Abstract
Many herbicide-resistant weed species are polyploids, but far too little about the evolution of resistance mutations in polyploids is understood. Hexaploid wild oat (Avena fatua) is a global crop weed and many populations have evolved herbicide resistance. We studied plastidic acetyl-coenzyme A carboxylase (ACCase)-inhibiting herbicide resistance in hexaploid wild oat and revealed that resistant individuals can express one, two or three different plastidic ACCase gene resistance mutations (Ile-1781-Leu, Asp-2078-Gly and Cys-2088-Arg). Using ACCase resistance mutations as molecular markers, combined with genetic, molecular and biochemical approaches, we found in individual resistant wild-oat plants that (1) up to three unlinked ACCase gene loci assort independently following Mendelian laws for disomic inheritance, (2) all three of these homoeologous ACCase genes were transcribed, with each able to carry its own mutation and (3) in a hexaploid background, each individual ACCase resistance mutation confers relatively low-level herbicide resistance, in contrast to high-level resistance conferred by the same mutations in unrelated diploid weed species of the Poaceae (grass) family. Low resistance conferred by individual ACCase resistance mutations is likely due to a dilution effect by susceptible ACCase expressed by homoeologs in hexaploid wild oat and/or differential expression of homoeologous ACCase gene copies. Thus, polyploidy in hexaploid wild oat may slow resistance evolution. Evidence of coexisting non-target-site resistance mechanisms among wild-oat populations was also revealed. In all, these results demonstrate that herbicide resistance and its evolution can be more complex in hexaploid wild oat than in unrelated diploid grass weeds. Our data provide a starting point for the daunting task of understanding resistance evolution in polyploids.
Keywords: ACCase mutation, Acc1 , herbicide resistance, hexaploid wild oat (Avena fatua), polyploidy, resistance evolution
Introduction
About 30–70% of plant species are of polyploid origin, and many herbicide-resistant weed species are polyploids. However, our understanding on the evolution of herbicide resistance in polyploid weeds is rather limited. Wild oats (A. fatua and A. sterilis) are self-pollinated allohexaploid species of the Poaceae (grass) family and are among the world's most common and economically damaging weeds in crop production. Acetyl-coenzyme A carboxylase (ACCase)-inhibiting herbicides have been widely used for wild oat control. ACCase (EC 6.4.1.2) is a key enzyme in most organisms, catalyzing the first step of fatty acid biosynthesis. Herbicidal inhibition of plant ACCase is also important in world agriculture for weed control. Three chemically distinct classes of herbicides that inhibit ACCase are the aryloxyphenoxypropionates, the cyclohexanediones and the more recent phenylpyrazolin class consisting of pinoxaden herbicide. These three chemical classes are hereafter collectively referred to as ACCase herbicides. In plants, ACCase in plastids is essential for the biosynthesis of primary fatty acids and cytosolic ACCase is involved in biosynthesis of long-chain fatty acids (Shorrosh et al., 1994). Homomeric ACCase in the cytosol of nearly all plant species, and heteromeric ACCase in the chloroplasts of dicots, are relatively insensitive to ACCase herbicides. In contrast, the plastidic homomeric ACCase found in nearly all grass species is ACCase herbicide sensitive, and thus grass species are generally controlled by ACCase herbicides.
Global and often intensive ACCase herbicide use has selected for resistance in many grass weeds (Heap, 2011), since the first report by Heap and Knight (1982). Biochemical mechanisms known to endow ACCase herbicide resistance in plants are target-site ACCase gene mutations and/or non-target-site enhanced rates of ACCase herbicide metabolism (reviewed by Devine and Shimabukuro, 1994; Délye, 2005; Powles and Yu, 2010). Thus far, among grass weeds, nine distinct amino-acid substitutions in the carboxyl transferase domain of plastidic ACCase genes have been identified as individually endowing target-site resistance to certain ACCase herbicides (reviewed by Délye et al., 2005; Powles and Yu, 2010; see also Collavo et al., 2011). In diploid grass species, these individual ACCase gene resistance mutations confer moderate to high-level resistance to ACCase herbicides (Délye et al., 2005; Yu et al., 2007). Depending on specific ACCase mutations, herbicides and herbicide rates, ACCase resistance alleles in diploid grass species have been found to be partially dominant or dominant over wild-type alleles (Christoffers et al., 2002; Tal and Rubin, 2004; Yu et al., 2007; Scarabel et al., 2011).
Many resistant wild-oat populations have been reported in at least 12 countries (Heap, 2011). Early studies of the mechanism(s) endowing resistance to ACCase herbicides in resistant Avena spp. populations revealed resistant ACCase (Maneechote et al., 1994, 1997; Seefeldt et al., 1996; Shukla et al., 1997; Cocker et al., 2000) with more recent reports identifying specific ACCase gene mutations conferring the resistance (Christoffers et al., 2002; Liu et al., 2007; Cruz-Hipolito et al., 2011; Beckie et al., 2012). However, hexaploidy in Avena and other polyploid species renders identifying and understanding herbicide resistance difficult. For herbicide resistance, these previous studies investigating the biochemical and molecular basis of ACCase herbicide resistance in Avena spp. did not investigate the complexities of gene transcription/expression in a polyploid species. Whereas in diploids both gene copies are usually transcribed and expressed, expression may be far more complicated in polyploid species. Hexaploid wild oat (2n=6 × =42) has a genome structure of AACCDD originating through allopolyploidy involving three diploid species (Rajhathy and Morrison, 1960; Jellen et al., 1994; Yang et al., 1999). A. longiglumis has been suggested as a possible A-genome donor in the evolution of hexaploid Avena spp. (Nikoloudakis et al., 2008), while the likely C-genome donor is A. ventricosa (Nikoloudakis and Katsiotis, 2008). The origin of the D genome is less clear, but it is more similar to the A genome than the C genome (Yang et al., 1999). All hexaploid Avena spp. show bivalent chromosome pairing with disomic inheritance (Thomas, 1992). Therefore, each gene is predicted to have three homoeologous copies, one from each of the three original genomes. While there is increasing understanding of homoeolog genetics and expression/silencing in allotetraploid Arabidopsis and polyploid crop species such as Brassica and wheat (reviewed by Wendel, 2000; Adams, 2007; Chen, 2007), all that is currently known for A. fatua plastidic ACCase genes is that there are three gene copies (Acc1;1, Acc1;2 and Acc1;3, Christoffers et al., 2002), and it is rarely known whether all homoeologous genes are expressed or how they interact and are inherited. In studying the molecular and biochemical basis of herbicide resistance in four hexaploid wild-oat (mainly A. fatua) populations, we identified individuals with one, two or three distinct resistance-endowing ACCase gene mutations. These multiple-mutation individuals provided a valuable opportunity to study gene expression and inheritance relating to natural selection in polyploids. Using resistance gene mutations as markers, we investigated homoeologous ACCase gene inheritance, transcription and in vitro enzyme activity in hexaploid Avena. Using herbicide resistance evolution as an example, we provide insights into resistance evolution in hexaploid wild oat.
Materials and methods
Plant material
Herbicide-resistant wild-oat populations M3/4, M3/5, M3/27 (A. fatua) and M5W/7 (A. sterilis), hereinafter called R1, R2, R3 and R4, respectively (Ahmad-Hamdani et al., 2011), were used in this study along with a susceptible A. fatua population (Owen and Powles, 2009). Plants surviving high rates of commercial ACCase herbicides diclofop-methyl fenoxaprop-p-ethyl, or sethoxydim, (all from Bayer CropScience, Melbourne, VIC, Australia) from each population enabled identification of specific ACCase resistance mutations among individual plants. These individuals with a specific ACCase mutation were grown to maturity in a glasshouse and allowed to self-pollinate and produce seeds. To ensure no chance of cross-pollination, individual plants were insulated in micro-perforated plastic bags and seeds from single plants were harvested separately. These single-plant-derived progeny lines with known ACCase resistance mutation(s) were used for subsequent studies.
Plant growth and herbicide treatment
Scarified seeds of the resistant progeny lines were germinated on agar (0.6% w/v) and seedlings transplanted into potting mix and grown in a controlled environment room with a day/night temperature of 20/15 °C and a 12-h photoperiod (500 μmol photons m−2 s−1). When seedlings reached the 3- to 4-leaf stage, one leaf from each plant was sampled for ACCase sequencing and mutation genotyping before the plants were herbicide treated for resistance phenotyping. The susceptible population was treated in the same manner. Individuals of the R progeny lines specifically identified to possess a known resistance-endowing single-ACCase mutation (Ile-1781-Leu, Asp-2078-Gly or Cys-2088-Arg) and the susceptible population were treated with a commercial formulation of the ACCase herbicide diclofop-methyl (delivered in 112 l ha−1 water at a pressure of 200 kPa). Diclofop-methyl was used at 2000, g ha−1, which fully controlled the S population. Plants were returned to the controlled environment room after treatment and visual assessment for resistance and susceptibility was made 3 weeks after treatment. Plants were recorded as resistant if they displayed strong growth after herbicide treatment and as susceptible if they displayed severe symptoms of leaf chlorosis, desiccation, retarded growth or plant death, similar to the S population.
