Abstract
Lineage commitment during embryonic stem cell (ESC) differentiation is controlled not only by a gamut of transcription factors but also by epigenetic events, mainly histone deacetylation and promoter DNA methylation. The DNA demethylation agent 5′-aza-2′-deoxycytidine (AzadC) has been widely described as an effective promoter of cardiomyogenic differentiation in various stem cell types. However, its toxicity and instability complicate its use. Therefore, the purpose of this study was to examine the effects of zebularine (1-(β-𝒟-ribofuranosyl)-1,2-dihydropyrimidin-2-1), a stable and non-toxic DNA cytosine methylation inhibitor, on mouse ESC (mESC) differentiation. Herein, we report that treating embryoid bodies, generated from mESCs, with 30 μM zebularine for 7 days led to greater cell differentiation and induced the expression of several cardiac-specific markers that were detected using reverse transcription-polymerase chain reaction (RT-PCR), real-time PCR, immunostaining and flow cytometry. Zebularine enhanced the expression of cardiac markers and the appearance of beating cells that responded to cardiac drugs, including ion channel blockers (diltiazem) and β-adrenergic stimulators (isoproterenol). Gene promoter methylation status was assessed using methylation-specific PCR (MSP) and validated by bisulfite sequencing analysis. Global gene expression profiling using microarrays showed that zebularine-differentiated cells are distinct from control ESCs. Pathway analysis revealed an enhancement of cellular processes such as embryonic development, cardiovascular system development and function. In addition, the whole-cell proteins exhibited different profiles as analyzed by two-dimensional differential-in-gel-electrophoresis. Our results indicate that zebularine regulates mesodermal differentiation of mESCs, controls promoter methylation of crucial cardiac genes and may help to improve cardiomyogenic differentiation.
Keywords: embryonic stem cells, mesoderm, methylation, zebularine, AzadC, nitric oxide
Embryonic stem cells (ESCs) hold a great potential for cell replacement, despite a number of significant hurdles that still stand in the way of the realization of that potential. The most important among these are differentiation efficiency, safety concerns and overcoming immune rejection.1 Currently, ESCs and their resulting differentiated cell types provide a promising in vitro system for the study of early human embryonic development.2, 3 As such, they can be used as cell lines with genetic disorders4, 5 and are being used increasingly in drug screening.6, 7
Differentiation of ESCs requires the repression of transcription factors involved in maintaining pluripotency and the activation of developmental genes. Both processes are directed by specific epigenetic mechanisms. An example of the first process is the promoter-methylation-dependent repression of Nanog and Oct3/4 as cells differentiate.8 Thus far, the activation of developmental genes during DNA-demethylation-induced stem cell differentiation has been less thoroughly studied. Instead, these developmental genes have been reported as being in a repressed state during the early stages of development because of the establishment of specific patterns of histone modifications, which consist of large regions of H3-Lys27 methylation harbouring smaller regions of H3-Lys4 methylation.9 This repressive chromatin state is mediated by the Polycomb group of proteins.10, 11, 12
Besides nitric oxide (NO)13 many epigenetic compounds efficiently invert genes' methylation status and histone patterns; such compounds are currently in use for the treatment of cancer.14 Because treatment of ESCs with 5′-aza-2′-deoxycytidine (AzadC) initiates cardiac differentiation and gene reactivation,15, 16 we tested the potential effect of zebularine (1-(β-𝒟-ribofuranosyl)-1,2-dihydropyrimidin-2-1) on ESCs.
Zebularine is a cytidine analogue that is able to inhibit DNA methyltransferases (DNMTs) through covalent binding17, 18, 19 and is a robust inhibitor of cytidine deaminase, a central enzyme for the metabolism of cytosine nucleoside analogues such as AzadC,20 which is increased in tumour cells as a resistance mechanism to antineoplastic drugs. Zebularine acts during DNA replication, substituting cytosine residues following its phosphorylation and conversion to deoxynucleotide. Once incorporated into DNA, it forms covalent complexes with DNMTs leading to inhibition of its activity, thereby decreasing DNA methylation.19 Overall, zebularine is a genome-wide demethylating agent with increased activity in proliferative cells.
Unlike AzadC, which is toxic in vitro and in vivo and is unstable in aqueous solution, zebularine is chemically stable in neutral and basic media.21, 22 In addition, zebularine has a smaller myelosuppressive effect than AzadC; this leads to minimal side effects, making zebularine a candidate drug for long-term tumour treatment by oral administration.22
We tested the effect of zebularine on mouse ESCs (mESCs) and detected rhythmic and synchronized beating areas in embryoid bodies (EBs). We next studied the gene and protein expression of cardiac markers, finding that zebularine-treated cells highly expressed cardiac-restricted markers and expressed low levels of pluripotency factors. Moreover, when gene expression was compared between cells treated with zebularine, AzadC and NO, the cardiac expression patterns showed that zebularine preferentially drives the differentiation of mESCs towards a cardiomyocyte-like phenotype. These findings support the hypothesis that zebularine regulates mesodermal differentiation more efficiently than the other drugs tested. Next, we analyzed methylation gene promoter status and detected that Nkx2.5, an early marker of the cardiac lineage differentiation program, was clearly unmethylated and thus transcriptionally activated following zebularine treatment. To decipher the global effect of zebularine on gene expression, we performed microarray analysis and found a significant number of differentially expressed genes with a B-statistic >1, demonstrating that the cellular transcriptome is clearly modified following zebularine treatment. The differentially expressed genes were selected using a linear model approach23 and implemented in the Linear Models for Microarray Bioconductor package and Ingenuity's Pathway Analysis (IPA) software. In addition, experiments having significant signatures or reverse/antisignatures were dissected using the entire mouse gene expression omnibus (GEO) experiments on the mouse Affymetrix platform deposited in the NCBI's GEO database. Moreover, proteins showed different patterns when subjected to two-dimensional differential-in-gel-electrophoresis (2D-DIGE). Finally, we studied the effect of zebularine on human ESCs (hESCs) and observed differences in the expression levels of some cardiac-specific genes after treatment.
Results
Zebularine preferably drives mESCs towards mesodermal lineage
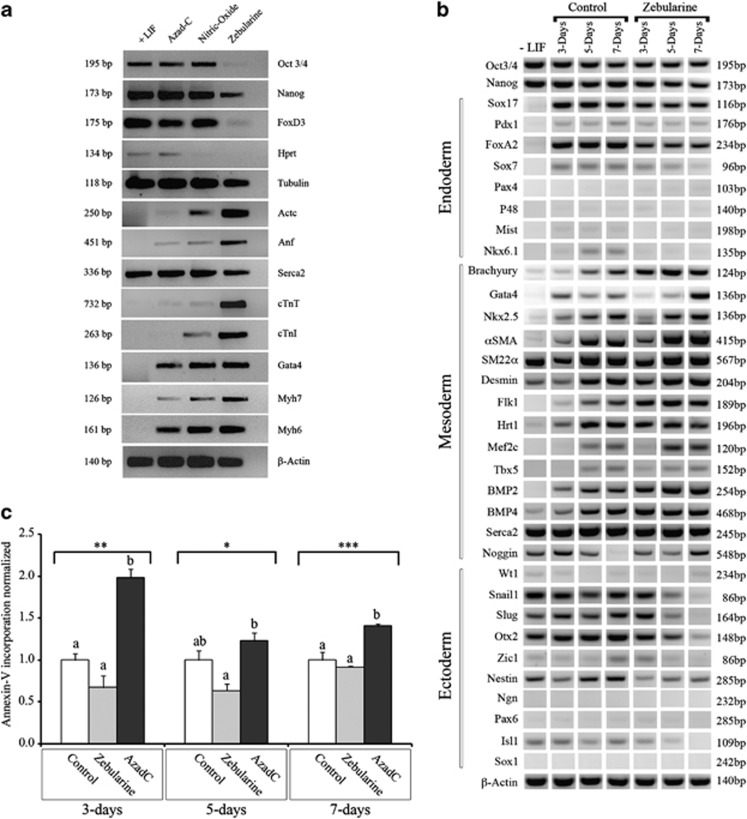
On the basis of the previous results demonstrating that AzadC and NO were able to promote cardiac differentiation of ESCs, we tested and compared the effect of zebularine on mESCs in standard culture conditions (+LIF (leukemia inhibitory factor)). Using reverse transcription-polymerase chain reaction (RT-PCR), we compared gene expression after treatment with each of these three molecules. When AzadC or NO was used, we observed that pluripotency markers were expressed at levels similar to control samples, whereas cardiac-specific genes were slightly increased. The expression of Gata4, Actc, Myh6, Myh7, cTnT and Anf was higher in AzadC- and NO-treated cells than in control cells. In contrast, Serca2 was expressed similarly in all samples. The only difference detected was the expression of cTnI in NO addition and Hprt in AzadC treatment. Interestingly, when we compared gene expression in zebularine-, AzadC- and NO-treated cells, we observed that zebularine caused a reduction in the expression of the pluripotency markers Oct3/4, Nanog and FoxD3 than did AzadC or NO. Moreover, zebularine-treated cells demonstrated higher expression of genes, especially those expressed in cardiac tissues, such as Actc, Anf, cTnT, cTnI, Myh6 and Myh7. Serca2 was expressed at a level similar to the other samples (Figure 1a).
Figure 1.
