Abstract
Head and neck squamous cell carcinoma (HNSCC) is an aggressive life-threatening disease associated with high mortality rates. While efforts have been made to explore the molecular mechanisms that contribute to the initiation and progression of HNSCC, most studies focus on protein-coding genes. Understanding of the genomic aberrations associated with noncoding genes (such as microRNAs) and their effects on HNSCC is still relatively limited. Recent evidence suggests that deregulation of microRNA genes (such as downregulation of miR-138) plays an important role in HNSCC. While deregulation of miR-138 has been frequently observed in HNSCC and other cancer types, the precise roles of miR-138 in tumorigenesis remain elusive. Recent bioinformatics analyses and functional studies using in vitro and in vivo systems have identified a number of functional targets for miR-138. These include genes that participate in essential biological processes that are highly relevant to the initiation and progression of HNSCC, including cell migration, epithelial to mesenchymal transition, cell cycle progression, DNA damage response and repair, senescence, and differentiation. However, the biological systems, study design, and data interpretation from these studies are highly variable, which hinder our understanding of the role of miR-138 in tumorigenesis at molecular level. In this review, we will first introduce the significance of microRNA deregulation in HNSCC. We will then provide a comprehensive review and integrative analysis of the existing studies on miR-138, and aim to define its molecular mechanisms that contribute to the initiation and progression of HNSCC.
1. INTRODUCTION
Head and neck/oral cancer (HNOC) is an understudied disease. While efforts have been made to identify molecular mechanisms that contribute to the initiation and progression of HNOC, most studies focus on protein-coding genes. MicroRNA deregulation and its role(s) in HNOC are still not fully elucidated. Recent studies on other types of cancers indicated that microRNAs play critical roles in tumorigenesis, including regulating cell migration and other cellular processes that contribute to metastasis, such as epithelial to mesenchymal transition (EMT) and extra-cellular matrix (ECM) remodeling. For example, miR-200 family and miR-205 have been confirmed as EMT modulators in several different cell types through the regulation of ZEB1 and ZEB2 (Burk et al., 2008; Korpal and Kang, 2008; Korpal et al., 2008; Park et al., 2008). A number of recent studies from our laboratory demonstrated that deregulation of miR-138 in HNOC also contribute to enhanced cell migration and EMT in HNOC ( Jiang et al., 2010; Jin et al., 2011; Liu et al., 2009b, 2011). Studies of other cancer types demonstrated that miR-138 regulates the cell cycle progression (Liu et al., 2012; Wang et al., 2012b), DNA-damage response (Wang et al., 2011b), and senescence (Mitomo et al., 2008; Rivetti di Val Cervo et al., 2012). These reports highlight the critical roles of miR-138 deregulation in tumorigenesis. Here, we presented a comprehensive review on the existing studies on miR-138 aiming to define the role of miR-138 deregulation in HNOC.
2. BACKGROUND
2.1. Head and Neck Squamous Cell Carcinoma
HNOC is the sixth most common cancer worldwide, accounting for approximately 6% of all cancer cases. According to the American Cancer Society, new oral cancer cases increased over 40% during the past 10 years, while the overall new cancer cases in the United States increased by about 20% in the same time period. Moreover, deaths associated with oral cancer increased by 10%, compared to the 2.8% increase in deaths for all cancer cases. The severity of HNOC is even worse worldwide, with over 263,000 new cases being diagnosed each year. For example, in South-Central Asia, home of approximately 20% of the world’s population, HNOC is the second most common cancer and the second leading death-causing disease in males (Global Cancer Facts & Figures, 2nd Edition; ACS, 2011).
Head and neck squamous cell carcinoma (HNSCC), which originates from the epithelium lining of this region, makes up the majority of HNOC (over 90%). As an invasive epithelial neoplasm, HNSCC most commonly arises in the tongue, floor of the mouth, gingival, buccal mucosa, pharynx, and larynx. It typically presents as a painless ulcer with raised borders, firm mass or indurated nodule, and may show early and extensive lymph-node metastases. The stages (tumor, node and metastasis) of HNSCC at diagnosis have a strong influence on survival and prognosis. Lymph-node metastasis decreases the survival rate by about 50%. Treatment for HNSCC usually includes surgery, often a radical en bloc resection of the tumor, lymph nodes, and involved soft tissue and bone. Surgical treatment is often combined with pre- and/or postoperative chemotherapy and radiotherapy, based on clinical judgment and histopathological results. Despite these interventions, more than 50% of patients with HNSCC will experience local relapse and distant metastasis. Recurrences and distant metastases are associated with poor prognoses. Furthermore, surgical intervention causes facial contour defects and can lead to functional impairment and psychological trauma in HNSCC patients. Unfortunately, the survival rates of patients with HNSCC have not significantly improved over the past several decades.
Heavy smoking and alcohol consumption impact the occurrence of HNSCC (Murata et al., 1996), with the intensity and duration of tobacco consumption directly correlated to the risk of developing HNSCC. However, in recent years, more HNSCC cases have been found in nonsmokers and nonalcohol consumers, which implies that environmental, immunologic, and/or genetic factors also contribute to the initiation and progression of HNSCC. For example, viral infection has been found in some cases of HNSCC. Human papillomavirus (HPV) has been widely studied and demonstrated to play an important role in the development of cervical cancer (Strati et al., 2006; Termine et al., 2008). Recent molecular and epidemiologic studies showed that about 15–25% of HNSCC contain genomic DNA from HPV, especially those arising in oropharyngeal sites, including tonsillar cancers. In fact, HPV is implicated in the increased incidence of HNSCC in several countries over the last few decades (Chaturvedi et al., 2011; Hong et al., 2010; Nasman et al., 2009). In the United States, the incidence of HPV− negative HNSCC declined by about 50% from 1988 to 2004, while the incidence of HPV-positive HNSCC increased by over 200% during the same period. Most impacted by this increase were young individuals, Caucasian individuals, and men (Chaturvedi et al., 2011), which happen to be the same groups of individuals that are associated with a higher percentage of oral HPV infection in the United States (Gillison et al., 2012). Lack of certain dietary factors such as vitamin E may also contribute to HNSCC tumorigenesis.
2.2. MicroRNA and HNSCC
One of the most significant achievements in the biological science in the last decade is the discovery of RNA interference (RNAi), a process within living cells that regulates gene expression at posttranscriptional levels. Historically, this process was described by other more generic names, such as cosuppression and posttranscriptional gene silencing. Only after the molecular mechanisms underlying these apparently unrelated processes were fully understood did it become apparent that they all described the RNAi phenomenon. RNAi is an RNA-dependent gene silencing process that is controlled by the RNA-induced silencing complex (RISC) and is initiated by two types of small RNA molecules—microRNA and small interfering RNA. However, the function of microRNA appears to be far beyond RNAi alone, including a direct interaction with gene promoters and epigenetic regulation of the DNA methylation and histone modification. By affecting gene regulation, microRNAs are involved in diverse biological activities, from cell differentiation, proliferation, apoptosis to the endocrine system, immune response, neurotransmitter synthesis, and circadian rhythm, to name a few.
MicroRNAs are the 21–23 nucleotide single-stranded RNA molecules found in eukaryotic cells. These tiny molecules are newcomers to the biological research. In the early years, the progress on microRNA research was slow and experienced substantial growing pains. While the first microRNA, lin-4, was characterized in Caenorhabditis elegans in the early 1990s (Lee et al., 1993), it was not until 2000 that researchers knew that microRNAs existed in humans. The short length and uniqueness of each microRNA rendered many conventional hybridization-based methods ineffective; very small RNAs are difficult to reliably amplify or label without introducing bias. In addition, hybridization-based methods for microRNA profiling relied on probes designed to detect known microRNAs or known microRNA species previously identified by sequencing or homology search. Moreover, the wide range of microRNA expression, from tens of thousands to just few molecules per cell, complicated the detection of microRNAs expressed at low copy numbers. Hence, many novel microRNAs may exist even in well-explored species. Nevertheless, recent advances in genomic technologies, data analysis, and bioinformatics approaches have made a significant impact on microRNA research. The microRNA field has experienced a major explosion in recent years. For example, the next generation deep-sequencing platforms are ideal for detecting and quantifying both known and novel microRNA sequences with high sensitivity and for a relatively low cost (Morin et al., 2008). The microRNA gene family is continuously growing with novel members discovered in association with rapid advances in genomic technologies, and reports on the functional characterizations of specific microRNA genes have dominated the recent literature.