ACCase gene sequencing
Genomic DNA (gDNA) was extracted from the leaf tissues of resistant and susceptible plants according to Yu et al. (2008) and the regions in the wild-oat plastidic ACCase carboxyl transferase domain with known and potential ACCase resistance mutation sites were amplified using published primer pairs SQCT-β1 F/SQCT-β1 R and SQCT-α1 F/SQCT-α1 R for wild oat (Liu et al., 2007 and Table 1) under modified PCR conditions. The PCR was conducted in a 25-μl volume that consisted of 1–2 μl of gDNA, 0.5 μℳ of each primer, and 12.5 μl of 2 × GoTaq Green Master Mix (Promega, Madison, WI, USA). The PCR was run with the following profile: 94 °C for 4 min; 35–40 cycles of 94 °C for 30 s, X °C for 30 s, and 72 °C for 30–120 s; followed by a final extension step of 5–7 min at 72 °C. X refers to the respective annealing temperature used for each primer pair (Table 1). The PCR products were purified from agarose gels using the Wizard SV Gel and PCR Clean-up System (Promega), and sequenced using a commercial sequencing service. All sequence chromatograms were visually checked for quality and consistency before sequences were assembled and aligned.
Table 1. Primer pairs (resistance mutation site numbering corresponds to amino acid sequence of full-length plastidic ACCase in A. myosuroides).
| Primer pair | Sequence (5′-3′) | Annealing temperature used (°C) | Usage | Product size (bp) | Targeted mutation site |
|---|---|---|---|---|---|
| SQCT-β1F | CATCATCTTTCTGTATGCCAGTGGG | 65 | Sequencing | 999 | -1781- |
| SQCT-β1R | CTGTATGCACCGTATGCCAAG | ||||
| SQCT-α1F | AATACATGTGATCCTCGTGCAG | 60 | Sequencing | 602 | -1999–2096- |
| SQCT-α1R | TCCTCTGACCTGAACTTGATCTC | ||||
| NsiI1781F | CTGAATGAAGAAGACTATGGTCG | 60 | dCAPS | 165 | 1781 |
| NsiI1781R | AGAATACGCACTGGCAATAGCAGCACTTCCATGCA | ||||
| RsaI2078F | TTCTCTGGTGGGCAAAGAGACCTTTTTGAAGG | 62 | dCAPS | 181 | 2078 |
| RsaI2078R | CATAGCACTCAATGCGATCTGGATTTATCTTGGTA | ||||
| SQCT-α1F | See above | 60 | CAPS | 602 | 2088 |
| SQCT-α1R | See above | ||||
| NsiI1781F | See above | 60 | cDNA cloning | 1122 | -1781–2096- |
| SQCT-α1R | TCCTCTGACCTGAACTTGATCTC | ||||
| NsiI1781F | See above | 60 | cDNA sequencing | 1050 | -1781–2096- |
Abbreviations: ACCase, acetyl-coenzyme A carboxylase; cDNA, complementary DNA; dCAPS, derived cleaved amplified polymorphic sequence.
ACCase resistance mutation genotyping and segregation analysis using PCR-based, non-homoeolog-specific molecular markers
Published derived cleaved amplified polymorphic sequence (dCAPS) markers for mutations at positions 1781 (Kaundun and Windass, 2006) and 2078 (Kaundun, 2010) were used in this study with modified primers (Table 1) and PCR conditions as described above. In addition, based on the ACCase sequence information obtained from the bulked susceptible (20 plants) and a number of individual R samples (at least 35), we developed a CAPS marker for the 2088 mutation (Table 2). The nucleotide T to C mutation at codon 2088, causing a Cys to Arg amino-acid substitution, creates a HaeII restriction site (Table 2). Sequence results revealed no other single-nucleotide polymorphisms around this restriction site. Therefore, the primer pair SQCT-α1F and SQCT-α1R (Table 1) was used to amplify a 602-bp fragment, which was then digested with HaeII. Wild-type 2088-Cys alleles would generate a single undigested 602 bp band, while mutant resistant 2088-Arg alleles would generate a 529 bp band, plus an undetectable 73 bp fragment. Heterozygous ACCase loci would produce both the 602 and 529 bp bands. Restriction digestions were carried out according to the manufacturer's recommendations (New England BioLabs, Ipswich, MA, USA) and digestion patterns were viewed on 2% agarose gels (electrophoresis at 90–100 V for 120–140 min) stained with ethidium bromide. These three (d)CAPS markers, for codons 1781, 2078 and 2088, were used to analyze segregation patterns for ACCase resistance mutation(s) in the R2-resistant progeny lines. While these (d)CAPS markers are codominant with wild-type alleles, they do not discriminate among ACCase gene copies and, therefore, were considered dominant markers (unable to detect true allelic heterozygosity) for the purposes of analyzing plant genotypes and segregation patterns (see discussions of homoeoallelic heterozygosity below).
Table 2. (d)CAPS markers used for identification of ACCase mutant alleles in wild-oat populations. Ambiguity nucleotide codes: R=A/G, Y=C/T.
| Target mutation | Primer pair used | Type | Restriction enzyme | Recognition site |
Fragment size (bp)
|
|
|---|---|---|---|---|---|---|
| Susceptible (S) | Resistant (R) | |||||
| 1781 | NsiI1781F/ NsiI1781R | dCAPS | NsiI | 5′-ATGCA^T-3′ | 130, 35 | 165 |
| 2078 | RsaI2078F/ RsaI2078R | dCAPS | Rsal | 5′-GT^AC-3′ | 181 | 147, 34 |
| 2088 | SQCT-α1 F/ SQCT-α1 R | CAPS | HaeII | 5′-RGCGC^Y-3′ | 602 | 529,73 |
Abbreviations: ACCase, acetyl-coenzyme A carboxylase; dCAPS, derived cleaved amplified polymorphic sequence.
ACCase complementary DNA (cDNA) fragment cloning and sequencing
Total RNA was extracted from leaf tissue of individual plants possessing all three ACCase mutations (1781+2078+2088) using the Plant RNeasy Mini Kit (Qiagen, Hilden, Germany). gDNA contamination was removed using the TURBO-DNA free kit (Ambion, Austin, TX, USA), which is highly effective in removing trace quantities of DNA contamination. Oligo(dT)-primed first-strand cDNA was prepared from 2 μg total RNA using the Omniscript Reverse Transcription system (Qiagen). The primers NsiI1781F (modified from Kaundun and Windass, 2006) and SQCT-α1R were paired to amplify a 1122 bp cDNA fragment containing all known resistance-endowing ACCase gene mutation sites (1781, 1999, 2027, 2041, 2078, 2088 and 2096), as well as polymorphic sites for the three ACCase gene copies in A. fatua (Christoffers et al., 2002). This fragment was cloned into the pGEM-T vector (Promega) and transformed into E. coli. White colonies with putative inserts were used as templates for PCR re-amplification of the 1122 bp fragment and screened for the 1781, 2078 or 2088 mutations using PCR-based markers (see below). Fragments that contained putative 1781, 2078 or 2088 mutations were selected for confirmatory sequencing using two forward primers, SQCT-α1F and NsiI1781F (Table 1), with overlapping amplification regions.
In vitro inhibition of ACCase activity by ACCase herbicides
The leaf tissue from R2 progeny lines determined to be homozygous (no dCAPS/CAPS marker segregation) for one or two ACCase mutation(s), or from plants of segregating progeny lines heterozygous for two or three mutations were used for in vitro ACCase activity assays. Leaf material at the 3 to 4-leaf seedling stage (unless otherwise specified) was harvested, snap frozen in liquid nitrogen and stored at −80 °C until use. ACCase extraction, partial purification and in vitro enzyme inhibition by ACCase herbicides was performed according to Yu et al. (2004). Two subsamples from each extraction were assayed, and there were at least two extractions per population per herbicide treatment. Technical grade diclofop acid, fenoxaprop acid and clethodim (94–98% purity, Bayer CropScience, and Sumitomo Chemical Australia, Epping, NSW, Australia) and sethoxydim (52%, Bayer CropScience) were used in assays to determine the herbicide concentration causing 50% inhibition of ACCase activity (I50). Herbicide stock solutions were prepared in 100 mM Tricine buffer. Stock solutions of diclofop or fenoxaprop contained 10% acetone with final concentrations of <1% acetone in each assay mix. Tissue protein content was determined by the Bradford method (Bradford, 1976).
The in vitro ACCase assay in A. fatua was reported to be affected by experimental conditions such as the composition of the extraction buffer and (NH4)2SO4 precipitation of the protein (Shukla et al., 1997). In order to test if different experimental conditions affected our resistant: susceptible I50 ratios, we determined I50 values when (1) ACCase was assayed with and without desalting, (2) when crude protein extract was precipitated with 10–40% versus 40% ammonium sulfate saturation and (3) when a different extraction buffer (buffer D in Shukla et al., 1997) was used. The herbicide I50 values were found to vary in each case for resistant and susceptible samples, but the I50 resistant/susceptible ratios (referred to as R/S ratios) remained similar. In addition, plants from purified L. rigidum mutant lines homozygous for ACCase mutations 1781, 2078 or 2088 (Yu et al., 2007) were included as positive controls for the assay.
Statistical analysis
The I50 was estimated by non-linear regression using the logistic model:
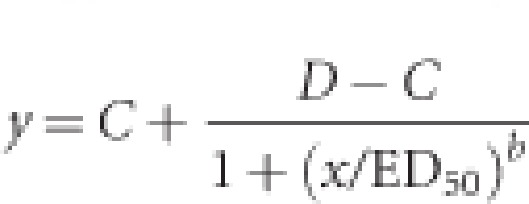 |
where C=lower limit, D=upper limit, ED50=dose giving 50% response and b=slope around ED50. Estimates were obtained using Sigmaplot (version 12.0, Systat Software Inc., San Jose, CA, USA). Data was subjected to one-way analysis of variance and significant differences in ACCase activities and I50 values among genotypes were determined by Duncan's test using SPSS software (version 11.5, SPSS Inc., Chicago, IL, USA). Data sets from different experiments were analyzed separately if significant differences were found because of differences in experimental set up or plant developmental stages.
