Abstract
Covalent attachment of small ubiquitin-like modifier (SUMO) to proteins is highly dynamic, and both SUMO-protein conjugation and cleavage can be regulated. Protein desumoylation is performed by SUMO proteases, which control cellular mechanisms ranging from transcription and cell division to ribosome biogenesis. Recent advances include the discovery of two novel classes of SUMO proteases, insights regarding SUMO protease specificity, and revelations of previously unappreciated SUMO protease functions in several key cellular pathways. These developments, together with new connections between SUMO proteases and the recently discovered SUMO-targeted ubiquitin ligases (STUbLs), make this an exciting period for the study of these enzymes.
Introduction
Cellular proteins are often covalently modified following their translation, providing organisms with added control of protein activity, localization or stability. Among the many known post-translational protein modifiers are other proteins, most commonly ubiquitin or one of several ubiquitin-like proteins (UBLs). Ubiquitin and UBLs share a common fold, known as the β-grasp fold, and a conserved enzymatic strategy for their activation and attachment to substrates1. The UBL known as small ubiquitin-related modifier (SUMO) is attached to a multitude of proteins in all species that have been analyzed, including Saccharomyces cerevisiae2–5, Arabidopsis thaliana6, 7 and humans8, 9. Correspondingly, sumoylation is known to affect numerous cellular processes. Although SUMO attachment to proteins can have distinct physiological consequences, its primary molecular function is to control interactions of the modified proteins with other proteins. Many cellular proteins bear SUMO-interacting motifs (SIMs) that bind SUMO non-covalently10 [Box 1].
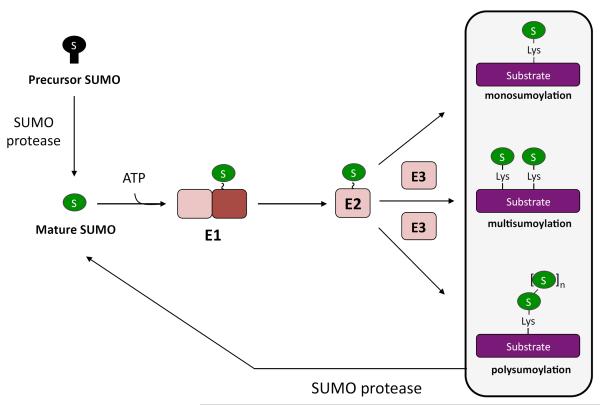
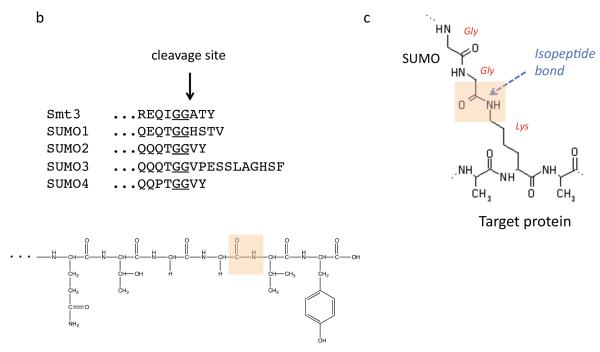
SUMO attachment to proteins requires ATP and typically involves three enzymes: a heterodimeric SUMO-activating enzyme (E1) that activates the SUMO C-terminal carboxyl group and then forms a thioester with the SUMO C-terminus, a SUMO-conjugating enzyme (E2), and one of several SUMO ligases (E3s)11 [FIG. 1]. Like ubiquitin, SUMO is usually ligated to substrate lysine side chains, forming amide (isopeptide) linkages. It may modify a single substrate lysine or multiple lysines (multisumoylation) or form SUMO chains on its substrates (polysumoylation). Yeast has only one form of SUMO (encoded by SMT3), but most vertebrates express at least three SUMO isoforms, SUMO1, SUMO2, and SUMO3, each encoded by a distinct gene. SUMO2 and SUMO3 are nearly identical and will hereafter be referred to as SUMO2/3 when isoform type needs to be specified. SUMO1 is ~48% identical to SUMO2/3. All SUMO isoforms are synthesized from C-terminally extended precursors that first need to be processed to their active forms, which terminate with a pair of glycine residues [FIG 1b]. A fourth SUMO paralogue (SUMO4) gene is present in humans, but it might not encode a molecule that is covalently coupled to proteins because its precursor does not seem to be processed in vivo12.
Figure 1. SUMO conjugation and deconjugation cycle.
a | SUMO (S) is translated in a precursor form (black) that must be processed by a SUMO protease before SUMO can be conjugated to other proteins. Mature SUMO is conjugated to proteins in an ATP-dependent manner in reactions catalyzed by three enzymes, E1, E2, and E3, in sequence. SUMO can be conjugated to a single lysine of a protein (monosumoylation), multiple lysines of a single protein (multisumoylation), or to SUMO that is already conjugated to a protein, forming a SUMO chain (polysumoylation). SUMO proteases deconjugate SUMO or SUMO chains from proteins. See Table 1 for a summary of the ability of each of the known SUMO protease to process and/or deconjugate the different forms of SUMO. b | The C-termini of S. cerevisiae Smt3 and the four mammalian SUMO isoforms are shown, with the di-glycine motif underlined. The site of cleavage is indicated by an arrrow. The chemical structure of the C-terminal tail of SUMO2 is also shown, with the scissile bond boxed in orange (bottom). c | The chemical structure of the isopeptide bond between the terminal glycine of mature SUMO and the lysine of a substrate is also shown, with the scissile bond boxed in orange.
Deconjugation of SUMO from proteins is carried out by enzymes referred to generically as SUMO proteases [FIG. 1]. These enzymes cleave precisely between the terminal glycine of SUMO and the substrate lysine [FIG. 1c]. Certain SUMO proteases are also responsible for SUMO precursor maturation and thus indirectly affect SUMO conjugation. The first SUMO protease to be described was the S. cerevisiae protein Ulp1 (UBL-specific protease 1)13. By comparing the sequence of the Ulp1 catalytic domain to sequence databases, a second budding yeast SUMO protease (Ulp2)14 and putative SUMO proteases in many other organisms were identified13, 15. The first human protein to be described as similar to Ulp1 was confirmed as a SUMO-specific protease and named sentrin-specific protease 1 (SENP1; SUMO is also known as sentrin)16. More extensive database searches identified additional putative human SENPs17, and subsequent studies have shown that humans express six SUMO proteases of the SENP class: SENP1–SENP3 and SENP5–SENP7. Moreover, recent studies have identified three new SUMO proteases in humans, desumoylating isopeptidase 1 (DeSI1), DeSI218 and ubiquitin-specific protease-like 1 (USPL1)19, which share little sequence similarity with the Ulp/SENP protease class.
Recent investigations into the biochemical activities and subcellular localization of SUMO proteases have contributed much towards our understanding of the specificity of these enzymes. New functions of SUMO proteases continue to be unraveled and intriguing examples of SUMO protease regulation have recently been uncovered. In this review we incorporate discussion of the newly discovered classes of SUMO proteases and the novel features found for the established Ulp/SENP enzymes into a broader appraisal of the SUMO protease field.
Unexpected Variety in SUMO proteases
The Ulp/SENP enzymes were the only known structural class of SUMO proteases until the very recent characterization of DeSI1 and DeSI218 and USPL119. All three groups are cysteine proteases with a papain-like proteinase fold. The identification of three distinct classes of SUMO proteases is reminiscent of the diversification seen for deubiquitylating enzymes (DUBs), which fall into five known structural classes20. The expansion of paralogues within the DUB superfamily, however, is far greater, with ~100 DUBs predicted for humans, none of which resembles the Ulp/SENP proteases. USPL1, however, as its name suggests, does distantly resemble the USP class of DUBs19. USPL1 is found throughout the vertebrates and in several invertebrates. The characterization of USPL1 in zebrafish (Danio rerio) provides evidence that the SUMO protease function of USPL1 is conserved in evolution. DeSI proteins are distinct from the known DUBs and Ulp/SENPs. Instead, they belong to the PPPDE (permuted papain fold peptidases of ds-RNA viruses and eukaryotes) class of proteases21, which do not have a representative in S. cerevisiae but are found in other fungi, including Schizosaccharomyces pombe. Whether non-mammalian PPPDE proteases function as SUMO-specific proteases has not been tested.