MicroRNA deregulation is a frequent event in HNSCC. We recently performed a meta-analysis based on 13 independent microRNAs profiling studies on HNSCC (Chen et al., 2012). Among the 432 differentially expressed microRNAs reported in these studies, 264 were up-regulated and 168 down-regulated microRNAs. The downregulation of miR-138 was initially reported in 4 cases of HNSCC of tongue (Wong et al., 2008), and thyroid carcinoma cell lines (Mitomo et al., 2008). In a recent study, to identify microRNAs associated with HNSCC metastasis, Liu et al. (2009b) examined the differential expression of microRNAs in 6 paired HNSCC cell lines with different metastatic potential (UM1/UM2, 1386Tu/1386Ln and 686Tu/686Ln). Reduced expression of miR-138 was observed in all 3 high invasive aggressive cell lines (UM1, 1386Ln and 686Ln). Restoring miR-138 expression led to suppression of cell invasion, cell cycle arrest and induced apoptosis. In contrast, the knockdown of miR-138 expression enhanced cell invasion and suppressed apoptosis. The fact that highly metastatic cells often showed reduced expression of miR-138 suggests the role of microRNA-138 as a tumor suppressor as well as a therapeutic target for HNSCC patients at risk of metastasis. The same group of investigators later validated the downregulation of miR-138 in HNSCC tissue samples ( Jiang et al., 2011).
2.3. MicroRNA-138 Biosynthesis
MicroRNA biogenesis has been well characterized (for detailed discussion on microRNA biogenesis, we refer you to earlier reviews (Bartel, 2004; Carthew and Sontheimer, 2009)). Two miR-138 genes were initially predicted in the mouse genome termed miR-138-1 and miR-138-2 (Lagos-Quintana et al., 2002; Weber, 2005), and their human homologs were mapped to chromosome 3p21.33 and 16q13, respectively. Interestingly, losses of heterozygosity (LOHs) at both chromosome loci have been frequently detected in HNSCC and appears to correlate with tumor progression (i.e. cervical lymph-node metastasis) (Hogg et al., 2002; Piccinin et al., 1998; Wang et al., 1999). Nevertheless, based on a series of analysis of the gene transcripts, it is believed that only one of the predicted miR-138 genes (miR-138-2) is functionally transcribed in humans (Obernosterer et al., 2006).
The miR-138-2 gene is an intergenic microRNA gene. While the precise genomic organization of miR-138-2 gene is not clear (the gene promoter has not been characterized and the length of the pri-miR-138-2 is unknown), it is believed that miR-138-2 gene utilizes the canonical pathway for its biogenesis (Obernosterer et al., 2006). In the canonical pathway, microRNAs are first transcribed as primary transcripts (pri-miR-138-2) with a local-hairpin structure, which also possess many characteristics of a typical messenger RNAs (mRNA), such as 5′ cap and 3′ poly-A tail. The pri-miR-138-2 is processed into 69-nucleotide stem-loop structures (known as pre-miR-138-2) in the cell nucleus by a protein complex consisting of the nuclease Drosha and the double-stranded RNA binding protein Pasha. The pre-miR-138-2 is then transported to the cytoplasm by Exportin-5 (Exp5; a member of the Ran transport receptor family). Once in the cytoplasm, pre-miR-138-2 is further cleaved by Dicer (a second RNase III endonuclease) to form a short double strand microRNA:microRNA* duplex. Finally, the microRNA:microRNA* duplex is unwound into 23-nucleotide mature microRNA and microRNA* by a helicase. The mature microRNAs are then incorporated into the RISC.
The genes for miR-138 have been used as a model system to investigate the posttranscriptional regulation of microRNA biogenesis (Obernosterer et al., 2006). Obernosterer et al. showed that while the precursor (pre-miR-138-2) is expressed ubiquitously in all the tissues they examined, the mature microRNA (miR-138) is found only in specific tissue/cell types. As such, they concluded that the tissue-specific expression of miR-138 is achieved in part by regulatory mechanism at posttranscriptional level (maturation steps).
2.4. MicroRNA-138-mediated Posttranscriptional Regulation
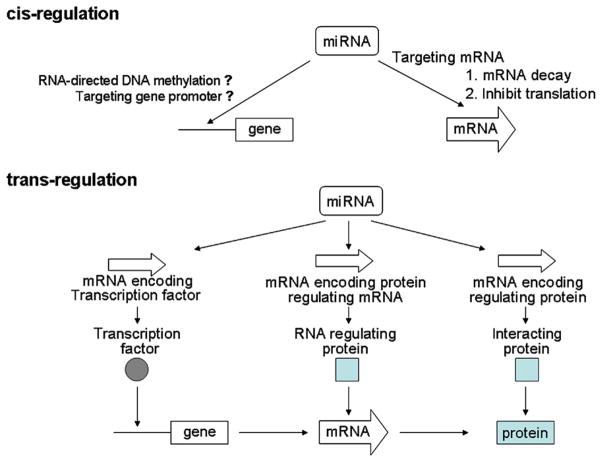
MicroRNAs are not involved directly in protein synthesis, but are believed to control the expression of more than one-third of the protein-coding genes in the human genome (Lewis et al., 2005, 2003; Xie et al., 2005). Each microRNA can target many mRNA transcripts and regulate hundreds of genes downstream. One microRNA can have multiple target sites in the mRNA transcript of a downstream gene. Therefore, microRNAs contribute a newly recognized level of gene expression regulation. As illustrated in Fig. 9.1, the potential mechanisms of microRNA-mediated gene regulation are multifactorial and encompass interaction(s) among different mechanisms. It has been demonstrated that microRNA binds to the target mRNA and regulates gene expression at the posttranscriptional levels (e.g. enhancing mRNA degradation and inhibiting translation). This cis-regulation occurs by binding the ~22 nucleotide mature microRNA to an imperfectly matched sequence in the target mRNA. Following the expression changes of specific microRNA-targeted genes (e.g. genes coding for transcription factors and genes coding for RNA regulating proteins), subsequent effects may alter the levels of other mRNAs (or protein interactions), and thus microRNA may exert its effects on the expressed genome through trans-regulatory mechanism(s). For more details on microRNA biogenesis, basic functions, and their roles in normal physiology and diseases, numerous excellent reviews are recommended (Ambros, 2004; Bartel, 2004; Bushati and Cohen, 2007; Chang and Mendell, 2007; Filipowicz et al., 2008; Kloosterman and Plasterk, 2006; Stefani and Slack, 2008).
Figure 9.1. Potential microRNA regulation mechanisms.
(Adapted from Comparative and functional genomics; Liu et al. (2009a)). (For color version of this figure, the reader is referred to the online version of this book.)
The microRNA-mediated posttranscriptional regulation occurs by binding the ~22 nucleotide mature microRNA to an imperfectly matched sequence in the target mRNA, where perfect matching of the seed region (typically encompasses the 5′ bases 2–7 of the microRNA) appears to be essential. The microRNA-138-FOSL1 regulatory module has been used as a model system to investigate the microRNA-targeting mechanism ( Jin et al., 2011). Using 6 commonly available bioinformatics tools (4-way PicTar, 5-way PicTar, TargetScanS, TargetScanHuman 5.1, miRanda at microrna.org, and miRanda at miRBase), only one canonical miR-138-targeting site was consistently identified in the 3′-UTR of the Fos-like antigen 1 (FOSL1) mRNA in a region that is not highly conserved ( Jiang et al., 2011). It is worth noting that the microRNA-targeting prediction tools described above are limited to the 3′-UTR of the mRNA sequence and do not consider possible noncanonical-targeting sites (e.g. allowing for G:U wobble base pairing). If the bioinformatics analysis was extended to the entire mRNA molecule, two additional canonical-targeting sites were located in the CDs of the FOSL1 mRNA. When G:U wobble base pairing was considered, three additional high-affinity (based on predicted minimum free energy) noncanonical-targeting sites were identified: one each in the 5′-UTR, CDs and 3′-UTR regions of the FOSL1 mRNA. As such, a total of six miR-138-targeting sites were identified (Table 9.1), one in the 5′-UTR, three in the CDs, and two in the 3′-UTR of the FOSL1 mRNA. These targeting sites were then experimentally validated in the HNSCC cell lines using luciferase reporter gene assays and ribonucleoprotein-immunoprecipitation (RIP-IP) assays (Jin et al., 2011).