Independent assortment of mutations was evaluated using logarithm of the odds score analysis to detect genetic linkage (Strachan and Read, 1999). This analysis compares the likelihood of two loci being linked versus unlinked (independent assortment). An expected Mendelian disomic inheritance segregation ratio of 27:9:9:9:3:3:3:1 was predicted from the self-pollination of a parent heterozygous for three different and unlinked dominant (d)CAPS markers. Likewise to test for polysomic inheritance, an expected segregation ratio of 147:69:69:69:15:15:15:1 was predicted for self-pollination of a hexaploid parent carrying three different (d)CAPS markers and three wild-type loci, where all six gene loci are unlinked and segregate randomly. In the polysomic model, the (d)CAPS markers were codominant among themselves but dominant over wild-type loci, and no chromatid segregation was considered (no double reduction).
Owing to relatively small sample sizes in dCAPS/CAPS marker segregation experiments, the exact test for goodness of fit for disomic and polysomic inheritance models was performed using the SAS software (version 9.3 for Windows, SAS Institute Inc., Cary, NC, USA) (program according to online Handbook of Biological Statistics) to analyze segregation ratios of different molecular genotypes in the R2 progeny lines.
Results
ACCase gene sequencing revealed that individual plants can possess from one to three different ACCase resistance mutations
At least 38 individual R plants from each of the four parent R populations were sequenced, resulting in the identification of three known ACCase gene resistance-endowing mutations; Ile-1781-Leu, Asp-2078-Gly and Cys-2088-Arg (Table 3). No other known ACCase resistance mutations were found. The 2078 mutation was the most common as it was found in resistant individuals within all four populations. The 2088 mutation was present only in individuals of two resistant populations (R2 and R3). The 1781 mutation was found only in one population (R2). Importantly in this R2 population, many individuals displayed more than one ACCase gene mutation, with 32% of individuals possessing two mutations, and one plant with all three resistance mutations (Table 3). While the majority of the plants in populations R2 and R3 had ACCase mutations, 89% of the resistant plants in population R1, and 26% in population R4, did not contain any of the known ACCase gene mutations (Table 3). These plants lacking ACCase mutations were found to possess non-target-site based resistance mechanisms (enhanced rates of herbicide metabolism) (Ahmad-Hamdani et al., 2012) unrelated to ACCase genes.
Table 3. ACCase carboxyl transferase domain sequencing results showing one, two or three different mutations in resistant individual plants of four wild-oat (Avena spp.) populations.
| Resistant Population | Number of plants analyzed |
Number of individuals in which specific mutation(s) were identified
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1781 (Ile-Leu) | 2078 (Asp-Gly) | 2088 (Cys-Arg) | 1781+2078 | 1781+2088 | 2078+2088 | 1781+2078+2088 | Nil | ||
| R1 | 38 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 34 |
| R2 | 38 | 2 | 10 | 12 | 1 | 8 | 3 | 1 | 1 |
| R3 | 38 | 0 | 34 | 1 | 0 | 0 | 0 | 0 | 3 |
| R4 | 43 | 0 | 32 | 0 | 0 | 0 | 0 | 0 | 11 |
Abbreviation: ACCase, acetyl-coenzyme A carboxylase.
Plants without identified mutations are suspected as having non-target-site resistance mechanism(s).
(d)CAPS marker analysis revealed homoeoallelic heterozygosity for ACCase resistance mutations in individual parent resistant plants
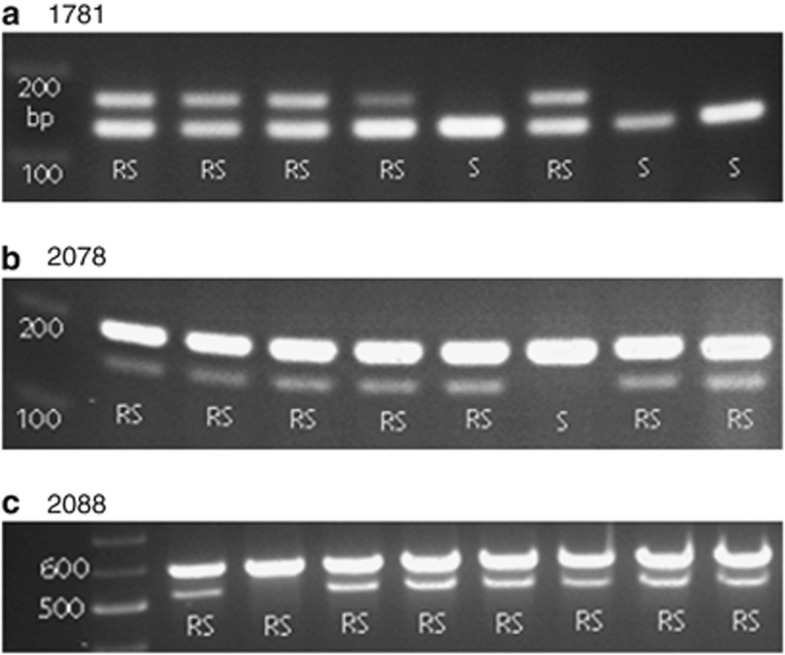
ACCase gene sequencing showed that individual parent resistant plants possessing resistance-endowing mutation(s) always displayed double peaks, that is, both mutant and wild-type nucleotides, at mutation sites in the sequence chromatograms (data not shown). Similarly, when these sequenced individuals were analyzed using the three (d)CAPS markers, plants either displayed both resistant mutant and wild-type bands (RS) or only susceptible wild-type bands (S) (Figure 1). This is in contrast to the genotype patterns (R, RS and S) usually detected in diploid species by the same markers (Kaundun and Windass, 2006; Kaundun, 2010). The most likely reason for consistent presence of wild-type nucleotides/bands at resistance mutation sites is the amplification of more than one homoeologous ACCase genes by the primers used in our assays, that is, homoeologous heterozygosity. Sequencing and (d)CAPS marker analysis would be expected to always reveal wild-type nucleotides/bands unless the same mutation was found in a homozygous state among all homoeologs. Homoeologous heterozygotes may or may not have allelic heterozygosity. We consider fixed heterozygosity, where there is polymorphim among homoeologs but each homoeolog itself is homozygous (no allelic heterozygosity), to be a specific type of homoeologous heterozygosity.
Figure 1.
Genomic (d)CAPS marker analysis of ACCase mutations showing homozygous susceptible (S) and heterozygous resistant (RS) (allelic and homoeologous) genotypes for the (a) 1781, (b) 2078 and (c) 2088 mutations detected in the parent resistant plants. Note: no homozygous resistant genotypes (RR) are evident, most likely due to fixed homoeologous heterozygosity.
(d)CAPS marker segregation analysis of progeny plants revealed three homoeologous ACCase gene loci, each segregating and assorting independently
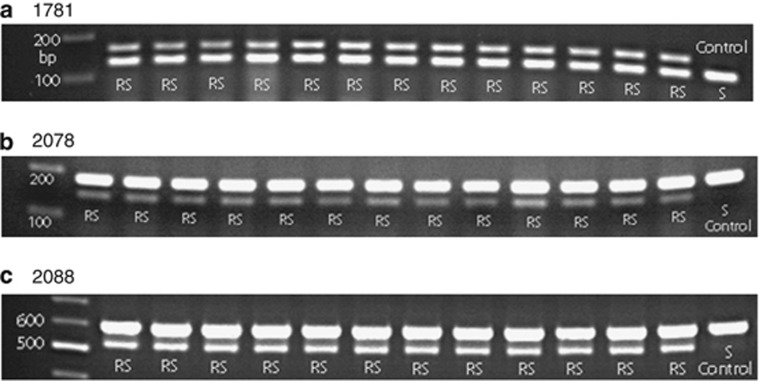
To explore parental genotypes and segregation patterns in more detail, individual R2 population plants possessing one mutation (1781, 2078 or 2088), two mutations (1781+2088 or 2078+2088) or three mutations (1781+2078+2088) were selfed and seeds from single plants harvested for subsequent study. If the parent plants identified by marker analysis were true allelic heterozygotes for a single resistance mutation, that is, having both wild-type and mutant alleles at the same locus (homoeologous copies are considered different loci); and the two alleles segregated at a single ACCase gene locus, then heterozygous RS (allelic and homoeologous) and homozygous S genotypes would be marker-detected among the progeny of RS-selfed plants. When this analysis was done for the single-mutation progeny lines carrying 1781, 2078 or 2088 mutation, only the RS (but no S) genotype was detected among the progeny (Table 4, Figure 2). As at least 18 (n⩾18) individual progeny from each line were analyzed, the probability of missing a homozygous S genotype among the progeny is low (P=0.75n=0.006). These results established that the single-mutation hexaploid Avena parent plants were homozygous for their particular resistance mutation (that is, AA_ _ _ _, BB_ _ _ _ or CC_ _ _ _ genotypes in Table 4, if A=1781, B=2078 and C=2088). The fixed heterozygosity (RS) detected in these plants by the molecular markers is due to the susceptible allele(s) or wild-type nucleotide being amplified from other ACCase gene loci (homoeoallelic heterozygosity). The growth of all RS genotypes was unaffected by diclofop treatment (2000, g ha−1), and no susceptible plants were found, establishing the RS genotypes with allelic R mutation homozygosity as diclofop resistant (Table 4).