That USPL1 most closely resembles a family of deubiquitylating enzymes but is in fact a SUMO-specific protease underscores the importance of biochemical characterization for putative proteases. A similar surprise was observed with the characterization of DEN1, which was originally annotated SENP8 due to its sequence similarity to the Ulp/SENP proteases, but was later found to be specific for the UBL Nedd8 rather than SUMO22–24.
Structures of SUMO proteases
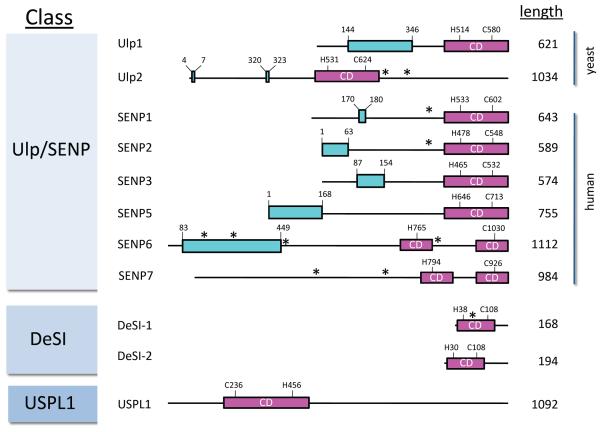
Most of the known SUMO proteases have substantial noncatalytic regions [FIG. 2a]. For the well-characterized Ulp/SENP proteases, N-terminal regions, although poorly conserved, regulate their intracellular localization25–30 [TABLE 1]. Yeast Ulp2 and multiple metazoan SENPs also contain putative SIMs in their non-catalytic regions [FIG. 2a]. A putative SIM can also be found in DeSI1, although in this case within the catalytic domain. If they are indeed functional, these SIMs might increase enzyme affinity for SUMO or SUMO chains or they might help orient the sumoylated protein on the SUMO protease. The catalytic domains of Ulp/SENP enzymes bind SUMO via surfaces that are distinct from SIMs31 [FIG. 2b, c].
Figure 2. Structures of SUMO proteases.
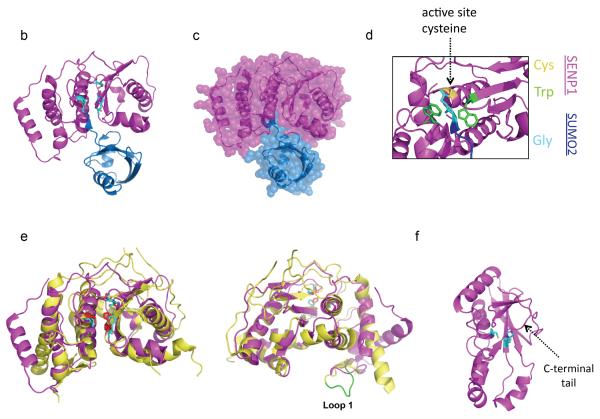
The domain organizations and lengths (in amino acids) of the SUMO proteases discussed in this article are shown. Each catalytic domain (CD) is colored magenta, with key histidine (H) and cysteine (C) residues shown. Putative SUMO-interacting motifs (SIMs) are indicated by asterisks. Regions shown to be important for intracellular localization of the protease are shown in cyan. For DeSI-1 and DeSI-2, the mouse proteins are shown. For USPL1, the human protein is depicted. Representatives from each of the three known classes of SUMO proteases exist in other organisms15, 21; however, in some cases it is unclear whether these proteins are indeed SUMO-specific proteases. b | Crystal structure of the catalytic domain of SENP1 (magenta) in complex with SUMO2 (blue) (Protein Databank (PDB) code 2CKH) shows the orientation of SUMO2 with respect to SENP1. This complex contains a covalent thiohemiacetal linkage between the active site cysteine of SENP1 and the C-terminal glycine of SUMO2. The catalytic residues are shown in stick format, with the atoms colored according to element (carbon = cyan; nitrogen = blue; oxygen = red; sulfur = orange). c | A space-filling representation of the structure shown in part b, which highlights the intimate contacts made between SENP1 and SUMO2. d | A higher magnification view of SENP1 in complex with SUMO2 (2CKH), showing the two Trp residues of SENP1 that surround the two Gly residues of SUMO2. e | The structure of the catalytic domain of SENP7 (yellow) (PDB code 3EAY) aligned to the structure of the catalytic domain of SENP1 (magenta) highlights the similarities and differences between SENP1/2 and SENP6/7. The catalytic residues are displayed in cyan for SENP1 and red for SENP7. Loop regions, including Loop-1 (right, green), of SENP7 are unique to SENP6 and SENP7. f | The structure of a monomer of DeSI-1 (PDB code 2WP7) shows the proposed catalytic dyad (Cys-His), which differs from the catalytic triad (Cys-His-Asp) utilized by the Ulp/SENP enzymes. The catalytic residues are displayed as in b
Table 1.
Properties of the SUMO proteases.
| Species | Name | Subcellular Localization | SUMO Isoform Preference | Precursor processing | Deconjugation | Chain Editing** |
|---|---|---|---|---|---|---|
| S. cerevisiae | Ulp1 | nuclear pore | Smt3 | Yes | Yes | No |
|
| ||||||
| Ulp2 | nucleoplasm | Smt3 | No | Yes | Yes | |
|
| ||||||
| Mammals | SENP1 | nuclear pore and nuclear foci | SUMO1 and SUMO2/3 | Yes | Yes | No |
|
| ||||||
| SENP2* | nuclear pore and nuclear foci, cytoplasm | SUMO2/3 > SUMO1 | Yes | Yes | No | |
|
| ||||||
| SENP3 | nucleolus | SUMO2/3 | Unknown | Yes | No | |
|
| ||||||
| SENP5 | nucleolus and mitochondria | SUMO2/3 | Yes | Yes | No | |
|
| ||||||
| SENP6 | nucleoplasm | SUMO2/3 | No | Yes | Yes | |
|
| ||||||
| SENP7 | nucleoplasm | SUMO2/3 | No | Yes | Yes | |
|
| ||||||
| DeSI-1 | cytoplasm and nucleus | SUMO1 and SUMO2/3 | Weak | Yes | Yes | |
|
| ||||||
| DeSI-2 | cytoplasm | Unknown | No | Unknown | Unknown | |
|
| ||||||
| USPL1 | Cajal bodies | SUMO2/3 > SUMO1 | Weak | Yes | Yes | |
SENP2 has at least 3 splice variants that localize differentially in cells. The 589 amino acid version localizes to the nuclear pore.
Chain editing refers to the ability of a protease to remove SUMO moieties from a SUMO chain without removing the entire SUMO chain from sumo-modified proteins.
Insight into the specificity and function of SUMO proteases has been gained through high-resolution structural investigations of the proteases, in some cases in complex with SUMO or SUMO-conjugated substrates. The first SUMO protease structure solved, that of the catalytic domain of yeast Ulp1 in a covalent transition state-like complex with SUMO31, revealed that the overall architecture of the Ulp1 catalytic domain resembles that of other cysteine proteases, as had been predicted from the sequence similarity of this domain to the adenovirus processing protease AVP13. Specificity for SUMO is achieved through an extensive interface involving unique hydrophilic interactions and numerous salt bridges. The active site pocket orients the catalytic triad residues (order: His-Asp-Cys) for substrate cleavage and positions the SUMO C-terminal diglycine motif in a shallow tunnel formed by two Trp residues — side chains of residues other than glycine would sterically clash with the narrow Trp tunnel. Structures for the catalytic domains of the mammalian SUMO proteases SENP1 and SENP2 are similar to that of Ulp1 and share the Trp tunnel and SUMO-binding surfaces noted for Ulp1 [FIG. 2b–d]32–35. As reviewed by Huang and Schulman36, SENP1 and SENP2 (and likely other members of the Ulp/SENP family) orient the scissile bond in a cis configuration, which leads to a kink in the SUMO C-terminal tails for SUMO precursors or in the isopeptide linkage to sumoylated proteins33, 34. These nonprolyl cis peptide bonds are rare in proteins and are thought to destabilize the scissile bond and promote cleavage.
