Abstract
Leprosy continues to be detected at near stable rates in China even with established control programs, necessitating new knowledge and alternative methods to interrupt transmission. A molecular epidemiology investigation of 190 patients was undertaken to define M. leprae strain types and discern genetic relationships and clusters in endemic and non-endemic regions spanning seventeen provinces and two autonomous regions. The findings support multiple locus variable number of tandem repeat (VNTR) analysis as a useful tool in uncovering characteristic patterns across the multiethnic and divergent geographic landscape of China. Several scenarios of clustering of leprosy from township to provincial to regional levels were recognized, while recent occupational or remote migration showed geographical separation of certain strains. First, prior studies indicated that of the four major M. leprae subtypes defined by single nucleotide polymorphisms (SNPs), only type 3 was present in China, purportedly entering from Europe/West/Central Asia via the Silk Road. However, this study revealed VNTR linked strains that are of type 1 in Guangdong, Fujian and Guangxi in southern China. Second, a subset of VNTR distinguishable strains of type 3, co-exist in these provinces. Third, type 3 strains with rpoT VNTR allele of 4, detected in Japan and Korea were discovered in Jiangsu and Anhui in the east and in western Sichuan bordering Tibet. Fourth, considering the overall genetic diversity, strains of endemic counties of Qiubei, Yunnan; Xing Yi, Guizhou; and across Sichuan in southwest were related. However, closer inspection showed distinct local strains and clusters. Altogether, these insights, primarily derived from VNTR typing, reveal multiple and overlooked paths for spread of leprosy into, within and out of China and invoke attention to historic maritime routes in the South and East China Sea. More importantly, new concepts and approaches for prospective case finding and tracking of leprosy from county to national level have been introduced.
Keywords: VNTR, Strain typing, Mycobacterium leprae, Leprosy, Transmission, Phylogeography, SNP typing, Maritime silk routes
1. Introduction
China is a large country, historically involved in international trade and migration through many centuries. Though the leprosy epidemic has been recorded since the time of Confucius (551-479 BCE), how leprosy spread into, within and from China are not completely resolved. Furthermore, as China has claimed elimination status for leprosy at the national level, active case finding efforts by mass, contact or group clue surveys have been either reduced or eliminated and a decentralized control program largely based on voluntary reporting and dermatological clinics has been adopted (Chen et al., 2001). The World Health Organization reported that 1324 new cases of leprosy cases detected in 2010, 85% of them being of the multibacillary form (WER, 2011). While, leprosy patients are still diagnosed nationwide, pockets of endemicity in the three ethnically diverse, mountainous and underdeveloped southwest provinces of Yunnan (YN), Guizhou (GZ) and Sichuan (SC) account for the major proportion of leprosy in China (Yu et al., 2010, Shen et al., 2008). In addition, migration of patients from highly endemic areas to other non-endemic areas with faster social-economic development prospects is ongoing (Shen et al., 2008). The case detection rates and delay in diagnosis are dependent on multiple factors such as age, occupation, nationality, endemicity, leprosy type and detection method (Chen et al., 2000, Shen et al., 2010, 2011). Therefore, newer tools and information can be incorporated to fully understand incidence and transmission of the various strains of leprosy in low and high endemic areas. In this regard, a molecular epidemiology approach utilizing strain typing of the pathogen Mycobacterium leprae linked to geographic, social, cultural and economic factors can provide useful complementary information. For instance, it will be helpful in case finding, in that once a cluster is identified by genotyping, it may be a clue to detect other undiagnosed leprosy patients by focusing on the geographical distribution or specific communities where the clusters are found, and taking into consideration their familial, social and occupational interactions. On the other hand, strain types that are novel to a region, may be explained by occupational migration, which provides opportunities for further spread of disease.
The present study attempts to expand on the knowledge of strain type diversity in China. Currently, a number of variable number of tandem repeat (VNTR) and single nucleotide polymorphisms (SNPs) have been discovered and applied to describe M. leprae strains for different geographical scales (Groathouse et al., 2004, Zhang et al., 2005, Monot et al., 2005, 2009, Truman et al., 2011). Strains within different regions and countries have been distinguished by applying VNTRs (Weng et al., 2007, Cardona-Castro et al., 2009, Fontes et al., 2009, Kimura et al., 2009, Phetsuksiri et al., 2012, Sakamuri et al., 2009, Srisungnam et al., 2009). Utilizing the SNP typing markers, in China, a single SNP type, i.e., 3K which is one of sixteen major types (Weng et al., 2007, Monot et al., 2009) has been detected. VNTR typing enabled further resolution of such type 3 strains at township, village, ethnic, and family scales to detect clusters of transmission in an endemic county in Yunnan Province (Weng et al., 2007, 2011). The present study extends our analyses and investigations in China and discusses strain types and genetic markers of similarity and differentiation amongst Chinese M. leprae strains covering a wider geographical range informative to provincial and national leprosy control programs.