Table 9.1.
Distribution of miR-138-targeting sites
| Number of genes | Location | Number of canonical sites per geneb | Number of noncanonical sites per genec | |
|---|---|---|---|---|
| FOSL1 | 1 | 5′-UTR | 0 | 1 |
| CDS | 2 | 1 | ||
| 3′-UTR | 1 | 1 | ||
|
| ||||
| FOS gene family | 4 | 5′-UTR | 0 | 0.5 |
| CDS | 1 | 4 | ||
| 3′-UTR | 0.25 | 1.5 | ||
|
| ||||
| Genes regulated by miR-138a | 194 | 5′-UTR | 0.079*** | 0.619* |
| CDS | 0.521** | 3.448 | ||
| 3′-UTR | 0.342*** | 1.501** | ||
|
| ||||
| Genes not regulated by miR-138 | 28,942 | 5′-UTR | 0.033 | 0.537 |
| CDS | 0.394 | 3.408 | ||
| 3′-UTR | 0.125 | 1.259 | ||
Genes regulated by miR-138 were determined as described previously [ Jiang et al. (2011) Hum Genet 129(2):189–97]. In brief, UM1 cells were transfected with miR-138 mimic and control mimic, and the transcript profiling was performed by microarrays. A total of 194 transcripts were downregulated upon ectopic transfection of miR-138 as measured by Affymetrix U133 + 2.0 arrays, which contain 29,136 unique and mapped transcripts used in our analysis. The significance of possible enrichment of predicted miR-138-targeting sites in the down-regulated transcripts was tested by Fisher’s exact test (
p < 0.1;
p < 0.05;
p < 0.001).
The canonical targeting site of miR-138 in the mRNA sequence is defined by the presence of seed sequence CACCAGC.
The noncanonical targeting site allows G:U wobble base pairing. The seed sequence on the mRNA molecule allows the substitution of C by U, and substitution of A by G.
In addition to FOSL1, Jin et al. also suggested that other members of the Fos-gene family (Fos, FosB, and FOSL2) may be regulated by miR-138. As showed in Table 9.1, in addition to FOSL1, miR-138-targeting sites were also identified on mRNAs of Fos, FosB, and FOSL2. It is worth noting that two canonical and seven noncanonical miR-138-targeting sites were identified in the CDs of the FosB mRNA, and three additional noncanonical sites were identified in the 3′-UTR of the FosB mRNA. This is consistent with the observation that ectopic transfection of miR-138 led to reduced FosB expression, and the knockdown of miR-138 led to an apparent increase in FosB levels in HNSCC cell lines ( Jin et al., 2011). Based on the sequence search and a previous study that identified 194 genes that were significantly down-regulated by miR-138 Jiang et al. (2011) also demonstrated significant enrichment of canonical microR-138-targeting sites in the 5′-UTR, CDs, and 3′-UTR of the genes that were regulated by miR-138. Significant enrichment of noncanonical microR-138-targeting sites was observed in the 3′-UTR of the genes that were regulated by miR-138. Apparent enrichment of noncanonical microR-138-targeting sites was also observed in 5′-UTR, but the difference is not statistically significant. These observations support the hypothesis that both canonical and noncanonical microRNA-targeting sites, and the microRNA-targeting sites located in all areas of mRNA molecule (e.g. 5′-UTR, CDs, 3′-UTR), may contribute to microRNA-mediated posttranscriptional regulation.
3. MICRORNA-138 DEREGULATION IN HNSCC AND ITS FUNCTIONAL RELEVANCE
The downregulation of miR-138 has been consistently observed in HNSCC (Jiang et al., 2011; Mitomo et al., 2008; Wong et al., 2008), as well as in other cancer types (Liu et al., 2012; Song et al., 2011; Wang et al., 2012b). Bioinformatics analysis revealed that miR-138 regulates a number of molecular pathways associated with tumorigenesis (Table 9.2). A number of miR-138-targeting genes have been experimentally validated in various cancer types (Table 9.3). These validated miR-138-target genes play essential roles in the initiation and progression of HNSCC, including cell migration, EMT, cell cycle regulation, DNA damage response and repair, and senescence.
Table 9.2.
Molecular pathways regulated by microRNA-138a
| KEGG Pathway | Pathway ID | Number of genes targeted in the pathway | −ln (p-value)b |
|---|---|---|---|
| Axon guidance | hsa04360 | 12 | 23.65 |
| Wnt-signaling pathway | hsa04310 | 7 | 4.63 |
| Bladder cancer | hsa05219 | 3 | 3.22 |
| Chronic myeloid leukemia | hsa05220 | 4 | 2.87 |
| mTOR-signaling pathway | hsa04150 | 3 | 2.73 |
| Small cell lung cancer | hsa05222 | 4 | 2.35 |
| ErbB-signaling pathway | hsa04012 | 4 | 2.25 |
| Nonsmall cell lung cancer | hsa05223 | 3 | 2.19 |
| Acute myeloid leukemia | hsa05221 | 3 | 2.13 |
| Glioma | hsa05214 | 3 | 1.68 |
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 | 3 | 1.63 |
| p53-signaling pathway | hsa04115 | 3 | 1.49 |
| Cell cycle | hsa04110 | 4 | 1.34 |
| Melanoma | hsa05218 | 3 | 1.33 |
| Pancreatic cancer | hsa05212 | 3 | 1.29 |
The microRNA-targeted KEGG pathways were searched from the DIANA-mirPath database. TargetScan 5 was used as target prediction software.
Computed using DIANA-mirPath [Papadopoulos et al. (2009) Bioinformatics 25:1991–3].
Table 9.3.
Functionally validated molecule targets of miR-138
| Gene | Gene name | Number of target site | Location of target site | Validation method | Putative function in tumorigenesis | Cancer type | Ref. |
|---|---|---|---|---|---|---|---|
| RhoC | Ras homolog gene family, member C | 1 | 3′-UTR | Reporter assay, RIP-IPc | Cell migration | HNSCC | (Jiang et al., 2010) |
| ROCK2 | Rho-associated, coiled-coil-containing protein kinase 2 | 1 | 3′-UTR | Reporter assay | Cell migration | HNSCC | (Jiang et al., 2010) |
| HIF-1α | Hypoxia-inducible factor 1, alpha | 1 | 3′-UTR | Reporter assay | Cell migration | Renal cell carcinoma | (Song et al., 2011) |
| VIM | Vimentin | 3 | 2 in CDs, 1 in 3′-UTR | Reporter assay | EMT | HNSCC | (Liu et al., 2011) |
| ZEB2 | Zinc finger E-box-binding homeobox 2 | 3 | 2 in CDs, 1 in 3′-UTR | Reporter assay | EMT | HNSCC | (Liu et al., 2011) |
| EZH2 | Enhancer of zeste homolog 2 | 2 | 1 in CDs, 1 in 3′-UTR | Reporter assay | EMT | HNSCC | (Liu et al., 2011) |
| Snai2a | Homolog of Snail 2 | 0 | Indirect target of miR-138 | N.A. | EMT | HNSCC | (Jin et al., 2011) |
| GNAI2 | G-protein alpha inhibiting activity polypeptide 2 | 2 | 2 in 3′-UTR | Reporter assay | Proto-oncogene, cell cycle | HNSCC | (Dhanasekaran et al., 1998) |
| FOSL1b | Fos-related antigen 1 | 6 | 1 in 5′-UTR, 3 in CDs, 2 in 3′-UTR | Reporter assay, RIP-IP | Proto-oncogene, cell cycle, EMT | HNSCC | (Dhanasekaran et al., 1998; Jin et al., 2011) |
| CCND1 | Cyclin D1 | 1 | 3′-UTR | Reporter assay | Proto-oncogene, cell cycle | Nasopharyngeal carcinoma | (Liu et al., 2012) |
| CCND3 | Cyclin D3 | 2 | 2 in 3′-UTR | Reporter assay | Proto-oncogene, cell cycle | Hepatocellular carcinoma | (Wang et al., 2012b) |
| H2AX | Histone H2A | 1 | 3′-UTR | Reporter assay | DNA damage response | Osteosarcoma | (Wang et al., 2011b) |
| XRCC1 | X-ray repair cross-complementing 1 | 1 | CDs | Reporter assay | DNA damage repair | Breast cancer | (Nicoloso et al., 2010) |
| TERT | Telomerase | 1 | 3′-UTR | Reporter assay | Senescence | Thyroid carcinoma | (Mitomo et al., 2008) |
| Sirt1 | Sirtuin 1 | 1 | 3′-UTR | Reporter assay | Senescence | N.A.d | (Rivetti diVal Cervo et al., 2012) |
Snai2 is an indirect target of miR-138. The miR-138-mediated Snai2 downregulation is achieved through the downregulation of FOSL1, which in turn leads to the reduced promoter activity of the Snai2 gene ( Jin et al., 2011).