Table 4. dCAPS and CAPS marker segregation of single-plant-derived (self-pollinated) progeny of the R2 population.
| Mutation(s) identified in parents | Progeny lines analyzed | No. of progeny plants |
Marker-identified genotypes in progeny
|
Inferred genotypes of the parents |
Phenotypic response to diclofop
a
|
||
|---|---|---|---|---|---|---|---|
| RS (mutation present) | S (mutation absent) | Resistant | Susceptible | ||||
| 1781 | 1 | 21 | 21 | 0 | AA_ _ _ _ | 21 | 0 |
| 2 | 18 | 18 | 0 | 18 | 0 | ||
| 2078 | 1 | 24 | 24 | 0 | BB_ _ _ _ | 24 | 0 |
| 2 | 20 | 20 | 0 | 20 | 0 | ||
| 2088 | 1 | 18 | 18 | 0 | CC_ _ _ _ | 18 | 0 |
| 2 | 18 | 18 | 0 | 18 | 0 | ||
| 1781+2088 | 1 | 24 | 24 (1781) | 0 | AACC _ _ | ||
| 24 (2088) | 0 | ||||||
| 2078+2088 | 1 | 32 | 32 (2078) | 0 | BBC_ _ _ | ||
| 27 (2088) | 5 | ||||||
| 1781+2078 +2088 | 1 | 71 | 55 (1781) | 16 | A_ B _ C _ | ||
| 50 (2078) | 21 | ||||||
| 49 (2088) | 22 | ||||||
Abbreviations: dCAPS, derived cleaved amplified polymorphic sequence; RS, resistant mutant and wild-type bands; S, the susceptible wild-type band.
Designations of R or S refer to the analyzed mutation-site nucleotides only. A=1781, B=2078 and C=2088 mutation, respectively.
When the genotyping finished, these plants with more than one mutation had already passed the 3- to 4-leaf stage and, therefore, were not treated with diclofop for phenotype assessment.
Figure 2.
gDNA (d)CAPS marker analysis of progeny plants derived from self-pollinated parents containing the (a) 1781, (b) 2078 or (c) 2088 mutation and showing no marker segregation in the progenies.
For the selfed plants containing the two mutations 1781+2088, all progeny plants examined (24) possessed both the 1781 and 2088 mutation and all were scored RS due to homoeologous heterozygosity (Table 4). The lack of S-genotype progeny indicates no allelic segregation for either of the two mutations, again likely because the parent plants were homozygous for the two mutations at different ACCase gene loci (that is, AACC _ _ in Table 4).
For the selfed plants containing the two mutations 2078+2088, 27 out of 32 progeny plants also had both mutations while five had only 2078 but not the 2088 mutation. This indicates that there was no allelic segregation for the 2078 mutation, but the plants did segregate for the 2088 mutation (as 2088 RS and 2088 S) (Table 4). The observed ratio of these 2088 segregates, RS:S=27:5, was a good fit (P=0.54) to the Mendelian 3:1 segregation ratio for a single locus. These results indicate that the parents were homozygous for the 2078 mutation at one ACCase gene locus but heterozygous for the 2088 mutation at another ACCase homoeolog (that is, BBC _ _ _ in Table 4).
For the selfed plant containing three mutations, 1781+2078+2088, the progeny plants segregated for each of the three mutations, indicating that the parent plant was heterozygous at all three mutant loci. Using the 1781 mutation as an example, 55 of 71 progeny plants were scored as RS with the remaining plants lacking the 1781 mutation (Table 4). Pairwise two-point logarithm of the odds score analyses were performed to detect any genetic linkage among mutations. Maximum logarithm of the odds score values (Zmax) were determined for all mutation pairs, and possible linkage was evaluated in both coupling and repulsion phases (with mutations as dominant markers). Results were Zmax=0.16 for 1781 and 2078 mutations linked in repulsion phase, 0.07 for 2078 and 2088 mutations linked in coupling phase and 0.03 for 1781 and 2088 mutations linked in coupling phase (data not shown). Recombination frequencies associated with each Zmax were 0.42, 0.45 and 0.47, respectively, and these values are near the 0.5 recombination frequency expected of unlinked loci. All Zmax values were also well below the threshold of 3.0 associated with genetic linkage (Strachan and Read, 1999), indicating that genetic linkage among the three homoeologous ACCase gene loci in wild oat is unlikely and that the three mutations in the parent plant assorted independently.
If the parent plants were heterozygous for each mutation, that is, AaBbCc where lower-case letters designate wild-type alleles, then selfing this parent would result in eight genotype categories among the progeny when genotyped with dominant d(CAPS) markers (listed in Table 5). The observed number of each genotype in the progeny of the self-pollinated 1781+2078+2088 mutant plant was found to be consistent with the predicted 27:9:9:9:3:3:3:1 Mendelian disomic segregation ratio for three unlinked loci genotyped using dominant markers (P=0.66) (Table 5). This observed segregation pattern is consistent with parent plants being heterozygous for each of the three mutations at each of three unlinked homoeologous ACCase gene loci (A_ B _C _ in Table 4).
Table 5. Exact test for goodness of fit of observed (d)CAPS marker segregation to a 27:9:9:9:3:3:3:1 ratio expected from disomic inheritance of three unlinked gene loci vs a 147:69:69:69:15:15:15:1 ratio expected from polysomic inheritance of six randomly segregating gene loci, assuming that there is no chromatid segregation (no double reduction).
| Mutation/allele |
Disomic model
|
Polysomic model
|
||||||
|---|---|---|---|---|---|---|---|---|
| Genotype |
Progeny plants
|
Genotype |
Progeny plants
|
|||||
| if A=1781, B=2078, C=2088 | Observed | Expected | P | if A=1781, B=2078, C=2088 | Observed | Expected | P | |
| 1781+2078+2088 | A_B_C_ | 27 | 29.9 | ABC_ _ _ | 27 | 26.1 | ||
| 1781+2078 | A_B_cc | 12 | 9.9 | AB_ _ _ _ | 12 | 12.2 | ||
| 1781+2088 | A_bbC_ | 12 | 9.9 | AC_ _ _ _ | 12 | 12.2 | ||
| 2078+2088 | aaB_C_ | 8 | 9.9 | BC_ _ _ _ | 8 | 12.2 | ||
| 1781 only | A_bbcc | 5 | 3.3 | A_ _ _ _ _ | 5 | 2.7 | ||
| 2078 only | aaB_cc | 4 | 3.3 | B_ _ _ _ _ | 4 | 2.7 | ||
| 2088 only | aabbC_ | 1 | 3.3 | C_ _ _ _ _ | 1 | 2.7 | ||
| Nil | aabbcc | 2 | 1.1 | wwwwww | 2 | 0.2 | ||
| Total | 71 | 71 | 0.66 | 71 | 71 | 0.009 | ||
Abbreviation: dCAPS, derived cleaved amplified polymorphic sequence.
Observations were made among progeny plants derived from a self-pollinated parent plant containing 1781, 2078 and 2088 mutations (A, B and C, respectively). In the disomic model, the parent would be heterozygous for each mutation (AaBbCc × AaBbCc). In the polysomic model, the parent would have all three mutations plus three wild-type alleles (ABCwww × ABCwww). Wild-type alleles are designated a, b or c in the disomic model and w in the polysomic model, and P is probability.
To further support disomic inheritance expected of Avena spp. hexaploids (Thomas, 1992) and associated with the 27:9:9:9:3:3:3:1 segregation ratio, progeny data were tested for goodness of fit to a polysomic inheritance model with random segregation of all six ACCase gene copies in hexaploid wild oat. Chromatid segregation (double reduction) was not considered. As the parent plant had all three mutations with no evidence of genetic linkage, and because two segregates did not carry any of the ACCase mutations (Table 5), the most likely parental genotype under a polysomic inheritance model would be ABCwww, where w is a wild-type ACCase gene. The genotype of the two segregates not found to have mutations would be wwwwww, derived from fertilization involving www gametes. Under polysomic inheritance, ABCwww is the most likely genotype of the parent plant because it could generate www gametes while still carrying all three mutant ACCase gene copies. With random segregation of all gene copies, self-pollination of the ABCwww parent would result in an expected segregation ratio of 147:69:69:69:15:15:15:1 when progeny are scored with dominant (d)CAPS markers. Observed progeny marker results were not a good fit to this polysomic inheritance model (P=0.009) (Table 5). However, the expected number of wwwwww segregants in the polysomic model was less than one (0.2), which may exaggerate the statistical significance of differences between observed and expected genotype frequencies. On the other hand, expecting 0.2 wwwwww segregants is most consistent with observing either nil or one wwwwww segregant, while we observed two (Table 5). Based on the binomial probability distribution (see Mendenhall, 1987), the probability of observing two or more wwwwww segregants among 71 progeny, where the wwwwww frequency is 1/400 (see the expected polysomic segregation ratio above), is only P=0.014. Observing the two wwwwww segregants is, therefore, improbable in the polysomic inheritance model.