The catalytic domains of SENP6, SENP7 and Ulp2 have low sequence identity to the catalytic domain of Ulp1 (less than 30%) but are more similar to one another and thus represent a distinct subgroup of Ulp and SENP enzymes13. SENP6 and SENP7 have four conserved loop insertions within the catalytic domain that are absent in the other mammalian SENPs. As revealed in the crystal structure of SENP7, the Loop-1 insertion – which is not conserved in yeast Ulp2 – projects towards the presumed SUMO-binding interface, suggesting a role in binding the SUMO-conjugated substrate [FIG. 2e]37. Consistent with this, in vitro activity assays with the mutant SENP7 catalytic domain lacking Loop-1 show that it is less efficient at deconjugating SUMO substrates than the wild-type version. Notably, SENP6 and SENP7 normally have higher activity toward SUMO2/3 than SUMO1 [Table 1]. Paralogue specificity of these proteases could be altered by mutating two key residues on SUMO2 that likely contact Loop-1 of SENP6 and SENP7 to those that are found in SUMO138. This SUMO2 mutant is no longer efficiently deconjugated by SENP6 or SENP7. Conversely, a mutant SUMO1 that has these two residues replaced by those of SUMO2 was a much better substrate for these SENPs. Thus, contacts between Loop-1 of SENP6 or SENP7 and SUMO2/3 contribute substantially to SUMO paralogue specificity.
Crystal structures of mouse DeSI1 [FIG. 2f] and a truncated human DeSI1 show several interesting features39, 40. The protease forms a dimer in which the active-site residues face a surface groove formed between the two monomers. Curiously, the C-terminal tails of the full-length DeSI1 fold into the groove and seem to block substrate access to the active sites. However, activity assays suggest truncation of the C-terminal tail has no effect on substrate cleavage rates39. SUMO-modified substrates might be able to displace the C-tails, or the occluded conformation may be a result of crystallization. Whether both active sites must be functional in the homodimer also remains to be determined.
In contrast to the catalytic triad in Ulps and SENPs, DeSI1 and DeSI2 have been suggested to have only a catalytic dyad (Cys-His)39. Cys108 and His38 are positioned as in other papain-fold proteases, and indeed, mutagenesis studies show that SUMO substrate cleavage is abolished when these residues are mutated18, 39. A comparative analysis of structures of the Nlp60/P60 cysteine protease superfamily, to which the DeSI enzymes belong, led to the suggestion that another residue in the DeSI enzymes binds a water molecule that in turn helps orient the catalytic His residue40. Structural analysis of DeSI1 in complex with a sumoylated substrate should help clarify these mechanistic issues. No high-resolution structure is currently available for USPL1, but its sequence similarity to DUBs of the USP class suggests its catalytic domain will have a comparable architecture19.
Substrate specificity of SUMO proteases
The specificity of the SUMO proteases has been extensively investigated both in vitro and in vivo and has been found to be strongly influenced by the localization of the protein [TABLE 1].
Yeast SUMO proteases
The yeast Ulp1 enzyme, unlike Ulp2, shows broad specificity in vitro, where it is capable of efficiently cleaving SUMO translational fusions and isopeptide-linked SUMO, including polySUMO modifications13, 31, 41. Ulp1 is largely responsible for SUMO precursor processing in vivo, and it also removes post-translational SUMO modifications from protein substrates13, 25. Ulp2 has particularly high activity towards the SUMO-SUMO linkages in polySUMO chains both in vitro and in vivo, although it also can cleave isopeptide linkages between SUMO and substrates14, 41. Genetic studies using yeast strains with mutations in Ulp1 or Ulp2 indicate that the two yeast SUMO proteases have distinct substrate specificities as unique sumoylated substrates accumulate in each protease-deficient strain, and the mutants have distinct phenotypic defects14.
Substrate specificity of Ulp1 and Ulp2 is strongly influenced by their restricted cellular localization25. Ulp2 is found throughout the nucleoplasm14, whereas Ulp1 localizes primarily to the inner surface of the nuclear pore complex (NPC). However, a fraction of Ulp1 is exported to the cytoplasm in a cell cycle-specific manner and desumoylates the heavily sumoylated septin proteins that form filaments at the bud neck of dividing cells42, 43. Localization is dictated by their noncatalytic N-terminal domains: indeed, an N-terminally deleted Ulp1 can localize throughout the cell. When expressed in an ulp2 null strain, this truncated Ulp1 enzyme partially suppresses the ulp2 mutant growth defects, such as irreversible arrest in response to DNA damage, and reduces the levels of sumoylated Ulp2 substrates, highlighting the importance of restricting Ulp1 localization in controlling its in vivo activity25. Moreover, high levels of Ulp1 activity mislocalized to the nucleoplasm are lethal25, 44. Nuclear localization of Ulp2, in contrast, is crucial for its function26.
Mammalian SUMO proteases
As observed with the yeast SUMO proteases, the in vivo specificities of the mammalian SENPs are governed in part by their localization. Most of the SENPs concentrate in the nucleus, with many of them localized to recognizable subnuclear compartments [Table 1]45. SENP3 and SENP5 localize to the nucleolus27, 46. SENP6 localizes throughout the nucleoplasm30, but a catalytically inactive SENP6 concentrates in nuclear foci47, a fraction of which seem to be PML (promyelocytic leukemia protein) nuclear bodies, consistent with PML being a substrate of SENP6. Interestingly, SENP2 encodes at least three alternatively spliced mRNA variants and the resulting proteins localize differently, including one variant that localizes to the cytoplasm45, 48. Since SENPs have been visualized in the cytoplasm to some extent48–50, and all are synthesized in the cytoplasm, SENP-mediated desumoylation is likely to occur at some frequency in the cytoplasm.
Deconjugation of sumoylated substrates by individual SENPs in vitro is generally not strongly SUMO paralogue-specific, although some of the SENPs exhibit greater activity towards one or another isoform [Table 1]. However, greater paralogue specificity in vivo is sometimes observed. Although the basis for the in vivo paralogue specificities of SENPs is not completely understood, competition for substrate binding by other cellular proteins is likely to be the explanation in most cases. An interesting example is the nearly exclusive modification in vivo of RanGAP1 by SUMO1 rather than SUMO2/3 even though in vitro the SUMO isoforms are conjugated to RanGAP1 with similar efficiencies51. Sumoylated RanGAP1 binds the NPC component Nup358, with SUMO1-RanGAP1 interacting more tightly with Nup358 than SUMO2/3-RanGAP1. This selectively protects the SUMO1-RanGAP1 conjugate from SUMO proteases.
By contrast to deconjugation of sumoylated proteins, SUMO precursor processing by SENPs is often paralogue-specific35, 46, 52. The basis for this discrimination lies in the predicted contacts made between the protease and propeptide residues following the C-terminal diglycine motif35. The propeptide sequences in the SUMO paralog precursors are quite divergent [FIG. 1b], and certain propeptide extensions fit better than others in binding pockets that lie adjacent to the catalytic residues of the protease35. These structure-based studies of precursor processing are currently limited to SENP1 and SENP2. Of the other SENPs, SENP5 has significant SUMO precursor processing activity for SUMO3 and limited activity toward SUMO146, SENP6 and SENP7 do not possess SUMO precursor processing activity37, and SENP3 has not been tested due to the inability to obtain purified, active SENP3 [TABLE 1].
DeSI1 was identified in a screen for proteins that bind to the transcriptional repressor BZEL (BTB-ZF protein expressed in effector lymphocytes)18. DeSI1 is diffusely localized throughout the cytoplasm and nucleus in 293T cells and desumoylation assays carried out both in 293T cells and with recombinant proteins revealed that DeSI1 specifically desumoylates BZEL. DeSI1 deconjugates both SUMO1 and SUMO2/3 from BZEL and weakly cleaves poly-SUMO2/3 chains18, 39. Importantly, the substrate specificity of DeSI1 is distinct from that of SENPs, as DeSI1 does not desumoylate known substrates of SENPs18. BZEL is currently the only known substrate of DeSI1 and knockdown of DeSI1 expression in 293T cells does not detectably alter bulk SUMO-conjugate profiles, suggesting a limited number of substrates. One caveat to this conclusion is that the very similar DeSI2, which also concentrates in the cytoplasm, might partially compensate for DeSI1. Therefore, it will be useful to evaluate cellular phenotypes in cells co-depleted for the two DeSI enzymes.