Of the 6 miR-138-targeting sites identified in FOSL1, 3 are canonical targeting sites and the other 3 are noncanonical targeting sites. The canonical targeting site for miR-138 is defined by the presence of seed sequence CACCAGC in the mRNA molecule. The noncanonical targeting site allows G:U wobble base pairing. The seed sequence on the mRNA molecule allows the substitution of C by U and the substitution of A by G.
RIP-IP assay demonstrating the miR-138-directed RISC binding to FOSL1 mRNA was performed in Jin et al. (2011) study as a positive control experiment.
The study was based on a replicative senescence model of human primary keratinocytes. It was not directly related to any specific cancer type.
3.1. MicroRNA-138 Effects on HNSCC Cell Migration
Cytoskeleton remodeling is essential for the cell invasion and migration, and plays a major role in the cancer cell metastasis. The Rho GTPases-signaling cascade is crucial in cytoskeleton remodeling, as well as regulating cell adhesion and migration. The overexpression of Rho family genes is frequently observed in cancer cells. The overexpression of RhoC and ROCK2, key genes in the Rho GTPases-signaling cascade, has been frequently linked to the enhanced metastatic potential in various cancer types. This is consistent with their functional roles. RhoC and ROCK2 are known to be involved in remodeling of cellular cytoskeleton, which in turn changes cell migratory behavior. A recent study by Jiang et al. (2010) suggested that miR-138 targets both RhoC and ROCK2. They showed that ectopic transfection of miR-138 reduced the expression of both RhoC and ROCK2 in HNSCC cells. This reduced expression consequently led to the reorganization of the stress fibers and the subsequent cell morphology transformation to a round bleb-like shape, and the suppression of cell migration and invasion. In contrast, the knockdown of miR-138 in HNSCC cells enhanced the expression of RhoC and ROCK2, which resulted in altered, elongated cell morphology, enhanced cell stress fiber formation, and accelerated cell migration and invasion. These observations are consistent with the notion that coordinated regulation of the actin cytoskeleton is central to cell motility, invasion, and metastasis. While miR-138 affects RhoC expression at both the mRNA and protein level, it only regulates ROCK2 expression at the protein level but not at the mRNA level, which suggests that miR-138 regulates ROCK2 gene expression primarily by inhibiting translation. Together with the previous finding that reduced miR-138 level is correlated with enhanced metastatic potential in HNSCC cells, the existing evidence established a novel regulatory paradigm in which miR-138 regulates RhoC-specific GTPase-signaling cascade by targeting both RhoC and ROCK2 mRNAs concurrently, and suppresses their expression at post-transcriptional levels. The previous studies demonstrated that the expression of RhoC is progressively increased as tumors become more aggressively metastatic, and that RhoC expression promotes metastasis (Fingleton, 2007; Ridley, 2004; Wu et al., 2004). The reduction of expression of RhoC and ROCK2 and subsequent increase in metastatic phenotypes (i.e. increase in migration and invasion) are in line with the previous findings.
In addition to its role in HNSCC, a recent study by Song et al. (2011) demonstrated that miR-138 regulates migration of clear cell renal cell carcinoma (ccRCC) cells. They showed that miR-138 controls the expression of Hypoxia-inducible factor-1alpha (HIF-1α), a critical regulator in cancer cells. The downregulation of miR-138 results in enhanced expression of HIF-1α, which enhanced the cell migration and reduced apoptosis in ccRCC cell lines. These results suggested that miR-138 may function as a tumor suppressor in different cancer types by inhibiting migration and invasion, and promote apoptosis.
3.2. MicroRNA-138 is a Multifunctional Molecular Regulator in EMT
Cancer initiation, progression, and metastasis require the activation of EMT. The activation of EMT-related biological pathways enables cancer cells to migrate and invade to adjacent tissues. EMT is a highly orchestrated series of events in which cell–cell and cell–ECM interactions are altered to release epithelial cells from the surrounding tissue, the cytoskeleton is reorganized to confer the ability to move through a three-dimensional ECM, and a new transcriptional program is induced to maintain the mesenchymal phenotype. It has been suggested that deregulation of microRNAs is involved in EMT and the progression of many different cancers. Previous studies revealed that miR-200 family and miR-205 regulate EMT by targeting the zinc finger E-box-binding homeobox (ZEB) family transcription repressors (ZEB1 and ZEB2), which consequently control the E-cadherin expression (Burk et al., 2008; Korpal and Kang, 2008; Korpal et al., 2008; Park et al., 2008). Similarly, the deregulation of miR-101 leads to the overexpression of polycomb group protein EZH2, which functions as an epigenetic regulator to inhibit the expression of E-cadherin and induce EMT (Kisliouk et al., 2011). A recent study showing that the downregulation of miR-138 in HNSCC cell lines is associated with marked morphological changes associated with EMT (e.g. loss of polarity and cell–cell adhesion, and the acquisition of mesenchymal-like cell morphology) suggested that miR-138 may play a role in EMT. To study the functional relevance of miR-138 in EMT, Liu et al. (2011) used an EMT-specific qRT-PCR array to examine the forced expression of miR-138-induced differential expression of EMT-related genes in 1386Ln cell line (an established HNSCC cell line with high metastasis potential). Among the 86 EMT-related genes examined, 23 genes were altered upon miR-138 overexpression including 9 down-regulated and 14 up-regulated genes. Subsequent bioinformatics-based analysis revealed that a number of these EMT-related genes are direct targets of miR-138, including VIM (vimentin), ZEB2, and EZH2 (enhancer of zeste homolog 2) (Liu et al., 2011), while others are potential indirect targets of miR-138, including Snai2 (homolog of Snail 2) ( Jin et al., 2011). Functional analyses demonstrated that miR-138 regulates the EMT via three distinct pathways: (i) direct targeting of VIM mRNA and controlling the expression of VIM at a posttranscriptional level, (ii) targeting the transcriptional repressors (ZEB2 and Snai2), which in turn regulates the transcription activity of the E-cad gene, and (iii) targeting the epigenetic regulator EZH2, which in turn modulates its gene-silencing effects on the downstream genes including E-cad. These results, together with our previously observed miR-138 effects on cell migration and invasion through targeting RhoC and ROCK2 concurrently, suggest that miR-138 is a multifunctional molecular regulator and plays major roles in EMT and in HNSCC progression (Fig. 9.2). The following sections will provide detailed discussion of the roles of miR-138 in EMT.
Figure 9.2.
