Abstract
New cases of leprosy are still being detected in Colombia after the country declared achievement of the WHO defined ‘elimination’ status. To study the ecology of leprosy in endemic regions, a combination of geographic and molecular tools were applied for a group of 201 multibacillary patients including six multi-case families from eleven departments. The location (latitude and longitude) of patient residences were mapped. Slit skin smears and/or skin biopsies were collected and DNA was extracted. Standard agarose gel electrophoresis following a multiplex PCR-was developed for rapid and inexpensive strain typing of M. leprae based on copy numbers of two VNTR minisatellite loci 27-5 and 12-5. A SNP (C/T) in gyrA (SNP7614) was mapped by introducing a novel PCR-RFLP into an ongoing drug resistance surveillance effort. Multiple genotypes were detected combining the three molecular markers. The two frequent genotypes in Colombia were SNP7614(C)/27-5(5)/12-5(4) [C54] predominantly distributed in the Atlantic departments and SNP7614 (T)/27-5(4)/12-5(5) [T45] associated with the Andean departments. A novel genotype SNP7614 (C)/27-5(6)/12-5(4) [C64] was detected in cities along the Magdalena river which separates the Andean from Atlantic departments; a subset was further characterized showing association with a rare allele of minisatellite 23-3 and the SNP type 1 of M. leprae. The genotypes within intra-family cases were conserved. Overall, this is the first large scale study that utilized simple and rapid assay formats for identification of major strain types and their distribution in Colombia. It provides the framework for further strain type discrimination and geographic information systems as tools for tracing transmission of leprosy.
Keywords: Leprosy, Mycobacterium leprae, SNP, gyrA, SNP7614, VNTR
1. Introduction
Leprosy is an infectious disease caused by the pathogen Mycobacterium leprae whose genome sequence was elucidated in 2001 (Cole et al., 2001). The narrow host range may be a consequence of the massive reductive genome evolution (Cole et al., 2001). Molecular methodologies for genotyping clinical strains of M. leprae have rapidly advanced in recent years. Two types of strain typing markers- variable number of tandem repeats (VNTRs) and single nucleotide polymorphisms (SNPs) that provide a range of resolution have been discovered and applied for different epidemiological questions (Groathouse et al., 2004; Zhang et al., 2005; Shin et al., 2000; Kimura et al., 2009; Srisungnam et al., 2009; Monot et al., 2005; Truman et al 2011). These applications include identification of family and community transmission clusters of leprosy and their evolution in geographic space, time and ethnicity (Weng et al., 2011; Sakamuri et al., 2009), the origin and global spread of leprosy (Matsuoka et al., 2006; Monot et al. 2005), the surveillance of drug resistance (Maeda et al., 2001; Singh et al., 2011) and detection of clusters of resistant cases ( Li et al., 2011), distinction of relapse from re-infections and mixed infections (da Silva Rocha et al., 2011; Sakamuri et al., 2009; Weng et al., 2011), and for obtaining evidence for zoonosis ( Truman et al., 2011) and ‘reverse zoonosis or anthoponosis’ (Suzuki et al., 2010).
At a global level, it has been hypothesized that leprosy originated in Central Asia/East Africa and migrated with humans over the centuries (Monot et al., 2005). This model was based on three M. leprae SNPs that produce four genotypes (SNP type 1, 2, 3 and 4). SNP type 2 is thought to be the progenitor of SNP type 1 strains that migrated to the East, and types 3 and 4 that are found in the New World in the west, dispersed from Europe and Africa respectively. Further global sampling and SNP discovery have led to the revisions of this model to account for discrepancies such as the predominant identification of type 3 and not type 1 in China and Japan in Asia; the four SNP types have since been expanded to 16 subtypes (Weng et al., 2007; Monot et al., 2009). While examining M. leprae VNTRs strain types in Colombia and Brazil (Cardona-Castro et al., 2009; Fontes et al., 2009), and their association with SNP types, we observed that the allele combination for the loci 27-5 and 12-5 was 4-5 in type 3 strains, while the combination was 5-4 most everywhere else (in all four SNP types ) except in China and Japan where it is predominantly 5-3 in SNP type 3 strains (Weng et al., 2007). This reversal of 27-5 and 12-5 alleles suggested that the SNP type 4s in S. America were not direct descendants of the local SNP type 3, but diversified at some point from the rest of the global strains (Cardona-Castro et al., 2009; Fontes et al., 2009) of types 1, 2 and 3. In the Philippines, two VNTR markers (GGT) 5 and 21-3 were sufficient to separate the SNP type 1 and type 3 strains (Sakamuri et al, 2009). Thus even a few VNTR markers can be informative of local subtypes.
In accordance with the possible separation of the American type 3 indicated by alternative 27-5 and 12-5 alleles, a second type of genomic marker which was identified during the course of drug resistance surveillance by us and others (da Silva Rocha et al., 2011; Truman et al., 2011) appeared to be associated with such type 3 strains. This marker is a C/T SNP in the gyrA gene in codon 99; the ‘T’ allele was found only in American SNP type 3 strains while the ‘C’ allele was found in all other regions and types including SNP type 4. This gyrA SNP along with many other SNPs was originally classified as ‘non-informative’ for the purposes of delineating phylogenetic relationships (Monot et al., 2005). However, in more recent work, the same gyrA SNP (denoted henceforth as SNP7614 to indicate position in M. leprae genome) and other SNPs were mapped which demonstrated that indeed the American type 3s are a branch of M. leprae which separated from all other extant global strains (Truman et al., 2011).