2. Materials and Methods
2.1. Patient inclusion, sample collection and M. leprae strain typing
During leprosy diagnosis, the collection of skin biopsies is routine and is performed by doctors of the Province or County level Skin Disease Control Stations (SDCS) of Centers of Disease Control (CDC) across China. A portion of the biopsies was placed in 70% ethanol for molecular studies. The use of these samples for research was approved by the ethical committee of the Beijing Tropical Medicine Research Institute. A subset of the patients belonged to earlier study cohorts (Weng et al., 2006, 2007, 2011; Xing et al., 2009).
2.2. M. leprae VNTR strain typing
M. leprae were strain typed using previously described methods (Kimura et al., 2009; Sakamuri et al., 2009; Weng et al., 2007, 2011). DNA extraction from biopsies followed by multiplex-PCR was performed. Fragment length analysis was applied to determine the VNTR patterns of each isolate based on the multiplex-PCR products (Main Research Center at Beijing Academy of Agriculture and Forestry Sciences or Proteomics and Metabolomics Facility, Colorado State University). Capillary electrophoresis was performed on an ABI 3730XL DNA Analyzer. Samples were typed at the following 17 VNTR loci: (AC)8b, (GTA)9, (GGT)5, (AT)17, 6-3a (rpoT), 21-3, (AC)9, (AT)15, (AC)8a, 27-5, 6–7, (TA)18, (TTC)21, 18-8, 23-3, 12-5 and (TA)10 (Groathouse et al., 2004; Kimura et al., 2009).
2.3. SNP typing of M. leprae
The SNP type (1–4) was identified for a subset of these samples (n=101). A PCR-RFLP based procedure was used for differentiation of SNP types 1–4 as described by Sakamuri et al. (Sakamuri et al., 2009); sequencing of SNPs was performed for samples that could not be typed by the RFLP assay (Monot et al., 2005). SNP subtyping of M. leprae was performed by sequencing of the PCR products using published primer sequences (Monot et al., 2009) for a subset of samples. Subtype 3K strains were identified by first sequencing SNPs at positions 2312059 (distinguishes SNP subtypes A–J from subtypes K–P) and 413902 (distinguishes subtypes A–K from subtypes L–P); if the strains were of 3K, there was no requirement to map the H/I and I/J SNPs to resolve the subtype. 1A–D were identified by sequencing SNPs at positions 8453, 313361 and 61425. PCR products were sequenced using ABI BigDye® Terminator v3.1 and an ABI 3730XL DNA Analyzer (Sangon Biotech, Shanghai Co., Ltd.)
2.4. Analysis of strain types
The population structure of 144 samples having complete data (shown in bold in Supplementary Table 1) was explored using principal component analysis (PCA), using the complete panel of VNTR loci. The SNP type of a subset of typed samples was not used as a locus in the analysis methods, but was used as a categorical variable. This analysis was performed at Colorado State University in the U.S. PCA was performed on individuals using the correlation matrix within the software Minitab 16.
Maximum parsimony analysis was performed using the software PAUP* 4.0b (Swofford, 2003) and the methods described by Sakamuri et al. (Sakamuri et al., 2009). The heuristic search method was used, with tree bisection-reconnection and 1,000 maximum trees. Loci were assigned weights corresponding to the reciprocal of Nei's unbiased gene diversity (Nei, 1987). A step matrix was implemented to apply a step-wise mutation model; a difference of one repeat equated to one step. The out group was defined as the set of SNP type 1 samples. From the 1,000 trees generated by PAUP* a consensus network was built using SplitsTree4 v4.11.3 (Huson and Bryant, 2006).