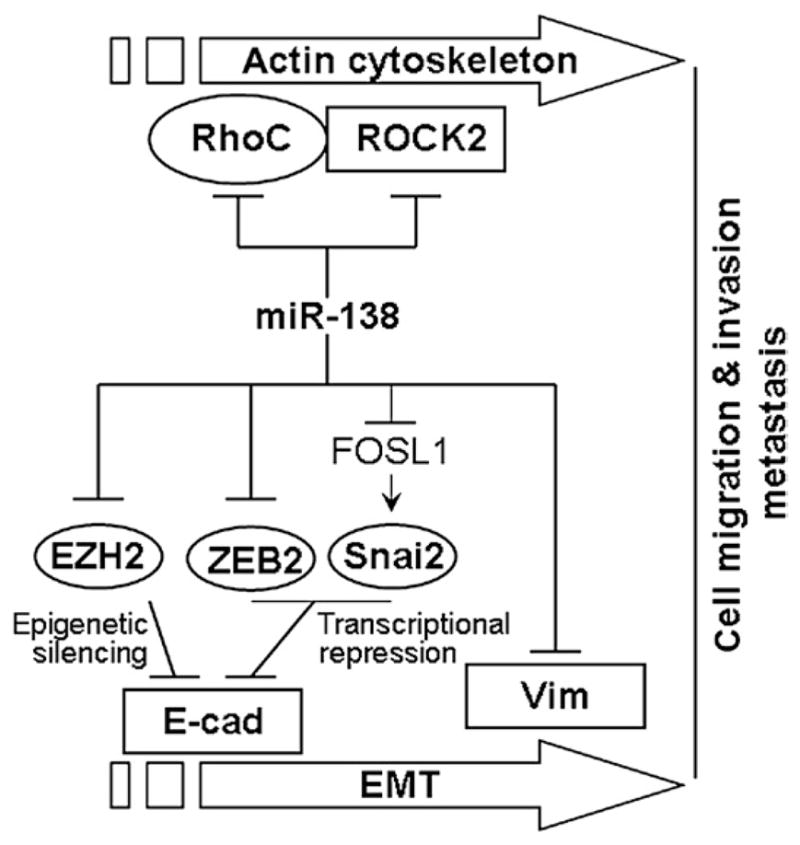
Potential roles of miR-138 in EMT and cancer cell metastasis.
Vimentin is a type III intermediate filament (IF) protein and the major cytoskeletal component of mesenchymal cells. Vimentin is often considered as a marker of mesenchymal cells or cells undergoing an EMT during both normal development and metastatic progression. In the process of EMT, the cellular IF status switches from a keratin-rich network (connecting to adherens junctions and hemidesmosomes) to a Vim-rich network (connecting to focal adhesions). Three miR-138-targeting sequences were identified in the VIM mRNA sequence. The first- and second-targeting sequences are located in the coding region and the third sequence is located in the 3′-UTR of the VIM mRNA. Direct targeting of miR-138 to specific sequences located in the mRNA of the VIM gene was confirmed using the luciferase reporter gene assays (Liu et al., 2011). Ectopic transfection of miR-138 in HNSCC cell lines resulted in reduced Vim expression, which is accompanied with increased E-Cad expression and reduced cell migration and invasion. The knockdown of miR-138 in HNSCC cell lines with anti-miR-138 LNA resulted in enhanced Vim expression, which is accompanied with decreased E-Cad expression and enhanced cell migration and invasion (Liu et al., 2011).
Downregulation of E-cadherin expression is one of the hallmarks of EMT. The expression of E-cadherin gene is tightly regulated by a number of transcriptional repressors, including members of ZEB gene family (ZEB1 and ZEB2) and Snai gene family (Snai1 and Snai2). The members of ZEB gene family mediate the EMT triggered by the key-signaling cascades such as TGFβ/BMP, NFκB, Ras-ERK2, and HIF-1. Three miR-138-binding sites were predicted in the ZEB2 mRNA—two located in the coding region and one in the 3′-UTR. However, the luciferase reporter gene assays showed that the site located in the 3′-UTR is largely responsible for the miR-138-mediated downregulation of ZEB2. It is worth knowing that the ZEB family repressors (both ZEB1 and ZEB2) are targeted by a number of different microRNAs, including the miR-200 family (Gregory et al., 2008). Interestingly, ZEB1 has been showed to suppress the expression of miR-200 family members, indicating that miR-200 members and ZEB factors reciprocally control each other in a negative feedback loop that maintain cells in either an epithelial or mesenchymal state (Bracken et al., 2008; Burk et al., 2008). It is not clear whether miR-138 is also regulated by ZEB factors (or its other target genes) by similar feedback mechanism(s). Further studies are needed to explore this potential regulatory mechanism.
In addition to ZEB2, the study by Liu et al. (2011) suggested that miR-138 also downregulates Snai2, a member of Snail family of zinc-finger transcriptional repressors that play an important role in the regulation of E-cad expression and EMT (Peinado et al., 2007). Overexpression of Snai2 is frequently observed in HNSCC, and is associated with lymph-node metastasis in the HNSCC patients (Wang et al., 2012a). However, no miR-138-targeting site was identified in the Snai2 mRNA. At the time, Liu et al. (2011) hypothesized that miR-138 indirectly regulates Snai2 by targeting factor(s) that control the Snai2 gene expression. Interestingly, in a subsequent study, Jin et al. (2011) identified FOSL1 as an additional target of miR-138. FOSL1 is a member of Fos gene family and is a known proto-oncogene. One of the established downstream genes of FOSL1 is Snai2 (Chen et al., 2009). Jin et al. (2011) then confirmed that the effect of this microRNA-138-FOSL1 regulatory module on Snai2 expression, as well as the Snai2-mediated repression of E-cadherin expression and the induction of EMT in HNSCC cells. It is worth noting that the results by Jin et al. (2011) also provided evidence suggesting that miR-138 may also target other members of the Fos-gene family (e.g. Fos, FosB, and FOSL2), which may in turn regulate Snai2 expression in a similar manner as FOSL1.
Epigenetic modifications such as histone methylation play an important role in EMT. EZH2 is a critical component of the polycomb repressive complex 2 (PRC2) that includes noncatalytic subunits Suz12 and Eed. It catalyzes trimethylation on Lysine 27 of histone 3 protein (H3K27Me3), which in turn leads to chromatin condensation and epigenetic silencing of the downstream genes (Sparmann and van Lohuizen, 2006). One of the well-established downstream target genes of EZH2 is E-cad, and the EZH2-mediated repression of E-cad is associated with EMT in several cancer types (Cao et al., 2008; Herranz et al., 2008; Huang et al., 2011). A recent study reported that overexpression of EZH2 in cancer cells downregulates the expression of E-cadherin gene through histone H3K27 trimethylation at the E-cadherin gene promoter (Cao et al., 2008). The knockdown of EZH2 in vitro has been shown to restore E-cadherin expression (Fujii and Ochiai, 2008; Rao et al., 2010). The results by Liu et al. (2011) demonstrated that the miR-138 downregulates the expression of EZH2 gene by binding to a conserved targeting site located in the 3′-UTR of the EZH2 mRNA. This miR-138-mediated EZH2 downregulation is reversely correlated with E-cad expression and EMT in HNSCC cells. Interestingly, miR-138 has previously been shown to target EZH2 in chickens (Kisliouk et al., 2011). Two miR-138-targeting sites were identified in the chicken EZH2 mRNA: an evolutionally conserved site located in the 3′-UTR, which is also presented in 3′-UTR of the human EZH2 mRNA, and a poorly conserved site that overlaps with the translational stop codon in the chicken EZH2. In humans, the seed region of this poorly conserved site has 2 base-substitutions (at position 4 and 6), which makes it nonfunctional. Nevertheless, the fact that EZH2 expression is downregulated by miR-138 in both chickens and humans suggested that the miR-138-medicated suppression of EZH2 gene is an evolutionally conserved molecular event. Liu et al. (2011) also observed an apparent increase in Suz12 level in an HNSCC cell line that was treated with anti-miR-138 LNA. However, no miR-138-targeting site was identified in the Suz12 mRNA, and currently the biological significance of these observed changes in Suz12 expression is not clear. It is worth noting that miR-200 has been shown to directly target Suz12 and control the E-cad expression by regulating PRC2 complex (Iliopoulos et al., 2010). Additional studies will be needed to fully explore the potential concordant effect(s) of anti-EMT microRNAs (e.g. miR-138 and miR-200) on PRC2-mediated repression of E-cad.