Effect of zebularine on mESC differentiation. (a) Detection and comparison of cardiac markers by RT-PCR in Control, AzadC, NO and zebularine treatment, undifferentiation markers are less detected and cardiac markers showed an enhanced expression after treatments. (b) Time course of zebularine treatment on lineage formation: mESCs cells were cultured as hanging drop during 3 days to form EBs and differentiate to derivatives of the three germ layers and then leaved in culture without treatment (Control) or treated with 30 μM zebularine. Gene expression was assessed in parallel at differentiating days 3, 5 and 7. Cells cultured without LIF (−LIF) are used as negative control for lineage formation. RT-PCR was performed with primers to detect pluripotency markers (Oct3/4 and Nanog), endoderm-specific markers (Sox17, Pdx1, FoxA2, Pax4, P48, Mist and Nkx6.1), mesoderm-specific markers (Brachyury, Gata4, Nkx2.5, αSMA, SM22α, Desmin, Flk1, Hrt1, Mef2c, tbx5, BMP2, BMP4, Serca2 and Noggin) and ectoderm-specific markers (Wt1, Snail1, Slug, Otx2, Zic1, Nestin, Ngn, Pax6. Islet1 and Sox1). β-Actin was used as the input control. (c) Apoptosis comparison for control-, zebularine- and AzadC-treated EBs. Time-course analysis and comparison of Annexin-V binding at differentiating days 3, 5 and 7, indicating that cells treated with zebularine are not undergoing apoptosis. Values were normalized from the means of the controls. Data are representative of three independent experiments statistically analyzed with analysis of variance (ANOVA) I (*P<0.05, **P<0.01 and ***P<0.001). Different letters at the top of the bars represent statistically different values; same letters represent statistically same values in discriminatory analysis Tukey's B test
As widely known, EB-mediated differentiation is the most common method to induce spontaneous cardiac differentiation; however, the results of this differentiation have been fairly successful. Athough we postulated that zebularine could be considered a critical parameter for optimizing early in vitro mesodermal differentiation, we treated EBs with 30 μM of zebularine, and then we assessed the gene expression at differentiating days 3, 5 and 7 for the three associated lineage markers (Figure 1b). We found that markers from different lineages are detected, interestingly, in zebularine-treated EBs, mesoderm markers (Brachyury, Gata4, Nkx2.5, αSMA, Desmin, Flk1, Hrt1, Mef2c BMP2, BMP4 and Noggin), which are markedly expressed starting at day 5; however, SM22α and Serca2 are equally detected in all experimental conditions. Moreover both endoderm markers (Sox17, Pdx1, FoxA2, Sox7, Pax4, P48, Mist and Nkx6.1) and ectoderm markers (Wt1, Snail1, Slug, Otx2, Zic1, Nestin, Ngn, Pax6, Isl1 and Sox1) are less or not detected at differentiating days 3, 5 and 7 when compared with control EBs, and in addition, Oct4 and Nanog are slightly less detected in zebularine-treated EBs compared with control and (−LIF). These results imply that zebularine is more efficient than AzadC or NO at promoting mesodermal gene expression.
Because of its lower toxicity, zebularine could potentially be used as a substitute for AzadC. To validate this hypothesis, we measured and compared Annexin-V incorporation at differentiating days 3, 5 and 7 (Figure 1c) and found that apoptosis was significantly reduced in zebularine compared with AzadC, and even more at day 3, where significant increase of apoptosis occurs as for AzadC, thus confirming that zebularine has no cytotoxic effect on EBs.
Zebularine promotes the expression of cardiac-specific genes and proteins
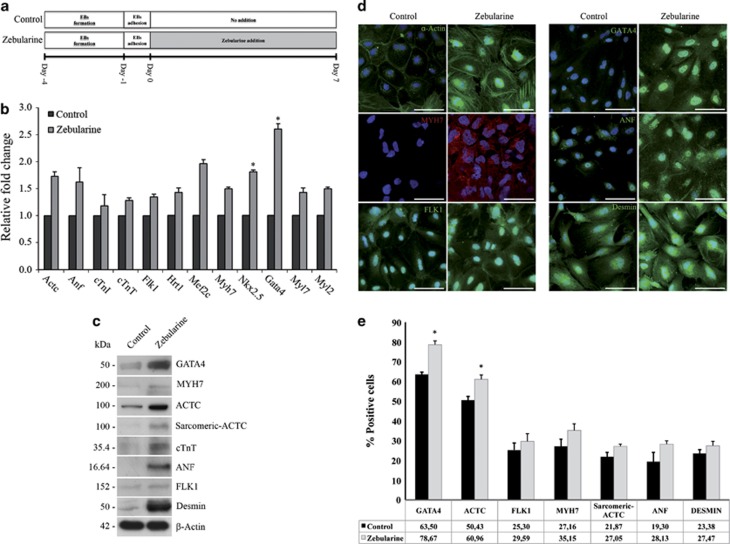
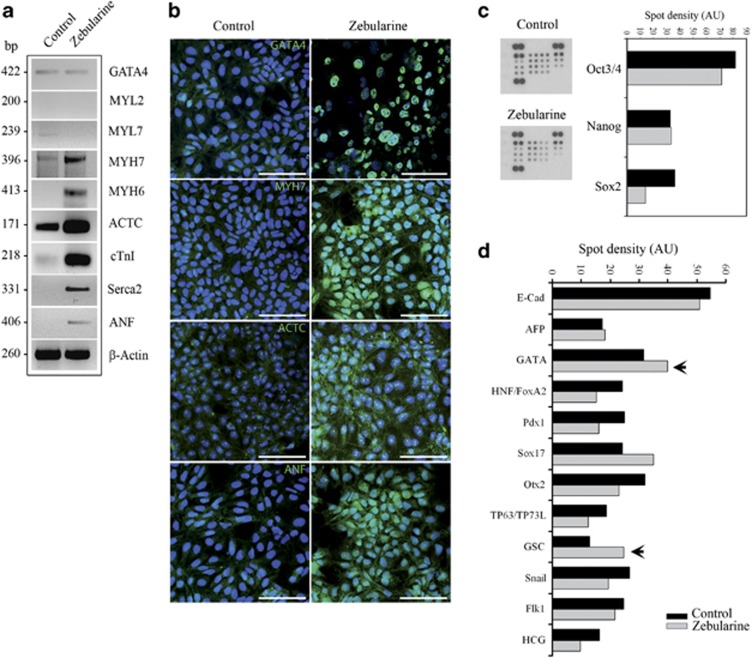
We used real-time PCR to analyze the mRNA levels of genes related to cardiac differentiation, and genes involved in the functional machinery of cardiac cells in EBs leaved to spontaneous differentiation during 7 days (from now on defined as Control) and EBs treated with zebularine (from now on defined as Zebularine) (Figure 2a). We detected elevated mRNA levels of cardiac-precursor genes Nkx2.5 (enhanced 1.8-fold (P>0.05)) and Gata4 (enhanced 2.6-fold (P>0.05)), both essential for cardiac cell commitment, elevated the expression of genes whose encoded proteins are essential components of contractile cardiac machinery, such as Actc, cTnI, cTnT, Myh7, Myl7 and Myl2, and increased the expression of other genes essential for cardiac development and function, such as Anf, Flk1, Hrt1 and Mef2c (Figure 2b).
Figure 2.
Zebularine promotes an overexpression of cardiac markers. (a) Timeline approach for differentiation of mESCs, showing the time periods for EBs formation, EBs adhesion, untreated control cells (no addition) and zebularine application. (b) Real-time PCR showing an overexpression of genes essential for cardiac development and function after zebularine treatment. The histograms represented the fold increase of each gene after treatment compared with the basal level of untreated control cells. Results are means±S.D. for three different experiments. (c) Western blot detection of cardiac protein expression; β-actin was used as a loading control. (d) Immunodetection corroborated the cardiac lineage markers after zebularine treatment; nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars; 50 μm. (e) Results from flow cytometric analysis showing the percentage of cells expressing cardiac-specific proteins in control and zebularine conditions. Data are represented as mean±standard error (±S.E.) for three different experiments. Statistical significance (*P<0.05) was determined by t-test
Using western blotting (Figure 2c), we found that zebularine promotes the expression of proteins that are restricted to mesoderm and cardiac phenotypes, such as Gata4, Actc, Myh7, cTnt, Anf, Desmin, Flk1 and α-actin. Immunostaining analysis (Figure 2d) showed that the precursor Gata4 was expressed at higher levels in zebularine-treated EBs and had a primarily nuclear distribution, whereas no expression was detected in control cells. Moreover, Actc and Myh7 proteins were markedly increased in cells treated with zebularine. Actc was clearly distributed in the cellular cytoskeleton and cellular prolongations in comparison to control cells, which only expressed low levels of this protein. Myh7 demonstrated a similar cytoskeletal distribution in treated cells and was not detected in control. Anf was expressed after zebularine treatment but not in control cells. Furthermore, Flk1 and Desmin, two proteins involved in cardiovascular system development, had increased expression following zebularine treatment (Figure 2d). Finally, by flow cytometry analysis we detected that cells treated with zebularine were more positive to proteins involved in cardiac differentiation and functionality (Figure 2e). Zebularine-treated cells are 15.17% more positive to Gata4, 10.53% to Actc, 4.29% to Flk1, 7.99% to Myh7, 5.18% to sarcomeric-Actc, 8.83% to Anf and 4.09% to Desmin. These results suggest that zebularine is able to induce the expression of cardiac-specific genes and proteins.