USPL1 was identified as a protein in HeLa cell extracts that crosslinked to either SUMO1 or SUMO3 that had been derivatized at its C-terminus by chemical ligation to vinyl methylester, a thiol reactive functional group that covalently modifies active site cysteines of enzymes19. USPL1 localizes to Cajal bodies within the nucleus of the cell, which is unique among the known SUMO proteases. Like Ulp1 and certain SENPs, USPL1 shows broad SUMO protease activity in vitro. Furthermore, bulk SUMO-conjugates are reduced upon USPL1 overexpression in HEK293T cells. In contrast, knockdown of USPL1 expression does not cause a detectable change in bulk SUMO-conjugate profiles, but it does impair cell growth. Interestingly, supplying an exogenous, non-catalytic form of USPL1 is able to suppress the growth defects caused by knockdown of endogenous USPL1. Thus, although USPL1 is clearly a SUMO protease, it appears to have an essential role in cell division that is independent of its SUMO protease activity.
Regulation of SUMO proteases
In addition to the constraints placed on SUMO protease activity by their localization or by substrate shielding, other mechanisms of regulating SUMO protease function, particularly in response to environmental stimuli, have been described.
Regulation of transcription
Altered transcription of specific mammalian SENP genes is a common mechanism affecting the sumoylation of specific proteins [FIG. 3a]. SENP1 expression is induced by androgen receptor signaling, and androgen response elements are present in the promoter region of SENP153. The SENP1 promoter also contains a hypoxia response element, and SENP1 levels increase upon oxygen deprivation (hypoxia) in a manner dependent on HIF1α54. SUMO proteases can in turn regulate transcription of other genes (see below), and in some cases, they control their own transcription. During both hypoxia and androgen signaling, SENP1 is proposed to participate in positive feedback loops wherein SENP1 has a stimulatory effect on its own transcription in addition to stimulating the transcription of the other targets of these pathways53–55. For example, SENP1 is proposed to desumoylate HIF1α (which is sumoylated, ubiquitylated and degraded in response to hypoxia) and enhance its stability, which is consistent with increased sumoylated HIF1α and decreased expression of HIF1α target genes in cells lacking SENP155. The exact mechanism of feedback control for SENP1 and androgen signaling remains to be determined53.
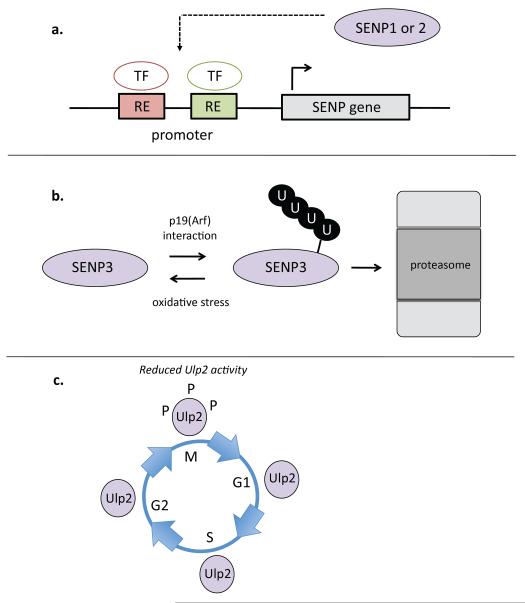
Figure 3. Regulation of SUMO proteases.
a | SENP1 and SENP2 levels are known to be affected by changes in their transcription under various conditions (see text). Fittingly, the promoters of SENP1 and SENP2 contain response elements (REs) that bind specific transcription factors (TFs). Both SENP1 and SENP2 can affect their own transcription by desumoylating transcription factors or factors that affect the activity or abundance of relevant transcription factors. b | SENP3 levels are regulated by the ubiquitin-proteasome system. SENP3 is constitutively ubiquitylated under normal growth conditions, leading to its proteasome-mediated degradation. Under conditions of mild oxidative stress, SENP3 ubiquitylation is reduced and SENP3 levels increase. When the tumor suppressor p19(Arf) is expressed, it interacts with SENP3, enhancing its ubiquitylation and degradation, leading to decreased levels of SENP3. c | Yeast Ulp2 is phophorylated during the mitosis phase of the cell cycle. Phosphorylation of Ulp2 is thought to inhibit its activity.
SENP2 is also known to regulate its own transcription, but in this case it operates in a negative feedback loop56. Following an early increase in SENP2 mRNA levels in response to genotoxic insults, SENP2 desumoylates the NF-κB activator NEMO (NF-κB essential modulator), which in turn attenuates SENP2 transcription as the response progresses. As NF-κB signaling promotes cell survival, this negative feedback loop is one mechanism used by cells to prevent the survival of cells that are damaged beyond repair.
Regulation by post-translational modifications
SUMO proteases are subject to post-translational modifications that regulate their activity and/or stability. SENP3 is modified by ubiquitin and subsequently degraded by the proteasome [FIG. 3b]. SENP3 turnover by the ubiquitin-proteasome system (UPS) is accelerated by its interaction with the tumor suppressor p19(Arf)57 and dampened by oxidative stress58. Under normal growth conditions, SENP3 is continuously ubiquitylated by the ubiquitin ligase CHIP59. Mild oxidative stress leads to association of the chaperone protein Hsp90 with SENP3, protecting it from CHIP-mediated ubiquitylation. Interestingly, stabilization of SENP3 leads to SENP3 accumulation in the nucleoplasm (outside of the nucleolus, where it is normally localized), and hence SENP3 can gain access to additional substrates, such as the general transcriptional activator p300, under these conditions58. p300 is a co-activator of HIF1α, and desumoylation of p300 enhances the expression of HIF1α target genes, which is a known cellular response to mild oxidative stress.
In yeast, Ulp2 accumulates in a (hyper)phosphorylated form specifically during mitosis [FIG. 3c], and this depends on the kinases Cdk1 and Cdc5 (Polo)60. Ulp2 is phosphorylated on its noncatalytic C-terminal domain, although the modified residues have not been definitively mapped. Interestingly, two known targets of Ulp2-dependent desumoylation that participate in chromatid cohesion and segregation, Top2 and Pds5, fail to accumulate in sumoylated form in a cdc5 mutant, consistent with Polo kinase acting as a negative regulator of Ulp260.
Regulation of SUMO protease activity
The specific activities of selected SUMO proteases are directly affected by certain cellular stresses. One example is during heat shock, which leads to a rapid increase in the sumoylation of many proteins in organisms from yeast to humans61. A very recent study found that several SENPs are irreversible inactivated by heat shock, both in cultured cells and in vitro62. Experiments using purified SENP catalytic domains suggest that this thermal inactivation is due to protein denaturation. Interestingly, SENP6 was more thermotolerant than the other SENPs, perhaps indicating a unique role for SENP6 during heat shock. It is unlikely that the thermal inactivation of SENPs is the only mechanism for cellular sumoylation alterations during heat shock. Another recent report suggests that the increase in sumoylation during heat shock involves changes in the ratio of sumoylated versus unmodified SAE2, one subunit of the dimeric SUMO E1 enzyme63. Heat shock leads to an increase in unmodifed SAE2 and a corresponding decrease in sumoylated SAE2, which is less active than unmodified SAE2 in the transfer of thioester-linked SUMO to E2. As suggested by Truong et al., a SUMO protease is likely to remove the inhibitory SUMOs from SAE2 during heat shock63, but this hypothesis remains to be tested. Since only a fraction of SAE2 exists in the sumoylated (less active) form under standard growth conditions, this mechanism may only account for a portion of the cellular sumoylation response to heat shock.
Another potential example of regulatory SUMO protease inactivation is the reversible formation of an intersubunit disulfide between two molecules of SENP1 (and potentially Ulp1 in yeast) in response to oxidative stress64. The active site cysteine participates in disulfide formation, and in vitro evidence suggests that reversible disulfide formation may protect the enzyme from irreversible oxidative inactivation of the catalytic cysteine.
