Crystals of recombinantly expressed ρ-Da1a toxin from eastern green mamba that were suitable for structure determination were discovered on defrosting a partially lyophilized sample.
Keywords: lyophilization, three-finger fold toxin, weak data
Abstract
ρ-Da1a toxin from eastern green mamba (Dendroaspis angusticeps) venom is a polypeptide of 65 amino acids with a strong affinity for the G-protein-coupled α1A-adrenoceptor. This neurotoxin has been crystallized from resolubilized lyophilized powder, but the best crystals grew spontaneously during lyophilization. The crystals belonged to the trigonal space group P3121, with unit-cell parameters a = b = 37.37, c = 66.05 Å, and diffracted to 1.95 Å resolution. The structure solved by molecular replacement showed strong similarities to green mamba muscarinic toxins.
1. Introduction
Snake toxins are routinely crystallized from resolubilized lyophilized samples (Rees et al., 1987 ▶). Typically, the powder is resuspended in acetate buffer pH 5.5 (Pawlak et al., 2006 ▶) since some toxins are poorly soluble at neutral pH (Rajashankar et al., 1999 ▶). This procedure gave crystals of synthesized ρ-Da1a toxin from eastern green mamba venom, but the best crystals were obtained from the Gly-ρ-Da1a-K34A mutant expressed in Escherichia coli; these crystals grew spontaneously during the lyophilization phase and survived freezing and defrosting, soaking in cryoprotectant and flash-cooling in liquid nitrogen.
Freeze-drying can induce structural changes in proteins, but these changes are often reversible (Griebenow & Klibanov, 1995 ▶) and therefore do not prevent crystallization when the protein is resuspended in buffer. Understanding how protein crystals can grow during freeze-drying could be important in developing new protein crystallization approaches. Freeze concentration can occur during lyophilization, leading to liquid–liquid phase separation between the components in the non-ice portion of the frozen sample (Heller et al., 1999 ▶), conditions that may be conducive to rapid nucleation and fast crystal growth. High protein concentrations are often needed to produce large crystals rapidly (Stura, 1999 ▶). Aspects of how the high concentration of the segregated protein can lead to better crystals are discussed in this paper. The experimental details of the production, purification, lyophilization, crystallization, crystal handling, data collection and structure determination of ρ-Da1a, both chemically synthesized and expressed in E. coli, are reported here in order to understand the parameters that allowed the crystallization of this toxin under these unusual circumstances.
2. Materials and methods
2.1. Toxin production
Recombinant Gly-ρ-Da1a-K34A was expressed in E. coli BL21 as a fusion with the ZZ domain of protein A, a high-level expression system developed for three-finger fold toxins (Drevet et al., 1997 ▶). A synthetic gene corresponding to the ENLYFQG-ρDa1a protein, flanked by the attB1 and attB2 sequences, was incorporated into the pENTRE plasmid following the manufacturer’s protocol (Invitrogene, Cergy-Pontoise, France). The toxin was recombined with ZZ into the pEXP plasmid; a tobacco etch virus (TEV) protease-cleavage sequence (NLYFQ↓G) was introduced at the N-terminus to give the final Gly-ρ-Da1a-K34A construct. The toxin was produced in inclusion bodies and was solubilized for 2 h in 4 M guadinium chloride, 100 mM Tris pH 8.0, 10 mM tris-(2-carboxyethyl)phosphine (TCEP) to prevent random oxidation of ρ-Da1a. After centrifugation at 18 000g for 60 min, the supernatant was diluted 1/10 in TEV buffer (50 mM Tris pH 8, 150 mM NaCl, 10 mM TCEP). TEV cleavage was performed overnight at 277 K with 10 mg TEV per 100 mg total protein. Gly-ρ-Da1a was then purified on nickel resin; it eluted at 50 mM imidazole, while the TEV and fusion protein eluted at 300 mM imidazole (Fig. 1 ▶ a).
Figure 1.