In this context, due to the evolving nature of the field of molecular epidemiology of leprosy, a number of genotyping platforms can be utilized to gain insight into the M. leprae genetic landscape. There are as many as 40 VNTR loci, and several hundred SNPs that are available for mapping. However, there would certainly be redundant information in the application of all markers, which is also neither feasible nor affordable in most settings, and may not be required until specific questions germane to local settings and control programs are to be addressed. The incidence rates, the origins of populations, and the history of leprosy differ from region to region and these can be reflected in the VNTR and SNP allelic diversities. Therefore, in this study, a simple low–cost three locus strain typing system composed of the SNP7614 in gyrA and two minisatellite VNTRs 27-5 and 12-5 was explored to verify previous findings, identify additional variants, and when dove-tailed with an ongoing drug resistance surveillance initiative (WHO, 2009), it produced the first formal molecular epidemiology database for a large set of geo-referenced leprosy patients from eleven endemic departments of Colombia. The approaches, findings and implications are presented.
2. Materials and Methods
2.1. Study sites, patients and clinical samples
The national and department level leprosy control programs of Colombia maintain a register of treated and in-treatment patients including those undergoing diagnostic procedures for leprosy. The officials in a number of departments were consulted to identify patients for inclusion in various research studies conducted by Instituto de Colombiano de Medicina Tropical (ICMT), from 2004 until 2011. Attempts were made to contact such individuals for possible inclusion in research projects. Individuals above age of 4 years (in some projects the minimal age was higher; minors required consent of guardians) and those consenting to participate or provide specimens for the research studies were included. These studies were approved by the local human research ethical committee in accordance with the Helsinki Ethical principles. Volunteers who participated in each study signed a consent form and authorized the use of biological samples for other future analyses.
In this molecular study, patients from the departments in the Andean region, and the Atlantic Coast were included. Bacteriological index (BI) measurements and Ridley-Jopling (RJ) clinical classifications were available at the time of initial diagnosis in some but not all patients as these procedures are not standard diagnostic protocols at all primary health centers (Ridley and Jopling, 1966). For the purposes of chemotherapy, the WHO, (WHO, 1982) system of clinical classification determined the patients as paucibacillary (PB) or multibacillary (MB) (http://www.who.int/lep/). Skin biopsies and/or lymph (a 50-200 μl volume of fluid exuded from pricking the ear lobes, lesion sites, elbows, and/or knees with a sterile lancet) were obtained from consenting patients by ICMT researchers at the time of enrollment. Smears were prepared for acid fast staining towards measuring BI and residual material was stored in 70% ethanol. The BI measurements were conducted at ICMT. BI positive patient specimens were selected for DNA extraction for two purposes, i.e., drug resistance surveillance and strain typing.
2.2. DNA extraction
Total DNA was extracted from skin biopsies or lymph by DNeasy kit (Qiagen,Valenicia, CA) as described previously (Kimura et al., 2009).
2.3. Multiplex PCR for drug resistance determining regions (DRDRs) and Restriction Fragment Length Polymorphism (RFLP) mapping of SNP7614 in gyrA
Four sets of primers (Table 1) were combined in a multiplex PCR (20 μl final volume; 0.25μM final concentration of each of the eight primers) in order to amplify drug resistance determining regions (DRDRs) in rpoB, folP1, gyrA and gyrB (Li et al., 2011). To distinguish the SNP7614 ‘C’ or ‘T’ allele within the gyrA DRDR amplicon, but not related to drug resistance phenotype, a portion (5 μl) of the multiplex DRDR PCR was incubated with FspI (New England Biolabs, USA), and NEBuffer 4 in a reaction volume of 10 μl. The sample was incubated at 37°C for 2 hours.
Table 1.
Primers for amplification of drug resistance targets and minisatellite VNTR loci
| Target | Amplicon size (bp) | Primer | Primer location | Primer sequence (5′ to 3′) |
|---|---|---|---|---|
| rpoB | 386 | rpoB-U | 2275587–2275568 | CAGGACGTCGAGGCGATCAC |
| rpoB-L | 2275202–2275218 | TCGTCAGCGGTCAAGTA | ||
| folP1 | 281 | folP1-U | 296746–296765 | TTCGTTCTCAGATGGCGGAC |
| folP1-L | 297026–297007 | GCCCACCAGACACATCGTTG | ||
| gyrA | 158 | gyrA-U | 7521–7539 | CCGTAGCCACGCTAAGTCA |
| gyrA-L | 7678–7660 | CCGGCGAACCGAAATTGCC | ||
| gyrB | 186 | gyrB-U | 6579–6602 | ACTGATCCTCGAAGTTCTGAACTG |
| gyrB-L | 6764–6749 | CAATGCCGTAATAATTTGCTTGAA | ||
| 27-5 | 327 (5 copies) | 27-5-U | 686939–686957 | ATTGAGCAGATGGCCGGTC |
| 27-5-L | 687265–687248 | AGCAGTCGGCACGCCCTT | ||
| 12-5 | 289 (5 copies) | 12-5-U | 1381580–1381598 | CTGGTCCACTTGCGGTACGAC |
| 12-5-L | 1381868–1381849 | GGAGAAGGAGGCCGAATACA |
The amplicon size and primer nucleotide positions are from the M. leprae TN strain as found in the Leproma website (http://genolist.pasteur.fr/Leproma/) and per Li et al., 2011 and Kimura et al., 2009.
