‘accidentally’, only presenting when a genuine ligand binds. This reduces the probability of incorrect signal transmission. It can also be used to create further binding sites. Data presented here suggest that autoinhibition may be occurring in eukaryotic homologs of Fdp, with the binding site being the C-terminal domain, and its N-terminal partner serving as an inhibitor (Fig. 5). This may explain why the affinity of FAS1 proteins for their ligands is apparently weak; it also suggests that single C-terminal FAS1 constructs may bind more tightly. It is therefore likely that antagonists based on the C-terminal FAS1 domain would bind more tightly to their ligands than the full-length protein, and could form the basis for useful drug targets.
Acknowledgements
We thank Mary Phillips-Jones, Eun-Lee Jeong and Sam Broad (University of Leeds) for advice and plasmids, Peter J.F. Henderson (University of Leeds, UK) for helpful discussions and for reading an early version of the manuscript, Samuel Kaplan (University of Texas Health Science Center, USA) for discussions and provision of unpublished information, Richard Tunnicliffe for help with structure calculation and Arthur Moir (University of Sheffield) for protein N-terminal sequencing. This work was funded through a White Rose Studentship (RGM).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Burroughs A.M., Balaji S., Iyer L.M., Aravind L. Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elkins T., Zinn K., McAllister L., Hoffmann F.M., Goodman C.S. Genetic analysis of a Drosophila neural cell-adhesion molecule: interaction of fasciclin-I and Abelson tyrosine kinase mutations. Cell. 1990;60:565–575. doi: 10.1016/0092-8674(90)90660-7. [DOI] [PubMed] [Google Scholar]
- 3.McAllister L., Goodman C.S., Zinn K. Dynamic expression of the cell adhesion molecule fasciclin-I during embryonic development in Drosophila. Development. 1992;115:267–276. doi: 10.1242/dev.115.1.267. [DOI] [PubMed] [Google Scholar]
- 4.McAllister L., Rehm E.J., Goodman C.S., Zinn K. Alternative splicing of micro-exons creates multiple forms of the insect cell-adhesion molecule fasciclin-I. J. Neurosci. 1992;12:895–905. doi: 10.1523/JNEUROSCI.12-03-00895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zinn K., McAllister L., Goodman C.S. Sequence analysis and neuronal expression of fasciclin-I in grasshopper and Drosophila. Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 6.Hortsch M., Goodman C.S. Drosophila fasciclin-I, a neural cell adhesion molecule, has a phosphatidylinositol lipid membrane anchor that is developmentally regulated. J. Biol. Chem. 1990;265:15104–15109. [PubMed] [Google Scholar]
- 7.Horiuchi K., Amizuka N., Takeshita S., Takamatsu H., Katsuura M., Ozawa H., Toyama Y., Bonewald L.F., Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor β. J. Bone Miner. Res. 1999;14:1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.E., Kim S.J., Lee B.H., Park R.W., Kim K.S., Kim I.S. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, βig-h3. J. Biol. Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki H., Auclair D., Kaji M., Fukai I., Kiriyama M., Yamakawa Y., Fujii Y., Chen L.B. Serum level of the periostin, a homologue of an insect cell adhesion molecule, in thymoma patients. Cancer Lett. 2001;172:37–42. doi: 10.1016/s0304-3835(01)00633-4. [DOI] [PubMed] [Google Scholar]
- 10.Gillan L., Matei D., Fishman D.A., Gerbin C.S., Karlan B.Y., Chang D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for avb3 and avb5 integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 11.Takeshita S., Kikuno R., Tezuka K., Amann E. Osteoblast-specific factor II: cloning of a putative bone adhesion protein with homology with the insect protein fasciclin-I. Biochem. J. 1993;294:271–278. doi: 10.1042/bj2940271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi H., Tsujimoto M. FEEL-1, a novel scavenger receptor with in vitro bacteria-binding and angiogenesis-modulating activities. J. Biol. Chem. 2002;277:34264–34270. doi: 10.1074/jbc.M204277200. [DOI] [PubMed] [Google Scholar]
- 13.Ruan K., Bao S., Ouyang G. The multifaceted role of periostin in tumorigenesis. Cell. Mol. Life Sci. 2009;66:2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo Y., Siriwardena B.S.M.S., Hatano H., Ogawa I., Takata T. Periostin: Novel diagnostic and therapeutic target for cancer. Histol. Histopathol. 2007;22:1167–1174. doi: 10.14670/HH-22.1167. [DOI] [PubMed] [Google Scholar]