Production of ρ-Da1a. The recombinant protein (molecular mass 27 487 Da) consisted of a fusion with the ZZ domain of protein A, a histidine tag and a TEV protease cleavage site. The fusion protein is produced in inclusion bodies, with a yield of 30–50 mg per litre of culture. (a) SDS–PAGE of the different fractions obtained during the production and purification of Gly-ρ-Da1a-K34A. Lanes 1 and 6, molecular-weight markers (labelled in kDa). Lanes 2 and 3, band at molecular mass 30 kDa corresponding to the fusion protein obtained after induction by 1 mM IPTG for 3 h at 310 K. Lane 4, elution of Gly-ρ-Da1a-K34A after TEV protease cleavage. Lane 5, elution of the TEV and fusion protein with 300 mM imidazole. (b) HPLC purification of Gly-ρ-Da1a-K34A after oxidation in folding buffer. (c) HPLC purification of the oxidized s-ρ-Da1a obtained from solid-phase peptide synthesis. Relevant peaks are marked with arrows.
The synthetic homologue of ρ-Da1a (s-ρ-Da1a) was assembled on an automatic peptide synthesizer (ABI 433, Applied Biosystems, USA) using Fmoc solid-phase chemistry (Quinton et al., 2010 ▶) and was purified by reverse-phase HPLC (Xbridge C18, 21 mm × 25 cm, 15 µm; P600 system; Waters, St Quentin en Yvelines, France) using a 15–30% acetonitrile/0.1% trifluoroacetic acid (TFA) gradient (Fig. 1 ▶ c).
Synthetic and recombinant toxins produced in inclusion bodies require refolding before they are functional. Refolding was carried out in a buffer consisting of 25% glycerol, 1 mM oxidized and reduced glutathione, 50 mM Tris pH 8.0 for 36–40 h at 277 K and the toxin was repurified by reverse-phase HPLC with a gradient of 10–40% acetonitrile with 0.1% TFA. Thus, the toxin solution before lyophilization contained acetonitrile and TFA but only traces of glycerol. After being placed in a freeze-drying flask the eluted protein was flash-frozen in a liquid-nitrogen bath and the flask was connected to a freeze-dryer (Christ Alpha 1-4 LD, Osterode, Germany) at a pressure of 1.5 Pa and with the cooling coil at 188 K. The final product was characterized by mass analysis (Quattro Micro, Micromass, Altrincham, England; Atheris Laboratories, Geneva, Switzerland) and quantified by amino-acid composition analysis.
2.2. Crystallization
For the synthetically produced toxin, the lyophilized protein was resuspended to yield a 10 mg ml−1 solution in 50 mM sodium acetate, 0.02% sodium azide pH 5.5. Crystallization experiments were carried out by the sitting-drop vapour-diffusion method at 293 K using CrysChem plates (Hampton Research, USA). To determine the solubility pattern, initial crystallization experiments consisting of six conditions from Stura Footprint Screen #1 (Stura et al., 1992 ▶; conditions 1D, 2C, 3D, 4C, 5C and 6D; Molecular Dimensions, UK) were set up. Larger crystals were obtained by optimization of condition 2C: 40% PEG 4000, 200 mM imidazole malate pH 7.5 with and without additives. For data collection, crystals were transferred into cryoprotectant solution (Table 1 ▶) using a polyimide litholoop (Molecular Dimensions, UK) mounted on a SPINE cap, briefly soaked, picked up and plunged directly into a carousel in liquid nitrogen for transport to a synchrotron facility. Crystals of recombinant Gly-ρ-Da1a-K34A that were used for structural studies were discovered in the defrosted sample when the protein solution was dispensed to set up crystallization trials. The presence of crystals in the Eppendorf tube was confirmed by defrosting a second aliquot. These crystals were stabilized in a solution consisting of 25% PEG 4000, 200 mM imidazole malate pH 7.5, 50 mM sodium acetate pH 5.5 before being transferred to the cryoprotectant solution (Table 1 ▶).