3. Results
3.1 Genetic diversity of M. leprae in China
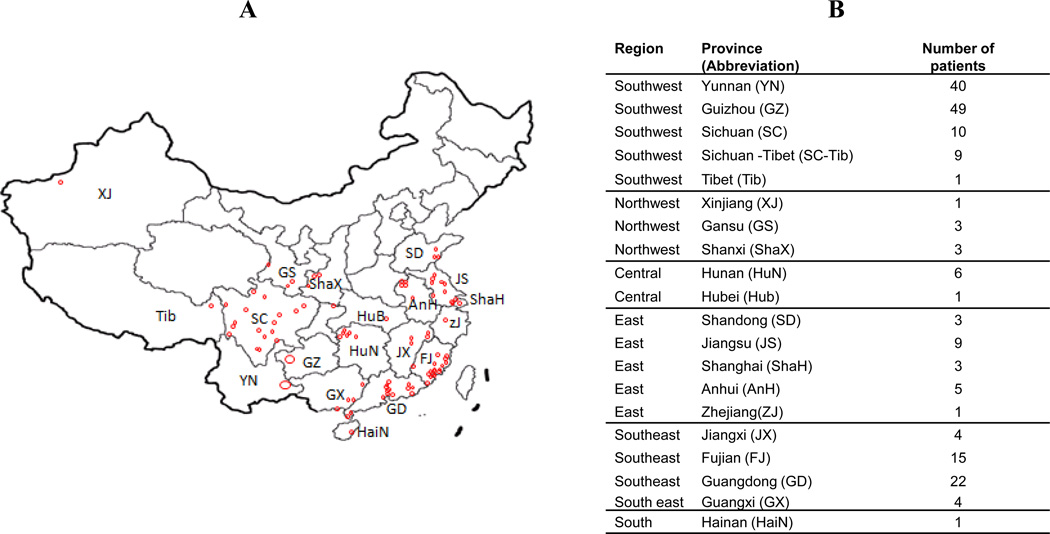
The M. leprae strain types in the skin biopsies of 190 leprosy patients diagnosed in 17 different provinces (including Shanghai) and two autonomous regions (Tibet and Xinjiang), shown in Figure 1 (Figure 1A and 1B) were determined. A panel of 17 VNTR loci was mapped by multiplex PCR -fragment length analyses methodology. Complete M. leprae VNTR profiles available for 144 patients were further analyzed. Nearly all DNA samples except those from Yunnan and Guizhou patients were also typed according to the SNP 1–4 system. The available demographic, clinical and M. leprae genotyping information for all the 190 patients has been compiled in Supplementary Table 1.
Figure 1. The geographical origins of the leprosy patients in the molecular epidemiology investigations.
A: An outline map of the provinces of China. Only the provinces where patients were identified for the study are labeled. Each patient is represented by a circle except in Yunnan and Guizhou where the single large circle represents all the patients. B: The geographical region, province names, abbreviations and number of patients are listed (Refer to Figure 5 for more detailed maps of Sichuan, Yunnan and Guizhou)
As a first approach, principal component analysis (PCA) was performed to explore the genetic variation within the VNTR dataset. Post analysis labeling of individuals according to the province, SNP type and selected VNTR loci alleles reveal several novel trends regarding the strain types and their geographical distributions in China (Figure 2). As seen in Figure 2A, the VNTR profiles of strains from different provinces separate into at least three subpopulations which are depicted within dotted lined circles (labeled as Ch1, Ch2 and Ch3) and a smaller group (labeled as Ch4) straddling Ch1 and Ch3.
Figure 2. M. leprae strain type variation in China.
A: A principal component analysis plot derived from VNTR mapping at 17 loci shows associations between geography and genotypes. The VNTR profiles of strains from different provinces separate into at least three subpopulations which are depicted within dotted lined circles (denoted as Ch1, Ch2 and Ch3) and a smaller group (Ch4) straddling Ch1 and Ch3. B: Association between VNTR derived population structure and SNP types. The same VNTR based PCA plot shown in panel A is labeled according to the SNP types of the individuals.
It was observed that several isolates obtained from patients diagnosed in the neighboring southeastern provinces of Guangdong (GD) and Fujian (FJ) were quite distinct from those isolated from elsewhere in China (Figure 2A, Ch1). Manual visualization of the VNTR alleles across the loci (Supplementary Table 1) indicates that these strains have alternative alleles at several of the typed loci. These readily noticeable VNTR allele differences prompted us to perform SNP typing to see if the Guandong and Fujian isolates were a subgroup of SNP type 3. This analysis revealed that the unique Guangdong and Fujian M. leprae strains are of SNP type 1; the first evidence for the existence of this type in China (Figure 2B). Instead of the more common 7 and 3 copy alleles detected thus far in China for loci 18-8 and 12-5 respectively, the alleles tended to be 8, and 4 or 5. Other contributing loci are (GGT)5, (AC)8b and (TA)10; the PCA loading plot and score plots of the allelic distribution patterns of these five and three other loci are shown in Figure 3 (A–I).
Figure 3. VNTR Allele distribution in M. leprae strains from different provinces.
A: The principal component analysis score loading plot of the variation in M. leprae strain types shown in Figure 2A. Panels B–I show the same plot and individuals except that the color coding indicates different alleles per the legend shown at each of eight loci that contribute to the population structure. B: 18-8, C: 12-5, D: (GGT)5, E: (AC)9, F: (AC)8a, G: (AC)8b, H: (TA)10 and I: 6-3a (rpoT)
A second subgroup is predominantly represented by patients of Jiangsu (JS) and Anhui (AnH), and patients of Tibetan ethnicity who reside in the western portion of the Sichuan province (Sichuan-Tibet, SC-Tib). These strains separate from all the other SNP type 3 individuals due to the 6-3a (rpoT) allele of 4 copies (Supplementary Table 1, Figure 2A, Ch2 and Figure 3I); the characteristic allele of the first polymorphic locus described for M. leprae strain typing seen only in parts of Japan, Korea, Indonesia and Mexico (Matsuoka et al., 2005, 2009).