The beating areas resulting from zebularine treatment respond to chronotropic drugs
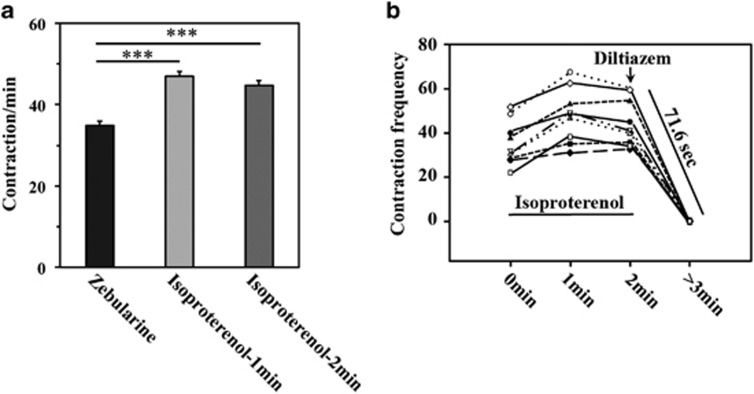
Zebularine treatment resulted in the appearance of beating areas that showed rhythmic and coordinated contractions (Supplementary Movie 1). To determine whether these contractions were functional, we used cardiotropic drugs to alter beating frequency. We administrated isoproterenol, which acts on β-adrenergic receptors, and diltiazem, a calcium channel blocker that produces a decrease in cardiac rhythm, provoking negative ionotropic and chronotropic function. After the addition of 2 μM isoproterenol (Figure 3a), we observed a 135.00% enhancement in contraction frequency after 1 min and a 126.25% enhancement after 2 min of exposure, demonstrating that isoproterenol provokes a very fast response in treated cells and that these cells possibly possess functional β-adrenergic receptors. On the other hand, and as we expected, 50 μM diltiazem (Figure 3b) led to complete inhibition of contraction 71.6 s after its addition (Supplementary Movies 1–3). Hence, zebularine promotes the differentiation of mESCs towards contractile cells that respond to cardiotropic regulators.
Figure 3.
Effects of chronotropic drugs in zebularine-treated cells. (a) Median of beating frequency in samples 1 and 2 min after treatment with 2 μM isoproterenol. Results are means±S.D. for 50 different areas from three experiments. ***A statistically significant difference of isoproterenol administration compared with zebularine (paired Student's t-test; P<0.001). (b) Contraction frequency registration of nine contractile areas from three different experiments showing that 2 μM isoproterenol increased beating frequency and 50 μM diltiazem was able to stop contractions 71.6 s after administration
Zebularine modifies the promoter methylation state of Nkx2.5
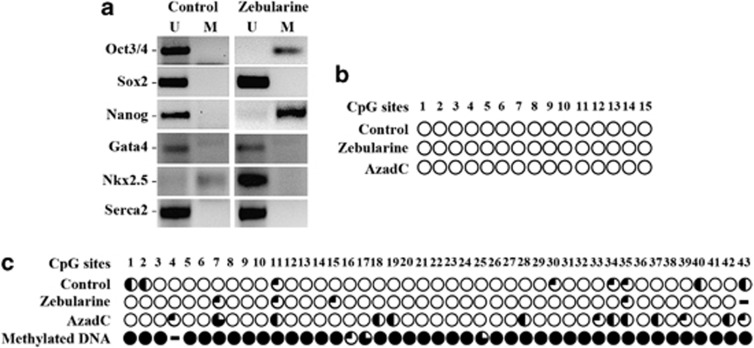
To determine whether zebularine produces changes in the methylation patterns, we studied gene promoter methylation status using methylation-specific PCR (MSP). As Figure 4a depicts, the pluripotency markers Oct3/4 and Nanog were transcriptionally active in control samples (unmethylated). This contrasts with zebularine-treated EBs, which had methylated promoters, suggesting that zebularine induced methylation-based gene silencing. In contrast, the Sox2 promoter was unmethylated in both samples, indicating that Sox2 expression was unaffected by zebularine. Moreover, the promoters of Gata4 and Serca2 were in an unmethylated state in both control and treated cells, suggesting that the expression of these genes was unaffected by zebularine.
Figure 4.
Analysis of gene promoter's methylation under zebularine treatment. (a) MSP of pluripotency and cardiac lineage markers showing that the promoters of both Oct3/4 and Nanog shifted from unmethylated (U) to methylated (M) state; the promoter of Nkx2.5 becomes unmethylated after zebularine treatment and no promoter methylation was detected in any of the other cardiac markers analyzed. (b) Bisulfite sequencing from five clones of Gata4 CpG island for Control, zebularine and AzadC-treated EBs, indicating that the promoter remains unmethylated in 15 CpG sites. (c) Bisulfite sequencing from five clones of Nkx2.5 promoter revealed that zebularine unmethylated almost all of the 43 CpG sites analyzed. The percentage of methylation is presented as the percentage of filled circles over total circles for a given CpG site from each clone analyzed:  =0%
=0%  =40%
=40% 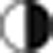 =60% =80%
=60% =80% 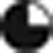 =100%
=100%
Interestingly, Nkx2.5 was epigenetically repressed in control cells, but appeared unmethylated in zebularine-treated cells. As Nkx2.5 associates with members of the GATA family, activates cardiac structural genes and is essential to initiate the cardiac differentiation, these data indicate that zebularine was able to induce demethylation of Nkx2.5, which leads to its expression, thereby promoting the expression of other fundamental genes during the cardiac differentiation of ESCs.
To confirm and validate the promoter methylation status of Nkx2.5 and Gata4, bisulfite-treated genomic DNAs obtained from Control, zebularine and AzadC samples were amplified, cloned and sequenced. As shown in Figure 4b, we found that the methylation pattern did not change and the Gata4 promoter remains completely unmethylated, however; and in concordance with the MSP results, Nkx2.5 promoter was almost completely unmethylated for zebularine and predominately methylated for AzadC (Figure 4c).
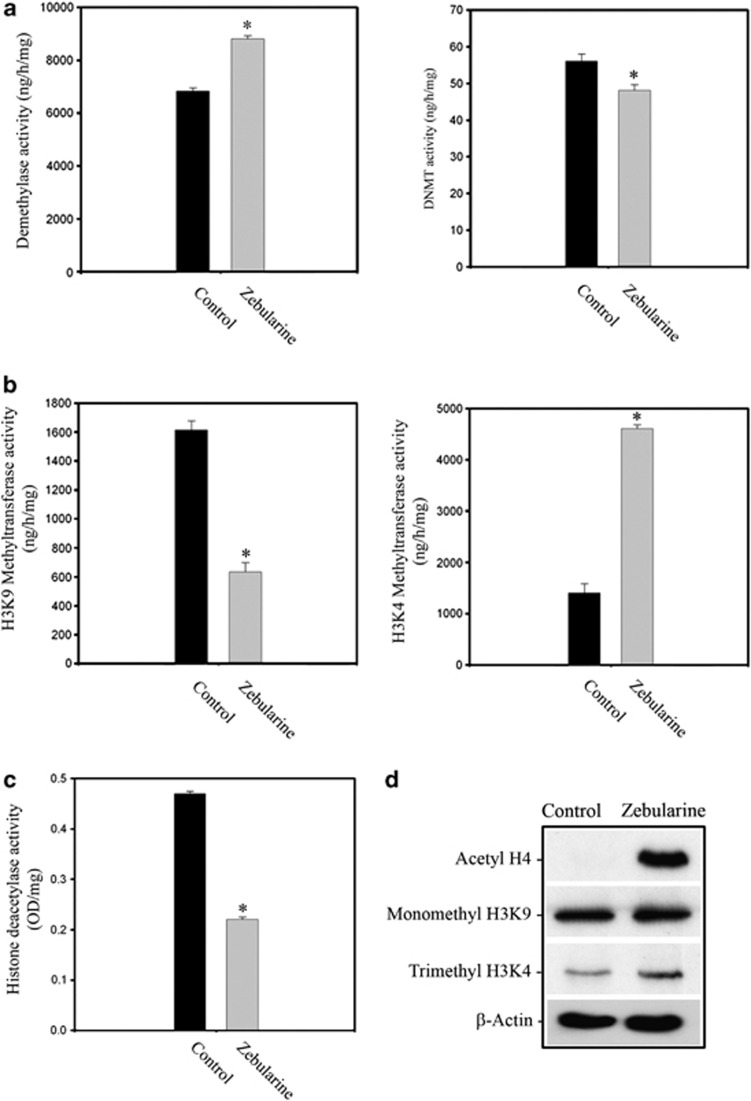
Consequently, we analyzed the activity of DNA demethylases and DNMTs. Following zebularine addition, DNA demethylase activity was significantly enhanced by 129.01% (P<0.01), whereas DNMT activity was reduced 14.20% (P=0.01) compared with control cells (Figure 5a). These data suggest that promoter methylation is reduced when cells are treated with zebularine and subsequently zebularine acts on DNMTs, decreasing their activity.
Figure 5.
Zebularine treatment caused (a) an increment of global demethylase activity and a decrease of DNTMs activity (b) a decrease of HMT (H3K9) and increase of H3K4 (c) reduction of global HDAC activity. Blots are representative of 4 independent experiments. (d) Western blot detection after zebularine treatment of acetylated state of histone H4, and methylated state of histones H3K4me3 and H3K9me1, β-actin was used as loading control
We also studied histone methyltransferase (HMT) activity for histone H3 lysine 9 (H3K9) and histone H3 lysine 4 (H3K4). After zebularine treatment, we found a 60.77% (P=0.017) reduction of HMT activity at H3K9, which is considered a marker for heterochromatin. In addition, the HMT activity at H3K4, a hallmark of euchromatin, was increased by 32.8% (P=0.01) after zebularine treatment (Figure 5b). We also detected that histone deacetylase (HDAC) activity was reduced by 52.74% (P=0.017) compared with control samples, indicating that chromatin exists in a more relaxed conformation in treated samples (Figure 5c). Although zebularine enhanced histone H4 acetylation, the acetylation state of histones plays a central role in determining gene expression and its overexpression could indicate a transcriptional upregulation; H3K4 and H3K9 methylation are detectable in zebularine-treated cells, the level of H3K9 methylation appears no different compared with control and H3K4 methylation appears slightly increased (Figure 5d), indicating the establishment of a specific pattern of histone modification.