The products were separated by electrophoresis in a 2% agarose gel in 1x TAE buffer. The product sizes were compared against those obtained from the NHDP63 reference DNA which has the SNP7614 ‘T’ allele instead of ‘C’ and is thus not restricted by FspI whose recognition sequence is TGC-GCA (the SNP ‘C’ allele is indicated in bold). The 20 bp DNA sizing standards were used for DNA length estimation (Bio-Rad, USA). Single locus PCRs were attempted if one of the products was not generated.
2.4. Multiplex PCR for VNTRs 27-5 and 12-5
The multiplex PCR (20 μl) included primers (Table 1) for 27-5 and 12-5 loci (0.2 μM final concentration each) and Qiagen Multiplex PCR master mix (Qiagen, USA). The VNTR multiplex PCR conditions were 15 min at 94°C for enzyme activation, and 40 thermal cycles of 94°C for 30 sec, 60°C for 120 sec and 72°C for 90 sec. A final extension step of 72°C for 10 min was included (Kimura et al., 2009). The NHDP63 strain of M. leprae was used as a PCR positive control and for detecting the reference alleles for 27-5 and 12-5 loci (4 and 5 copies respectively) (Kimura et al., 2009). The alleles of the VNTR loci could be determined after separation of the PCR products by agarose gel electrophoresis and ethidium bromide staining of DNA. VNTR stuttering (generation of -1 or +1 repeats) during replication can occur during PCR or in vivo (Kimura et al., 2009). For allele determination, the dominant product was selected.
2.5. Geo-referencing methods
Patient residence location (latitude and longitude) was recorded by Garmin eTrex Vista HCx instrument in decimal degrees units system. The coordinates were plotted on freely accessible Google Earth software (earth.google.com) and visualized using Google Maps (maps.google.com). Maps depicting patient locations and their M. leprae genotypes were created in Google Maps.
2.6. Statistical Analysis
Statistical Package for the Social Sciences, SPSS software version 15.0.1 (SPSS Inc, Chicago, IL) was used for data processing. Chi-squared distribution test was used for comparisons of qualitative variables between groups.
3. Results and Discussion
3.1. Geo-referenced patient cohort
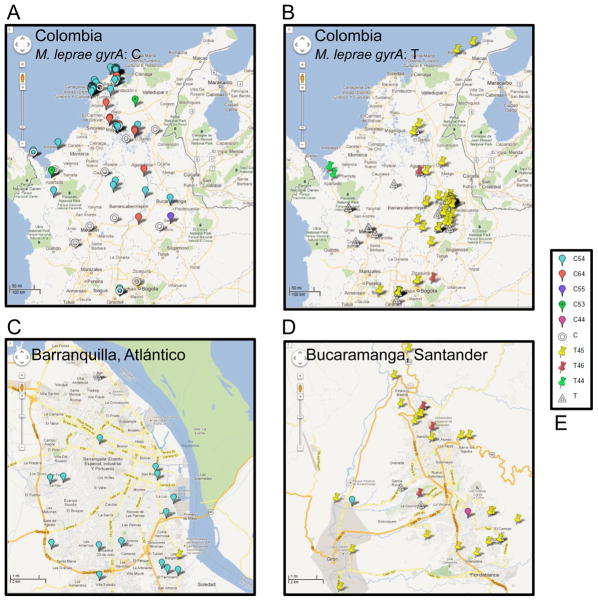
The new case detection rate per year is in the range of 300-400 nationally. ICMT has contacted the local leprosy control offices in one or more departments at different periods over a period of 7 years. The control program does not routinely collect samples from each patient for research. There are finite resources for research personnel to travel to the departments, arrange for patient recruitment and sample collection. For molecular studies, biological specimens such as lymph (i.e., from slit skin samples) or biopsies were collected in ethanol. In order to conserve molecular reagents and clinical materials, a subset of these samples (from BI positive patients at the time of sample collection) was selected for DNA extraction and M. leprae specific target amplifications because PCR positivity is related to the detection of bacilli in skin samples. Considering the logistics, and remaining biological sample availability in the repository from BI positive patients, the sample set of 201 from patients in eleven departments (Guajira, Cesar, Magdalena, Atlántico, Bolívar, Norte de Santander, Santander, Cundinamarca, Tolima, Antioquia and Chocó) that was compiled for the current molecular studies to represent the Colombian M. leprae diversity is reasonable. The geographical locations of the patients mapped on Google Earth and Google Maps are represented in Figure 1 (1A and 1B). The southern departments are under-represented as the leprosy control program does not report cases in these regions and are not shown in Figure 1. Furthermore there was limited access to public health posts and contacts in these departments with clinicians from ICMT.
Figure 1. The geographical origins of the patients, the differential distribution of the gyrA SNP7614 ‘C’ and ‘T’ alleles, and the fine mapping of the three locus genotypes in Colombia.
A: Map depicting the distribution of patients with the M. leprae gyrA ‘C’ allele. B: Map depicting the distribution of patients with the M. leprae gyrA ‘T’ allele. The Panels A and B have a scale bar of 50 miles. C: Zoomed in view of the strain types in Barranquilla, Atlántico. D: Zoomed in view of the strain types in Bucaramanga, Santander. The Panels C and D have a scale bar of 1 mile. E. The key for the place-marks representing different genotypes. The empty donut and empty triangle indicate that alleles for one or both of the VNTRs could not be determined for the gyrA C or T type isolates.