Cellular roles of SUMO proteases
SUMO proteases affect numerous cellular processes and when mutated, cause a corresponding diversity of defects, including lethality13, 55, 65. Because of their diverse roles, we cannot review all known or suspected SUMO protease functions here, but instead focus on a few illustrative examples. In many cases, the relevant sumoylated substrate or substrates are not known with certainty. This uncertainty relates to two general observations regarding in vivo sumoylation. First, at steady state only a small percentage of any given protein substrate is present in its sumoylated form. Second, it is often the case that multiple proteins involved in a particular physiological process or pathway are sumoylated and/or interact noncovalently with SUMO. Therefore, blocking or enhancing the sumoylation of any one protein often does not yield a strong phenotypic deficiency.
Transcription
Numerous transcriptional programmes are regulated by the SUMO pathway, and multiple recent investigations implicate SUMO proteases in the regulation of specific transcription factor activities18, 58, 66–75. Most commonly, Ulp- or SENP-mediated removal of SUMO results in increased transcriptional activity of the target factor. This is presumably due to the variety of factors involved in transcriptional repression, such as histone deacetylases (HDACs), polycomb-related repressors and other co-repressors, that are more effectively recruited by sumoylated transcription factors than their unmodified forms76. In the case of the transcription factor Elk1, its sumoylation recruits HDAC277. Sumoylated Elk1 is desumoylated mainly by SENP1 and SENP1 depletion dampens transcriptional activation mediated through Elk168.
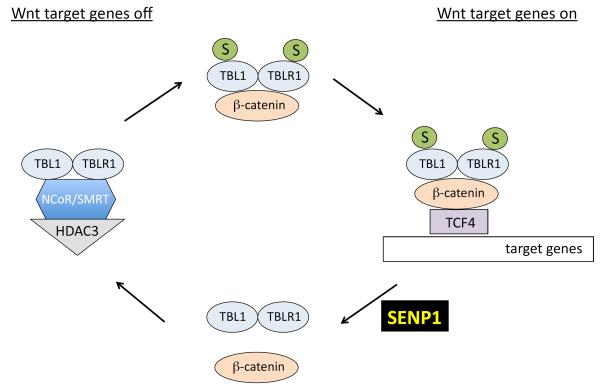
However, exceptions to this generalization are known, as in the case of SENP1-mediated regulation of the Wnt signaling pathway [FIG. 4]72. A major regulator of developmental programmes, the Wnt pathway is tightly coordinated to prevent aberrant cell growth and proliferation. The transcriptional regulators TBL1 and TBLR1 were previously reported as activators of the Wnt pathway through their interaction with β-catenin, a transcriptional coactivator78. TBL1 and TBLR1 are sumoylated following activation of the Wnt pathway, and sumoylation-deficient mutants of these proteins are unable to activate a Wnt signaling response72. Sumoylation of TBL1-TBLR1 was found to promote TBL1-TBLR1 dissociation from the NCoR-HDAC3 corepressor complex and to promote formation of a TBL1SUMO-TBLR1SUMO-β-catenin complex, which is recruited to Wnt target genes by TCF4, a sequence-specific DNA-binding protein. Of the SENP proteases tested, only SENP1 interacts with TBL1-TBLR1, and ectopic expression of SENP1 in 239T cells resulted in desumoylation of TBL1-TBLR1 and decreased activation of Wnt pathway target genes. This effect was abolished with a SENP1 catalytic mutant (SENP1C603S), indicating that the SUMO-deconjugating activity of SENP1 is required. Lastly, shRNA knockdown of SENP1 in 293T cells led to the accumulation of sumoylated TBL1-TBLR1 and enhanced activation of Wnt target genes. These data identify SENP1 as a negative regulator of the Wnt signaling pathway that seems to work by triggering disassembly of a transcriptional co-activator complex.
Figure 4. The function of SENP1 in transcription during Wnt signaling.
Activation of Wnt signaling leads to the sumoylation of TBL1 and TBLR1, which function together as a transcriptional co-activator complex. Sumoylation of TBL1-TBLR1 promotes the dissociation of TBL1-TBLR1 from the NcoR/HDAC3 corepressor complex and promotes the association of TBL1-TBLR1 with β-catenin. The TBL1SUMO-TBLR1SUMO-β-catenin complex is recruited to Wnt target genes by the sequence-specific DNA-binding protein TCF4. SENP1 is responsible for desumoylation of TBL1SUMO-TBLR1SUMO, which leads to the disassembly of the TBL1SUMO-TBLR1SUMO-β-catenin complex and decreased activation of Wnt target gene expression.
Cell cycle progression
Ulp1 is essential for progression past the G2/M phase of the cell cycle in budding yeast13. Ulp1 is required for SUMO precursor processing, but providing mature SUMO does not reverse the growth defects of a temperature-sensitive ulp1 mutant. This suggests that these cell cycle defects are caused by the persistent or excess sumoylation of Ulp1 target proteins. The identities of the key sumoylated proteins linked to the cell cycle function(s) of Ulp1 remain unknown, although proteins with roles in cell division, such as the spindle-orientation protein Kar9, have been identified as Ulp1 substrates42, 79, 80.
Yeast Ulp2 also has roles in cell division14, 81. At elevated temperatures and following DNA damage14, Ulp2 is essential for cell division, functioning in the resumption of the cell cycle after the checkpoint has been turned off or the DNA damage has been repaired82, 83. Under normal conditions, yeast can divide in the absence of Ulp2, but growth is poor, and ulp2-deficient cells have abnormal cell size and morphology14. Poor growth in these cells has been linked to improper centromere cohesion dynamics84, in part due to persistent or excess sumoylation of DNA topoisomerase II (Top2). Indeed, ulp2-deficient cells that express Top2 that cannot be sumoylated display reduced centromere cohesion defects and show improved growth at elevated temperature84. Poor growth in ulp2-deficient cells is also thought to be due in part to enhanced spontaneous homologous recombination (HR); these HR defects could be due to altered sumoylation of several substrates, as many proteins within nuclear recombination foci are sumoylated85.
Poor growth in yeast cells lacking Ulp2 has also been linked to abnormal mitotic spindles14, but the basis of this is not clear. However, studies of human SENP6, which resembles Ulp2 in structure and function, have shed light on a role for SUMO2/3 deconjugation during kinetochore assembly86. Kinetochores are large protein assemblies that physically link spindle microtubules to the centromeres of chromosomes during mitosis. A kinetochore subcomplex known as the CENP-H/I/K complex is less abundant in cells depleted of SENP6, and these cells mis-segregate chromosomes at an elevated rate compared with control cells. Interestingly, co-depletion of a ubiquitin ligase (RNF4; see below) that ubiquitylates polysumoylated proteins stabilizes the CENPH/I/K complex and partially restores accurate chromosome alignment during metaphase. Since polysumoylated CENPH and CENPI accumulate in these SENP6- and RNF4-depleted cells, the authors propose that under normal conditions RNF4 targets CENPH- and CENPI-containing complexes that have deviated from their correct assembly pathway. Thus, SENP6 is thought to ensure proper kinetochore assembly on chromosomes by only desumoylating correctly assembled kinetochores leaving off-pathway intermediates to be ubiquitylated by RNF4 and ultimately degraded.
Human SENP5 also has a role in cell division, as knockdown of SENP5 results in decreased cell proliferation and abnormal nuclear morphology46. Understanding the mechanistic basis of these defects awaits further investigation.
Ribosome biogenesis
Eukaryotic ribosome biogenesis is a tightly regulated and coordinated process, and current data highlight a connection between ribosome maturation and the SUMO pathway. The connection was first established in S. cerevisiae, in which numerous SUMO pathway mutants, including a temperature-sensitive Ulp1, were shown to have defects in ribosomal RNA (rRNA) processing87.
In mammalian cells, SENP3 has been found to physically interact with multiple proteins involved in ribosome maturation, including nucleophosmin (NPM1)88, 89 and a protein complex including PELP1, TEX10 and WDR1890. The early steps of ribosome biogenesis occur in the nucleolus, and NPM1, SENP3 and SENP5 co-localize in subdomains of the nucleolus89. Depletion of SENP388, 89 or SENP589 results in rRNA processing defects, as does depletion of NPM191, 92. Although the substrates of SENP3 and SENP5 that are pertinent to ribosome biogenesis remain unclear, sumoylated forms of NPM1, GNL2 (a putative 60S preribosomal export factor) and the 60S ribosomal subunit RPL37A have been reported to accumulate upon depletion of SENP3, SENP5, or SENP3 and SENP5 in combination88, 89. In the case of NPM1, the results of two reports are conflicting. While one study provides evidence that NPM1 is desumoylated by SENP3 and that constitutive sumoylation of NPM1 impairs rRNA processing88, the other study presents data arguing against NPM1 as a substrate of SENP3 (or SENP5) but supporting a role for NPM1 in regulating the abundance of SENP3 and SENP589.