Reverse transciptase-PCR (d)CAPS analysis revealed that three different ACCase mutations can all be transcribed in a single plant
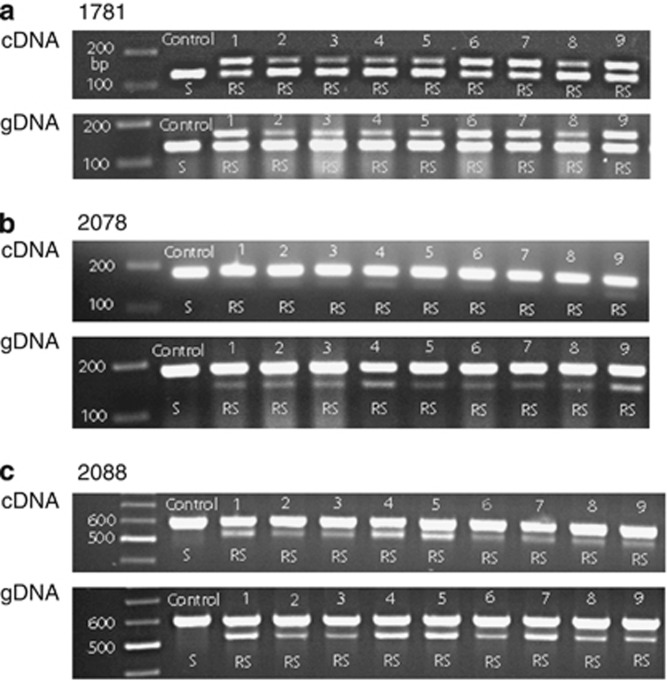
To confirm that individual plants express all three ACCase resistance mutations, leaf material from nine progeny plants (at the 3 to 4-leaf stage) that were each confirmed to carry all three mutations were used for cDNA-based marker analysis. It was found that the genotype patterns for each of the three mutations were similar when determined using either cDNA or gDNA (note that cDNA and gDNA amplicons have the same sizes as there are no introns within the amplified fragments) (Figure 3), indicating that genes carrying any of the three ACCase mutations are transcribed. As the three mutations in these plants are each harbored in a different homoeolog, the results also indicate transcription of all three homoeologous ACCase gene copies. However, a much weaker R band was observed relative to the strong S band in both the cDNA and gDNA restriction patterns, especially for the 2078 mutation (Figure 3), probably due to relatively low PCR amplification efficiency for this mutant allele.
Figure 3.
(d)CAPS marker analysis (a, 1781; b, 2078; c, 2088) of progeny plants carrying three ACCase mutations (1781+2078+2088) using cDNA versus gDNA and showing the same genotype pattern for each of the mutant alleles.
cDNA cloning further confirmed transcription of the three ACCase mutant alleles in three different Acc1 gene copies
Two individual progeny plants that were confirmed to have all three different ACCase resistance mutations in genomic and reverse transciptase-PCR genotyping (plant 1, 4 in Figure 3) were used for cDNA cloning. The 1122 kb fragment encompassing the 1781, 2078 and 2088 mutation sites was cloned into a plasmid vector and 28 clones with putative inserts from the two plants were picked at random and screened for the 1781, 2078 and 2088 mutations using the three (d)CAPS markers. Thirteen clones had the 1781 mutation, seven had the 2088 mutation, one had the 2078 mutation and seven were wild type. Among the 21 clones that contained mutations, each individual clone was found to have only one of the three mutations (Supplementary Figure S4). Sequencing of the 1781, 2078 and 2088 individual clones further confirmed the three mutations, and alignment of polymorphic sequence regions with the three known Acc1 homoeologous sequences revealed three resistant alleles (haplotypes). The 2078 haplotype sequence is clearly homologous to Acc1;3 and the 1781 and 2088 haplotypes are similar to Acc1;2/Acc1;1 (Figure 4). Therefore, it is clear from molecular marker segregation, reverse transciptase-PCR marker analysis and cDNA cloning that hexaploid wild oat has three ACCase gene copies, each of which can harbor resistance mutation(s). Importantly, all three copies are expressed as mRNA.
Figure 4.
Partial sequence alignment (from cloned fragments) showing the three haplotype sequences of the 2088, 1781 and 2078 mutations, corresponding, respectively, to the ACCase gene copies Acc1;1, Acc1;2 and Acc1;3 in wild oat (Christoffers et al., 2002). Note that the sequence region containing the 1781, 2078 or 2088 codon was not included due to infrequent informative single-nucleotide polymorphisms (SNPs). Boxed regions indicate SNPs discriminating the three haplotypes (GenBank accession AF231335 for Acc1;1-1, AF231334 for Acc1;1-2, AF231336 for Acc1;2 and AF464875 for Acc1;3).
Assay of in vitro ACCase activity revealed that the 1781, 2078 or 2088 mutation in wild oat confers lower-level resistance to ACCase herbicides than in diploid grass species
To determine if the 1781, 2078 or 2088 resistance mutations individually express as resistant ACCase in wild-oat populations, ACCase activity in the presence or absence of ACCase herbicides was assayed in partially-purified leaf ACCase extracts from susceptible versus resistant plants homozygous, respectively, for each of the three resistance mutations (progeny derived from those in Table 4). The ACCase I50 was estimated for each herbicide and each ACCase resistance mutation (R) versus wild type (S), to give a R/S ratio.
As expected, ACCase from the susceptible plants was confirmed to be sensitive to ACCase herbicides, with I50 values (Table 6) in the range of reported plastidic ACCase values for susceptible grass weed species (Délye, 2005). In contrast, ACCase from resistant plants was significantly less sensitive to ACCase herbicides than the susceptible plants. This reduced sensitivity to ACCase herbicides further confirmed expression of ACCase resistance mutation(s) at the protein level. Importantly, the level of resistance conferred by each mutation as indicated by R/S ratios (1.7–10.8 fold), was generally low (Table 6). Two additional herbicide-susceptible wild-oat populations (A. fatua and A. sterilis) were used to assess population variation in ACCase herbicide sensitivity, and the diclofop I50 values determined for these two susceptible populations (6.38–6.46 μℳ, data not included in Table 6) were found similar to that of the susceptible plants originally used. In addition, ACCase isolated from resistant plants (populations R1 and R4) without known ACCase resistance mutations was found to be sensitive to fenoxaprop and sethoxydim herbicides, with I50 values of 1.8 and 2.2 μℳ, respectively (data not included in Table 6).
Table 6. ACCase I50 values and R/S ratios determined for wild oat (A. fatua) homozygous progeny lines (see Table 4) from this study in comparison with the R/S ratios reported for the diploid grass species Lolium rigidum and A. myosuroides using purified lines that are homozygous for the specific ACCase mutations.
| Mutation/mutant allele |
Diclofop acid
|
Fenoxaprop acid
|
Sethoxydim
|
Clethodim
|
||||
|---|---|---|---|---|---|---|---|---|
| I50 (μℳ) | R/S ratio | I50 (μℳ) | R/S ratio | I50 (μℳ) | R/S ratio | I50 (μℳ) | R/S ratio | |
| Avena spp (hexaploid) | ||||||||
| 1781-Leu | 16.05 a | 2.0 | 9.79 a | 5.3 | 18.64 a | 10.8 | 2.08 a | 3.9 |
| 2078-Gly | 15.98 a | 2.0 | 7.24 b | 3.9 | 9.79 b | 5.7 | 1.02 b | 1.9 |
| 2088-Arg | 13.54 a | 1.7 | 8.94 ab | 4.8 | 6.10 c | 3.5 | 1.02 b | 1.9 |
| Susceptible | 7.96 b | 1.86 c | 1.73 d | 0.53 c | ||||
| Lolium rigidum a (diploid) | ||||||||
| 1781-Leu | 6 | >100 | 18 | |||||
| 2078-Gly | 32 | >100 | 41 | |||||
| 2088-Arg | 38 | 37 | 48 | |||||
| A. myosuroides b (diploid) | ||||||||
| 2078-Arg | 23 | 53 | 36 | |||||
Abbreviations: ACCase, acetyl-coenzyme A carboxylasel; R/S ratios, resistant/susceptible ratios.
Means with different letters in a column are significantly different (α=0.05).
The diclofop and clethodim I50 R/S data are from Yu et al. (2007) and the I50 values are not included here because the absolute I50 values of R and S plants can vary because of experimental and assay conditions, while the I50 R/S ratios are more relevant and comparable across species and laboratories. The sethoxydim data is from the current study.
The data are from Délye et al. (2005) and the I50 values not included here.
Reported ACCase I50 R/S ratios for purified resistant diploid L. rigidum (Yu et al., 2007) and resistant diploid Alopecurus myosuroides (Délye et al., 2005) for the four herbicides (where available) are also cited in Table 6 for comparison. It is evident that the 1781, 2078 or 2088 mutation homozygous in diploid resistant L. rigidum or A. myosuroides results in much higher ACCase I50 R/S ratios than those displayed by the same mutation(s) in hexaploid wild oat; especially for the 2078 or 2088 mutation.
In order to assess if multiple ACCase gene mutations endow ACCase with higher resistance than single mutations, wild-oat plants having two or more mutations (see Table 4) were assessed for their ACCase sensitivity to diclofop in a separate experiment (experiment II) and the results were compared with the data for the single mutations from experiment I. In general, the ACCase I50 R/S ratios were found to be higher for multiple mutations relative to single mutations (Table 7). It is noteworthy that the 1781+2088 mutants had an I50 R/S ratio of 3.6 μℳ, which equals the sum of the individual R/S values of the 1781 (2.0 μℳ) and 2088 (1.7 μℳ) mutants. This indicates an additive resistance interaction among mutant alleles across genomes. Clearly, as will be discussed, the 1781, 2078 or 2088 ACCase mutation alone in hexaploid wild oat does not confer high-level in vitro or in vivo resistance to ACCase herbicides. It is also notable that in the absence of herbicides, the specific activity of ACCase was much lower for the 2078 or 2088 mutation compared with that of susceptible plants, while the activity of ACCase was unaffected by the 1781 mutation (Table 7). Reduced ACCase activity was also recorded from plants containing two or three resistance mutations (Table 7).