Table 1. Data-collection, processing and refinement statistics for Gly--Da1a-K34A.
| PDB code | 4iye |
| Crystallization | During lyophilization |
| Cryoprotectant | 27% PEG 8000, 15% PEG MME 550, 10% glycerol, 99mM TrisHCl pH 8.0 |
| Data collection | |
| Synchrotron source | ID14-1, ESRF |
| Wavelength () | 0.9334 |
| Space group | P3121 |
| Unit-cell parameters () | a = b = 37.37, c = 66.05 |
| Molecules in asymmetric unit | 1 |
| Resolution () | 32.41.95 (2.071.95) |
| R merge (%) | 17.1 (108.8) |
| R p.i.m. (%) | 16.0 (94.7) |
| Mean I/(I) | 11.43 (1.12) |
| CC1/2 | 99.4 (65.9) |
| Completeness (%) | 93.9 (69.1) |
| Multiplicity | 8.44 (2.89) |
| Refinement | |
| Resolution () | 32.41.95 (2.001.95) |
| No. of reflections | 3738 (172) |
| R work (%) | 17.0 (29.0) |
| R free (%) | 25.2 (51.0) |
| R.m.s. deviations | |
| Bond lengths () | 0.016 |
| Bond angles () | 1.648 |
| Ramachandran plot, No. of residues† | |
| Favoured | 62 [98.48%] |
| Allowed | 1 [1.52%] |
| Outliers | 0 |
Values from Coot.
2.3. Data collection and analysis
Data-collection attempts were carried out using single crystals on beamlines ID29 and ID14-1 at the European Synchrotron Radiation Facility, Grenoble, France at 100 K. Only one of the seven crystals of s-ρ-Da1a grown by sitting-drop vapour diffusion gave diffraction data with reflections extending beyond 3 Å resolution. The spots could be indexed in XDS (Kabsch, 2010 ▶) in a primitive lattice with unit-cell parameters a = 37.6, b = 37.7, c = 65.9 Å, α = 90.2, β = 90.0, γ = 119.8°, but the data could not be merged with reasonable statistics. Both crystals retrieved from the defrosted sample of the Gly-ρ-Da1a-K34A mutant diffracted well, with visible data to better than 2.4 Å resolution. Indexing carried out with MOSFLM (Leslie, 2006 ▶) and XDS gave identical unit-cell parameters for the recombinant form. Data reduction with XDS for these crystals was consistent with the trigonal space group P3121, with unit-cell parameters a = b = 37.37, c = 66.05 Å (Table 1 ▶), suggesting that the recombinant and the synthesized forms of ρ-Da1a crystallize in similar lattices.
2.4. Structure determination and refinement
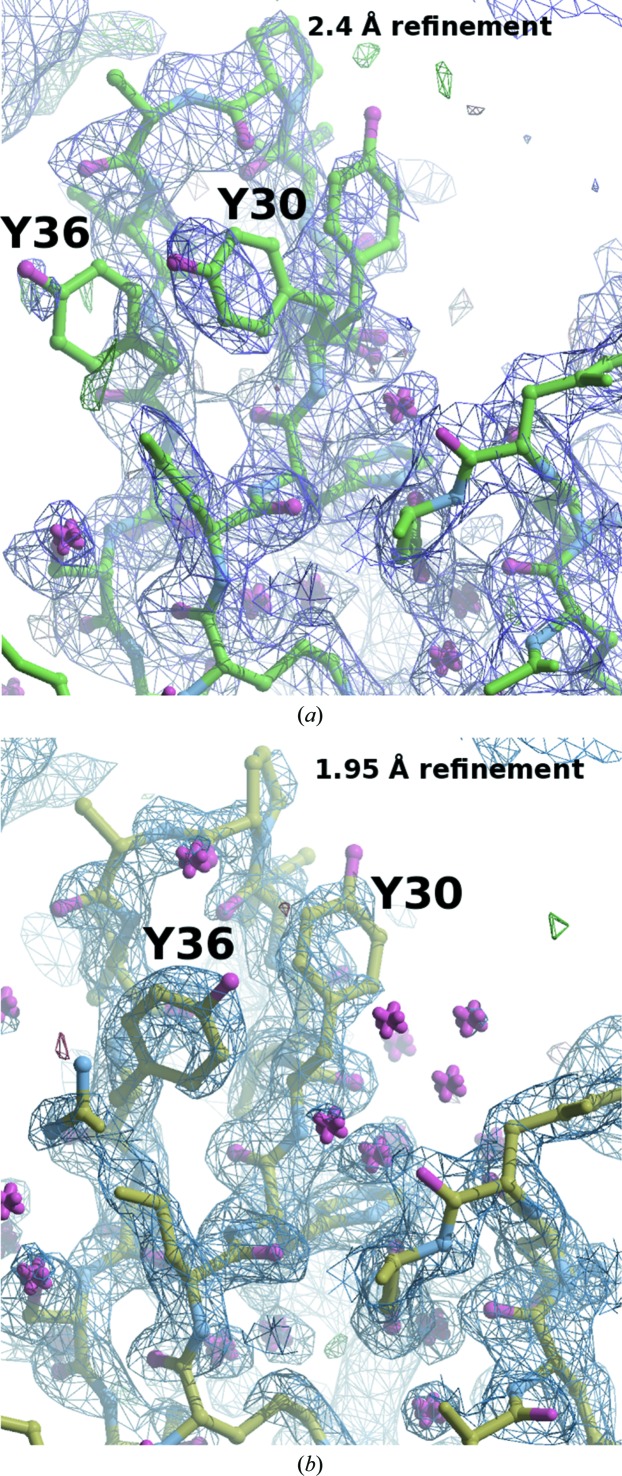
The structure was solved by molecular replacement in MOLREP (Vagin & Teplyakov, 2010 ▶) using the structure of the three-finger muscarinic toxin 2 (MT2; PDB entry 1ff4, space group P3221, unit-cell parameters a = b = 63.0, c = 37.0 Å; R. Ménez, M.-H. Le Du, J.-F. Gaucher & A. Ménez, unpublished work) as a model. A solution with one molecule in the asymmetric unit was only obtained after removing the tip of loop 1 (residues 6–11) and using data in the range 15–2.7 Å. The molecular-replacement solution confirms the attribution to space group P3121 rather than its enantiomorph P3221. This corresponds to a Matthews coefficient of 1.83 Å3 Da−1 and a solvent content of 33% for the crystals; this is a tighter packing compared with MT2, which has a solvent content of 59%. Only 2% of the entries in the PDB have such a low solvent content. Rebuilding was carried out with Coot (Emsley et al., 2010 ▶) and was followed by refinement with REFMAC (Murshudov et al., 2011 ▶) and phenix.refine (Adams et al., 2010 ▶) at 2.4 Å resolution. The electron density was clearly interpretable in all regions, with the only ambiguities being