PELP1, TEX10 and WDR18 interact with SENP3 and one another90. PELP1, TEX10 and WDR18 show distant homology to Rix1, Ipi1 and Ipi3, which form a complex involved in ribosome biogenesis in yeast93, 94. PELP1 desumoylation by SENP3 is thought to control the localization of PELP1, which is partitioned between the nucleolus and the nucleoplasm90. However, the importance of PELP1 localization can only be speculated at this time. Future studies on the roles of SUMO dynamics in ribosome biogenesis are expected to shed light on this exciting biological connection.
Mitochondrial dynamics
Even though most SUMO-regulated processes are localized to the nucleus, instances of SUMO functioning outside of the nucleus continue to emerge. One example is the control of mitochondrial dynamics95–97. Many mitochondrial proteins are modified by SUMO1 in cultured mammalian cells, and SUMO1 overexpression leads to mitochondrial fragmentation95. Concurrent overexpression of SENP5 can prevent this SUMO1-induced mitochondrial fragmentation and additional evidence supports roles for SENP5 in both mitochondrial fission and fusion96. Although SENP5 localizes to the nucleus for most of the cell cycle, it is found at mitochondria during mitosis97. SENP5 is required for cell division46, 97 and the localization of SENP5 to mitochondria at mitosis is suggested to facilitate mitochondrial inheritance during cell division97. Although SENP5 is expected to have many (unidentified) substrates on mitochondria, one established substrate is Drp1 (dynamin-related protein), a GTPase involved in mitochondrial fission96. SUMO1 modification of Drp1 enhances its association with mitochondria, but subsequent desumoylation by SENP5 is thought to facilitate the assembly of Drp1 into active, fission-promoting polymers97. However, the role of SENP5 in mitochondrial dynamics is not limited to regulating the function of Drp1 in fission since SENP5 knockdown also affects mitochondrial fusion and leads to a complex mitochondrial morphology that cannot be explained by the effects of SENP5 on Drp1 alone96. Deciphering the exact roles of SUMO1 and SENP5 in mitochondrial dynamics will require the identification of more of the mitochondrial SUMO1/SENP5 substrates and further assays of mitochondrial fusion and fission.
SUMO proteases and the STUbL pathway
The surfaces of the SUMO and ubiquitin proteins differ markedly in their physical properties, and, unlike ubiquitin, SUMO does not directly target proteins to the proteasome. However, the recent discovery of a functionally conserved family of enzymes known as SUMO-targeted ubiquitin ligases (STUbLs), which catalyze the addition of ubiquitin to proteins that have been previously sumoylated, illuminated a pathway in which SUMO can indirectly target proteins for proteasome-mediated destruction or non-proteolytic but ubiquitin-dependent regulation98–101 [FIG. 5]. There are likely to be multiple STUbLs, but the best described are the related human RNF4 homodimer, the budding yeast Slx5–Slx8 heterodimer, and the fission yeast Rfp1/2-Slx8 heterodimer. These proteins all have multiple SIMs, and each subunit of the complex bears a RING domain that is important for dimerization and ubiquitin ligase activity102, 103.
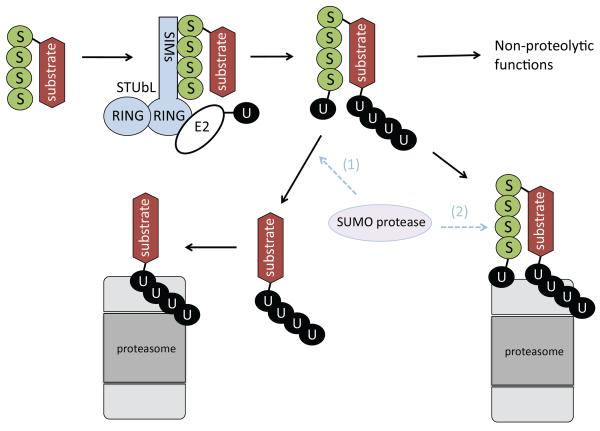
Figure 5. SUMO-targeted ubiquitin ligases (STUbLs).
STUbLs are enzymes that catalyze the addition of ubiquitin to proteins that have been previously sumoylated. SUMO (S), likely in the form of SUMO chains, attached to STUbL substrates is recognized by SIMs of the STUbL. The ubiquitin (U)-charged E2 enzyme (E2-U) is recruited by the dimeric RING domains of the STUbL, and ubiquitin is transferred to the substrate. Potential sites of attachment for ubiquitin on the substrate include the substrate itself and/or the attached SUMO moieties. STUbL activity results in proteins that are modified by both SUMO and ubiquitin. Ubiquitin addition can target proteins to the proteasome for destruction or non-proteolytic fates. It is currently unclear whether SUMO proteases act on STUbL substrates destined for the proteasome. If SUMO proteases act on these substrates, it could be before arrival at the proteasome (1) and/or at the proteasome (2). Alternatively, SUMO could be destroyed along with the substrate at the proteasome. Ubiquitin on STUbL substrates is likely recycled by deubiquitylating (DUB) activity of the proteasome (not shown).
STUbL-mediated ubiquitylation is currently known to play key roles in regulating specific transcriptional programs104 and DNA repair pathways105, 106, processes also regulated by SUMO proteases (see above)107. Since STUbLs and SUMO proteases can both affect cellular sumoylation dynamics, there are likely to be many interactions between these enzymes, and recent studies have indeed connected these two types of enzymes86, 100, 108, 109. An outstanding example derives from a genetic screen using the mot1-301 budding yeast mutant, which has a defect in the general transcriptional regulator Mot1108. Genetic suppressors of the temperature-sensitive growth defect revealed mutations in SLX5, SLX8, SUMO E1, SUMO E2, or either of the two SUMO proteases (ULP1 and ULP2). The mutant Mot1-301 protein is sumoylated, unlike the normal version of the protein, and loss of Slx5 or Slx8 leads to metabolic stabilization of the mutant protein, consistent with its potential STUbL-dependent ubiquitylation110. How mutations in ULP1 and ULP2 suppress the mutant mot1-301 phenotype is less clear. Loss of SUMO protease activity might indirectly impair SUMO conjugation to Mot1-301 as a result of defects in precursor maturation and SUMO recycling.
Ulp1 and Ulp2 have been connected to Slx5–Slx8 in two other studies. Slx5 overexpression can partially compensate for loss of Ulp1, but only if mature SUMO is provided100. These data suggest that although excess Slx5 cannot satisfy the SUMO precursor processing function of Ulp1, it can prevent growth defects caused by persistent or excessive sumoylation of certain proteins13. Enhanced Slx5–Slx8 STUbL activity may inactivate these proteins, either by targeting them to the proteasome for destruction or by simply removing them from their sites of action. Given what is known about STUbLs and SUMO proteases, one might reason that reduced STUbL activity would be exacerbated by reduced SUMO protease activity. However, defects associated with loss of Slx5–Slx8 are suppressed by mutation of ULP2109, suggesting that enhanced (poly)sumoylation of certain proteins can suppress STUbL deficiency. Since there are currently few known Ulp2 substrates and no confirmed natural substrates of Slx5–Slx8, these genetic interactions await molecular characterization. The genetic interaction between Ulp2 and Slx5–Slx8 is even more complex in that deletion of SLX5 or SLX8 suppresses the defects caused by lack of ULP2. Interestingly, an analogous situation has been observed in human cells, in that knockdown of RNF4 partially suppresses the mitotic defects caused by SENP6 knockdown (see above)86.
Conclusions and perspectives
From recent work, Ulp and SENP enzymes are becoming better understood at the biochemical, cell biological and physiological levels. Many Ulp and SENP substrates have been identified and in some cases the mechanism by which desumoylation affects the activity or function of the target protein has been established. Still, each of the Ulp/SENP enzymes is likely to have many substrates, most of which await identification. Future studies are likely to be directed towards more global identification of specific SUMO protease substrates, including substrates of the newly identified DeSI enzymes and USPL1. Given that many sumoylated proteins are part of large macromolecular complexes, it will be important to determine how each SUMO protease substrate fits into networks of interacting proteins.