3.2. Detection of a polymorphism in M. leprae gyrA in Colombian isolates
Drug resistance surveillance was one of the research objectives that contributed to the development of the patient cohort and sample repository. In this regard, four gene targets were amplified by single and/or multiplex PCR- rpoB, folP1, gyrB and gyrA regions involved in resistance to rifampicin, dapsone and ofloxacin respectively (Figure 2A) and sequenced (Hernandez et al., 2008, WHO 2010). During the course of these investigations, we noted that NHDP63, an armadillo-derived human isolate of M. leprae DNA that was routinely used as positive DNA control for PCR was found to have a nucleotide change in codon 99; CGT instead of the CGC seen in the sequenced TN strain (genolist.pasteur.fr/Leproma/), resulting in a silent mutation (Arg to Arg). The same gyrA ‘T’ variant was being detected in many of the Colombian M. leprae isolates.
Figure 2. Genetic mapping of Colombian M. leprae isolates.
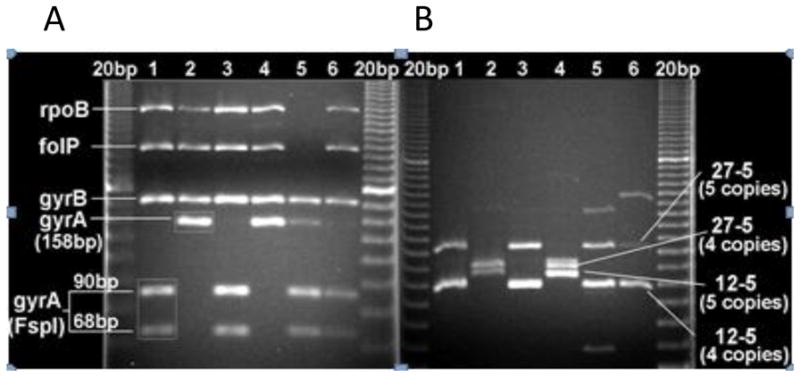
A: PCR-RFLP (FspI) for gyrA C/T (SNP 7614) after multiplex amplification of drug resistance target regions within rpoB, folP1, gyrB and gyrA genes. 20 bp DNA ladder is in the first and last lanes. gyrA in samples 1, 3, 5 and 6 were cut (90 and 68 bp products) indicating the presence of the ‘C’ allele, but remained intact (158bp) in samples 2 and (‘T’ allele). B: Multiplex PCR for the 27-5 and 12-5 VNTR loci. Samples 1, 3, 5 and 6 have 5 copies respectively, while 2 and 4 have 4 copies.
3.3. A Novel rapid PCR-RFLP assay for the gyrA SNP
To utilize this new gyrA SNP as a genetic typing marker and to expedite the screening, a PCR-RFLP assay was designed. The restriction enzyme FspI recognizes the ‘C’ but not the ‘T’ allele; (Figure 2A) and fortuitously does not cut within the rpoB, folP1 and gyrB amplicons. Therefore FspI digestion could be performed directly on the multiplexed PCR products (Figure 2A). The gyrA amplicons were sequenced before we developed the FspI methodology for SNP7614. Seventy one of the 95, ‘C’ type samples were determined by DNA sequence and 57 by FspI. Thirty three samples were mapped by both methods and shown to be concordant. Seventy four of the 118, ‘T’ type samples were sequenced and 56 were determined by FspI-RFLP. Twelve samples were evaluated by both methods showing concordance. Based on these results, the PCR-RFLP is useful for rapid mapping of the SNP7614 and in resource limited settings. In most samples assigned the ‘C’ allele, more than 90% of the amplicon was restricted. Partial digestion may be related to amplicon excess or insufficient enzyme or incubation time. Only two samples showed partial cutting (less than 5-10%of the starting amplicon); these were assigned the ‘T’ allele. Low level of co-infections and PCR inhibitions are possible; the dominant RFLP pattern was considered for allele assignment. Users of this method can consider including an unrelated template with an FspI site in the PCR as an internal control.
3.4. Relationship between SNP7614 with geographical location, and the development of a three locus genetic mapping system including VNTR alleles of 27-5 and 12-5 loci
With just one marker, the SNP 7614, and a simple, first level classification of patients as belonging to Andean or Atlantic departments, a robust snap shot of the strain distribution pattern in Colombia was revealed (Supplementary Tables 1A, 1B and 1C). The strains with the allele ‘C’ are significantly associated with the Atlantic region compared to those with the ‘T’ allele (odds ratio [OR], 32.97; P, 1X10-7; confidence interval [CI],13.7-81.52; Chi2 Yates corrected, 93.62). There were only few exceptions to the trend as shown in Supplementary Table 1C (shown in grey shading). A review of the residential histories along with the exact location of the municipalities where the patients were enrolled, verified that 16 of 26 had lived in the alternative region where infection could have occurred before diagnosis or in municipalities bordering the two regions. In some departments, such as Antioquia, there are cities along the small Atlantic coastline portion, which account for three of the Turbo patients with the ‘C’ type SNP. Likewise, there are municipalities in the Atlantic departments that have contact with the Andean departments, such as along the ports of the river Magdalena which allows for movement of people and products, where the ‘T’ type allele was detected. Besides the 16, there are two patients from the leprosarium in the city of Agua de Dios, Cundinamarca, who have the ‘C’ type instead of the common ‘T’ type. For one such patient, the origin five years prior to residence in this city was not known, while the other lived in this city for 30 years.
Another interesting illustration showing benefit from first line SNP7614 typing relates to two patients (aunt and niece) in Guajira. These patients belong to an ethnic group named Wayúu, which accounts for a significant percent of the Guajira population. The members of this Amerindian ethnicity are known to migrate back and forth in Colombia and seasonally across the Colombian-Venezuelan borders. Incidentally, the SNP7614 was ‘T’ for these patients, which is the dominant allele in Venezuela (Singh et al., 2011).