Taken together, this evidence demonstrates that miR-138 regulates the EMT through 3 distinct pathways in HNSCC cells (Fig. 9.2). Together with the previous observation that miR-138 regulates cell migration and invasion by concurrently targeting RhoC and ROCK2 ( Jiang et al., 2010), this evidence suggests that miR-138 is a multifunctional molecular regulator and plays major roles in EMT. Additional studies are required to explore its potential as a novel therapeutic target for cancer patients at the risk of metastasis.
3.3. MicroRNA-138 Regulating Cell Cycle by Targeting Multiple Proto-oncogenes
To understand the roles of microRNA in complex biological processes (i.e. tumorigenesis), it is important to identify the functional modules involved in complex interactions between microRNAs and their targets (e.g. mRNA). As part of a top-down approach to identify the miR-138-targeting genes, Jiang et al. (2011) carried out a genome-wide expression profiling experiments on HNSCC cell lines that transfected with either miR-138 or negative control. A panel of 194 unique transcripts was significantly down-regulated in cells transfected with miR-138. A comprehensive screening using six different sequence-based microRNA-target prediction algorithms revealed that 51 out of these 194 down-regulated transcripts are potential direct targets for miR-138. These targets include chloride channel, nucleotide-sensitive, 1A (CLNS1A), G-protein alpha-inhibiting activity polypeptide 2 (GNAI2), solute carrier family 20, member 1 (SLC20A1), eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), FOSL1, and Rho-related GTP-binding protein C (RhoC). The authors then focused on the effect of miR-138 on GNAI2, a known proto-oncogene that is involved in the initiation and progression of several different types of tumors (Dhanasekaran et al., 1998). GNAI2 belongs to the family of Gi alpha proteins that includes 3 polypeptides: Gi alpha 1 (GNAI1), Gi alpha 2 (GNAI2), and Gi alpha 3 (GNAI3). They form heterotrimers with beta and gamma subunits, and are involved in a wide variety of signaling events mediated by G-protein-coupled receptor (GPCR). Among the Gi alpha family members, GNAI2 gene is the only one that contains the conserved miR-138-targeting sequence. Jiang et al. (2011) confirmed the direct targeting of miR-138 to the two candidate-binding sequences located in the 3′-untranslated region of GNAI2 mRNA using luciferase reporter gene assays. The knockdown of miR-138 in HNSCC cells enhanced the expression of GNAI2 at both mRNA and protein levels. In contrast, ectopic transfection of miR-138 reduced the expression of GNAI2, which consequentially led to reduced proliferation, cell cycle arrest, and apoptosis. Interestingly, a recent report showed that GNAI2 is also a functional target of miR-30d in hepatocellular carcinoma (HCC) cells (Yao et al., 2010). It is worth noting that downregulation of miR-30d has also been observed in OSCC (Kozaki et al., 2008). Taken together, this evidence suggests a novel paradigm in which microRNAs regulate GPCR signaling by targeting GNAI2 mRNA and suppressing its expression at posttranscriptional levels.
The cell cycle is regulated by a family of the cyclin-dependent kinases (CDKs) and their activating partners (cyclins). The G1 to S phase transition is regulated primarily by members of cyclin D family (CCND1, CCND2, and CCND3) in complex with CDK4/CDK6 and cyclin E family (CCNE1 and CCNE2) in complex with CDK2. These complexes cooperate in phosphorylating and preventing Rb from binding to E2F, thus activating E2F-mediated transcription and driving cells from the G1 into the S phase. Two independent recent studies demonstrated that miR-138 targets CCND1 and CCND3 in nasopharyngeal carcinoma (NPC) and HCC, respectively (Liu et al., 2012; Wang et al., 2012b). Both studies showed that miR-138 is frequently downregulated in NPC and HCC. The levels of CCND1 and CCND3 were inversely correlated with miR-138 expression in NPC and HCC respectively. Both studies showed that forced expression of miR-138-suppressed proliferation and colony formation in vitro and inhibited tumorigenesis in xenograft nude mice (Liu et al., 2012; Wang et al., 2012b). Direct targeting of miR-138 to CCND3 mRNA was further confirmed using luciferase reporter gene assays (Wang et al., 2012b). These studies, together with the evidence described earlier showing that miR-138 targets other oncogenes (e.g. FOSL1, RhoC, ROCK2) and regulates cell migration and EMT, suggest that miR-138 functions as a tumor suppressor, and may serve as a useful therapeutic agent for microRNA-based therapy.
3.4. Role of microRNA-138 in DNA Damage and Chemoresistance
Several recent studies suggested that microRNAs play important roles in DNA damage response and DNA repair (Hu et al., 2010; Lal et al., 2009; Pothof et al., 2009; Zhang et al., 2010). This microRNA-mediated regulation of DNA damage response has the potential to improve the efficacy of cancer therapies such as chemotherapy and radiotherapy, which rely on the induction of DNA damage. To gain a systematic understanding of the role of microRNAs in DNA damage response, Wang et al. (2011b) conducted a cell-based microRNA library screen to identify the effects of microRNAs on ionizing radiation (IR)-induced DNA damage. They found that over-expression of miR-138 enhanced cellular sensitivity to IR, as well as other DNA-damaging agents (cisplatin and camptothecin), in an osteosarcoma cell line. They further demonstrated that miR-138 directly targeted the histone H2AX gene, one of the major players in the DNA damage response pathways (Wang et al., 2011b). It is worth noting that H2AX is also targeted by miR-24 (Lal et al., 2009), suggesting that H2AX is a major target of the microRNA-mediated posttranscriptional regulation, and may ultimately govern the microRNA-mediated DNA damage response. The role of miR-138 on the DNA damage response was independently validated recently in a cisplatin-resistant nonsmall cell-lung cancer cells (A549/DDP), where the upregulation of miR-138 increased the sensitivity of A549/DDP cells to cisplatin in in vitro drug sensitivity assays (Wang et al., 2011a). The authors also found that excision repair cross-complementation group 1 (ERCC1) was negatively regulated by miR-138 and that downregulation of ERCC1 at the protein level was correlated with elevated levels of miR-138 in A549/DDP cells. These results suggested that miR-138 is a potential therapeutic agent for cancer treatment as overexpression of miR-138 will sensitize tumor cells to DNA-damaging agents. Nicoloso et al. (2010) identified a functional noncanonical miR-138-targeting site in the DNA repair gene X-ray repair cross-complementing 1 (XRCC1). Their case control study suggested that a specific single-nucleotide polymorphism in this miR-138-targeting site is associated with increased risk for breast cancer.
While the accumulating evidence suggests critical roles of miR-138 in DNA damage response, the effect of miR-138 on the cellular responsiveness to other chemotherapy is still unclear. A few preliminary reports suggested that miR-138 deregulation may contribute to specific chemo-resistance. For example, miR-138 was found to be upregulated in the vincristine-induced multidrug resistance leukemia cell line (Zhao et al., 2010). The downregulation of miR-138 has been observed in gefitinib-resistance lung adenocarcinoma cell line (Qin et al., 2011). Nevertheless, these reports are based on single cell line and have not been independently validated. It is critically important to define the effect of miR-138 on multiple chemotherapy drugs, since most types of cancer (including HNSCC) are subjected to multimodality treatment. Future studies will be needed to test the feasibility of utilizing miR-138 as a chemosensitizer/radiosensitizer in therapy for certain types of cancer.
3.5. Role of miR-138 in Senescence
Cellular senescence (also known as replicative senescence) is the phenomenon by which normal cells lose the ability to divide. This is often triggered by DNA damage (including shortened telomeres). If the damage cannot be easily repaired, cells will then self-destruct (apoptosis, programmed cell death). Senescence not only contributes to the natural aging, but also inhibits malignant progression. As such, senescence is considered as a tumor-suppressive mechanism. Malignant cells that bypass this arrest become immortalized by telomere extension due mostly to the activation of telomerase reverse transcriptase (TERT) responsible for synthesis of telomeres. Mitomo et al. (2008) showed that the downregulation of miR-138 is associated with the overexpression of TERT in the anaplastic thyroid carcinoma (ATC) cell lines they examined. Forced expression of miR-138 led to reduced TERT expression in ATC cell lines. The direct targeting of miR-138 to the 3′-UTR of the TERT mRNA was confirmed by luciferase reporter gene assays in HEK-293 cells. In addition to miR-138, other microRNAs have also been shown to regulate telomerase activity. For example, miR-150 expression resulted in reduced telomerase activity, shortened telomeres, and induction of senescence in lymphoma cells (Watanabe et al., 2011). Unlike miR-138, which regulates telomerase activity by direct targeting TERT mRNA, miR-150 regulates the telomerase activity by indirect mechanisms (suppressing the AKT activity and inducing the p53 expression). These results indicated that TERT is a key target of microRNA-induced senescence, which can be regulated directly or indirectly by a senescence-associated microRNA (e.g. miR-138).