These data indicate that zebularine acts as a chromatin-modifying agent, inducing methylation and silencing of Oct3/4 and Nanog, both of which are essential to maintain pluripotency. In addition, this drug is able to demethylate and increase the expression of Nkx2.5, which directs cells to enter a cardiomyocyte differentiation program. Moreover, zebularine indirectly alters the activity of enzymes that are implicated in the epigenetic regulation of gene transcription and chromatin structure.
Zebularine alters global gene expression profiling
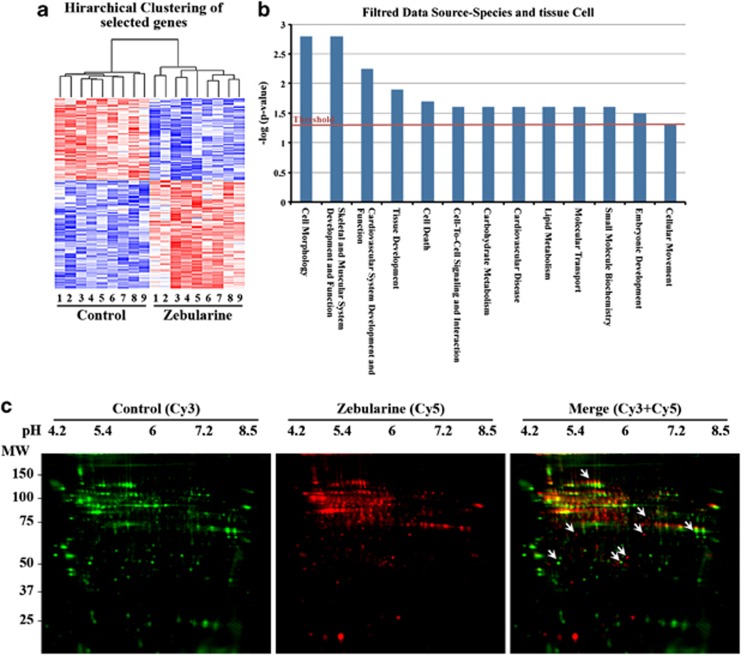
To characterize the genome-wide gene expression profile, we used Affymetrix microarray platform. A set of differentially expressed genes is presented as a heatmap to illustrate groups of coregulated genes; hierarchical clustering (Figure 6a) indicated that the gene expression profile of zebularine is distinct from that of control. The data were then globally normalized, filtered and deposited in NCBI's GEO database (GSE29072). Using Gene Ontology categories, 266 genes (119 upregulated and 147 downregulated) were identified from a total of 45 102 genes as having an expression level that differed significantly (P<0.001) upon zebularine treatment. The potential functions and known biological roles of these genes are listed in Supplementary Table 2. Data were most associated with biological process, system development, organ and multiorgan development, anatomical structure development, anatomical structure morphogenesis, cellular components and molecular functions. Some of the genes upregulated by zebularine matched the mesoderm markers, these include BMP4 and Brachyury.
Figure 6.
Microarrays, IPA analysis and proteomics assays for zebularine-treated cells. (a) Hierarchical clustering of the gene expression changes between zebularine and Control. The columns represent different replica of each condition replica. A red color represents upregulation, whereas a blue color represents downregulation. (b) Plot for biological functions generated by IPA analysis showing enhanced threshold for those related to cardiomuscular development. Enrichment score (y axis) is reported as the minus log transformation on the geometric mean of P-values from the enriched annotation terms associating with one or more of the gene group members. The genes are clustered into significantly enriched groups for specific biological functions. (c) Comparison of control- and zebularine-treated cells proteins using 2D-DIGE. Protein fractions from control were labeled with Cy3 (green) and zebularine ones with Cy5 (red), and then mixed. The mixed protein samples were separated by isoelectric focusing, and then SDS-PAGE. Merge picture from the superposition of fluorescence images shows a pool of proteins with differential mobility in zebularine condition (arrows)
We compared our microarray results with the results from other studies by analyzing all mouse GEO studies that used the mouse Affymetrix platform in the GEO database. A high positive score implies that the mouse GEO studies shares over- and underexpressed signatures with our experiment, whereas a high negative score indicates a reverse or antipattern compared with our study. The scores were calculated using weighted Kolmogorov–Smirnov statistics, which were then used to generate a Connectivity Map.24 These computations were carried out using the GEO query package of R Bioconductor and the statistical packages in R. Out of the microarrays we analyzed, 3750 showed a significantly similar or significantly reverse signature to our experiment; as a first approximation, we focused on similarity/dissimilarity scores with a P<0.05 and found 71 studies with high similarity to our results (P-values from 0.695 to 1) and 54 studies with low similarity (P-values from −0.695 to −1) (Table 1).
Table 1. (A) High similarity (P<0.05) and (B) low similarity (P<0.05).
|
(A) | ||
|---|---|---|
| GEO study | Similarity score | P-value |
| DNA demethylation effect on dermal fibroblasts: Aza-dC<=>untreated (agent) (GSE4696) | 1.04 | <0.0001 |
| Transcription factors Nanog and Oct4 knockdown effect on ESCs: Nanog knockdown<=>Control (protocol) (GSE4189) | 1 | <0.0001 |
| ESC line response to the conditional expression of neurogenin 3: 3 days<=>0 days (time) (GSE3653) | 0.95 | <0.0001 |
| NA demethylation effect on terminally differentiated cells: 5-aza-2dC<=>control (agent) (GSE4768) | 0.885 | <0.0001 |
| Cell culture model of extraembryonic endoderm: R1<=>IM8A1-I (ratio) (cell line) | 0.883 | <0.0001 |
| Hair follicle and skin molecular signatures: matrix<=>dermal papilla (cell type) | 0.846 | <0.0001 |
| Transcription factor Nrl deficiency effect on photoreceptor development: P2<=>E16 (development stage) | 0.839 | <0.0001 |
| Glucosamine and high glucose effect on renal mesangial cells: 25 mM glucose<=>control (agent) | 0.837 | <0.0001 |
| Phosgene effect on lungs: time course 12 h<=>0 h (time) | 0.835 | <0.0001 |
| Blastocyst response to various MAPK pathway inhibitors (MOE430A): JNK inhibitor<=>untreated (ratio) (agent) | 0.827 | <0.0001 |
| DNA demethylation effect on terminally differentiated cells: osteoblast<=>fibroblast (cell type) | 0.822 | <0.0001 |
| Blastocyst response to various MAPK pathway inhibitors (MOE430 2.0): JNK inhibitor<=>untreated (agent) | 0.818 | <0.0001 |
| Trans-10, cis-12-conjugated linoleic acid delipidiative effect on white adipose tissues: time course trans-10, cis-12-conjugated linoleic acid<=>control (agent) | 0.807 | <0.0001 |
| Estrogen effect on lung: time course 6 h<=>3 h (time) | 0.801 | <0.0001 |
| Mechanical stress effect on fibroblasts from various fetal tissues (MG-430A): tendon fibroblast<=>skin fibroblast (cell line) | 0.797 | <0.0001 |
| Spinal cord injury model: time course site of trauma<=>caudal to trauma (specimen) | 0.794 | <0.0001 |
| DNA demethylation effect on terminally differentiated cells: osteoblast<=>chondrocyte (cell type) | 0.791 | <0.0001 |
| Transcription factor Oct1-deficient fibroblast response to ionizing radiation (430A): γ-radiation<=>control (agent) | 0.79 | <0.0001 |
| Pancreatic development (MG-U74B): E14.5<=>E12.5 (age) | 0.79 | <0.0001 |
| Topoisomerase IIβ deficiency effect on the developing embryonic brain: topIIβ null<=>wild type (genotype/variation) | 0.79 | <0.0001 |
| Phosgene effect on lungs: time course 24 h<=>0 h (time) | 0.788 | <0.0001 |
| Oocyte development (MOE430B): secondary follicle<=>primary follicle (development stage) | 0.785 | <0.0001 |
| Oocyte development (MOE430B): 12 days<=>6 days (age) | 0.785 | <0.0001 |
| CH1 domain deletion, p300 and CBP heterozygous-null mutant fibroblasts response to hypoxia: normoxia<=>hypoxia (stress) | 0.78 | <0.0001 |
| Hair follicle and skin molecular signatures: matrix<=>dermal fibroblasts (cell type) | 0.772 | 0.006818 |
| Conjugated linoleic acid delipidative effect on obese line: time course (dye-swap) 14 days (ratio) (time) | 0.771 | 0.006818 |
| Cardiac development in embryo: E14.5<=>E11.5 (development stage) (GSE1479) | 0.771 | 0.006818 |
| Pancreatic development (MG-U74B): E15.5<=>E12.5 (age) | 0.763 | 0.006818 |
| Differential repression of TLR responses by PPARg and LXRs: GW7845<=>none (treatment) | 0.76 | 0.006818 |
| Corneal stromal cell differentiation: adult<=>10 days postnatal (protocol) | 0.758 | 0.006818 |
| Fibroblast growth factor 2 effect on embryonic fibroblast: dose response 40 ng/ml<=>4 ng/ml (ratio) (dose) | 0.75 | 0.01271 |
| Epididymis development: postnatal day 0.5<=>gestational day 18 (age) | 0.75 | 0.01815 |
| Pancreatic development (MG-U74B): E13.5<=>E12.5 (age) | 0.748 | 0.01815 |
| ERK MAP kinase inactivation during G0/G1-to-S-phase transition: time course FGF<=>control (protocol) | 0.748 | 0.01852 |
| Testis developmental time course: embryonic (MG-U74A): 18.5 d.p.c.<=>11.5 d.pc. (development stage) | 0.746 | 0.01852 |
| Phosgene effect on lungs: time course 8 h<=>0 h (time) | 0.741 | 0.01852 |
| Testis development: gestation day 16<=>gestation day 12 (time) | 0.741 | 0.01852 |
| Cardiac development in embryo: E16.5<=>E11.5 (development stage) (GSE1479) | 0.74 | 0.01852 |
| Ovary development: gestational day 14<=>gestational day 12 (age) | 0.738 | 0.01852 |
| Phosgene effect on lungs: time course 48 h<=>0 h (time) | 0.736 | 0.01852 |
| Acute myocardial infarction model: time course (MG-U74B) 48 h<=>4 h (time) | 0.736 | 0.01852 |
| Heart failure and NO eNOS knockout<=>wild type (strain) | 0.736 | 0.01852 |
| Embryonic kidney: ureteric bud and metanephric mesenchyme (MOE430 2.0): ureteric bud tip<=>ureteric bud stalk (tissue) | 0.734 | 0.01852 |
| MAP kinase activation effect on heart: time course MKK3bE transgenic<=>control (genotype/variation) | 0.729 | 0.01852 |
| Glycerol kinase knockout effect on liver glycerol kinase knockout<=>wild type (genotype/variation) | 0.729 | 0.01852 |
| Myotube response to PGC-1α-induced mitochondrial biogenesis: time course (MG-U74B) 3 days<=>0 days (time) | 0.728 | 0.02131 |