- 15.Nummela P., Lammi J., Soikkeli J., Saksela O., Laakkonen P., Holtta E. Transforming growth factor beta-induced (TGFBI) is an anti-adhesive protein regulating the invasive growth of melanoma cells. Am. J. Pathol. 2012;180:1663–1674. doi: 10.1016/j.ajpath.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.E., Jeong H.W., Nam J.O., Lee B.H., Choi J.Y., Park R.W., Park J.Y., Kim I.S. Identification of motifs in the fasciclin domains of the transforming growth factor-β-induced matrix protein βig-h3 that interact with the avb5 integrin. J. Biol. Chem. 2002;277:46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- 17.Park S.W., Bae J.S., Kim K.S., Park S.H., Lee B.H., Choi J.Y., Park J.Y., Ha S.W., Kim Y.L., Kwon T.H., Kim I.S., Park R.W. βig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with a3b1 integrin. Exp. Mol. Med. 2004;36:211–219. doi: 10.1038/emm.2004.29. [DOI] [PubMed] [Google Scholar]
- 18.Huber O., Sumper M. Algal-cams – isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to Drosophila fasciclin-I. EMBO J. 1994;13:4212–4222. doi: 10.1002/j.1460-2075.1994.tb06741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson K.L., Jones B.J., Bacic A., Schultz C.J. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borner G.H.H., Sherrier D.J., Stevens T.J., Arkin I.T., Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in arabidopsis. A genomic analysis. Plant Physiol. 2002;129:486–499. doi: 10.1104/pp.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H.Z., Kim Y., Guo Y., Stevenson B., Zhu J.K. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai S., Matsumoto J., Nagasuga T. Specific skin-reactive protein from culture filtrate of Mycobacterium bovis BCG. Infect. Immunity. 1981;31:1152–1160. doi: 10.1128/iai.31.3.1152-1160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsrud P., Lindblad P. Fasciclin domain proteins are present in Nostoc symbionts of lichens. Appl. Environ. Microbiol. 2002;68:2036–2039. doi: 10.1128/AEM.68.4.2036-2039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds W.S., Schwarz J.A., Weis V.M. Symbiosis-enhanced gene expression in cnidarian–algal associations: cloning and characterization of a cDNA, sym32, encoding a possible cell adhesion protein. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2000;126:33–44. doi: 10.1016/s0742-8413(00)00099-2. [DOI] [PubMed] [Google Scholar]
- 25.Ulstrup J.C., Jeansson S., Wiker H.G., Harboe M. Relationship of secretion pattern and MPB70 homology with osteoblast-specific factor-2 to osteitis following Mycobacterium bovis BCG vaccination. Infect. Immunity. 1995;63:672–675. doi: 10.1128/iai.63.2.672-675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oke V., Long S.R. Bacterial genes induced within the nodule during the Rhizobium–legume symbiosis. Mol. Microbiol. 1999;32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 27.Thapa N., Lee B.-H., Kim I.-S. TGFBIp/βig-h3 protein: a versatile matrix molecule induced by TGFβ. Int. J. Biochem. Cell. Biol. 2007;39:2183–2194. doi: 10.1016/j.biocel.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Munier F.L., Frueh B.E., Othenin-Girard P., Uffer S., Cousin P., Wang M.X., Héon E., Black G.C.M., Blasi M.A., Balestrazzi E., Lorenz B., Escoto R., Barraquer R., Hoeltzenbein M., Gloor B., Fossarello M., Singh A.D., Arsenijevic Y., Zografos L., Schorderet D.F. BIGH3 mutation spectrum in corneal dystrophies. Invest. Ophthalmol. Vis. Sci. 2002;43:949–954. [PubMed] [Google Scholar]
- 29.Kannabiran C., Klintworth G.K. TGFBI gene mutations in corneal dystrophies. Hum. Mut. 2006;27:615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 30.Clout N.J., Tisi D., Hohenester E. Novel fold revealed by the structure of a FAS1 domain pair from the insect cell adhesion molecule fasciclin I. Structure. 2003;11:197–203. doi: 10.1016/s0969-2126(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 31.Yoo J.-H., Kim E., Kim J., Cho H.-S. Crystallization and preliminary crystallographic analysis of the fourth FAS1 domain of human βig-h3. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:893–895. doi: 10.1107/S1744309107039358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr M.D., Bloemink M.J., Dentten E., Whelan A.O., Gordon S.V., Kelly G., Frenkiel T.A., Hewinson R.G., Williamson R.A. Solution structure of the Mycobacterium tuberculosis complex protein MPB70 – from tuberculosis pathogenesis to inherited human corneal disease. J. Biol. Chem. 2003;278:43736–43743. doi: 10.1074/jbc.M307235200. [DOI] [PubMed] [Google Scholar]
- 33.Hunter C.N., Turner G. Transfer of genes coding for apoproteins of reaction center and light-harvesting LH1 complexes to Rhodobacter sphaeroides. J. Gen. Microbiol. 1988;134:1471–1480. [Google Scholar]