Sumoylation of many proteins greatly increases following certain cellular stresses, such as heat shock (see above), treatment with ethanol or hydrogen peroxide111 and inositol starvation112, but the direct involvement of SUMO proteases in these phenomena has been subject to only limited investigation. Notably, yeast cells exposed to high levels of alcohol, but not other tested stresses, relocalize Ulp1 to the nucleolus113. Whether nucleolar sequestration of Ulp1 is important for cells to survive high alcohol levels is not known. Identifying changes in SUMO protease activity, levels or localization in response to these stresses, in addition to isolating clusters of substrates that are differentially processed by these proteases, will be necessary to understand the physiological significance of these responses.
The catalytic domain of the Ulp and SENP proteases is confined to a ~200-residue stretch, but all of these enzymes are substantially larger. Besides their roles in cellular localization, the functions of the remaining domains are poorly defined. One study showed that a noncatalytic region of SENP2 is the most important region for SENP2 interaction with its substrate NEMO56, suggesting that noncatalytic regions can play important roles in substrate recognition. Another study showed that the N-terminus of SENP2 influences the SUMO paralogue specificity of the enzyme, as full-length SENP2 displayed different activities than its catalytic domain45. Most in vitro studies of Ulp and SENP enzymes have employed only catalytic domain fragments, and although challenging, more studies using full-length proteins are needed. The noncatalytic regions of Ulp and SENP enzymes may also bind accessory proteins that influence substrate specificity, activity and/or localization of the protease. Similarly, the noncatalytic regions of the DeSI enzymes and USPL1 are also likely to have unappreciated functions.
Many questions remain about STUbLs and their relationship to SUMO proteases. Does sumoylation of STUbL substrates have a function prior to their recognition and ubiquitylation by STUbLs? One attractive idea is that changes in the length of a substrate-linked SUMO chain can convert a protein for which sumoylation yields a particular functional activity into a STUbL substrate, triggering the end of the process that depended on the initial SUMO modification. Changes in SUMO protease activity could affect such a system. The existence of proteasome substrates that are modified by both SUMO and ubiquitin raises several interesting questions [FIG. 5]. For example, does a SUMO protease act on these proteins before or during proteasome recognition of the substrate? Do any SUMO proteases physically associate with the proteasome? Although the site of ubiquitin conjugation on most STUbL substrates is unclear, there is evidence that STUbLs can add ubiquitin directly to the SUMO or SUMO chain moieties of these substrates114, 115. Processing of such substrates might involve a protease that cleaves between SUMO and ubiquitin, and such an activity has been suggested for the yeast protein Wss1, a metalloprotease that has been genetically linked to both the SUMO and STUbL pathways115, 116. We can expect that many of these central questions will begin to be answered over the next few years.
Box 1 | SUMO-interacting motifs.
A SUMO-interacting motif (SIM) is a short sequence element (less than 10 amino acids), in some cases including phosphorylated amino acids, that is found in many proteins and that interacts with a specific surface groove of SUMO10. Although the properties of all SIMs are conserved, consensus sequences for given SUMO paralogue-SIM interactions are only beginning to emerge117. Most SIMs that have been identified to date have two important elements, a core of 3–4 hydrophobic residues (usually valine or isoleucine) and a nearby acidic region, which can be glutamate or aspartate side chains or phosphorylated serines or threonines. The hydrophobic residues of the SIM bind to the β2 strand of SUMO and extend the β-sheet of the β-grasp fold; the acidic residues bind a nearby basic patch on the SUMO surface, reinforcing the interaction and orientation of the SIM (parallel or anti-parallel to the β2 strand)118, 119. Although the SIM is the best-studied motif that binds SUMO, a variant SUMO-binding motif has recently been described120, and others are likely to be discovered.
Acknowledgements
We thank Christian Schlieker and Jennifer Gillies for their helpful comments on the manuscript. We also acknowledge support from NIH grants GM046904 and GM053756 to M.H., an NIH NRSA postdoctoral fellowship (F32 GM097794) to C.M.H., and an NSF predoctoral fellowship to N.R.W.
Glossary
- nuclear pore complex
Large multiprotein complex that forms a channel in the nuclear envelope of an eukaryotic cell, joining the inner and outer nuclear membranes and allowing transport of proteins to and from the nucleus.
- Septin
Highly conserved protein family first identified in yeast and more recently found in a wide range of animal cells. They are thought to function primarily in the control of cytokinesis in yeast, where they form a 10-nm filamentous ring that encircles the bud neck.
- ubiquitin-proteasome system
A system of selective, ATP-dependent protein degradation, in which target proteins that have been conjugated by ubiquitin are degraded by the 26S proteasome.
- histone deacetylase
An enzyme that removes the acetyl groups of core histones; its activity has an important function in transcriptional regulation and cell-cycle progression through alterations in chromatin structure.
- polycomb-related repressors
A family of proteins that can remodel chromatin and silence genes. This protein family was first discovered in Drosophila melanogaster.
- homologous recombination
A DNA recombination pathway, which includes the repair of double-strand DNA breaks, that uses a homologous double-stranded DNA molecule as a template for the repair of the broken DNA.
- Centromeres
Region of a chromosome that is attached to the spindle during nuclear division.
- mitochondrial fission
Mitochondrial membrane constriction and scission that promotes fragmentation of the mitochondrial network. The process is highly regulated and together with the opposing process of mitochondrial fusion (joining) is responsible for the dynamics observed for the mitochondrial network.
- response element
A short sequence of DNA within the promoter region of a gene that binds specific DNA-binding transcription factors, thus impacting transcription of the (typically adjacent) gene.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–9. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–51. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 3.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–8. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 4.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–10. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 5.Wykoff DD, O'Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci U S A. 2010;107:17415–20. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:16512–7. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matic I, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39:641–52. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Bruderer R, et al. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–8. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–71. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517–20. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 13.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–51. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 14.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20:2367–77. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–95. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–9. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 17.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 18.Shin EJ, et al. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 2012;13:339–46. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz S, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012 doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 21.Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3:1440–50. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza HM, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–43. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 23.Gan-Erdene T, et al. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 24.Wu K, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–91. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 25.Li SJ, Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol. 2003;160:1069–81. doi: 10.1083/jcb.200212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroetz MB, Su D, Hochstrasser M. Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol Biol Cell. 2009;20:2196–206. doi: 10.1091/mbc.E08-10-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–77. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 28.Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–6. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- 29.Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423–7. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D, et al. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J Cell Biol. 2006;174:939–49. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–76. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 32.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12:1519–31. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Reverter D, Lima CD. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol. 2006;13:1060–8. doi: 10.1038/nsmb1168. [DOI] [PubMed] [Google Scholar]
- 34.Shen L, et al. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol. 2006;13:1069–77. doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J. 2006;397:279–88. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang DT, Schulman BA. Breaking up with a kinky SUMO. Nat Struct Mol Biol. 2006;13:1045–7. doi: 10.1038/nsmb1206-1045. [DOI] [PubMed] [Google Scholar]
- 37.Lima CD, Reverter D. Structure of the human SENP7 catalytic domain and poly-SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem. 2008;283:32045–55. doi: 10.1074/jbc.M805655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alegre KO, Reverter D. Swapping small ubiquitin-like modifier (SUMO) isoform specificity of SUMO proteases SENP6 and SENP7. J Biol Chem. 2011;286:36142–51. doi: 10.1074/jbc.M111.268847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suh HY, et al. Crystal structure of DeSI-1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins. 2012;80:2099–104. doi: 10.1002/prot.24093. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, et al. Structural analysis of papain-like NlpC/P60 superfamily enzymes with a circularly permuted topology reveals potential lipid binding sites. PLoS One. 2011;6:e22013. doi: 10.1371/journal.pone.0022013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–20. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi Y, Mizoi J, Toh EA, Kikuchi Y. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J Biochem. 2000;128:723–5. doi: 10.1093/oxfordjournals.jbchem.a022807. [DOI] [PubMed] [Google Scholar]