| Cardiac development in embryo: E18.5<=>E11.5 (development stage) (GSE1479) | 0.725 | 0.02131 |
| Pancreatic development (MG-U74B): E16.5<=>E12.5 (age) | 0.725 | 0.02131 |
| Lymphotoxin-β receptor signaling inhibition effect on lymph nodes: time course (MG-430B) 35 days<=>27 days (time) | 0.72 | 0.02394 |
| Spermatogonial stem cell activity in testis laminin binding germ cells (MG-U74C): laminin binding<=>interstitial (cell type) | 0.72 | 0.02394 |
| Phosgene effect on lungs: time course 72 h<=>0 h (time) | 0.72 | 0.02394 |
| Pyruvate effect on muscle cells: sodium pyruvate<=>untreated (agent) | 0.72 | 0.02394 |
| Hematopoietic cells at various stages of differentiation: granulocyte<=>ST-HSC (ratio) (development stage) | 0.719 | 0.02394 |
| Hippocampus of various inbred strains: DBA/2J<=>129S1/SvImJ (strain) | 0.717 | 0.02706 |
| Phosgene effect on lungs: time course 8 h<=>4 h (time) | 0.716 | 0.02706 |
| Gonadal somatic cells during the critical period of sex determination: time course 11.5 d.p.c.<=>10.5 d.p.c. (age) | 0.714 | 0.0303 |
| Embryonal carcinoma cell line response to retinoic acid or trichostatin A: trichostatin A<=>control (agent) | 0.711 | 0.03571 |
| Uniparental duplication of chromosome 7 regions effect on various tissues (MG-U74A): 13.5 d.p.c. placenta<=>13.5 d.p.c. embryo (tissue) | 0.709 | 0.03892 |
| MAP kinase activation effect on heart: time course male<=>female (gender) | 0.708 | 0.04206 |
| Acute myocardial infarction model: time course (MG-U74A) 24 h<=>12 h (time) | 0.706 | 0.04239 |
| Transcription factor Oct1 deficient fibroblast response to ionizing radiation (430B): γ-radiation<=>control (agent) | 0.705 | 0.04239 |
| Phosgene effect on lungs: time course phosgene<=>none (agent) | 0.705 | 0.04239 |
| Osteoblast differentiation (MG-U74B): 17 days<=>7 days (time) | 0.704 | 0.04239 |
| Ovary development: gestational day 14<=>gestational day 11 (age) | 0.702 | 0.04239 |
| Hyperoxic lung injury (U74Cv2): Nrf2 mutant<=>wild type (strain) | 0.701 | 0.04487 |
| Testis development: gestation day 14<=>gestation day 12 (time) | 0.7 | 0.04487 |
| Embryonic kidney: ureteric bud and metanephric mesenchyme (MOE430 2.0): ureteric bud tip<=>metanephric mesenchyme (tissue) | 0.699 | 0.04611 |
| Oocyte development (MOE430B): 22 days<=>6 days (age) | 0.696 | 0.04611 |
| Cystic fibrosis transmembrane conductance regulator deficiency in different genetic backgrounds: CFTR deficient<=>control (genotype/variation) | 0.695 | 0.04611 |
| Androgen effect on spermatogenesis: time course 10 days<=>8 days (age) |
0.695 |
0.04878 |
|
(B) | ||
|
GEO study |
Similarity score |
P-value |
| Oocyte development (MOE430B): primary follicle<=>large antral follicle (development.stage) | −0.695 | 0.04611 |
| Male and female salivary gland comparison: submandibular gland<=>meibomian gland (tissue) | −0.698 | 0.04611 |
| Corneal stromal cell differentiation: keratocyte<=>cornea (cell type) | −0.702 | 0.04239 |
| Male and female salivary gland comparison: submandibular gland<=>parotid gland (tissue) | −0.706 | 0.04239 |
| Uniparental duplication of chromosome 7 regions effect on various tissues (MG-U74A): newborn heart<=>13.5 d.p.c. placenta (tissue) | −0.707 | 0.04239 |
| PGC-1α-null brown adipocyte response to cAMP: PGC-1α null<=>wild type (genotype/variation) | −0.711 | 0.03571 |
| Hair follicle development: time course 1 day<=>1 year (age) | −0.711 | 0.03571 |
| Smooth muscle cell differentiation induced by retinoic acid: time course control (agent) (GSE1506) | −0.712 | 0.03309 |
| Smooth muscle cell differentiation induced by retinoic acid: time course 0 h (time) (GSE1506) | −0.712 | 0.03309 |
| Testis development: postnatal day 2<=>gestation day 14 (time) | −0.713 | 0.03309 |
| Extraocular and hindlimb skeletal muscle cell differentiation: time course (MG-430B): 24 h<=>0 h (time) | −0.714 | 0.0303 |
| Ovary development: gestational day 18<=>gestational day 14 (age) | −0.716 | 0.02706 |
| Male and female salivary gland comparison:submandibular gland<=>lacrimal gland (tissue) | −0.718 | 0.02394 |
| Guided differentiation into dopaminergic neurons and random differentiation into EBs neural precursors (stage 3) (development stage) | −0.721 | 0.02131 |
| Myogenic transcription factor MyoD mutant expression effect on embryonic fibroblast: time course 24 h<=>6 h (time) | −0.721 | 0.02131 |
| Spermatogonial stem cell self-renewal: time course 18 h<=>control (time) | −0.722 | 0.02131 |
| Spermatogonial stem cell self-renewal: time course GDNF, GFRα1 withdrawal<=>GDNF, GFRα1 pre-withdrawal (protocol) | −0.722 | 0.02131 |
| Male and female salivary gland comparison: sublingual gland<=>parotid gland (tissue) | −0.729 | 0.01852 |
| Embryonic stem cell line response to the conditional expression of neurogenin 3 undifferentiated: ESCs<=>EBs (development stage) (GSE3653) | −0.73 | 0.01852 |
| Pubertal mammary gland development: 4 weeks<=> 3 weeks (age) | −0.734 | 0.01852 |
| Medullary thymic epithelial cells medullary TEC<=>dendritic (cell type) | −0.738 | 0.01852 |
| Chondrocyte differentiation: time course 9 days<=>3 days (time) | −0.741 | 0.01852 |
| Hematopoietic stem cell proliferation after 5-fluorouracil treatment: time course 30 days<=>0 days (time) | −0.744 | 0.01852 |
| Spermatogonial stem cell self-renewal: time course 4 h<=>control (time) | −0.747 | 0.01852 |
| Corneal stromal cell differentiation cell culture<=>adult (protocol) | −0.75 | 0.01815 |
| Brown fat cell response to PGC-1α and PGC-1β deficiency: PGC-1β siRNA<=>control siRNA (agent) | −0.751 | 0.01271 |
| Embryonic stem cell differentiation induced by various chemicals: time course HMBA (agent) | −0.754 | 0.01271 |
| Hair follicle and skin molecular signatures: melanocytes<=>matrix (cell type) | −0.755 | 0.01271 |
| Transcription factor Egr3 expression effect on primary myotubes (MG-430A): truncated Egr3<=>wild-type Egr3 (genotype/variation) | −0.761 | 0.006818 |
| Testosterone effect on female lacrimal, meibomian and submandibular glands: submandibular gland<=>meibomian gland (tissue) | −0.765 | 0.006818 |
| Hair follicle and skin molecular signatures: outer root sheath<=>matrix (cell.type) | −0.766 | 0.006818 |
| Chondrocyte differentiation: time course 9 days<=>6 days (time) | −0.767 | 0.006818 |
| Guided differentiation into dopaminergic neurons and random differentiation into EB-guided differentiation (growth protocol) | −0.77 | 0.006818 |
| Testis development: postnatal day 2<=>gestation day 16 (time) | −0.77 | 0.006818 |
| Cardiac development in embryo: whole heart<=>both ventricles (tissue) | −0.773 | 0.006818 |
| ESC differentiation induced by various chemicals: time course RA (agent) (Platform GPL1261) | −0.774 | 0.006818 |
| Corneal stromal cell differentiation: myofibroblast<=>cornea (cell type) | −0.778 | 0.006818 |
| Embryonic stem cell line response to the conditional expression of neurogenin 3: 10 days<=>0 days (time) (GSE3653) | −0.779 | 0.006818 |
| Peripheral myelin protein 22 gene dosage and point mutation effect on sciatic nerve: PMP22 knockout<=>wild type (genotype/variation) | −0.78 | 0.006818 |
| Corneal stromal cell differentiation: fibroblast<=>cornea (cell type) | −0.789 | <0.0001 |
| Endothelial progenitor cells in fetal liver: feeder cell layer<=>LEPs_Sca1 minus (cell type) | −0.789 | <0.0001 |
| ESC line response to the conditional expression of neurogenin 3: 10 days<=>3 days (time) | −0.791 | <0.0001 |
| Hematopoietic stem cell proliferation after 5-fluorouracil treatment: time course 6 days<=>0 days (time) | −0.794 | <0.0001 |
| ESC differentiation induced by various chemicals: time course 6 days (time) | −0.798 | <0.0001 |
| Cell culture model of extraembryonic endoderm: ICR<=>129/Sv (ratio) (strain) | −0.807 | <0.0001 |
| Cell culture model of extraembryonic endoderm: XEN1–3<=>R1 (ratio) (cell line) | −0.807 | <0.0001 |
| Cell culture model of extraembryonic endoderm: PO<=>129/Sv (strain) | −0.81 | <0.0001 |
| Antenatal steroid effect on the placenta: dexamethasone<=>control (agent) | −0.823 | <0.0001 |
| Protein tyrosine kinase Src R388A mutant rescue by imidazole in living cells: time course Src R388A Y527F<=>Src D386N Y527F (cell line) | −0.836 | <0.0001 |
| Acute myocardial infarction model: time course (MG-U74B) 4 h<=>24 h (time) | −0.855 | <0.0001 |
| ESC differentiation induced by various chemicals: time course 2 days (time) (Platform GPL1261) | −0.932 | <0.0001 |
| Embryonic stem cell differentiation induced by various chemicals: time course 4 days (time) (Platform GPL1261) | −0.963 | <0.0001 |
| ESC differentiation induced by various chemicals: time course DMSO (agent) (Platform GPL1261) | −1.1 | <0.0001 |
Bold text denotes studies that involve the use of different agents (AzadC, Retinoic Acid, DMSO) for ESC differentiation, cardiac development and DNA demethylation, and the GEO accession number for these studies are in italics within parentheses.