Furthermore, when alleles of the three loci gyrA SNP7614, 27-5 and 12-5 were combined, a number of genotypes (denoted by an alphanumeric code for the alleles of the loci in that order) were detected as shown (Supplementary Table 2A). In conjunction with geographical mapping, a pattern of strain type distribution is obvious: the Atlantic coastal states Bolivar and Atlántico predominantly have the genotype C54 (Figure 1A), while the central Andean states (Santander, Antioquia, Cundinamarca, Tolima, Cesar and Norte de Santander) carry the T45 type of strains (Figure 1B). The association between the genotypes and geography is significant (Supplementary Tables 2B and 2C). The strains with T45 genotype have 21.9 times the odds as those of all other known genotypes of belonging to the Andean region of residence (P, 1x10-7; Chi2, 56.77; confidence interval [CI], 2.11-4.07). On the other hand, the strains with C54 genotype have 21.48 times the odds as those of all other known genotypes of belonging to the Atlantic region (P, 1x10-7, Chi2, 54.41; confidence interval [CI], 8.04-62.54). The river Magdalena that runs south to north appears to separate these major strain types, while a novel C64 genotype is found along this boundary. The department of Magdalena has all three genotypes. Of 13 samples with the C64 genotype, 9 were available for further analysis. Using the recently developed HRM assay for SNP typing they were found to be of type 1 (Li et al., 2012).
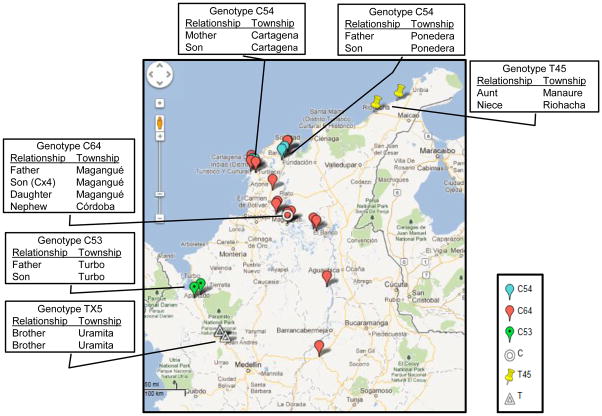
3.6. Genotypes of multifamily cases
Patients with known family relationships were linked by their three- locus M. leprae genotypes. The locations and compositions of six multi-case family clusters are shown in Figure 3. The multi-case family of C64 type is interesting because there appear to be additional cases outside the family in the same or neighboring townships (Figure 3). Besides the 27-5 with 6 copy allele, eight of the C64 isolates carried the one copy allele at 23-3, which is also rare in the VNTR typed global strains of M. leprae. One family in Turbo had the minor C53 genotype, while two of the four cases of another minor group C55 were identified in the town of Soledad near Barranquilla, Atlántico. The 27-5 and 12-5 alleles of 5 and 3 have been detected in Japan and China.
Figure 3.
Map of multi-case families and C64 genotypes
4. Conclusions
Leprosy is a neglected topical disease, with near stable global incidence rates even after implementation of multidrug therapy regimens and decentralized leprosy control programs. Therefore further investigations using informed surveillance strategies are required to curb transmission chains and networks as opposed to the passive case detection and treatment systems currently in practice. M. leprae genotyping technologies are rapidly advancing, but have yet to be formally integrated into control programs in many countries. In this regard, to gain a first broad level of insight into the distribution and types of the M. leprae strains, the molecular epidemiology effort described here, demonstrates that a simple three locus system is instructive and powerful for Colombia. It can be achieved by multiplex PCR-agarose gel electrophoresis mapping systems that do not require costly laboratory facilities or operator training. The sizes of the 27-5 and 12-5 amplicons on the gels reveal their alleles; while for the SNP7614 mapping, the Fsp1 based PCR-RFLP is a time saving and inexpensive technique. Strong associations between the SNP 7614 C/T and previously reported 27-5 and 12-5 loci were detected, while a number of other genotypes were revealed.
The modern Colombian human racial and ethnic composition reflects events of colonization by European (primarily Spanish) conquistadors, the introduction of African slaves (Klein et al., 2007, Rojas et al., 2010) and of late, immigration by Asians. These exogenous races admixed with and/or replaced the Native American Indians since the 15th century (Bedoya et al., 2006, Wang et al., 2008). Leprosy is believed to have been brought to the New World in the post –Colombian period by the recent historic events of colonialism and slavery. It can be seen that the two major strain types in Colombia, defined as C54 and T45 are segregated across the coastal and Andean states. This may mirror the primary routes of entry and settlement of the various populations in these regions. The strains in Colombia with minisatellite 27-5 and 12-5 alleles of 5 and 4 are frequently of the SNP type 4 as shown previously (Cardona-Castro et al., 2009; Fontes et al., 2009) and may be derived from African slaves, while those with 4 and 5 are of SNP type 3 as found in America which may have been brought by the Europeans (Spaniards, Portuguese and Italians) and diverged from those elsewhere in the world. To demonstrate further support, a set of 15 random samples with the C54 genotype were mapped at SNP14,676 (one of the three SNP typing loci that distinguishes the type 4 from types 1,2 and 3) (Monot, et al., 2005) by PCR sequencing. The SNP14,676 ‘T’ allele which is restricted to the type 4, as first described for strains found in West Africa (Monot et al., 2005) was found in 14 of 15 samples. Monot et al proposed that type 4 spread to the new world by slave trade. Further subtyping of type 4 strains was not conducted at this stage. Note that, those of the C64 genotype were of type 1. On the other hand, eight strains with SNP7614 with ‘T’ allele were found to have the ‘C’ allele of SNP14,676, meaning they could be type 1, 2 or 3. Based on the literature that SNP7614 ‘T’ has been seen only in strains of North and South America associated with type 3 strains of subtype 3I (Monot et al., 2009, Truman et al.2011, Singh et al., 2011) and other data (global drug resistance surveillance, and our own sampling of Asian strains). In a study in S. America, 172 strains with the ‘T’ allele were shown to be of SNP 3 (subtype 3I), while those with ‘C’ allele were of type 4 (N, O and P) and 1D (Singh et al., 2011). The type 3 strains from Asia and other type strains (1, 2, and 4) have the ‘C’ allele, Thus, SNP7614 ‘T’ may be a strong predictor for the type 3 strains found in Colombia; in future studies, this relationship can be formally established. It would be interesting to explore demographic and historic relationships of the 3I, 1D and type 4 strains in Venezuelan patients with those found in Colombia.