A more recent study focusing on senescence of normal keratinocytes provided additional evidence supporting the role of miR-138 in replicative senescence. Using a replicative senescence culture model of keratinocytes, the authors showed that miR-138 is significantly upregulated in senescent keratinocytes as compared to proliferating cells (Rivetti di Val Cervo et al., 2012). The upregulation of miR-138 was further confirmed in aged skin from healthy subjects (>60 years) as compared to skin from young subjects (<10 years). They further demonstrated that forced expression of miR-138, as well as miR-181, is sufficient to induce premature senescence. Their functional analysis established a feedback regulatory loop involving p63, Sirt1, and several microRNAs (including miR-138, miR-181, and miR-130b) (Rivetti di Val Cervo et al., 2012). It is worth noting that p53, a major player in senescence, apoptosis, as well as many other essential cellular functions, has been shown to be targeted by miR-138 in induced pluripotent stem (iPS) cell (Ye et al., 2012). However, the role of miR-138-mediated downregulation of p53 in senescence is not clear. Taken together, while the precise mechanism(s) is still under investigation, the studies described above clearly suggested a role of miR-138 in replicative senescence.
4. BIOLOGICAL PROCESSES REGULATED BY MICRORNA-138
As described above, the involvement of miR-138 in tumorigenesis is through its regulation of a number of biological pathways, of which many of these pathways are also involved in developmental process. Not surprisingly, a number of recent studies demonstrated that miR-138 also plays major roles in development. Morton et al. (2008) showed that miR-138 is required for cardiac morphogenesis during embryonic development in the zebrafish. Siegel et al. (2009) showed that miR-138 is required in dendritic spine morphogenesis in the rat. miR-138 also plays a role in thermotolerance acquisition in chicks, which involves neuronal network remodeling and sensory development (Kisliouk et al., 2011). miR-138 is differentially expressed during the development of mammary gland in mice (Wang and Li, 2007). A case–control study showed that a specific polymorphism in the miR-138 gene (miR-138-2) is associated with panic disorder (Muinos-Gimeno et al., 2011).
The observed effects of miR-138 on various developmental events (and the corresponding defects) may tie to its role in the stem cell differentiation. iPS cells can be obtained by reprogramming somatic cells with the forced expression of Oct4, Sox2, and Klf4, with or without c-Myc (named as OSKM or OSK, respectively). However, the efficiency of this reprogramming/induction is generally low. An increasing number of studies found that iPS cell-induction efficiency can be enhanced by modulating several microRNAs during the reprogramming of somatic cells ( Judson et al., 2009; Melton et al., 2010). In a recent study, Ye et al. (2012) showed that miR-138 promotes the generation of the iPS cells by facilitating the reprogramming of the somatic cells. Their study showed that the forced expression of miR-138 significantly enhanced the efficiency of both OSKM- and OSK-initiated somatic cell reprogramming to iPS cells, without sacrificing the pluripotent characteristics of the resulted iPS cells. Ye et al. (2012) also showed that miR-138 is endogenously expressed in embryonic stem cells. As such, miR-138 may be responsible for maintaining the stem cells in the dedifferentiated state. This is supported by 2 independent studies investigating the role of miR-138 on mesenchymal stem cell (MSC) differentiation. Eskildsen et al. (2011) demonstrated that miR-138 inhibits osteogenic differentiation of human MSCs. Forced expression of miR-138 inhibited osteoblast differentiation of MSCs in vitro, and reduced ectopic bone formation in vivo, whereas the knockdown of miR-138 by anti-miR-138 promoted expression of osteoblast-specific genes, and enhanced bone formation in vivo. Similarly, Yang et al. (2011) demonstrated that miR-138 regulates adipogenic differentiation of adipose tissue-derived MSCs (hAD-MSCs). Their data showed that forced expression of miR-138 in hAD-MSCs inhibited the adipogenic gene expression program (including key adipogenic transcription factors and other adipogenic marker genes), and suppressed lipid droplets accumulation. Thus, these studies demonstrated that miR-138 play an important role in pluripotency maintenance, and retain the stem cells in dedifferentiated state.
5. CONCLUSION
In summary, we provided a comprehensive review and integrative analysis of the existing studies on miR-138, and presented evidence supporting its roles in cell migration, EMT, and cell cycle progression, as well as its potential involvement in DNA damage response and repair, senescence, and differentiation. It is clear that miR-138 is a multifunctional molecular regulator, and the precise regulation of this gene is critically important. More studies will be needed to fully understand the mechanism that regulates the expression of this noncoding gene. It is worth noting that miR-138 is just one of the deregulated microRNAs in HNSCC. Other microRNA genes may have redundant functions (or opposite effects) as miR-138. For example, miR-200 family members also regulate EMT (Burk et al., 2008; Korpal and Kang, 2008; Korpal et al., 2008; Park et al., 2008), and they may exhibit synergistic effect with miR-138 in regulating EMT. As such, functional investigations of multiple microRNAs (together with their target genes) are required. This systematic approach is essential to fully assess the relevance of microRNA in the complex biological system (e.g. HNSCC).
HNSCC is one of the undertreated and understudied cancer types. Historically, investigation on this disease has always been lagging behind studies on other cancer types. However, a number of recent studies on HNSCC contributed to major improvement in our overall understanding of cancer biology, including the investigation on genomic aberrations associated with microRNA genes in HNSCC. To the best of our knowledge, miR-138 is one of the best studied noncoding genes in HNSCC. The relationships of miR-138 expression to tumorigenesis, HNSCC cell migration, and EMT suggest that the measurement of miR-138 expression may provide prognostic information for HNSCC patients and their care providers. This information could help to craft more clinically appropriate and cost-effective therapeutic management strategies in a variety of healthcare settings. As the biologic changes within tumors become more precisely described at a molecular level for a particular patient, more effective and better directed radiotherapy and chemotherapy regimens can be planned. Such enhancements to the characterization and treatment of HNSCC would add beneficial elements to a personalized medicine model that could aid in maximizing disease-specific treatment options, minimizing collateral tissue damage, and opening the door for improved outcomes for a greater number of patients. Nevertheless, a large-scale patient-based study will be needed to fully explore the potential value of miR-138 as a biomarker.
MicroRNA-138 is a multifunctional molecular regulator. In addition to its well-documented role in cell proliferation, migration and EMT, the understanding of the relevance of miR-138 in senescence and stem cell differentiation is still in its infancy. It will be interesting to determine whether miR-138 regulates aspects of telomere signaling and DNA damage response. Deregulation of miR-138 may be a surrogate marker for telomere dysfunction in HNSCC, which may have important ramifications for cellular senescence and treatment outcomes. In addition to its roles in HNSCC and tumorigenesis of other cancers, the functional study of miR-138 gene (e.g. using transgenic models) may also provide new insights into the developmental process and the pathogenesis of other diseases (obesity, diabetes, etc.)