As this classification is very approximate and gives only basic information, we also analyzed the data using IPA to classify specific groups of genes involved in cellular functions related to cardiogenesis (Figure 6b). Interestingly, the most often recorded biological functions were related to skeletal, muscular and cardiovascular system development and function, tissue development, cell-to-cell signaling and embryonic development. In addition to these gene expression changes, 2D-DIGE proteomics assay results showed that zebularine also affects protein mobility patterns, altering the acidic shift in several proteins (Figure 6c).
Zebularine enhances the expression of cardiac lineage markers in hESCs
We speculated whether zebularine was able to produce the same effect on hESCs. To address this issue, we used HS-181 – an hESC cell line. After 7 days of treatment, no beating areas were detected. To determine whether zebularine-treated hESCs possessed cardiac expression markers, we analyzed the gene expression using RT-PCR (Figure 7a). As with mESCs, we detected Gata4 expression in control and treated samples, and increased expression of Actc, Myh7, Myh6, cTnI, Serca2 and Anf in zebularine-differentiated cells; however, no expression of Myl2 and Myl7 was detected, possibly explaining the absence of contractile areas in treated cells. We then studied the protein expression using immunostaining (Figure 7b) and detected that zebularine slightly enhanced the protein levels of Gata4, Myh7, Actc and Anf. In addition, results from an hESC-specific protein array demonstrated a slight inhibition of pluripotency marker expression and increased levels of mesodermic proteins after zebularine treatment (Figures 7c and d). As a result, zebularine is able to induce cardiac-specific gene and protein expression in HS-181, although less effectively than in mESCs. Future comparative investigations will shed light on whether zebularine is able to induce complete cardiac differentiation in various cell types.
Figure 7.
Characterization of HS-181 cell line after treatment with zebularine. (a) RT-PCR of cardiac markers after treatment with zebularine. Myh7, Myh6, Actc, cTnI and Serca2 present more expression after treatment. (b) Immunostaining of treated cells to detect cardiac-specific proteins. Scale bars; 50 μm. (c) Blots of Proteome Profiler Array and the resulting quantification histograms demonstrating inhibition of pluripotency marker expression and (d) increased levels of mesodermic proteins after zebularine treatment (black arrows)
Discussion
It has been widely demonstrated that ESCs are able to give rise to cardiomyocytes when treated with growth factors and chemical compounds.25, 26, 27, 28 Among the many different protocols used to differentiate ESCs towards cardiomyocytes, NO addition to ESC culture was able to upregulate cardiomyogenesis processes and inhibit cell death.29, 30, 31 AzadC has also been demonstrated to be an activator of the cardiac differentiation,15 although 1 μM of AzadC leads to cell death and disappearance of the cardiac phenotype. Thus, AzadC possesses a significant limitation in producing cardiac-like cells from ESCs because of its cytotoxicity and inability to maintain a cardiac phenotype.
In this study, we demonstrated that zebularine is able to induce a mesodermal differentiation in mESCs and found that its application led to the increased expression of cardiac-restricted genes. Beyond the overexpression of cardiomyocyte-specific markers, we found that the pluripotency markers were downregulated. Interestingly, based on gene expression, zebularine appears to be more efficient at promoting the development of cardiac phenotypes than AzadC and NO, particularly in light of some reports suggesting that AzadC fails to differentiate stem cells to a cardiac lineage.32, 33
Furthermore, protein expression analysis confirmed that proteins involved in cardiac differentiation and functionality were highly expressed in the treated cells. These data were supported by immunostaining and flow cytometry analysis. To our knowledge, this is the first time that zebularine has been shown to differentiate ESCs towards cardiac-like cells more efficiently than other tested drugs.
Also, we observed that zebularine addition induced rhythmic and coordinated contractions and the ability to respond to cardiotropic drugs, suggesting that zebularine-treated cells possess cardiac-like functionality.
During cell differentiation, it has been suggested that the accumulation of cell lineage-specific methylation patterns could give rise to tissue-dependent and differentially methylated regions, such as those found in some CpG islands.34 This indicates that promoter methylation of specific genes involved in cellular differentiation may occur and thereby commit cells towards a specific lineage. Analysis of promoter methylation status of some genes involved in pluripotency and cellular differentiation showed that after zebularine administration, Oct3/4 and Nanog were transcriptionally inactivated via DNA methylation, suggesting that these cells would be biased towards differentiation. Meanwhile, the Nkx2.5 gene was clearly more activated after zebularine treatment, as its promoter became unmethylated. This indicates that cells treated with zebularine not only decrease the expression of pluripotency markers but also start to express genes that have pivotal roles in cardiac differentiation. In addition, we have demonstrated that zebularine is able to enhance histone H4 acetylation and modify the activity of HDAC and DNMT, in addition to reducing the methylation of H3K9. Zebularine also enhanced DNA demethylase activity and increased the methylation of H3K4. Hence, these global modifications induced by zebularine open and relax the structure of chromatin, increasing the accessibility of promoters to transcriptional machinery.
It has been recently suggested that DNMTs, in addition to incorporating methyl groups into gene promoter regions, may also enforce gene silencing through the recruitment of transcriptional repressor protein complexes that carry proteins with HDAC activity.35 This would create a scenario where DNA methylation is correlated with the compaction of chromatin structure. This suggestion is in concordance with our data that demonstrate a decrease of DNMT activity in conjunction with a decrease in HDAC activity.
Furthermore, using genome-wide expression array and 2D-DIGE protein analyses, we have demonstrated that zebularine strongly modifies gene and protein expression. Interestingly, our microarray data matched existing GEO studies (with a high similarity score). However, further analyses should be performed, at least at the protein level.
Finally, zebularine treatment of hESCs enhanced cardiac-restricted genes and protein expression, although no beating cells were detected. The absence of beating areas could be explained by the fact that zebularine is incorporated into DNA during the S phase of the cell cycle. As the doubling time of hESCs is longer than that of mESCs, this would explain why zebularine was less efficient in hESCs than in mESCs. Moreover, a protein array panel demonstrated a quantitative increase in mesoderm markers and a slight decrease in pluripotency markers.
These results provide new mechanistic insights regarding the function of zebularine during early ESC differentiation. The link between zebularine and its epigenetic effects opens a new field of investigation aimed at defining the rules of differentiation processes. Indeed, the possibility of priming normal cells with zebularine ex vivo may represent a strategy to obtain specialized cell populations potentially useful for cell therapy.
Materials and Methods
Cell culture and drug administration
The mouse ESC cell line (ES-D3 (D3); ATCC Number: CRL-1934) was cultured in DMEM (Dulbecco's medium) with 15% FBS, 10 U/ml penicillin, 10 μg/ml streptomycin, 2 mM glutamine, 10−4 M β-mercaptoethanol, 1 × non-essential amino acids and 103 U/ml LIF, and then maintained at 37 °C and 5% CO2. The drug administration to mESCs was prepared in three different experimental ways: (1) standard culture conditions in presence of LIF to compare the effect of different agents: the cells were divided into control group, NO-treated group, Azad-C-treated group and zebularine-treated group, control cultures (+LIF) were left untreated and experimental cultures were treated with 1 μg/ml AzadC, 100 μM of NO and 30 μM of zebularine for 7 days. (2) In vitro differentiation to generate derivatives of the three primary germ layers: EBs were generated using the hanging drop method during 3 days in the absence of LIF, and then plated and incubated for 24 h in a 10-cm plastic dish in DMEM medium without LIF to adhere, control cultures were left untreated and experimental cultures were treated with 30 μM of zebularine for 3, 5 and 7 days; in parallel cells cultured in monolayer without LIF and without generating EBs, (−LIF) were used as negative control. (3) Effect of zebularine on mESC differentiation: EBs were generated as in experimental point 2 and the cells were treated with 30 μM of zebularine for 7 days (see Figure 2a).