at the tip of loop 2 (residues 28–36; Fig. 2 ▶ a). The X-ray diffraction frames were later reprocessed to 1.95 Å resolution to extract the weak high-resolution data to help in map interpretation. Mean I/σ(I) and CC1/2 values (Karplus & Diederichs, 2012 ▶) were used as a guide to determine an appropriate resolution cutoff for the data (Table 1 ▶). This resulted in improved maps in which the positions of Phe30 and Phe36 could unambiguously be determined after two cycles of rebuilding and refinement (Fig. 2 ▶ b).
Figure 2.
Electron density in the tip region of loop 2. (a) Refinement at 2.4 Å resolution leaves certain ambiguities in the positioning of Phe30 and Phe36. (b) By incorporating the weak data to 1.95 Å resolution the two phenylalanine side chains could unambiguously be positioned in the electron density. The X-ray diffraction data could be used to 1.95 Å resolution. The figures were produced using PyMOL (DeLano, 2002 ▶) and the Raster3D (Merritt & Murphy, 1994 ▶) screenshot option in Coot.
3. Results and discussion
3.1. Crystals of ρ-Da1a
Crystals of recombinant E. coli-expressed Gly-ρ-Da1a-K34A were discovered when the protein was dispensed from the defrosted sample into an Eppendorf tube. X-ray diffraction data could be collected from these crystals after they had been transferred into a cryoprotectant solution consisting of 27%(w/v) PEG 8K, 15%(v/v) PEG MME 550, 10%(v/v) glycerol, 99 mM Tris–HCl pH 8.0 and flash-cooled in liquid nitrogen. The data collected on beamline ID14-1 at the European Synchrotron Radiation Facility could be indexed and merged with good statistics in the trigonal space group P3121 (Table 1 ▶). To determine the process that could have led to the formation of crystals in the frozen and defrosted sample aliquoted in Eppendorf tubes, we looked at the nonconcentrated samples. We did not find crystals in any aliquot at 0.7 mg ml−1, suggesting that crystal formation requires a higher protein concentration. Toxin samples redissolved in Milli-Q water at 10 mg ml−1 using protein that had been completely lyophilized to give a fluffy powder did not show the presence of crystals, suggesting that proper lyophilization does not lead to protein crystallization. The crystals were uniquely present in aliquots originating from a lyophilization process that did not terminate with a powder. When this particular sample was removed from the apparatus a small piece of ice was still present. The increase in pressure from 1.5 Pa to atmospheric pressure caused the ice to melt. After lyophilization at low pressure for about 1 h, the liquid-nitrogen-cooled peptide solution, which was initially at 0.7 mg ml−1, reached a concentration of about 50 mg ml−1 in the ice–protein slurry. Within a time period of 30 min, the sample was aliquoted in Eppendorf tubes at room temperature and the aliquots were stored in a freezer at 253 K. Crystal growth must have occurred during this time period of not longer than 30 min during which the residual ice melted and the sample was aliquoted. Nucleation could have occurred earlier at the triple point, since liquid water exists in minute amounts in the frozen sample, but any significant crystal growth is unlikely at 188 K. Nucleation could also have occurred as the ice melted, when the sample would have been at a very high protein concentration.