The third subgroup includes the bulk of the SNP type 3 isolates and represents the highly endemic Yunnan (YN), Guizhou (GZ) and Sichuan (SC) provinces and the majority of the provinces studied (Figure 2A, Ch3). Although the number of patients from Jiangxi and Guangxi is small, it is interesting that genotypes of three strains from each province (shown as Ch4 in Figure 2A) are between those of Guandong-Fujian and Yunnan-Sichuan-Guizhou. This pattern corresponds to the geographical relationships between these provinces.
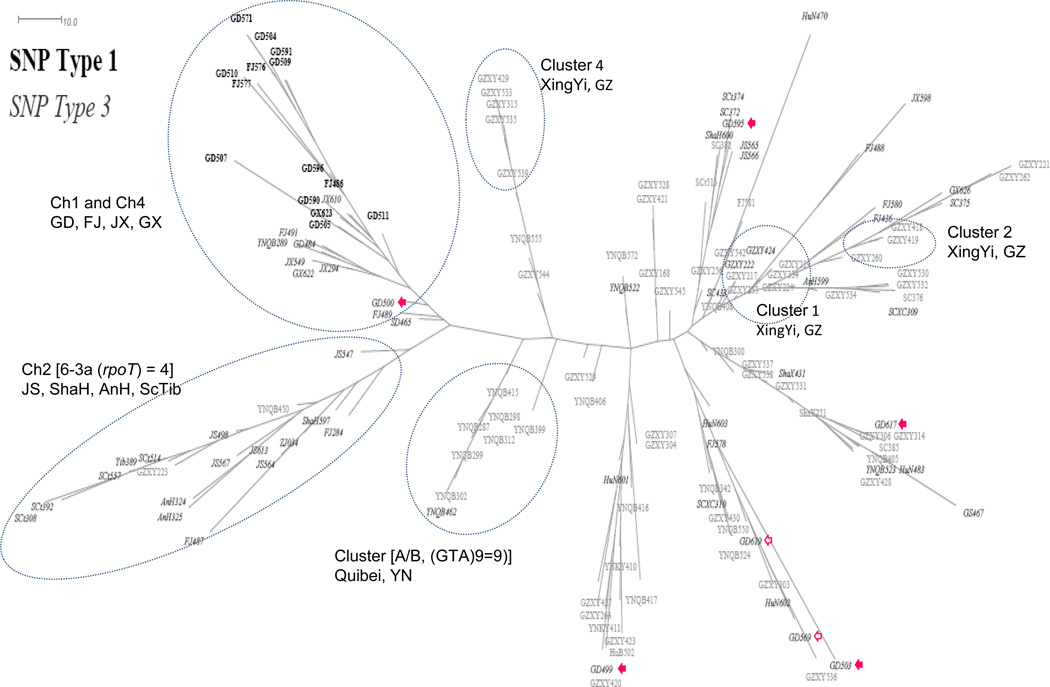
The prominent PCA groupings described above were evident when the VNTR data were analyzed by a parsimony algorithm, using a weighted step-wise mutation model (Figure 4).
Figure 4. A maximum parsimony tree based on multilocus VNTR M. leprae strain typing.
The individuals are labeled (per Supplementary Table 1). The SNP type 1 individuals were used as the outgroup. The individuals shown within dotted circles represent groupings and transmission clusters with known epidemiological links (also see Figure 2A). Empty and filled arrows show patients enrolled in Guangdong clinics whose M. leprae genotypes differ from the majority of Guangdong patients. The filled arrows represent patients with known migration history from other provinces while no information is available for patients represented by empty arrows. GD500 is a migrant from Jiangxi; the M. leprae genotype falls within Ch4 where other Jiangxi patients cluster.
3.2 Implications of M. leprae strain typing
Aside from providing new insight into the broad strain types across a larger number of provinces across China, the current study is of practical relevance to leprosy control efforts in provincial, county and national scale.
3.2.1 Multi-case families as indicators of ongoing transmission
As in previous studies, multi-case families (MCFs) in different provinces were identified (Supplementary Table 1) with intra-MCF VNTR similarity attesting to shared infectious source. On the other hand, in two family cases in Guizhou (GZXY540 and GZXY421; GZXY220 and GZXY221), the VNTR patterns are divergent; the source of infection for one of the cases is due to a temporally remote infection or from outside the family, assuming the common view that the source of infection is M. leprae patients.
3.2.2 Detection of patient migration
Although the majority of the Guangdong strains are of SNP type 1 (13/22) or a version of SNP type 3 similar to those seen in Jiangxi and Guangxi (3/22), six isolates diverged (see PCA and parsimony tree, Figure 2A and Figure 4). When the histories of these six patients were reviewed, four were confirmed as being migrants from other provinces. The origin and the migration history of four patients are detailed but no follow-up investigation was performed, (see footnotes in Supplementary Table 1). Three of these patients were non-Han, while the majority of Guangdong patients are of Han ethnicity.