The human ESC cell line (HS-181) was derived in the Fertility Unit of Karolinska University Hospital, Huddinge at the Karolinska Institute after approval of a project entitled ‘Derivation and early differentiation and characterization of hESC lines' by the Karolinska Institute Research Ethics Board South, Drno 454/02. This line was derived from an embryo that could not be used for the infertility treatment of a couple. Both partners of the couple signed a consent form for donation of the embryo for derivation of a possible permanent stem cell line to be used in stem cell research. The HS-181 line is included in the EU hESC registry (http://www.hescreg.eu/) and cultured as described by Hovatta et al.36 To promote differentiation, cells were cultured on Matrigel, bFGF was withdrawn from the medium and cells were subjected to EB formation and 30 μM zebularine treatment for 7 days, as described for mESCs.
Analysis of apoptosis
Apoptosis was evaluated using an Annexin-V-FITC apoptosis detection kit II (BD Pharmingen, San Diego, CA, USA). Briefly, the cells were divided into Control group, zebularine-treated group and Azad-C-treated group. In each time point (3, 5 and 7 days), cells were trypsinized to prepare a single-cell suspension (detached cells) and centrifuged at 1800 r/min for 6 min at 4 °C. Cell pellets were washed two times with cold PBS and then stained with Annexin-V-FITC and propidium iodide (PI), following the manufacturer's instructions. For each sample, FITC-Annexin-V and PI staining were analyzed using Flow Cytometry (BD FACSCalibur cytometry System, San Jose, CA, USA). The data were analyzed using the CellQuest software (BD, San Jose, CA, USA).
Control samples were normalized to 1. The SPSS statistical package (v. 13.0, SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Data were analyzed for statistical significance using one-way ANOVA and Tukey's B test: an analysis of variance type I (ANOVA I) was used to determine the statistical significance differences between treatments. Once confirmed, Tukey's B discriminatory analysis was performed to determine the existence of significance differences between the groups.
Flow cytometry
Cardiac proteins expression was analyzed by flow cytometry. Briefly, EBs were trypsinized and fixed with 4% paraformaldehyde. The cells were treated with blocking solution (PBS containing 3% BSA, 0.1% Triton X-100), and incubated for 1 h at 4 °C with primary anti-Gata4, anti-Actc, anti-Flk1, anti-Myh7, anti-sarcomeric Actc, anti-Anf and anti-Desmin. After washing, the cells were incubated for 30 min with secondary antibodies. Unlabeled cells and cells labeled with secondary antibody were used as controls. The data were analyzed using the CellQuest software to calculate the percentage of positive cells.
RT-PCR analysis
Total RNA was extracted from control and treated cells using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. RNA concentration was quantified using a NanoDrop spectrophotometer, and 1 μg of total RNA was reverse transcribed using MMLV retrotranscriptase (Promega, Madison, WI, USA) and random hexamers.
cDNA was amplified using 0.05 U/μl of Eco-Taq polymerase (Ecogen, Barcelona, Spain) and 400 nM of specific primers. In all, 26 amplification cycles were used. Quantitative real-time PCR was performed using SYBR-Green and detected using an ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA). Primer information is described in Supplementary Table 1.
Western blotting assay
Cells were lysed in RIPA buffer (Sigma-Aldrich, St Louis, MO, USA) complete with protease inhibitors (Roche Diagnostics GmbH, Mannheim, Germany). Protein concentration was quantified by Bradford colorimetric assay (Sigma-Aldrich), and 20 μg of proteins were separated using SDS-PAGE, transferred to PDVF membrane and probed with anti-β-actin (Sigma-Aldrich; A5441), anti-Actc (Santa Cruz Biotechnology, Dallas, TX, USA; SC15335), anti-Gata4 (Abcam, Cambridge, UK; AB4132), anti-Myh7 (Santa Cruz Biotechnology; SC12117), anti-Anf (Abcam; AB5490), anti-cTnT (Abcam; AB10214), anti-Desmin (Sigma-Aldrich; D8281), anti-Flk1 (Millipore, Billerica, MA, USA; 07-716), anti-sarcomeric Actc (Sigma-Aldrich; A7811), anti-acetyl histone H4 (Upstate, Lake Placid, NY, USA; 06-598), anti-monomethyl Lys9 H3 (Upstate; 07-450) and anti-trimethyl Lys4 H3 (Upstate; 07-473).
Immunofluorescence staining
Cells were fixed and permeabilized with methanol, incubated at −20 °C for 20 min and blocked with donkey serum in PBS (pH 7.4), and then labeled with appropriate primary antibodies (1:1000). The primary antibodies used were mostly the same as those used for western blotting, and the secondary antibodies used were Alexa Fluor 488 goat anti-rabbit (Invitrogen; A11034), Alexa Fluor 488 goat anti-mouse (Invitrogen; A11029) and Alexa Fluor 568 donkey anti-goat (Invitrogen; A11057). Digital images were obtained using a Leica SP5 confocal microscope (Leica, Mannheim, Germany) or an Olympus IX71 inverted microscope (Olympus, Tokyo, Japan).
Proteome profile array
Protein expression profiles were assayed using the specific human pluripotent Stem Cell array kit ‘Proteome Profiler Array' (R&D Systems Europe, Abingdon, UK; ARY010) following the manufacturer's instructions. This array allowed us to detect simultaneously 15 different stem cell markers related to pluripotency and differentiation processes. The densitometry analysis of the resulting spots was carried out using the ImageJ 1.41 software (Wayne Rasband, NIH, Bethesda, MD, USA).
Chronotropic drugs treatment
Beating areas of cell culture treated with zebularine were subjected to 2 μM isoproterenol, and the frequency of beating was counted 1 and 2 min after treatment. The medium was changed and 50 μM diltiazem was added to the cultures, and contractions were counted until they stopped. Movies were captured on Olympus IX71 inverted microscope using Olympus DP70 digital camera system (Olympus) and the Olympus DP Controller software (Olympus).
Bisulfite modification and MSP
Genomic DNA was extracted by phenol–chloroform extraction and purified using the Wizard DNA Clean-up system (Promega). Bisulfite modification was performed using the EZ-DNA Methylation-gold kit (Zymo Research Corp., Irvine, CA, USA) following the manufacturer's instructions. Specific primers (Supplementary Table 1) were designed using Methyl Primer Express Software (Applied Biosystems), and MSP was performed as follows: 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min and 72 °C for 7 min. Samples were loaded on a 2% agarose gel.
Bisulfite sequencing
DNA was isolated using DNeasy kit (Qiagen, Hilden, Germany) from EBs (Control, Zebularine and Azad-C) grown for 7 days to allow differentiation. In all, 1 μg of DNA was then bisulphite converted using the Cells-to-CpGM Bisulfite Conversion kit (Applied Biosystems) according to the manufacturer's recommendation. Methyl Primer Express Software (Applied Biosystems) was used to design primers for CpG islands flanking the same sequence region used for MSP (Supplementary Table 1). Bisulfite-modified DNA was amplified using a hot start Taq polymerase (MyTaq HS Red DNA Polymerase; Bioline Reagents Ltd., London, UK), starting with denaturation at 95 °C for 5 min, and then 35 cycles of denaturation at 95 °C for 30 s, annealing at optimal Tm for 15 s, extension at 72 °C for 15 s, followed by an extra extension at 72 °C for 10 min. Amplicons were purified using the QIAquick PCR Purification kit (Qiagen) and were cloned in pGEM-T vector at a 3:1 insert:vector molar ratio. DH5α-competent cells (Invitrogen) were transformed by heat shock and grown overnight on LB-agar plates containing ampicillin, X-gal and IPTG. CpG primers described above was used to amplify bisulfite-modified DNA directly from 4 to 10 white colonies; amplicons were then purified enzymatically with ExoSAP-IT (USB, Cleveland, OH, USA) and 50 ng of PCR product were bidirectionally sequenced using the BigDye Terminator v.3.1 Cycle Sequencing kit (Applied Biosystems). Fragments were separated on an ABI3730xl DNA Analyzer (Applied Biosystems) and analyzed with Variant Reporter Software v.1.0 (Applied Biosystems). BiQ Analyzer software (Max-Planck-Institut, Saarbrüken, Germany) was used to analyze methylation patterns and generate diagrams. In all, 1 μg of DNA from Control EBs was methylated by MSssI CpG methyltransferase using the recommended protocol (New England Biolabs, Ipswich, MA, USA) and used in parallel with samples as control for bisulfite conversion and sequencing efficiency and accuracy.
Epigenetic enzyme activity analysis
The epigenetic enzyme activity measurements and calculations were performed according to the manufacturer's protocols and formulas. DNMTs activity (Catalog no.: P-3001), HMT activity H3K4 (Catalog no.: P-3002), HMT activity H3K9 (Catalog no.: P-3003) and demethylase activity (Catalog no.: P-3019) were measured using 10 μg of nuclear protein extracts in the EpiQuik Kits (Epigentek, Farmingdale, NY, USA). HDAC activity was analyzed using the HDAC Colorimetric Assay Kit (BioVision, Milpitas, CA, USA), and the colorimetric intensity was measured using a Varioskan Flash microplate reader (Thermo Electron, Vantaa, Finland). Nuclear proteins were isolated using the Subcellular Proteome Extraction Kit (Calbiochem, San Diego, USA), and 5 μg of extracts were used for this assay.