3.2. Structure determination
Molecular replacement using small proteins such as the three-finger toxins is very sensitive to structural deviations; in the case of Gly-ρ-Da1a-K34A the tip of loop 1 had to be removed before a valid solution could be found. The functional loops are defined as loop 1 (amino acids 4–16), loop 2 (amino acids 25–41) and loop 3 (amino acids 47–56) organized into two antiparallel β-sheets. Loop 1 forms the first β-sheet, which is composed of two strands of residues: Cys3–Ser6 and Ile12–Glu15 (Fig. 3 ▶). The second β-sheet comprises three antiparallel strands split into loops 2 and 3. These three strands are formed by residues Cys24–His29, Asp37–Gly41 and Cys58–Asn64. The four disulfide bridges characteristic of three-finger toxins are in the hydrophobic core between cysteines 3–24, 17–42, 46–57 and 58–63. The Gly-ρ-Da1a-K34A mutant displayed the same affinity as the synthetic toxin ρ-Da1a. Neither the additional N-terminal glycine nor Lys34 play any functional role.
Figure 3.
Superimposition of ρ-Da1a (yellow) with the muscarinic toxins MT1 (cyan), MT2 (green) and MT7 (Fruchart-Gaillard et al., 2008 ▶; pink) and two MT1–MT7 chimeras (shades of grey; PDB entries 3fev and 3neq; Fruchart-Gaillard et al., 2012 ▶). The greatest variation among the toxins is at the tip of loop 1, which is the section that was removed from the MT2 model during molecular replacement (PDB entry 1ff4). Overall, the structure of ρ-Da1a-K34A is closest to that of MT1. A major involvement of loop 1 in the interaction with the receptor is also supported by the fact that MT1, which has similar amino acids 1–15 to those of ρ-Da1a, binds the α1A-adrenoceptor with a K i of 20 nM.
3.3. Structural comparisons of ρ-Da1a
Venomous animals use the toxins present in their venoms to subdue their prey and for self-defence. To interact very efficiently and selectively with major physiological targets such as receptors and ion channels, the toxins have been subjected to an evolutionary process that has resulted in a large repertoire of small polypeptides with functional properties that are of interest for pharmacological and/or therapeutic applications.
Determination of their three-dimensional structures and their comparison with related toxins allows an understanding of the features that determine their pharmacological profiles with respect to a variety of human receptors. The solution of the crystallographic structure of ρ-Da1a is an important achievement because of its atypical pharmacological properties. Although structurally similar to the muscarinic toxins (MTs) present in the venom of African mambas, such as MT7 and MT1, the main target of ρ-Da1a is the α1A-adrenoceptor, for which it has an affinity of 350 pM. The amino-acid replacement present in ρ-Da1a with respect to MT7 and MT1 drastically changes the selectivity profile: MT7 and MT1 have affinities of 34 pM and 24 nM for muscarinic receptor M1 and 13 µM and 310 nM for M4, but greater than 20 µM and 62 nM, respectively, for the α1A-adrenoceptor. The structure of ρ-Da1a completes the study of engineered muscarinic chimeric toxins designed to have an altered pharmacological profile compared with the parent toxins (Fruchart-Gaillard et al., 2012 ▶). The amino-acid sequence similarity of ρ-Da1a to other three-finger toxins is matched by structural similarity (Fig. 3 ▶). Within the structural family of the three-finger fold toxins, small deviations can result in large changes in the selectivity, which makes crystallographic studies essential to direct engineering studies aimed at modifying the selectivity profiles of toxins that are not naturally selected for human targets. The crystal structures of the toxins can then be used in modelling and mutagenesis studies to understand the toxin–receptor interaction (Fruchart-Gaillard et al., 2002 ▶; Marquer et al., 2011 ▶).