The finding of the C64 genotypes as being SNP type 1 is novel. These C64 isolates are geographically contained along a tract and carry another unusual VNTR minisatellite marker which indicates that they are a phylogenetically and epidemiologically validated strain type. The type 1 detected mainly in Asia is presumed to be an off shoot of SNP type 2. Thus, the geographical and ethnic origins of the Colombian patients may be further explored, both retrospectively and prospectively, particularly for patients from the identified locations. It is interesting that the predominant strain types of M. leprae have not yet dispersed or mixed uniformly across Colombia. This may be related to the relatively recent modern history of Colombia and introduction of leprosy. Besides the Africans, immigrant populations have been noted to cities near the Magdalena river basin, such as from Lebanon from where SNP 1 may have been introduced to Colombia. SNP type 2s in the western hemisphere have been reported mainly in Mexico; restricted to the western states of Jalisco and Sinaloa (Matsuoka et al., 2009). The SNP7614 status of these strains is not known and the relationship of these SNP type 2s with the local Mexican type 3s and 4s has not been described. It was proposed that these strains may have a pre-Colombian introduction from Asia via the Bering Strait (Matsuoka et al., 2009). It would be interesting to understand this in the context of strains found in Mexico and S. America.
The southern and eastern states are under-represented in this study as very few leprosy cases are reported from these states into the database SIVIGILA (Sistema Nacional de Vigilancia en Salud Pública) managed by the government leprosy control program through the Colombian National Institute of Health (http://www.ins.gov.co/). However, proximity of several of these states to endemic Brazil and Venezuela suggest that there must be unreported leprosy in these areas as well. The common strain types detected in these neighboring countries are type 3 and 4, as in Colombia.
The current database is neither complete nor cumulative for the period studied since it is a product of several limited duration projects initiated by academic investigators. Due to the protracted incubation period of leprosy, prospective and long term efforts are necessary to unlock the full potential of molecular epidemiology. The identification of multi-case family and community clusters indicates delay in diagnosis and continued transmission; these situations can be addressed locally and nationally. Secondary and higher level genotyping should then be possible due to the growing number of VNTR and SNP markers (Kimura et al., 2009, Jensen et al., 2011). There are other alternatives for genotyping which are not dependent on DNA sequencing methods such as the high resolution melt-PCR (HRM-PCR); as applied here for the SNP typing of a subset of samples (Li et al., 2012) and SNP typing by PCR-RFLP (Sakamuri et al., 2009).
In Colombia, as in most countries, there has not been any formal effort by the leprosy control program to include molecular or spatial epidemiology in their operations. The utility of SNP7614 by itself, and in combination with minisatellites has provided a convenient primary and secondary level typing system, revealing interesting scenarios compatible with location and migration histories. The Colombian patients represent a cross-section of the leprosy cases, and there is no suggestion at this stage that all individuals with a shared three locus genotype are related through a single recent short range transmission scenario. However, it is reasonable that distant temporal or community associations exist because the history of recorded leprosy in Colombia is only a few centuries old. It is intended that this work, will foster prospective studies involving sample banking at the time of diagnosis. Technologies for strain typing such as next generation genome sequencing are evolving rapidly; however, these are not yet ready for implementation in regional clinical laboratories. On the other hand, it is possible to introduce the first line genotyping tests we have described. Applications include recognition of strain types from migrant individuals, better definition of primary contacts and distinction of relapse from reinfection. There is also, known human contact with armadillos in Colombia with possibility for zoonotic transmission, as has been demonstrated in N. America (Truman et al., 2011). In Colombia, wild armadillo strains have not been harvested, isolated or strain typed at multiple loci thus far. Locals are known to hunt the animals as a source of meat and blood for medicinal purposes, and for crafting containers and musical instruments. In more recent questionnaires, patients have been asked about their interactions with armadillos.
Overall, the hope is that molecular epidemiology will be recognized as a component of leprosy control programs at the population and individual levels to better understand the origin of new cases and retracing routes of transmission. More sophisticated genetic tests can be coordinated at national reference or research laboratories.
Supplementary Material
Supplementary Table 1: A: Distribution of SNP7614 ‘C’ and ‘T’ in Andean and Atlantic departments. B: Associations of SNP7614 with patient geographical region of origin, and C: Distribution of SNP7614 ‘C’ and ‘T’ according to city of origin.