Microarray analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen), and 1 μg was used to obtain the gene expression profile of each sample. The signals were quantified using spectrophotometry and verified using a microfluidics-based platform (Genomic core facility of CABIMER). All samples showed the characteristics of high-quality RNA and were subjected to subsequent analysis. cRNA was hybridized on an Affymetrix GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA, USA). The data presented in this publication have been deposited in NCBI's GEO37 and are accessible through GEO Series accession number GSE29072 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29072). In addition, data were subjected to Gene Ontology analysis and IPA software to determine the possible biological functions and pathways.
2D-DIGE proteomic analysis
Proteins were extracted with 30 mM Tris, 7 M urea, 2 M thiourea and 4% (w/v) CHAPS, pH 8.0. Cells were then sonicated in a Bioruptor and centrifuged at 12 000 r.p.m. at 4 °C for 10 min. The supernatant was collected, 1 μl of TCEP was added and samples were incubated for 30 min in the dark at 37 °C. Next, Cy3 or Cy5 dye was added (2 μl) and the staining reaction was stopped using 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 2% (v/v) pharmalyte and 130 mM DTT. Proteins were quantified using the Bradford colorimetric assay (Sigma-Aldrich), and 1D electrophoresis was performed with Inmobiline Drystrip pH 3-11 NL, 18-cm strips (GE Healthcare, Buckinghamshire, UK). In all, 5 μg of protein were used per strip. The strips were focused in an IPGphor system for 22 h. 2D-PAGE was performed with 10% duracryl acrylamide gels (Genomics Solutions, Ann Arbor, MI, USA), and electrophoresis was performed using an Ettan Dalt VI system (Amersham, Buckinghamshire, UK) according to the following protocol: 2.5 W per gel for 30 min, followed by a second step of 12.5 W per gel for 5 h. Fluorescence was visualized using a Typhoon Trio (Amersham).
Statistical analysis
Values are presented as mean±S.D., unless otherwise indicated, statistical significance was calculated using an unpaired Student's t-test and P<0.05 was considered significantly different.
Acknowledgments
The technical assistance and suggestions of N Mellado and EJ López are greatly appreciated. We thank A Krishan for his help with microarray analysis and K Kamenarova for her assistance with Sanger (direct) sequencing. We were supported by Fundación Progreso y Salud, Consejería de Salud, Junta de Andalucía (Grant PI-0022/2008); Consejería de Innovación Ciencia y Empresa, Junta de Andalucía (Grant CTS-6505; INP-2011-1615-900000); FEDER co-funded grants from Instituto de Salud Carlos III (RD06/0010/0025; PI10/00964) and the Ministry of Health and Consumer Affairs (Advanced Therapies Program Grant TRA-120). CIBERDEM is an initiative of the Instituto de Salud Carlos III. AH was awarded a ‘Ramón Areces' postgraduate scholarship.
Glossary
- AzadC
5′-aza-2′-deoxycytidine
- DNMT
DNA methyltransferase
- EBs
embryoid bodies
- ESC
embryonic stem cell
- GEO
gene expression omnibus
- HDAC
histone deacetylase
- hESC
human embryonic stem cell
- HMT
histone methyltransferase
- H3K4
histone H3 lysine 4
- H3K9
histone H3 lysine 9
- IPA
Ingenuity's Pathway Analysis
- LIF
leukemia inhibitory factor
- LIMMA
linear models for microarray
- mESC
mouse embryonic stem cell
- MSP
methylation-specific PCR
- NO
nitric oxide
- PI
propidium iodide
- T-DMR
tissue-dependent and differentially methylated region
- zebularine
1-(β-𝒟-ribofuranosyl)-1,2-dihydropyrimidin-2-1
- 2D-DIGE
two-dimensional differential-in-gel-electrophoresis
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by Y Shi
Supplementary Material
References
- Hmadcha A, Domínguez-Bendala J, Wakeman J, Arredouani M, Soria B. The immune boundaries for stem cell based therapies: problems and prospective solutions. J Cell Mol Med. 2009;13:1464–1475. doi: 10.1111/j.1582-4934.2009.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, et al. Developmental study of fragile X síndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Dvash T, Ben-Yosef D, Eiges R. Human embryonic stem cells as a powerful tool for studying human embryogenesis. Pediatr Res. 2006;60:111–117. doi: 10.1203/01.pdr.0000228349.24676.17. [DOI] [PubMed] [Google Scholar]
- Pickering SJ, Minger SL, Patel M, Taylor H, Black C, Burns CJ, et al. Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reprod Biomed Online. 2005;10:390–397. doi: 10.1016/s1472-6483(10)61801-9. [DOI] [PubMed] [Google Scholar]
- Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, et al. Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online. 2005;10:105–110. doi: 10.1016/s1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- Festag M, Sehner C, Steinberg P, Viertel B. An in vitro embryotoxicity assay based on the disturbance of the differentiation of murine embryonic stem cells into endothelial cells. I: Establishment of the differentiation protocol. Toxicol In Vitro. 2007;21:1619–1630. doi: 10.1016/j.tiv.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Fico A, Manganelli G, Simeone M, Guido S, Minchiotti G, Filosa S. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cells Dev. 2008;17:573–584. doi: 10.1089/scd.2007.0130. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med. 1999;190:1595–1604. doi: 10.1084/jem.190.11.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papait R, Monti E, Bonapace IM. Novel approaches on epigenetics. Curr Opin Drug Discov Devel. 2009;12:264–275. [PubMed] [Google Scholar]
- Choi SC, Yoon J, Shim WJ, Ro YM, Lim DS. 5-Azacytidine induces cardiac differentiation of P19 embryonic stem cells. Exp Mol Med. 2004;36:515–523. doi: 10.1038/emm.2004.66. [DOI] [PubMed] [Google Scholar]
- Yoon BS, Yoo SJ, Lee JE, You S, Lee HT, Yoon HS. Enhanced differentiation of human embryonic stem cells into cardiomyocytes by combining hanging drop culture and 5-azacytidine treatment. Differentiation. 2006;74:149–159. doi: 10.1111/j.1432-0436.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- Marquez VE, Kelley JA, Agbaria R, Ben-Kasus T, Cheng JC, Yoo CB, et al. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. Ann N Y Acad Sci. 2005;1058:246–254. doi: 10.1196/annals.1359.037. [DOI] [PubMed] [Google Scholar]
- Marquez VE, Barchi JJ, Jr, Kelley JA, Rao KV, Agbaria R, Ben-Kasus T, et al. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. The magic of its chemistry and biology. Nucleosides Nucleotides Nucleic Acids. 2005;24:305–318. doi: 10.1081/ncn-200059765. [DOI] [PubMed] [Google Scholar]
- Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321:591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos D, van Overveld W.Decitabine: a historical review of the development of an epigenetic drug Ann Hematol 200584(Suppl 13–8.Erratum in: Ann Hematol 2006; 85: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin J, Eisenhauer E. DNA methyltransferase inhibitors – state of the art. Ann Oncol. 2002;13:1699–1716. doi: 10.1093/annonc/mdf314. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Chuang JC, Byun HM, Egger G, Yang AS, Dubeau L, et al. Long-term epigenetic therapy with oral zebularine has minimal side effects and prevents intestinal tumors in mice. Cancer Prev Res. 2008;1:233–240. doi: 10.1158/1940-6207.CAPR-07-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Chen K, Wu L, Wang ZZ. Extrinsic regulation of cardiomyocyte differentiation of embryonic stem cells. J Cell Biochem. 2008;104:119–128. doi: 10.1002/jcb.21604. [DOI] [PubMed] [Google Scholar]
- Riebeling C, Schlechter K, Buesen R, Spielmann H, Luch A, Seiler A. Defined culture medium for stem cell differentiation: applicability of serum-free conditions in the mouse embryonic stem cell test. Toxicol In Vitro. 2011;25:914–921. doi: 10.1016/j.tiv.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Yan P, Otsuji TG, Narazaki G, Uosaki H, Fukushima H, et al. Induction and enhancement of cardiac cell differentiation from mouse and human induced pluripotent stem cells with cyclosporin-A. PLoS One. 2011;6:e16734. doi: 10.1371/journal.pone.0016734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Mujoo K, Sharin VG, Bryan NS, Krumenacker JS, Sloan C, Parveen S, et al. Role of nitric oxide signaling components in differentiation of embryonic stem cells into myocardial cells. Proc Natl Acad Sci USA. 2008;105:18924–18929. doi: 10.1073/pnas.0810230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumenacker JS, Katsuki S, Kots A, Murad F. Differential expression of genes involved in cGMP-dependent nitric oxide signaling in murine embryonic stem (ES) cells and ES cell-derived cardiomyocytes. Nitric Oxide. 2006;14:1–11. doi: 10.1016/j.niox.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Kanno S, Kim PK, Sallam K, Lei J, Billiar TR, Shears LL., II Nitric oxide facilitates cardiomyogenesis in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12277–12281. doi: 10.1073/pnas.0401557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song J, Liu W, Wan Y, Chen X, Hu C. Growth and differentiation of rat bone marrow stromal cells: does 5-azacytidine trigger their cardiomyogenic differentiation. Cardiovasc Res. 2003;58:460–468. doi: 10.1016/s0008-6363(03)00265-7. [DOI] [PubMed] [Google Scholar]
- Martin-Rendon E, Sweeney D, Lu F, Girdlestone J, Navarrete C, Watt SM. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox Sang. 2008;95:137–148. doi: 10.1111/j.1423-0410.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, et al. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- Estève PO, Chin HG, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci USA. 2005;102:1000–1005. doi: 10.1073/pnas.0407729102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta O, Mikkola M, Gertow K, Strömberg AM, Inzunza J, Hreinsson J, et al. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum Reprod. 2003;18:1404–1409. doi: 10.1093/humrep/deg290. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.