4. Conclusions
4.1. Low-temperature crystallization
Experimentally, the nucleation and crystal growth of Gly-ρ-Da1a-K34A occurred at low temperature during lyophilization starting with a sample at 0.7 mg ml−1 and large crystals grew in 30 min. With a reservoir of 45% PEG 4000 and protein at 5 mg ml−1, a few days were required to grow crystals of comparable size, but of inferior quality, at room temperature. The significance of this report lies in the observation that the crystals formed under uncontrolled extreme low temperatures survived freezing, defrosting and cryocrystallography.
We would like to understand what led to the nucleation and enlargement of these crystals as it could eventually form the basis for new methods to crystallize proteins. Unfortunately, our attempts to reproduce the event did not meet with success. The physics of lyophilization is well understood; it takes place at the triple point, where vapour, liquid and ice coexist, typically in a vacuum (1.5 Pa) and at a temperature around 188 K. The presence of liquid water makes it possible, but not probable, that crystal nucleation could have occurred under these extreme conditions. At temperatures of 185–195 K supercooled liquid-like ultraviscous solvent can flow through channels in protein crystals (Weik et al., 2005 ▶), probably driven by changes in temperature. However, the transport properties at these temperatures are unlikely to be conducive to substantial crystal growth. It is probably safe to assume that subsequent growth occurred as the ice melted until the protein solution reached a concentration at which it was no longer saturated or until the sample was aliquoted and frozen at 293 K. This may have involved dynamic processes driven by temperature and protein concentration gradients with strong convective flows, which are not easily reproduced.
Even these brief conjectures are sufficient to make us understand that current protein crystallization techniques do not explore the entire temperature range that could be profitably used to crystallize proteins. Most protein crystallization experiments have been carried out at temperatures above the value at which water reaches its maximum density (277.13 K), so little is known about the crystallization of proteins at temperatures close to or below the freezing point of water owing to a lack of suitable equipment to precisely control the temperature for protein crystal growth under such conditions. Since crystal contacts involve water-mediated protein–protein interactions via the formation of hydrogen bonds, the behaviour of water is of great interest in protein crystallization. Below 277.13 K pure water becomes anomalous, becoming less dense with decreasing temperature. This behaviour can be explained by water-cluster models (Ludwig, 2001 ▶) involving extensive hydrogen bonding. The results reported here suggest that crystals could be grown at subzero temperatures in a protein–ice slurry.
Without accurate temperature control in the range 258–277 K, some crystallization studies could be carried out in the presence of compounds typically used as cryoprotectants because of their ability to disrupt ice formation. To grow crystals in the absence of ice at subzero temperatures, cryomixtures could be formulated to act as precipitants (Vera & Stura, 2013 ▶). The degree of supersaturation could be controlled by using higher glycerol concentrations to aid protein solubility, in contrast to MPD which is more effective as a precipitant. Working in a slurry would be more complicated but could be an interesting development since as the water crystallizes to ice the protein concentration would increase, stimulating rapid crystal growth.
4.2. Impurity segregation
Lyophilization or freeze-drying of aqueous solutions of proteins is a gentle concentration process that makes use of low temperatures and low pressure at the triple point to avoid the direct liquid–gas transition and boiling of the sample. It becomes irreversible only if it involves the loss of structural water molecules and denaturation (Griebenow & Klibanov, 1995 ▶). This process can also lead to protein concentration after liquid–liquid separation and can mediate the segregation and precipitation of impurities and less soluble misfolded protein. Nucleation and crystal growth under these conditions can be very fast, as in the case presented here. From the indexing, the crystal lattices obtained for both the synthesized and expressed proteins appear to be identical, suggesting that there is a strong tendency to crystallize in a well defined crystal form. From the observation of the amount of undissolved aggregates in the lyophilized frozen and defrosted sample, the improvement in crystal quality could be linked to the elimination of less soluble contaminants.