Acknowledgments
This work was supported in part by NIH PHS grants (CA139596). Y.J. is supported by a T32 training grant (DE018381). We thank Ms. Katherine Long for her editorial assistance.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, Mehra R, Laxman B, Cao X, Yu J, Kleer CG, Varambally S, Chinnaiyan AM. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, Liu K, Huang S. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69:9228–9235. doi: 10.1158/0008-5472.CAN-09-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I, Wang C, Liu X, Zhou X. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol. 2012;48:686–691. doi: 10.1016/j.oraloncology.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran N, Tsim ST, Dermott JM, Onesime D. Regulation of cell proliferation by G proteins. Oncogene. 1998;17:1383–1394. doi: 10.1038/sj.onc.1202242. [DOI] [PubMed] [Google Scholar]
- Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fingleton B. Molecular targets in metastasis: lessons from genomic approaches. Cancer Genomics Proteomics. 2007;4:211–221. [PubMed] [Google Scholar]
- Fujii S, Ochiai A. Enhancer of zeste homolog 2 downregulates E-cadherin by mediating histone H3 methylation in gastric cancer cells. Cancer Sci. 2008;99:738–746. doi: 10.1111/j.1349-7006.2008.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L, Graubard BI, Chaturvedi AK. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA. 2012;307:693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg RP, Honorio S, Martinez A, Agathanggelou A, Dallol A, Fullwood P, Weichselbaum R, Kuo MJ, Maher ER, Latif F. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p. 21.3 in head and neck squamous cell carcinoma. Eur J Cancer. 2002;38:1585–1592. doi: 10.1016/s0959-8049(01)00422-1. [DOI] [PubMed] [Google Scholar]
- Hong AM, Grulich AE, Jones D, Lee CS, Garland SM, Dobbins TA, Clark JR, Harnett GB, Milross CG, O’Brien CJ, Rose BR. Squamous cell carcinoma of the oropharynx in Australian males induced by human papillomavirus vaccine targets. Vaccine. 2010;28:3269–3272. doi: 10.1016/j.vaccine.2010.02.098. [DOI] [PubMed] [Google Scholar]
- Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, Yang PM, Chang YF, Marquez VE, Chen CC. Methotrexate induces apoptosis through p53/p21-dependent pathway and increases E-cadherin expression through downregulation of HDAC/EZH2. Biochem Pharmacol. 2011;81:510–517. doi: 10.1016/j.bcp.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Dai Y, Liu X, Wang C, Wang A, Chen Z, Heidbreder CE, Kolokythas A, Zhou X. Identification and experimental validation of G protein alpha inhibiting activity polypeptide 2 (GNAI2) as a microRNA-138 target in tongue squamous cell carcinoma. Hum Genet. 2011;129:189–197. doi: 10.1007/s00439-010-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, Shi F, Zhou X. Down-regulation of the Rho GTPase signaling pathway is involved in the microRNA-138 mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–512. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang C, Liu X, Mu W, Chen Z, Yu D, Wang A, Dai Y, Zhou X. Molecular characterization of the microRNA-138-Fos-like antigen 1 (FOSL1) regulatory module in squamous cell carcinoma. J Biol Chem. 2011;286:40104–40109. doi: 10.1074/jbc.C111.296707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisliouk T, Yosefi S, Meiri N. MiR-138 inhibits EZH2 methyltransferase expression and methylation of histone H3 at lysine 27, and affects thermotolerance acquisition. Eur J Neurosci. 2011;33:224–235. doi: 10.1111/j.1460-9568.2010.07493.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen Z, Yu J, Xia J, Zhou X. MicroRNA profiling and head and neck cancer. Comp Funct Genomics. 2009a doi: 10.1155/2009/837514. Article ID: 837514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009b;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K, Wu M, Liang Y, Liu P, Tang J, Lu WH, Feng QS, Chen LZ, Qian CN, Bei JX, Kang T, Zeng YX. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, Dai Y, Zhou X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, Kato K, Komatsu H, Ikeda K, Wakabayashi G, Masuda T. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinos-Gimeno M, Espinosa-Parrilla Y, Guidi M, Kagerbauer B, Sipila T, Maron E, Pettai K, Kananen L, Navines R, Martin-Santos R, Gratacos M, Metspalu A, Hovatta I, Estivill X. Human microRNAs miR-22, miR-138-2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry. 2011;69:526–533. doi: 10.1016/j.biopsych.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Murata M, Takayama K, Choi BC, Pak AW. A nested case–control study on alcohol drinking, tobacco smoking, and cancer. Cancer Detect Prev. 1996;20:557–565. [PubMed] [Google Scholar]
- Nasman A, Attner P, Hammarstedt L, Du J, Eriksson M, Giraud G, Ahrlund-Richter S, Marklund L, Romanitan M, Lindquist D, Ramqvist T, Lindholm J, Sparen P, Ye W, Dahlstrand H, Munck-Wikland E, Dalianis T. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral-induced carcinoma? Int J Cancer. 2009;125:362–366. doi: 10.1002/ijc.24339. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirpath: Intergrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Piccinin S, Gasparotto D, Vukosavljevic T, Barzan L, Sulfaro S, Maestro R, Boiocchi M. Microsatellite instability in squamous cell carcinomas of the head and neck related to field cancerization phenomena. Br J Cancer. 1998;78:1147–1151. doi: 10.1038/bjc.1998.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Liu B, Li Y, You J, Zhou Q. Screening and identification of microRNAs related to acquired gefitinib-resistance in lung adenocarcinoma cell lines. Zhongguo Fei Ai Za Zhi. 2011;14:478–483. doi: 10.3779/j.issn.1009-3419.2011.06.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, Zeng YX, Kung HF, Xie D. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. Rho proteins and cancer. Breast Cancer Res Treat. 2004;84:13–19. doi: 10.1023/B:BREA.0000018423.47497.c6. [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C, Calin GA, Candi E, Melino G. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Zhang X, Wang C, Wu Y, Cai W, Gao J, Hong B. MiR-138 suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in clear cell renal cell carcinoma 786-O cells. Asian Pac J Cancer Prev. 2011;12:1307–1311. [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Strati K, Pitot HC, Lambert PF. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A. 2006;103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine N, Panzarella V, Falaschini S, Russo A, Matranga D, Lo Muzio L, Campisi G. HPV in oral squamous cell carcinoma vs head and neck squamous cell carcinoma biopsies: a meta-analysis (1988–2007) Ann Oncol. 2008;19:1681–1690. doi: 10.1093/annonc/mdn372. [DOI] [PubMed] [Google Scholar]
- Wang C, Li Q. Identification of differentially expressed microRNAs during the development of Chinese murine mammary gland. J Genet Genomics. 2007;34:966–973. doi: 10.1016/S1673-8527(07)60109-X. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu X, Huang H, Ma H, Cai W, Hou J, Huang L, Dai Y, Yu T, Zhou X. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer. 2012a;130:2249–2258. doi: 10.1002/ijc.26226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhong M, Liu W, Li J, Huang J, Zheng L. Alterations of microRNAs in cisplatin-resistant human non-small cell lung cancer cells (A549/DDP) Exp Lung Res. 2011a;37:427–434. doi: 10.3109/01902148.2011.584263. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012b doi: 10.1093/carcin/bgs113. [Epub ahead of print: Feb 23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gleich L, Pavelic ZP, Li YQ, Gale N, Hunt S, Gluckman JL, Stambrook PJ. Cervical metastases of head and neck squamous cell carcinoma correlate with loss of heterozygosity on chromosome 16q. Int J Oncol. 1999;14:557–561. doi: 10.3892/ijo.14.3.557. [DOI] [PubMed] [Google Scholar]
- Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011b;9:1100–1111. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, Shimizu N, Sawada K. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Wu M, Wu ZF, Kumar-Sinha C, Chinnaiyan A, Merajver SD. RhoC induces differential expression of genes involved in invasion and metastasis in MCF10A breast cells. Breast Cancer Res Treat. 2004;84:3–12. doi: 10.1023/B:BREA.0000018426.76893.21. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bian C, Zhou H, Huang S, Wang S, Liao L, Zhao RC. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011;20:259–267. doi: 10.1089/scd.2010.0072. [DOI] [PubMed] [Google Scholar]
- Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, Wan D, Yao M, Li J, Gu J, He X. MicroRNA-30d promotes tumor invasion and metastasis by targeting G alphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, Jia W, Deng AM, Liu H, Kang J. miR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells. 2012 doi: 10.1002/stem.1149. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang L, Hu J, Ruan J. miR-138 might reverse multidrug resistance of leukemia cells. Leuk Res. 2010;34:1078–1082. doi: 10.1016/j.leukres.2009.10.002. [DOI] [PubMed] [Google Scholar]