- 34.Keen N.T., Tamaki S., Kobayashi D., Trollinger D. Improved broad host range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 35.Potter C.A., Ward A., Laguri C., Williamson M.P., Henderson P.J.F., Phillips-Jones M.K. Expression, purification and characterisation of full-length histidine protein kinase RegB from Rhodobacter sphaeroides. J. Mol. Biol. 2002;320:201–213. doi: 10.1016/S0022-2836(02)00424-2. [DOI] [PubMed] [Google Scholar]
- 36.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 38.Linge J.P., Williams M.A., Spronk C., Bonvin A., Nilges M. Refinement of protein structures in explicit solvent. Proteins: Struct. Funct. Bioinf. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 39.Djordjevic D., Wiedmann M., McLandsborough L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002;68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter C.N., Coomber S.A. Cloning and oxygen-regulated expression of the bacteriochlorophyll biosynthesis genes bchE, bchB, bchA and bchC of Rhodobacter sphaeroides. J. Gen. Microbiol. 1988;134:1491–1497. [Google Scholar]
- 41.Duggan P.S., Parker S.D., Phillips-Jones M.K. Characterisation of a Rhodobacter sphaeroides gene that encodes a product resembling Escherichia coli cytochrome b561 and R. sphaeroides cytochrome b562. FEMS Microbiol. Lett. 2000;189:239–246. doi: 10.1111/j.1574-6968.2000.tb09237.x. [DOI] [PubMed] [Google Scholar]
- 42.Moody R.G., Phillips-Jones M.K., Williamson M.P. NMR assignment of the Rhodobacter sphaeroides fasciclin-1 domain protein (Fdp) Biomol. NMR Assign. 2007;1:11–12. doi: 10.1007/s12104-007-9011-6. [DOI] [PubMed] [Google Scholar]
- 43.Jefferson K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004;236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Roh J.H., Smith W.E., Kaplan S. Effects of oxygen and light intensity on transcriptome expression in Rhodobacter sphaeroides 2.4.1 – Redox active gene expression profile. J. Biol. Chem. 2004;279:9146–9155. doi: 10.1074/jbc.M311608200. [DOI] [PubMed] [Google Scholar]
- 45.Yu N.Y., Wagner J.R., Laird M.R., Melli G., Rey S., Lo R., Dao P., Sahinalp S.C., Ester M., Foster L.J., Brinkman F.S.L. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park S.J., Park S., Ahn H.C., Kim I.S., Lee B.J. Conformational resemblance between the structures of integrin-activating pentapetides derived from βig-h3 and RGD peptide analogues in a membrane environment. Peptides. 2004;25:199–205. doi: 10.1016/j.peptides.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 47.Clout N., Hohenester E. A model of FAS1 domain 4 of the corneal protein βig-h3 gives a clearer view on corneal dystrophies. Mol. Vis. 2003;9:440–448. [PubMed] [Google Scholar]
- 48.Feng D.F., Doolittle R.F. Progressive alignment of amino acid sequences and construction of phylogenetic trees from them. Methods Enzymol. 1996;266:368–382. doi: 10.1016/s0076-6879(96)66023-6. [DOI] [PubMed] [Google Scholar]
- 49.Elavazhagan M., Lakshminarayanan R., Zhou L., Ting L.W., Tong L., Beuerman R.W., Chaurasia S.S., Mehta J.S. Expression, purification and characterization of fourth FAS1 domain of TGFβIp-associated corneal dystrophic mutants. Protein Expr. Purif. 2012;84:108–115. doi: 10.1016/j.pep.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 50.Runager K., Basaiawmoit R.V., Deva T., Andreasen M., Valnickova Z., Sørensen C.S., Karring H., Thøgersen I.B., Christiansen G., Underhaug J., Kristensen T., Nielsen N.C., Klintworth G.K., Otzen D.E., Enghild J.J. Human phenotypically distinct TGFBI corneal dystrophies are linked to the stability of the fourth FAS1 domain of TGFBIp. J. Biol. Chem. 2011;286:4951–4958. doi: 10.1074/jbc.M110.181099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orecchia P., Conte R., Balza E., Castellani P., Borsi L., Zardi L., Mingari M.C., Carnemolla B. Identification of a novel cell binding site of periostin involved in tumour growth. Eur. J. Cancer. 2011;47:2221–2229. doi: 10.1016/j.ejca.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 52.Basaiawmoit R.V., Oliveira C.L.P., Runager K., Sørensen C.S., Behrens M.A., Jonsson B.H., Kristensen T., Klintworth G.K., Enghild J.J., Pedersen J.S., Otzen D.E. SAXS models of TGFBIp reveal a trimeric structure and show that the overall shape is not affected by the Arg124His mutation. J. Mol. Biol. 2011;408:503–513. doi: 10.1016/j.jmb.2011.02.052. [DOI] [PubMed] [Google Scholar]
- 53.Williamson M.P. Garland Science; New York: 2011. How Proteins Work. [Google Scholar]
- 54.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]