Table 7. ACCase activity (in the absence of diclofop) and diclofop I 50 values determined for plants containing different resistance mutations/mutant alleles.
| Mutation/mutant allele | I50 values | I50 R/S ratio | ACCase activity (nmol HCO−3 mg−1 per protein min−1) | Activity R/S ratio |
|---|---|---|---|---|
| Experiment I | ||||
| 1781-Leu | 16.05 a | 2.0 | 1.58 a | 0.97 |
| 2078-Gly | 15.98 a | 2.0 | 0.50 b | 0.31 |
| 2088-Arg | 13.54 a | 1.7 | 0.39 b | 0.24 |
| Susceptible | 7.96 b | 1.63 a | ||
| Experiment II | ||||
| 1781+2078+2088 | 15.92 c | 2.6 | 2.23 b | 0.72 |
| 2078+2088 | 18.08 b | 3.0 | 1.39 c | 0.45 |
| 1781+2088 | 21.68 a | 3.6 | 1.34 c | 0.43 |
| Susceptible | 6.06 d | 3.10 a | ||
Abbreviations: ACCase, acetyl-coenzyme A carboxylase; R/S ratios, resistant/susceptible ratios.
Experiment I was conducted using plant materials at the 3- to 4-leaf stage and experiment II at the 4- to 5-leaf stage. Means with different letters in a column within an experiment are significantly different (α=0.05).
In order to determine if herbicide treatment can induce higher ACCase activity and I50 values, the 2078 or 2088 mutants that were foliar-sprayed with 2000, g ha−1 diclofop-methyl were used for determination of ACCase activity and diclofop I50 values. The results (not shown) were similar to those without diclofop pre-treatment, indicating that herbicide treatment did not induce higher ACCase activity.
Discussion
Multiple ACCase resistance mutations in individual hexaploid wild-oat plants
Resistance mutation diversity within weed populations and individual plants is important in optimizing weed management strategies. One of the most important findings of this study with hexaploid ACCase herbicide-resistant wild oat is that individuals can possess multiple ACCase resistance mutations among homoeologous genes. While it is common for individuals of cross-pollinated diploid weed species such as A. myosuroides and L. rigidum (Délye et al., 2005; Yu et al., 2007) to have more than one different ACCase resistance mutation (compound heterozygosity), multiple mutations have not been reported, to our knowledge, in any field-evolved herbicide-resistant polyploid weed species. Indeed, we found that individuals with two distinct ACCase mutations are not uncommon (12 of 157 analyzed plants), and we found one individual with three mutations (1781+2078+2088) (Table 3). The accumulation of three resistance mutations in one plant could occur through independent mutations in all three homoeologous ACCase gene copies, or due to cross-pollination between individuals carrying different homoeologous ACCase mutations. Although wild oat is known as a preferentially self-pollinated species, outcrossing can range from 0 to 12% (Imam and Allard, 1965; Murray et al., 2002). Multiple herbicide target-site mutations in individuals of self-pollinated polyploid weed species have been rare thus far, and hence these plants are useful material for genetic and genomic studies for gene expression in polyploid species.
Genetics and expression of homoeologous ACCase gene loci in hexaploid wild oat
Hexaploid wild oat has a genome structure of AACCDD, and genome C is clearly distinguishable from A and D (Yang et al., 1999). A previous study of ACCase herbicide-resistant A. fatua identified three homoeologous plastidic ACCase gene copies, Acc1;1, Acc1;2 and Acc1;3, with the 1781 mutation in Acc1;1 (Christoffers et al., 2002). In comparison with Avena spp. diploids, Acc1;3 was found to likely represent the C genome, while Acc1;1 and Acc1;2 likely represent the similar A/D genomes (MJ Christoffers, unpublished data). Subsequent studies with other resistant A. fatua populations also found the 2078 or 2088 mutations in Acc1;1 (Christoffers and Peterson, 2007). However, in studies with resistant A. sterilis and A. fatua populations, Liu et al. (2007) and Beckie et al. (2012) indicated that any of the three Acc1 homoeologs can host ACCase resistance mutations. In our study, we identified an individual possessing three distinct ACCase resistance mutations and this provided a unique opportunity to investigate the inheritance of multiple resistance mutations in a polyploid species. We were able to show that individual wild-oat plants can be heterozygous for three different ACCase mutations in each of the three Acc1 homoeologs, with the 2078 mutation clearly residing in Acc1;3 (tentatively assigned to the C genome), and the 1781 and 2088 mutations in Acc1;1/Acc1;2 (likely the A and D genomes) in individuals of the R2 population (Figure 4).
Secondly, our results were consistent with the three homoeologous Acc1 loci segregating and assorting independently of each other, following classical disomic Mendelian inheritance due to restricted homoeologous pairing and recombination in meiosis (Table 5). Evidence against our data fitting an alternative polysomic inheritance model was highly dependent on the identification of two wild-type plants among segregating progeny. As we cannot be absolutely certain that a genotyping error was not made, we strictly cannot rule out polysomic inheritance based on our data alone. However, the disomic inheritance model favored by the exact test analysis (Table 5) is in line with the established mode of inheritance for hexaploid Avena spp. (Thomas, 1992). This disomic segregation pattern in hexaploid Avena observed in the current study is also consistent with previous reports on inheritance of ACCase resistance traits (Murray et al., 1995, 1996) and the segregation of wild-type Acc1;1-1 and resistant Acc1;1-2 alleles in A. fatua (Christoffers et al., 2002).
Thirdly, all three mutant alleles at the three Acc1 loci are expressed as mRNA (Figure 3, Supplementary Figure S4). Consistent with this conclusion is the recent addition to the GenBank database of 3′-end cDNA sequences for the Acc1;2 and Acc1;3 sequences of well-studied UM1 wild oat (HQ244401 and HQ244402, respectively), indicating the transcriptional activity of these genes in another wild-oat population.
It is currently unclear if the three homoeologous ACCase genes in A. fatua contribute equally to the transcript pool. In allopolyploid cotton, wheat and other plant species, numerous genes have shown strong expression bias toward one copy (reviewed by Wendel, 2000; Adams, 2007). Many weed species are polyploid and little is currently known about gene expression/silencing or epigenetic regulation in polyploid weeds. In the polyploid Schoenoplectus mucronatus, acetolactate synthase gene methylation was detected and epigenetic regulation implicated (Scarabel et al., 2009). Clearly, more research is needed to understand selection and evolution of herbicide resistance genes in polyploid species.
Fixed heterozygosity has also been observed in other polyploid weed species (for example, in Salsola tragus, Warwick et al., 2010) in regard to acetolactate synthase resistance mutations. Therefore, PCR-based gene-specific (but not copy-specific) co-dominant molecular markers or sequencing primers designed against conserved regions of herbicide target genes cannot discriminate true (allelic) heterozygotes from homoeoallelic heterozygotes in polyploid species like wild oat. Likewise, detection of true allelic heterozygosity may be similarly complicated in diploid species having multiple herbicide target gene copies. Fixed polymorphism detected by non-copy-specific markers can be indicative of polyploidy or multiple single-genome copies of the gene.
ACCase resistance mutations and resistance evolution in hexaploid Avena
Most resistance-endowing ACCase gene mutations have been characterized in diploid species (especially A. myosuroides and L. rigidum) by in vitro ACCase assay and whole-plant bioassay using purified lines homozygous for each of the mutations. Among them, the Ile-1781-Leu mutation was found to confer high-level resistance to the cyclohexanediones herbicide sethoxydim (Table 6) and moderate-level resistance to the aryloxyphenoxypropionates herbicide diclofop (Yu et al., 2007). The Asp-2078-Gly or Cys-2088-Arg mutations conferred high-level resistance to both aryloxyphenoxypropionates and cyclohexanediones herbicides (Délye et al., 2005; Yu et al., 2007; and Table 6). In contrast, in vitro inhibition of resistant wild-oat ACCase revealed that each of the mutations conferred much lower-level resistance to the selected ACCase herbicides (Table 6). A similar low-level (2- to 5-fold) resistance to ACCase herbicides was also recently reported for the same 2078 and the Ile-2041-Asn mutation in other A. fatua populations (Cruz-Hipoloto et al., 2011). The low resistance observed at the ACCase level for the 1781, 2078 or 2088 resistance mutation in hexaploid wild oat, compared with diploid grass weed species A. myosuroides and L. rigidum (Table 6), is likely due to polyploidy and dilution of resistant ACCase by susceptible ACCase in wild oat. Assuming equal expression of the three plastidic homoeologous ACCase genes, wild-oat plants homozygous for a given mutation at one ACCase locus would still have two-thirds of the total expressed ACCase as wild-type herbicide sensitive. Furthermore, because ACCase requires two monomers to form a homodimer to yield active ACCase, fully resistant homodimers would likely be further diluted in hexaploid species. Although not examined in this study, differential expression of the three homoeologous ACCase copies is possible and may influence the relative amount of resistant ACCase. If the expression of the ACCase gene copy harboring the ACCase mutation was low compared with other copies, the level of resistance conferred by the mutation would be even further suppressed in wild oat. In contrast, in a diploid homozygous for a given ACCase resistance mutation, all expressed ACCase would be the herbicide resistant isoform. Therefore, an ACCase mutation that confers resistance in diploids may not necessarily endow enough resistance in hexaploids to be selected for in the field (depending on the dominance of the resistance allele, herbicides and herbicide rates). For example, diploid resistant L. rigidum survived the Australian field rate of clethodim (60 g ha−1) or pinoxaden (30 g ha−1) due to the 2078 or 2088 resistance mutation (Yu et al., 2007), but hexaploid wild oat possessing the same mutation rarely survived the herbicide at the same or lower rates (Ahmad-Hamdani et al., 2011; AV Varanasi and MJ Christoffers, submitted).