Supplementary Table 2: A: Genotypes based on SNP7614 and VNTRs 27-5 and 12-5 in Andean and Atlantic departments, B: Association of the genotype T45 based on gyrA SNP7614 and VNTRs 27-5 and 12-5 with Andean region of origin, and C: Association of the genotype C54 based on gyrA SNP7614 and VNTRs 27-5 and 12-5 with Atlantic region of origin.
Highlights.
Affordable geographic and molecular tools for mapping leprosy strain types in Colombia.
With only three markers, multiple M. leprae genotypes were detected; two were predominant.
The dominant genotype in Atlantic region is gyrA SNP7614(C)/27-5(5)/12-5(4) [C54].
The dominant genotype is gyrA SNP7614 (T)/27-5(4)/12-5(5) [T45] in the Andean region.
A novel genotype gyrA SNP7614 (C)/27-5(6)/12-5(4) [C64] in Colombia is linked to SNP type 1
Spatial mapping shows limited overlap of transmission of major genotypes.
Acknowledgments
The work was possible through support from COLCIENCIAS Code 325651928980, COLCIENCIAS code 325649326207, and Instituto Colombiano de Medicina Tropical-Universidad CES (to NCC) and National Institutes of Health, USA, NIH AI063457 and AI-25469 (to VV and PJB). We sincerely appreciate the cooperation and coordination by staff and clinicians of the leprosy control programs and the support of the patients. We thank Rama Murthy Sakamuri for assistance with strain typing efforts in the laboratory and Jason Vander Heiden for assistance with geographic data analysis. Prof. Gabriel Bedoya is acknowledged for his tutorial support to NCC, and review of this work.
Abbreviations
- GIS
Global Information System
- GPS
Global Positioning System
- SNP
Single Nucleotide Polymorphism
- MLVA
Multiple Locus Variable Number of Tandem Repeat Analysis
- VNTR
Variable Number of Tandem Repeat Analysis
Footnotes
Competing interest
The authors declare they have no competing interests.
Authors’ contributions
JCBA and NCC are the first authors. The authors JCBA and IMRM performed the field work. JJCBA, IMRM, and WL performed the laboratory work and collected the data. NCC, PJB and VV designed the study. JCBA, NCC and VV analyzed the results. VV and NCC wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bedoya G, Montoya P, García J, Soto I, Gurgeois S, Carvajal L, Labuda D, Alvarez V, Ospina J, Hedrick PW, Ruiz-Linares A. Admixture dynamics in Hispanics: A shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona-Castro N, Beltrán-Alzate JC, Romero-Montoya IM, Meléndez E, Torres F, Sakamuri RM, Li W, Vissa V. Identification and comparison of Mycobacterium leprae genotypes in two geographical regions of Colombia. Lepr Rev. 2009;80:316–321. [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward SJ, Barrell BG. Massive gene decay in the leprosy bacillus. Nature. 2001;22:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Fontes AN, Sakamuri RM, Foschiani IM, Ura S, Rio MO, Brega A, Nunes E, Brennan PJ, Vissa DV, Suffys PN. Genetic diversity of Mycobacterium leprae isolates from Brazilian leprosy patients. Lepr Rev. 2009;80:302–315. [PubMed] [Google Scholar]
- Groathouse NA, Rivoire B, Kim H, Lee H, Cho SN, Brennan JP, Vissa VD. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J Clin Microbiol. 2004;42:1666–1672. doi: 10.1128/JCM.42.4.1666-1672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández E, Cardona-Castro N, Rodríguez G, Villegas S, Beltrán JC, Kimura M, Vissa V, Gómez Y. Estudio de resistencia a la rifampicina y la dapsona en tres pacientes con recurrencia de lepra. Rev Panam Salud Publica. 2008;23:73–77. doi: 10.1590/s1020-49892008000200001. [DOI] [PubMed] [Google Scholar]
- Jensen RW, Rivest J, Li W, Vissa V. DNA fingerprinting of Mycobacterium leprae strains using Variable Number Tandem Repeat (VNTR) – Fragment length Analysis (FLA) JoVE. 2011;53 doi: 10.3791/3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Sakamuri RM, Groathouse NA, Rivoire BL, Gingrich D, Krueger-Koplin S, Cho S, Brennan PJ, Vissa VD. Rapid Variable-Number Tandem-Repeat Genotyping for Mycobacterium leprae Clinical Specimens. J Clin Microbiol. 2009;47:1757–1766. doi: 10.1128/JCM.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HS, Vinson B. African Slavery in Latin America and the Caribbean. Oxford University Press; 2007. [Google Scholar]
- Li W, Sakamuri RM, Lyons DE, Orcullo FM, Shinde V, Dela Pena EL, Maghanoy AA, Mallari IB, Tan EV, Nath I, Brennan PJ, Balagon M, Vissa V. Transmission of dapsone-resistant leprosy detected by molecular epidemiological approaches. Antimicrob Agents Chemother. 2011;55:5384–5387. doi: 10.1128/AAC.05236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Matsuoka M, Kai M, Thapa P, Khadge S, Hagge DA, Brennan PJ, Vissa V. Real-time PCR and high-resolution melt analysis for rapid detection of Mycobacterium leprae drug resistance mutations and strain types. J Clin Microbiol. 2012;50:742–753. doi: 10.1128/JCM.05183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Matsuoka M, Nakata N, Kai M, Maeda Y, Hashimoto K, Kimura H, Kobayashi K, Kashiwabara Y. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob Agents Chemother. 2001;45:3635–3639. doi: 10.1128/AAC.45.12.3635-3639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Zhang L, Fafutis L, Legua P, Wiens C. Polymorphism in the rpoT gene in Mycobacterium leprae isolates obtained from Latin American countries and its possible correlation with the spread of leprosy. Fems Microbiol Lett. 2006;243:311–315. doi: 10.1016/j.femsle.