3.2.3 Leprosy within the endemic provinces of Yunnan, Guizhou and Sichuan
As Yunnan, Guizhou and Sichuan carry a significant proportion of the total number of new cases in China, these mountainous and economically underdeveloped southwestern provinces continue to be the focus of national leprosy control efforts. The three provinces have designated several autonomous prefectures for a variety of ethnic minority nationalities, whose proportion and distribution vary from province to province. In the present dataset, the majority of the population and leprosy cases were of Han ethnicity in Guizhou and parts of Sichuan, but not in Sichuan-Tibet and Yunnan (Supplementary Table 1). Movement of family members or entire families for transient or semi-permanent work is common, due to some connectivity between these provinces by roads and trains.
The prefecture and county level distribution of the Sichuan, Yunnan, and Guizhou patients of this study are shown schematically in Figure 5 (A–C). The Sichuan patients near the Tibetan province reside in the Garze (also known as Ganzi) Tibetan autonomous prefecture and Ngawa (also known as Aba) autonomous prefecture, while the remaining have been sampled from six other prefectures (Figure 5A). Yunnan patients were predominantly enrolled from the county of Qiubei (QB) in Wenshan prefecture (Figure 5B); only three were from the neighboring Kaiyuan County (KY) in Honghe prefecture. The Guizhou patients are from XingYi City (XY) in Qianxinan prefecture which is designated as the autonomous province for Buyei (or Buyi) and Miao nationalities (Figure 5C). The Qianxinan prefecture of Guizhou adjoins the Wenshan prefecture of Yunnan.
Figure 5. The spatial distribution of the Sichuan, Yunnan and Guizhou patients.
The relative size of the provinces is approximately to scale. Numbers embedded in the maps refer to specific prefectures per the key in the above referenced maps. D: A township level localization of Guizhou patients (the ID numbers per Supplementary Table 1) is shown in a map created with Google Maps application ((https://maps.google.com/maps?hl=en.). The placemarks were centered according to the latitude and longitude of the township. The superscripts placed after some of the individuals indicate distinct shared VNTR patterns. The geographical proximities of patients (within a range of 10–15 miles or less) with shared M. leprae VNTR genotypes (see Supplementary Table 1) suggests the presence of previously unknown transmission clusters. The arrow points to the town of BaJieZhen.
The patients from Sichuan are more geographically scattered among different prefectures than other provinces and distinct strain type clustering was not observed. However, the east-west distribution across the province of strains with rpoT 3 versus 4 copies is clear (Figure 5A), with the Zang/Tibetan nationality patients more likely to be infected with the latter strain type.
M. leprae genotypes along with the familial-ethno- spatial distribution of Yunnan-Qiubei (YN-QB) patients have been well characterized in prior longitudinal investigations (Weng et al 2007, 2011, Xing et al., 2009) which revealed highly clustered and unclustered strains. The present study indicates that the strain type responsible for the dominate cluster (identified as groups A and B in Weng et al, 2007), which includes a number of multi-case families in the northern townships of Qiubei, is not seen in Guizhou or Sichuan. This group has a (GTA)9 copy of 9, and is shown as a distinct branch (labeled A/B) in Figure 4. This indicates restricted spread of the strain outside of YN-QB. The strain type was associated with patients of the Zhuang ethnicity.
With regard to strain types within the prefecture level city of Xing Yi in Guizhou, as in YN-QB, clusters with similar strain types were detected (Supplementary Table 1, Figure 4 and Figure 5D). One such cluster of at least seven cases with an atypical allele of 7 for (AC)8b (cluster number 4, Supplementary Table 1) is interesting. Four of these cases are from the township of BaJieZhen. There were five Buyei minority cases in the entire cohort, and three belong to this cluster and reside in this township (Figure 5D).
Discussion
Although records of leprosy in China date back thousands of years, there is still limited information on its origins and routes of spread. Utilizing molecular epidemiological approaches, based on VNTR markers, we had previously shown that the current strain types are highly conserved in a survey of strains both within a highly endemic region and across China as a whole (Weng et al., 2006). A number of mini-satellite loci were found to be non-polymorphic, requiring microsatellite mapping for the fine resolution of community strains (Weng et al., 2006, 2007). The SNP based typing system showed that Chinese strains belong to the 3K lineage. To accommodate this finding, it was proposed that the SNP type 3 arrived in China, from Europe/ and Central Asia via the Silk Road (Monot et al., 2009). The 3K subtype per se has, to date, been detected in several southeast (Indonesia, Philippines, New Caledonia) and northeast Asian countries (Japan and Korea). These Asian countries also have SNP type 1 strains. However, in other endemic Asian countries such as India, Bangladesh and Nepal, SNP type 3 samples have not been described. Geographic boundaries such as mountains and oceans could have contributed to some of these restricted patterns of flow. However, it is difficult to reconcile the genetic homogeneity of M. leprae in China with its prehistoric and ancient involvement in routes of human migration, trade and religious links with many countries, including neighbors where SNP type 1 are dominant. Besides addressing this broader issue, there is a need for a better description of the genetic diversity of M. leprae within China for pragmatic applications of molecular strain typing which yields new information on endemic clusters and effect of migration, which can be exploited for implementation of informed contact tracing, active early detection screening programs and collaboration of national and provincial leprosy control programs anxious to further reduce or even eliminate leprosy from China.