- 43.Elmore ZC, et al. Sumo-dependent substrate targeting of the SUMO protease Ulp1. BMC Biol. 2011;9:74. doi: 10.1186/1741-7007-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panse VG, Kuster B, Gerstberger T, Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol. 2003;5:21–7. doi: 10.1038/ncb893. [DOI] [PubMed] [Google Scholar]
- 45.Kolli N, et al. Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J. 2010;430:335–44. doi: 10.1042/BJ20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Bacco A, et al. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–98. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hattersley N, Shen L, Jaffray EG, Hay RT. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol Biol Cell. 2011;22:78–90. doi: 10.1091/mbc.E10-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishida T, Kaneko F, Kitagawa M, Yasuda H. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J Biol Chem. 2001;276:39060–6. doi: 10.1074/jbc.M103955200. [DOI] [PubMed] [Google Scholar]
- 49.Kim KI, et al. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J Biol Chem. 2000;275:14102–6. doi: 10.1074/jbc.275.19.14102. [DOI] [PubMed] [Google Scholar]
- 50.Kadoya T, et al. Desumoylation activity of Axam, a novel Axin-binding protein, is involved in downregulation of beta-catenin. Mol Cell Biol. 2002;22:3803–19. doi: 10.1128/MCB.22.11.3803-3819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu S, et al. Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1. Mol Cell. 2009;33:570–80. doi: 10.1016/j.molcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Z, Au SW. Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem J. 2005;386:325–30. doi: 10.1042/BJ20041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bawa-Khalfe T, Cheng J, Wang Z, Yeh ET. Induction of the SUMO-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J Biol Chem. 2007;282:37341–9. doi: 10.1074/jbc.M706978200. [DOI] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Induction of SENP1 in endothelial cells contributes to hypoxia-driven VEGF expression and angiogenesis. J Biol Chem. 2010;285:36682–8. doi: 10.1074/jbc.M110.164236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–95. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MH, Mabb AM, Gill GB, Yeh ET, Miyamoto S. NF-kappaB induction of the SUMO protease SENP2: A negative feedback loop to attenuate cell survival response to genotoxic stress. Mol Cell. 2011;43:180–91. doi: 10.1016/j.molcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuo ML, den Besten W, Thomas MC, Sherr CJ. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 2008;7:3378–87. doi: 10.4161/cc.7.21.6930. [DOI] [PubMed] [Google Scholar]
- 58.Huang C, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–62. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan S, et al. Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90. EMBO J. 2010;29:3773–86. doi: 10.1038/emboj.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–19. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- 61.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 62.Pinto MP, et al. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochim Biophys Acta. 2012;1823:1958–66. doi: 10.1016/j.bbamcr.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Truong K, Lee TD, Chen Y. Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J Biol Chem. 2012;287:15154–63. doi: 10.1074/jbc.M112.353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Z, et al. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 2008;22:127–37. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 65.Kang X, et al. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hwang EJ, et al. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem Biophys Res Commun. 2009;378:513–7. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- 67.Alm-Kristiansen AH, Norman IL, Matre V, Gabrielsen OS. SUMO modification regulates the transcriptional activity of FLASH. Biochem Biophys Res Commun. 2009;387:494–9. doi: 10.1016/j.bbrc.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 68.Witty J, Aguilar-Martinez E, Sharrocks AD. SENP1 participates in the dynamic regulation of Elk-1 SUMOylation. Biochem J. 2010;428:247–54. doi: 10.1042/BJ20091948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindberg MJ, Popko-Scibor AE, Hansson ML, Wallberg AE. SUMO modification regulates the transcriptional activity of MAML1. FASEB J. 2010;24:2396–404. doi: 10.1096/fj.09-149401. [DOI] [PubMed] [Google Scholar]
- 70.Murata T, et al. Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J Biol Chem. 2010;285:23925–35. doi: 10.1074/jbc.M109.095356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaikkonen S, Makkonen H, Rytinki M, Palvimo JJ. SUMOylation can regulate the activity of ETS-like transcription factor 4. Biochim Biophys Acta. 2010;1799:555–60. doi: 10.1016/j.bbagrm.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Choi HK, et al. Reversible SUMOylation of TBL1-TBLR1 regulates beta-catenin-mediated Wnt signaling. Mol Cell. 2011;43:203–16. doi: 10.1016/j.molcel.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 73.Shimshon L, et al. SUMOylation of Blimp-1 promotes its proteasomal degradation. FEBS Lett. 2011;585:2405–9. doi: 10.1016/j.febslet.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 74.Cong L, Pakala SB, Ohshiro K, Li DQ, Kumar R. SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function. J Biol Chem. 2011;286:43793–808. doi: 10.1074/jbc.M111.267237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang TH, Xu S, Tailor P, Kanno T, Ozato K. The Small Ubiquitin-like Modifier-Deconjugating Enzyme Sentrin-Specific Peptidase 1 Switches IFN Regulatory Factor 8 from a Repressor to an Activator during Macrophage Activation. J Immunol. 2012 doi: 10.4049/jimmunol.1201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–92. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–7. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 78.Li X, et al. SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 2008;15:739–50. doi: 10.1038/sj.cdd.4402303. [DOI] [PubMed] [Google Scholar]
- 79.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–91. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 80.Leisner C, et al. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr Biol. 2008;18:1249–55. doi: 10.1016/j.cub.2008.07.091. [DOI] [PubMed] [Google Scholar]
- 81.Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwartz DC, Felberbaum R, Hochstrasser M. The Ulp2 SUMO protease is required for cell division following termination of the DNA damage checkpoint. Mol Cell Biol. 2007;27:6948–61. doi: 10.1128/MCB.00774-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee MT, Bakir AA, Nguyen KN, Bachant J. The SUMO isopeptidase Ulp2p is required to prevent recombination-induced chromosome segregation lethality following DNA replication stress. PLoS Genet. 2011;7:e1001355. doi: 10.1371/journal.pgen.1001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–82. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 85.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–7. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 86.Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–92. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panse VG, et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–21. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–9. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yun C, et al. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–95. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–78. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–15. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 93.Nissan TA, et al. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 94.Krogan NJ, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–39. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 95.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–5. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 96.Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–88. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 97.Zunino R, Braschi E, Xu L, McBride HM. Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J Biol Chem. 2009;284:17783–95. doi: 10.1074/jbc.M901902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Prudden J, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–12. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie Y, et al. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–84. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- 101.Uzunova K, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–75. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 102.Plechanovova A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–9. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012 doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abed M, et al. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011;30:1289–301. doi: 10.1038/emboj.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin Y, et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–95. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dou H, Huang C, Singh M, Carpenter PB, Yeh ET. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell. 2010;39:333–45. doi: 10.1016/j.molcel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Z, Jones GM, Prelich G. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics. 2006;172:1499–509. doi: 10.1534/genetics.105.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mullen JR, Das M, Brill SJ. Genetic Evidence that Poly-sumoylation Bypasses the Need for a SUMO-targeted Ub Ligase. Genetics. 2010 doi: 10.1534/genetics.110.124347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol. 2009;29:1694–706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–8. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Felberbaum R, Wilson NR, Cheng D, Peng J, Hochstrasser M. Desumoylation of the endoplasmic reticulum membrane VAP family protein Scs2 by Ulp1 and SUMO regulation of the inositol synthesis pathway. Mol Cell Biol. 2012;32:64–75. doi: 10.1128/MCB.05878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sydorskyy Y, et al. A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol Cell Biol. 2010;30:4452–62. doi: 10.1128/MCB.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mullen JR, Brill SJ. Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates. J Biol Chem. 2008;283:19912–21. doi: 10.1074/jbc.M802690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mullen JR, Chen CF, Brill SJ. Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. Mol Cell Biol. 2010;30:3737–48. doi: 10.1128/MCB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Biggins S, Bhalla N, Chang A, Smith DL, Murray AW. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–70. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Namanja AT, et al. Insights into high affinity small ubiquitin-like modifier (SUMO) recognition by SUMO-interacting motifs (SIMs) revealed by a combination of NMR and peptide array analysis. J Biol Chem. 2012;287:3231–40. doi: 10.1074/jbc.M111.293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–9. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 119.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–27. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 120.Rendtlew Danielsen J, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–87. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]