Whereas gene redundancy in hexaploids may minimize deleterious effects of gene mutations, it may also buffer the effect of resistance mutations (both at target and non-target sites) in herbicide resistance evolution. The dilution effect of resistance mutations by additional susceptible (wild type) homoeologous gene copies may explain in part why wild oat, in comparison with other diploids, is relatively slow to evolve levels of resistance that facilitate identification of resistant populations. In Australia, diclofop was released for Lolium and wild oat control in 1978 and despite the presence of both species in many treated fields, resistant L. rigidum was reported in 1982 (Heap and Knight, 1982), whereas resistance in wild oat was not reported until 1992 (Mansooji et al., 1992). Very intensive surveys revealed much greater frequency of herbicide resistance in L. rigidum than in wild oat in the same region (Owen et al., 2007; Owen and Powles, 2009). Of course, slower resistance evolution in wild oat relative to L. rigidum is influenced by several other factors including smaller population sizes, longer seed bank turnover time and extended seed germination (Owen and Powles, 2009). Although self-pollination in wild oat may increase selection for resistance by promoting allelic homozygosity, it may also slow the spread of resistance alleles. Nonetheless, molecular and genetic evidence from the current study indicates that herbicide resistance and evolution in hexaploid weed species can be considerably more complex than in diploid species.
Although single ACCase mutations in hexaploid wild oat provide relatively low-level resistance to the ACCase herbicides, resistance levels can be significantly increased if individual plants can accumulate multiple ACCase mutations. Based on diclofop I50 R/S ratios, individuals with two mutations had higher resistance than those with a single mutation (Table 7). However, three mutations did not have the expected higher resistance profile, probably because the progeny plants that were used for ACCase assay were mostly heterozygous at two to three Acc1 loci, with homozygotes for the three mutations at three loci (AABBCC genotype, Table 5) being extremely rare. Although the number of resistant alleles in each case cannot be accurately determined, especially when involving three mutations, the data clearly indicates an additive effect on resistance level across gene copies/genomes, especially when the individuals are homozygous for different mutations. Indeed, the additive effect of resistant acetolactate synthase genes from different genomes has been observed at the acetolactate synthase and whole-plant levels in a mutagenized imidazolinone-resistant wheat line (TealIMI15A) (Newhouse et al., 1992; Pozniak and Hucl, 2004; Anderson et al., 2004; Pozniak et al., 2004).
Effect of ACCase gene mutations on ACCase activity
Importantly, resistant plants possessing the 2078 or 2088 mutation had <40% of the extractable ACCase activity of susceptible plants (Table 7). Multiple mutations involving 2078 and/or 2088 also had reduced ACCase activity (40–70% of the S, Table 7). This is intriguing given that mutant ACCase isoforms likely account for only about 1/3 of the total ACCase isoforms in the assay mixture. Reduced ACCase activity of the 2078 or 2088 mutation has also been observed in diploid L. rigidum and A. myosuroides (Yu et al., 2007; Délye et al., 2005). While the single 2078 or 2088 mutation results in significant reduction of in vitro ACCase activity in both hexaploid wild oat and diploid L. rigidum (Yu et al., 2007), it is unknown if the mutations in wild oat have any fitness cost as they do in Lolium (MV Auib, Q Yu, S Powles, unpublished). In A. myosuroides, only plants homozygous for the 2078 mutation were found to have a fitness penalty (Menchari et al., 2008). As a polyploid with additional gene copies, wild oat may more readily tolerate mutations that decrease ACCase expression and/or ACCase activity.
Non-target-site resistance mechanisms are also evident in resistant wild-oat populations
For fenoxaprop, sethoxydim and clethodim, the ACCase I50 R/S ratios observed for the 1781, 2078 or 2088 mutation at the ACCase level (Table 6) were generally correlated with the LD50 R/S ratios observed at the whole-plant level (Ahmad-Hamdani et al., 2011). Thus, it's likely that resistance to these three herbicides in these wild-oat populations is largely due to the 1781, 2078 or 2088 mutation. In contrast, high-level resistance to diclofop at the whole plant level (LD50 R/S ratios >20-fold) (Ahmad-Hamdani et al., 2011), was not consistent with low diclofop ACCase I50 R/S ratios (Table 6), suggesting the possibility of additional non-target-site resistance mechanisms. Some resistant individuals had no known ACCase mutations (Table 3) and their ACCase was herbicide sensitive (see Results). Our recent work on these resistant plants without ACCase resistance mutations demonstrated enhanced herbicide metabolism as a non-target-site resistance mechanism (Ahmad-Hamdani et al., 2012). Both a resistant ACCase and enhanced rates of herbicide metabolism have been demonstrated in a resistant A. sterilis population (Maneechote et al., 1997). Thus, the high-level resistance to some ACCase herbicides (for example, diclofop or fenoxaprop) observed at the population level (Ahmad-Hamdani et al., 2011) is likely due to an ACCase resistance mutation plus metabolic resistance (Ahmad-Hamdani et al., 2012).
In summary, identifying and understanding herbicide resistance and its evolution in hexaploid plant species is more challenging than in diploids. The role of resistance-endowing gene mutations in hexaploid Avena species may not be simply inferred from other unrelated diploid species. More complex factors, such as dilution or gene dosage effects and intergenomic gene expression, must be considered in understanding herbicide resistance and evolution in polyploid plant species.
Data archiving
Partial ACCase sequence alignment showing the three haplotype sequences of the 2088, 1781 and 2078 mutants (Figure 4), and (d)CAPS marker analysis of ACCase resistance mutations in E. coli clones (Supplementary Figure S4) have been deposited at Dryad: doi:10.5061/dryad.04562.
Acknowledgments
This work was supported by the Grains Research and Development Corporation of Australia (grants to the Australian Herbicide Resistance Initiative) and by a PhD scholarship to MS Ahmad-Hamdani from the Ministry of Higher Education Malaysia. We are grateful to Drs Henri Darmency, Christophe Délye and Danica Goggin for valuable comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Adams KL. Evolution of duplicate gene expression in polyploid and hybrid plants. J Heredity. 2007;98:136–141. doi: 10.1093/jhered/esl061. [DOI] [PubMed] [Google Scholar]
- Ahmad-Hamdani MS, Owen MJ, Yu Q, Powles SB. ACCase-inhibiting herbicide resistant wild oat (Avena fatua L.) populations from the Western Australian grain belt. Weed Technol. 2011;26:130–136. [Google Scholar]
- Ahmad-Hamdani MS, Yu Q, Han H, Cawthray GR, Wang SF, Powles SB.2012Herbicide resistance endowed by enhanced rates of herbicide metabolism in wild oat (Avena spp.) Weed Sciin pressdoi: 10.1614/WS-D-12-00078.1 [DOI]
- Anderson JA, Matthiesen L, Hegstad J. Resistance to an imidazolinone herbicide is conferred by a gene on chromosome 6DL in the wheat line cv 9804. Weed Sci. 2004;52:83–90. [Google Scholar]
- Beckie HJ, Warwick SI, Sauder CA. Basis for herbicide resistance in Canadian populations of wild oat (Avena fatua) Weed Sci. 2012;60:10–18. [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffers MJ, Berg ML, Messersmith CG. An isoleucine to leucine mutation in acetyl-CoA carboxylase confers herbicide resistance in wild oat. Genome. 2002;45:1049–1056. doi: 10.1139/g02-080. [DOI] [PubMed] [Google Scholar]
- Christoffers MJ, Pederson SN. Response of wild oat (Avena fatua) acetyl-CoA carboxylase mutant to pinoxaden. Abstr Weed Sci Soc Am. 2007;47:258. [Google Scholar]
- Cocker KM, Coleman JOD, Blair AM, Clarke JH, Moss SR. Biochemical mechanisms of cross-resistance to aryloxyphenoxypropionate and cyclohexanedione herbicides in populations of Avena spp. Weed Res. 2000;40:323–334. [Google Scholar]
- Collavo A, Panozzo S, Lucchesi G, Scarabel L, Sattin M. Characterization and management of Phalaris paradoxa resistant to ACCase-inhibitors. Crop Prot. 2011;30:293–299. [Google Scholar]
- Cruz-Hipolito H, Osuna MD, Dominguez-Valenzuela JA, Espinoza N, De Prado R. Mechanism of resistance to ACCase-inhibiting herbicides in wild oat (Avena fatua) from Latin America. J Agr Food Chem. 2011;59:7261–7267. doi: 10.1021/jf201074k. [DOI] [PubMed] [Google Scholar]
- Délye C, Zhang XQ, Michel S, Matejicek A, Powles SB. Molecular bases for sensitivity to acetyl-coenzyme A carboxylase inhibitors in black-grass. Plant Physiol. 2005;137:794–806. doi: 10.1104/pp.104.046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Délye C. Weed resistance to acetyl coenzyme A carboxylase inhibitors: an update. Weed Sci. 2005;53:728–746. [Google Scholar]
- Devine MD, Shimabukuro RH.1994Resistance to acetyl coenzyme A carboxylase inhibiting herbicidesIn: Powles SB, Holtum JAM, (eds).Herbicide Resistance in Plants: Biology and Biochemistry Lewis Publishers: Boca Raton, FL; 83–139. [Google Scholar]
- Heap I, Knight R. A population of ryegrass tolerant to the herbicide diclofop-methyl. J Aust Inst Agric Sci. 1982;48:156–157. [Google Scholar]
- Heap IM.2011International Survey of Herbicide-Resistant WeedsAvailable at: http://www.weedscience.org . (Accessed March 2012).