To address these gaps, the current work expands the survey of strain types by including larger numbers of patients, representing 16 provinces and two autonomous regions, and additional VNTR loci, including mini-satellites previously thought to be non-polymorphic. Novel strain types in geographically well defined pockets were thus identified. First, SNP 1 type strains [SNP subtype 1D (n=17) and 1A (n=1)] hitherto not described in China, were found in the southeastern provinces. Second, SNP type 3 strains with the 4 copy rpoT locus were detected in two divergent locations: (ethnically Tibetan parts of Sichuan) and the eastern provinces (Jiangsu and Anhui). Third, there are distinguishable SNP type 3 strains in southeastern provinces of Fujian, Guangdong, Jiangxi and Guangxi. In Xinjiang (XJ), a patient carried the SNP type 2, in agreement with the geographic and ethnic proximity to those of central Asia and Nepal where this lineage is prevalent.
These observations lead us to propose that there were at least three routes and periods for leprosy introduction and/or transmission in China. The northern route via the Silk Road from central Asia has been implicated previously (Monot et al., 2005, 2009) The second is the southern Silk Road on the Sea (Maritime Silk Road) which involved the countries of southeast Asia, and was most influential during the Tang dynasty in the 8th century AD, after the decline of the land Silk Road, although its origins are more ancient dating back to the Han dynasty (202 BC-AD 220). The Maritime Silk Road started at Guangzhou, Guangdong. The third is another Maritime Silk Road originating from the port of Ningbo (in Zhejiang) linking China with Japan and Korea, to the east via the East China Sea (http://english.ningbo.gov.cn). Theories of a prehistoric southern route of human migration out of Africa and on the origins of the Asians have been discussed in the literature, so the possibility of the spread of leprosy along southern maritime route cannot be excluded and was considered, but was not investigated at this time.
The southern and eastern Maritime Silk Roads in the South China Sea and Indian Ocean originating from Guangzhou could be the link between China and its neighboring Asian countries (such as Thailand, Philippines, India, and Nepal) where SNP 1 strains currently dominate. The eastern Canton region of China, containing the provinces Fujian and Guangdong have a long maritime history and were actively involved in trade routes developed during the Tang dynasty for transport of silk, porcelain, tea and other products to the countries of southeast Asia (see http://en.wikipedia.org/wiki/File:Transasia_trade_routes_1stC_CE_gr2.png for a schematic map which depicts trading routes related to the ‘Silk Road’ during the period 500 BCE to 500 CE). More recently, 30,000 people immigrated to the Philippines at the beginning of 16 century. Overseas ethnic Chinese in Europe and the Americas are predominantly from these regions. Compared to SNP type 3, SNP type 1 is currently a minor population in China and the reasons for its restriction to this part of China would be interesting to examine. Further sampling from Guangdong, Fujian and neighboring provinces, particularly the northern province of Zhejiang, may indicate the hub(s), the frequency, the geographical gradient and range, and the ethnicities of the patients carrying the SNP type 1 and SNP type 3 strains. Zhejian is of particular interest due to its history as a shipbuilding site and producer of trade goods.
Globally, a small number of strains carry 4 copies of a hexamer sequence at the rpoT locus, rather than 3. The 4 copy allele was the first genetic polymorphism for M. leprae and could distinguish Japanese and Korean strains from those around the world (Matsuoka et al., 2000). Finding this allele within China confirms their relatedness to Japanese strains. Another shared feature is a 3-copy allele at the 12-5 locus; the 4 and 5 copy alleles are common in southeastern China, and other regions throughout the world. The Chinese patients with the rpoT 4 copy allele were located in two widely separated regions, one closer to Japan and Korea and the other on the western boundary between Tibetan and Sichuan; these strains were of type of 3K. It is unclear why the rpoT 4 copy allele was found in patients with such a wide separation, but considerations have been made of familial or religious connections. The patients or physicians did not have any knowledge of possible common ethnicity or familial histories. There may have been a historic separation of the 4-copy strains between these two regions. Buddhism, the role of monasteries and monks may have been involved in this phenomenon.