Many avenues that could be used to crystallize proteins have been left unexplored, with subzero crystallization being one of these. Without wishing to discourage further investigations, the obstacles to achieving this in a scientifically reproducible manner appear to be too great with current crystallization equipment and the rewards are too uncertain to suggest that it should be pursued without equipment that allows good control of temperature and pressure and the possibility of microscopic observation of the sample.
Supplementary Material
PDB reference: Gly-ρ-Da1a-K34A, 4iye
Acknowledgments
We are grateful to the ESRF and the staff of beamlines ID29 and ID14-1 for the allocation of beam time and for assistance during data collection.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Drevet, P., Lemaire, C., Gasparini, S., Zinn-Justin, S., Lajeunesse, E., Ducancel, F., Pinkasfeld, S., Courçon, M., Tremeau, O., Boulain, J. C. & Ménez, A. (1997). Protein Expr. Purif. 10, 293–300. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fruchart-Gaillard, C., Gilquin, B., Antil-Delbeke, S., Le Novère, N., Tamiya, T., Corringer, P.-J., Changeux, J.-P., Ménez, A. & Servent, D. (2002). Proc. Natl Acad. Sci. USA, 99, 3216–3221. [DOI] [PMC free article] [PubMed]
- Fruchart-Gaillard, C., Mourier, G., Blanchet, G., Vera, L., Gilles, N., Ménez, R., Marcon, E., Stura, E. A. & Servent, D. (2012). PLoS One, 7, e39166. [DOI] [PMC free article] [PubMed]
- Fruchart-Gaillard, C., Mourier, G., Marquer, C., Stura, E., Birdsall, N. J. M. & Servent, D. (2008). Mol. Pharmacol. 74, 1554–1563. [DOI] [PubMed]
- Griebenow, K. & Klibanov, A. M. (1995). Proc. Natl Acad. Sci. USA, 92, 10969–10976. [DOI] [PMC free article] [PubMed]
- Heller, M. C., Carpenter, J. F. & Randolph, T. W. (1999). Biotechnol. Bioeng. 63, 166–174. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Ludwig, R. (2001). Angew. Chem. Int. Ed. 40, 1808–1827.
- Marquer, C., Fruchart-Gaillard, C., Letellier, G., Marcon, E., Mourier, G., Zinn-Justin, S., Ménez, A., Servent, D. & Gilquin, B. (2011). J. Biol. Chem. 286, 31661–31675. [DOI] [PMC free article] [PubMed]
- Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869–873. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Pawlak, J., Mackessy, S. P., Fry, B. G., Bhatia, M., Mourier, G., Fruchart-Gaillard, C., Servent, D., Ménez, R., Stura, E., Ménez, A. & Kini, R. M. (2006). J. Biol. Chem. 281, 29030–29041. [DOI] [PubMed]
- Quinton, L. et al. (2010). Br. J. Pharmacol. 159, 316–325. [DOI] [PMC free article] [PubMed]
- Rajashankar, K. R., Tsai, I.-H. & Betzel, Ch. (1999). Acta Cryst. D55, 1064–1065. [DOI] [PubMed]
- Rees, B., Samama, J.-P., Thierry, J.-C., Gilibert, M., Fischer, J., Schweitz, H., Lazdunski, M. & Moras, D. (1987). Proc. Natl Acad. Sci. USA, 84, 3132–3136. [DOI] [PMC free article] [PubMed]
- Stura, E. A. (1999). Crystallization of Proteins: Techniques, Strategies and Tips. A Laboratory Manual, edited by T. Bergfors, pp. 113–124. La Jolla: International University Line.
- Stura, E. A., Nemerow, G. R. & Wilson, I. A. (1992). J. Cryst. Growth, 122, 273–285.
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vera, L. & Stura, E. A. (2013). Cryst. Growth Des., 10.1021/cg301531f.
- Weik, M., Schreurs, A. M. M., Leiros, H.-K. S., Zaccai, G., Ravelli, R. B. G. & Gros, P. (2005). J. Synchrotron Rad. 12, 310–317. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Gly-ρ-Da1a-K34A, 4iye