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Gonzalez AV, Estrada I, Carreno-Martinez C, Fafutis-Morris M. Various genotypes of Mycobacterium leprae from Mexico reveal distinct Geographic distribution. Lepr Rev. 2009;80:322–326. [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Araoz R, Coppée JY, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Vimmond M, Flageul B, Cho SN, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamesipour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- Ridley RS, Jopling WH. Classification of leprosy according to immunity—a five group system. Int J Lepr. 1966;34:225–273. [PubMed] [Google Scholar]
- Rojas W, Parra MV, Campo O, Caro MA, Lopera JG, Arias W, Duque C, Naranjo A, García J, Vergara C, Lopera J, Hernandez E, Valencia A, Caicedo Y, Cuartas M, Gutiérrez J, López S, Ruiz-Linares A, Bedoya G. Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers. Am J Phys Anthropol. 2010;143:13–20. doi: 10.1002/ajpa.21270. [DOI] [PubMed] [Google Scholar]
- Sakamuri R, Kimura M, Li W, Kim HC, Lee H, Kiran MD, Black WC, Balagon M, Gelber R, Cho SN, Brennan PJ, Vissa DV. Population-based molecular epidemiology of leprosy in Cebu, Philippines. J Clin Microbiol. 2009;47:2844–2854. doi: 10.1128/JCM.02021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YC, Lee H, Walsh GP, Kim JD, Cho SN. Variable numbers of TTC repeats in Mycobacterium leprae DNA from leprosy patients and use in strain differentiation. J Clin Microbiol. 2000;38:4535–4538. doi: 10.1128/jcm.38.12.4535-4538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Rocha A, Cunha Dos Santos AA, Pignataro P, Nery JA, de Miranda AB, Soares DF, Brum Fontes AN, Miranda A, Ferreira H, Boéchat N, Novisck Gallo ME, Sarno EN, De Oliveira ML, Suffys PN. Genotyping of Mycobacterium leprae from Brazilian leprosy patients suggests the occurrence of reinfection or of bacterial population shift during disease relapse. J Med Microbiol. 2011;60:1441–1446. doi: 10.1099/jmm.0.029389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Busso P, Paniz-Mondolfi A, Aranzazu N, Monot M, Honore N, Fernandes AF, Virmond M, Villareal-Olaya ME, Rivas C, Cole ST. Molecular drug susceptibility testing and genotyping of Mycobacterium leprae strains from South America. Antimicrob Agents Chemother. 2011;55:2971–2973. doi: 10.1128/AAC.00201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisungnam S, Rudeeaneksin J, Lukebua A, Wattanapokayakit S, Pasadorn S, Mahotarn K, Ajincholapan, Sakamuri RM, Kimura M, Brennan PJ, Phetsuksiri B, Vissa DV. Molecular epidemiology of leprosy based on VNTR typing in Thailand. Lepr Rev. 2009;80:280–289. [PubMed] [Google Scholar]
- Suzuki K, Udono T, Fujisawa M, Tanigawa K, Idani G, Ishii N. Infection during infancy and long incubation period of leprosy suggested in a case of a chimpanzee used for medical research. J Clin Microbiol. 2010;48:3432–3434. doi: 10.1128/JCM.00017-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman RW, Singh P, Sharma R, Busso P, Rougemont J, Paniz-Mondolfi A, Kapopoulou K, Brisse S, Scollard DM, Gillis TP, Cole ST. Probable Zoonotic Leprosy in the Southern United States. N Engl J Med. 2011;364:1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro LM, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, Ruiz-Linarez A. Geographic Patterns of Genome Admixture in Latin American Mestizos. PLoS Genet. 2008;4(3):e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Wang Z, Liu J, Kimura M, Black WC, 4th, Brennan PJ, Li H, Vissa VD. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J Clin Microbiol. 2007;45:1728–1734. doi: 10.1128/JCM.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Heiden JV, Xing Y, Liu J, Vissa VD. Transmission of leprosy in Qiubei County, Yunnan, China: insights from an 8-year molecular epidemiology investigation. Infect Genet Evol. 2011;11:363–374. doi: 10.1016/j.meegid.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Chemotherapy of leprosy for control programmes. WHO; Geneva, Switzerland: 1982. WHO Technical Report Series. 768. [PubMed] [Google Scholar]
- World Health Organization. Guidelines for global surveillance of drug resistance in leprosy. WHO; India: 2009. http://www.searo.who.int/LinkFiles/Situation_1-Guidelines_GSDRL_GLP-09.pdf. [Google Scholar]
- Zhang L, Budiawan T, Matsuoka M. Diversity of potential short tandem repeats in Mycobacterium leprae an application for molecular typing. J Clin Microbiol. 2005;43:5221–5229. doi: 10.1128/JCM.43.10.5221-5229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: A: Distribution of SNP7614 ‘C’ and ‘T’ in Andean and Atlantic departments. B: Associations of SNP7614 with patient geographical region of origin, and C: Distribution of SNP7614 ‘C’ and ‘T’ according to city of origin.
Supplementary Table 2: A: Genotypes based on SNP7614 and VNTRs 27-5 and 12-5 in Andean and Atlantic departments, B: Association of the genotype T45 based on gyrA SNP7614 and VNTRs 27-5 and 12-5 with Andean region of origin, and C: Association of the genotype C54 based on gyrA SNP7614 and VNTRs 27-5 and 12-5 with Atlantic region of origin.