Finally, this study identifies a few Chinese strains of a novel SNP type 3 in Guangxi and Jiangxi and in the neighboring Guangdong and Fujian provinces; the co-localization with the type 1 strains in this part of China is interesting. These were found to be of subtype 3K. While there is no specific evidence herein, there is a possibility that the 3K strains (and the 1D strains) in this region may have entered or originated first in China and/or other countries in Asia (Philippines, Indonesia, Japan and Korea) and disseminated from the south, perhaps by the Maritime Silk Road and the ensuing interactions amongst the people rather than from the northern Silk Road on land. These events would have preceded the modern European colonization events which we had been previously surmised to be responsible for the prevalence of the SNP type 3K in the Philippines (Sakamuri et al., 2009). Islam is a religion practiced in many countries that were linked by the ancient trading routes where the 3K strains are found. The European/American strains of type 3I have not been shown anywhere in Asia to-date (only one strain of 3J, found in New Caledonia has been definitively described). In Turkey and Iran, the strain types reported thus far are 2F and 3K, without the putative intermediates (2G–3J) (Monot et al., 2009). SNP type 2 strains have been seen in Indonesia, Myanmar, Japan and Korea also (Matsuoka et al., 2005). Taken together, it is not clear, when and where the SNP type 2, the presumed ancestor of both types 1 and 3, diverged. This may have occurred in Asia, from where it dispersed in all directions (with type 3 heading back toward Europe as well). One patient with SNP type 2 strain was identified in Xinjiang. Whether SNP type 2 strains moved further east or north of China is not yet known, and why these are not yet seen in endemic parts of Yunnan, Guizhou and Sichuan is intriguing. Perhaps, more sampling in the future, in areas along the three routes may reveal intermediates between SNP type 1, 2 and 3K.
The significance, origins and evolutionary history of leprosy in Asia may be revealed by higher resolution mapping, such as by whole genome sequencing of representative strains of all subtypes. Yunnan and Guizhou border the mainland southeast Asian countries of Myanmar, Laos, and Vietnam where leprosy is endemic. These regions including Cambodia and Thailand may share a common history of leprosy. With regard to leprosy control measures, it is clear that at many levels new insight was gained in our investigations. We revealed regional pockets of specific leprosy genotypes, including those with linkages to multi-case families and/or ethnicity. Implementing innovative social and spatial network tools in concert with molecular epidemiology, as described in this study, can aid in rational, cluster by cluster tracking and disruption of ongoing leprosy transmission.
Supplementary Material
Demographic, clinical and M. leprae genotyping information for the leprosy patients in the study
Highlights.
-
*
VNTR and SNP typing identified novel regional M. leprae strain types in China.
-
*
SNP type 1 and a variant of the Chinese SNP type 3 were found in the south.
-
*
rpoT with four short tandem repeats were found in the west and east.
-
*
Maritime routes in the south and east may be involved in entry/spread of leprosy.
-
*
Spatial-ethnic-genetic mapping aid in tracing clusters and spread of leprosy.
Acknowledgements
We are grateful to the patients who participated in this study and to the doctors and clinical staff from each of the provinces who provided one or more patient samples and information for this study. We graciously acknowledge Dr. Li Huan-Ying and Dr. Patrick Brennan for their leadership and support of leprosy research investigations in BTMRI, China and CSU, USA. Students in Dr. Weng’s and Dr. Vissa’s laboratory are acknowledged for their assistance. The work was supported by grants from the National Natural Science Foundation of China (30670111) and National Institutes of Health/NIAID (RO1-AI-63457, ARRA supplement RO1-AI-63457 S1 and contract NO1 AI-25469).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cardona-Castro N, Beltrán-Alzate JC, Romero-Montoya IM, Meléndez E, Torres F, Sakamuri RM, Li W, Vissa V. Identification and comparison of Mycobacterium leprae genotypes in two geographical regions of Colombia. Lepr. Rev. 2009;80:316–321. [PubMed] [Google Scholar]
- Chen XS, Li WZ, Jiang C, Ye GY. Leprosy in China: delay in the detection of cases. Ann. Trop. Med. Parasitol. 2000;94:181–188. doi: 10.1080/00034980057527. [DOI] [PubMed] [Google Scholar]
- Chen XS, Li WZ, Jiang C, Zhu CB, Ye GY. Studies on mode of detection of leprosy in China during the years 1981–1998. Lepr Rev. 2001;72:302–310. doi: 10.5935/0305-7518.20010037. [DOI] [PubMed] [Google Scholar]