- Imam AG, Allard RW. Population studies in predominantly self-pollinated species. VI. Genetic variability between and within natural populations of wild oats from differing habitats in California. Genetics. 1965;51:49–62. doi: 10.1093/genetics/51.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellen EN, Gill BS, Cox TS. Genomic in situ hybridization Differentiates between A/D- and C-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena) Genome. 1994;37:613–618. doi: 10.1139/g94-087. [DOI] [PubMed] [Google Scholar]
- Kaundun SS, Windass JD. Derived cleaved amplified polymorphic sequence, a simple method to detect a key point mutation conferring acetyl CoA carboxylase inhibitor herbicide resistance in grass weeds. Weed Sci. 2006;46:34–39. [Google Scholar]
- Kaundun SS. An aspartate to glycine change in the carboxyl transferase domain of acetyl CoA carboxylase and non-target-site mechanism(s) confer resistance to ACCase inhibitor herbicides in a Lolium multiflorum population. Pest Manag Sci. 2010;66:1249–1256. doi: 10.1002/ps.2003. [DOI] [PubMed] [Google Scholar]
- Liu WJ, Harrison DK, Chalupska D, Gornicki P, O'Donnell CC, Adkins SW, et al. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc Nat Acad Sci USA. 2007;104:3627–3632. doi: 10.1073/pnas.0611572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneechote C, Holtum JAM, Preston C, Powles SB. Resistant acetyl-CoA carboxylase is a mechanism of herbicide resistance in a biotype of Avena sterilis ssp. ludoviciana. Plant Cell Physiol. 1994;35:627–635. [Google Scholar]
- Maneechote C, Preston C, Powles SB. A diclofop-methyl-resistant Avena sterilis biotype with a herbicide-resistance acetyl-coenzyme A carboxylase and enhanced metabolism of diclofop-methyl. Pestic Sci. 1997;49:105–114. [Google Scholar]
- Mansooji AM, Holtum JA, Boutsalis P, Matthews JM, Powles SB. Resistance to aryloxyphenoxypropionate herbicides in 2 wild oat species (Avena fatua and Avena sterilis ssp. ludoviciana) Weed Sci. 1992;40:599–605. [Google Scholar]
- Menchari Y, Camilleri C, Michel S, Brunel D, Dessaint F, Le Corre V, et al. Weed response to herbicide: regional-scale distribution of herbicide resistance alleles in the grass weed Alopecurus myosuroides. New Phytol. 2006;171:861–874. doi: 10.1111/j.1469-8137.2006.01788.x. [DOI] [PubMed] [Google Scholar]
- Menchari Y, Chauvel B, Darmency H, Délye C. Fitness costs associated with three mutant acetyl-coenzyme A carboxylase alleles endowing herbicide resistance in black-grass Alopecurus myosuroides. J Appl Ecol. 2008;45:939–947. [Google Scholar]
- Mendenhall W.1987Introduction to Probability and Statistics7th edn.Duxbury Press: Boston; 173–181. [Google Scholar]
- Murray BG, Brûlé-Babel AL, Morrison IN. Two distince alleles encode for acetyl-CoA carboxylase inhibitor resistance in wild oat (Avena fatua) Weed Sci. 1996;44:476–481. [Google Scholar]
- Murray BG, Morrison IN, Frieses LF. Pollen-mediated gene flow in wild oat. Weed Sci. 2002;50:321–325. [Google Scholar]
- Murray BG, Morrison IN, Brûlé-Babel AL. Inheritance of acetyl-CoA carboxylase inhibitor resistance in wild oat (Avena fatua) Weed Sci. 1995;43:233–238. [Google Scholar]
- Newhouse K, Smith WA, Starrett MA, Schaefer TJ, Singh BK. Tolerance to imidazolinone herbicides in wheat. Plant Physiol. 1992;100:882–886. doi: 10.1104/pp.100.2.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloudakis N, Katsiotis A. The origin of the C-genome and cytoplasm of Avena polyploids. Theor Appl Genet. 2008;117:273–281. doi: 10.1007/s00122-008-0772-9. [DOI] [PubMed] [Google Scholar]
- Nikoloudakis N, Skaracis G, Katsiotis A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8-ITS2 and IGS Avena sp. sequences. Mol Phylogenet Evol. 2008;46:102–115. doi: 10.1016/j.ympev.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Ohsako T, Tominaga T. Nucleotide substitutions in the acetolactate synthase genes of sulfonylurea-resistant biotypes of Monochoria vaginalis (Pontederiaceae) Genes Genet Syst. 2007;82:207–215. doi: 10.1266/ggs.82.207. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Powles SB. Distribution and frequency of herbicide-resistant wild oat (Avena spp.) across the Western Australian grain belt. Crop Pasture Sci. 2009;60:25–31. [Google Scholar]
- Powles SB, Yu Q. Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol. 2010;61:317–347. doi: 10.1146/annurev-arplant-042809-112119. [DOI] [PubMed] [Google Scholar]
- Pozniak CJ, Birk IT, O'Donoughue LS, Menard C, Huci PJ, Singh BK. Physiological and molecular characterization of mutation-derived imidazolinone resistance in spring wheat. Crop Sci. 2004;44:1434–1443. [Google Scholar]
- Pozniak CJ, Hucl PJ. Genetic analysis of imidazolinone resistance in mutation-derived lines of common wheat. Crop Sci. 2004;44:23–30. [Google Scholar]
- Rajhathy T, Morrison JW. Genome homology in the genus Avena. Can J Genet Cytol. 1960;2:278–285. [Google Scholar]
- Scarabel L, Locascio A, Furini A, Sattin M, Varotto S. Characterisation of ALS genes in the polyploid species Schoenoplectus mucronatus and implications for resistance management. Pest Manag Sci. 2009;66:337–344. doi: 10.1002/ps.1883. [DOI] [PubMed] [Google Scholar]
- Seefeldt SS, Fuerst EP, Gealy DR, Shukala A, Irzyk GP, Devine MD. Mechanisms of resistance to diclofop of two wild oat (Avena fatua) biotypes from the Williamette Valley of Oregon. Weed Sci. 1996;44:776–781. [Google Scholar]
- Scarabel L, Panozzo S, Varotto S, Sattin M. Allelic variation of teh ACCase gene and response to ACCase-inhibiting herbicides in pinoxaden-resistant Lolium spp. Pest Manag Sci. 2011;67:932–941. doi: 10.1002/ps.2133. [DOI] [PubMed] [Google Scholar]
- Shorrosh BS, Dixon RA, Ohlrogge JB. Molecular cloning, characterization, and elicitation of acetyl-CoA carboxylase from alfalfa. Proc Natl Acad Sci USA. 1994;91:4323–4327. doi: 10.1073/pnas.91.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Dupont S, Devine MD. Resistance to ACCase-inhibiting herbicides in wild oat: evidence for target site-based resistance in two biotypes from Canada. Pestic Biochem Physiol. 1997;57:147–155. [Google Scholar]
- Strachan T, Read AP. Human Molecular Genetics 2. Wiley-Liss: New York; 1999. pp. 274–276. [Google Scholar]
- Tal A, Rubin B. Molecular characterization and inheritance of resistance to ACCase-inhibiting herbicides in Lolium rigidum. Pest Manage Sci. 2004;60:1013–1018. doi: 10.1002/ps.903. [DOI] [PubMed] [Google Scholar]
- Thomas T.1992Cytogenetics of AvenaIn: Marshall HG, Sorrells ME, (eds).Oat Science and Technology American Society of Agronomy and Crop Science Society of America: Madison, WI; 473–507. [Google Scholar]
- Uchino A, Ogata S, Kohara H, Yoshida S, Yoshioka T, Watanabe H. Molecular basis of diverse responses to acetolactate synthase-inhibiting herbicides in sulfonylurea-resistant biotypes of Schoenplectus juncoides. Weed Biol Manag. 2007;7:89–96. [Google Scholar]
- Warwick S, Sauder CA, Beckie HJ. Acetolactate synthase (ALS) target-site mutations in ALS inhibitor-resistant Russian thistle (Salsola tragus) Weed Sci. 2010;58:244–251. [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Yang Q, Hanson L, Bennett MD, Leitch IJ. Genome structure and evolution in the allohexaploid weed Avena fatua L. (Poaceae) Genome. 1999;42:512–518. [PubMed] [Google Scholar]
- Yu Q, Collavo A, Zheng MQ, Owen M, Sattin M, Powles SB. Diversity of acetyl-coenzyme A carboxylase mutations in resistant Lolium populations: evaluation using clethodim. Plant Physiol. 2007;145:547–555. doi: 10.1104/pp.107.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Friesen S, Zhang XQ, Powles SB. Tolerance to acetolactate synthase and acetyl-coenzyme A carboxylase inhibiting herbicides in Vulpia bromoides is conferred by two co-existing resistance mechanisms. Pestic Biochem Physiol. 2004;21:21–30. [Google Scholar]
- Yu Q, Han HP, Powles SB. Mutations of the ALS gene endowing resistance to ALS-inhibiting herbicides in Lolium rigidum populations. Pest Manag Sci. 2008;64:1229–1236. doi: 10.1002/ps.1624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.