- Fontes AN, Sakamuri RM, Baptista IM, Ura S, Moraes MO, Martínez AN, Sarno EN, Brennan PJ, Vissa VD, Suffys PN. Genetic diversity of Mycobacterium leprae isolates from Brazilian leprosy patients. Lepr. Rev. 2009;80:302–315. [PubMed] [Google Scholar]
- Groathouse NA, Rivoire B, Kim H, Lee H, Cho SN, Brennan JP, Vissa VD. Multiple polymorphic loci for molecular typing of strains of Mycobacterium leprae. J. Clin. Microbiol. 2004;42:1666–1672. doi: 10.1128/JCM.42.4.1666-1672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kimura M, Sakamuri RM, Groathouse NA, Rivoire BL, Gingrich D, Krueger-Koplin S, Cho SN, Brennan PJ, Vissa V. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J. Clin. Microbiol. 2009;47:1757–1766. doi: 10.1128/JCM.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Maeda S, Kai M, Nakata N, Chae GT, Gillis TP, Kobayashi K, Izumi S, Kashiwabara Y. Mycobacterium leprae typing by genomic diversity and global distribution of genotypes. Int. J Lepr. Other Mycobact. Dis. 2000;68:121–128. [PubMed] [Google Scholar]
- Matsuoka M, Zhang L, Morris MF, Legua P, Wiens C. Polymorphism in the rpoT gene in Mycobacterium leprae isolates obtained from Latin American countries and its possible correlation with the spread of leprosy. FEMS. Microbiol. Lett. 2005;243:311–315. doi: 10.1016/j.femsle.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Gonzalez AV, Estrada I, Carreno-Martinez C, Fafutis-Morris M. Various genotypes of Mycobacterium leprae from Mexico reveal distinct Geographic distribution. Lepr. Rev. 2009;80:322–326. [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Araoz R, Coppée JY, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Virmond M, Flageul B, Cho SN, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamesipour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 2009;41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York, NY, USA: Columbia University Press; 1987. [Google Scholar]
- Phetsuksiri B, Srisungngam S, Rudeeaneksin J, Buncho S, Lukebua A, Wongtrungkapun R, Paitoon S, Sakamuri RM, Brennan PJ, Vissa V. SNP genotypes of Mycobacterium leprae isolates in Thailand and their combination with rpoT and TTC genotyping for analysis of leprosy distribution and transmission. Jpn. J. Infect. Dis. 2012;65:52–56. [PubMed] [Google Scholar]
- Sakamuri R, Kimura M, Li W, Kim HC, Lee H, Kiran MD, Black WC, Balagon M, Gelber R, Cho SN, Brennan PJ, Vissa DV. Population-based molecular epidemiology of leprosy in Cebu, Philippines. J. Clin. Microbiol. 2009;47:2844–2854. doi: 10.1128/JCM.02021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhang GC, Chen XS, Zhou M, Yu MW, Yan LB. A long-term evolution on the epidemiological characteristics of leprosy, towards the goal of its elimination in 1949 – 2007 in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29:1095–1100. Chinese. [PubMed] [Google Scholar]
- Shen J, Zhou M, Xu X, Ray A, Zhang G, Yan L. A big challenge in case finding at low endemic situation: analysis on 1462 new leprosy patients detected in China in 2007. Lepr Rev. 2010;81:176–183. [PubMed] [Google Scholar]
- Shen JP, Yang RD, Wang J, Zhou M. Historical comparisons on related features among newly registered leprosy patients in the endemic areas of Wenshan district, Yunnan province. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:565–567. Chinese. [PubMed] [Google Scholar]
- Srisungnam S, Rudeeaneksin J, Lukebua A, Wattanapokayakit S, Pasadorn S, Mahotarn K, Ajincholapan, Sakamuri RM, Kimura M, Brennan PJ, Phetsuksiri B, Vissa DV. Molecular epidemiology of leprosy based on VNTR typing in Thailand. Lepr. Rev. 2009;80:280–289. [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Truman RW, Singh P, Sharma R, Busso P, Rougemont J, Paniz-Mondolfi A, Kapopoulou A, Brisse S, Scollard DM, Gillis TP, Cole ST. Probable zoonotic leprosy in the southern United States. N. Engl. J. Med. 2011;364:1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Wen Y, Tian X-J, Wang H-B, Tan X-J, Li H-Y. Preliminary study on the genotyping of Mycobacterium leprae on 50 isolates from China. Zhongua Liu Xing Bing Xue Za Zhi (Chinese Journal of Epidemiology) 2006;27:402–405. [PubMed] [Google Scholar]
- Weng X, Wang Z, Liu J, Kimura M, Black WC, 4th, Brennan PJ, Li H, Vissa VD. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J. Clin. Microbiol. 2007;45:1728–1734. doi: 10.1128/JCM.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Heiden JV, Xing Y, Liu J, Vissa VD. Transmission of leprosy in Qiubei County, Yunnan, China: insights from an 8-year molecular epidemiology investigation. Infect. Genet. Evol. 2011;11:363–374. doi: 10.1016/j.meegid.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekly epidemiological record. Leprosy update. No. 36. 2011;86:389–400. ( http://www.who.int/lep/situation/new_cases/en/index.html, http://www.who.int/wer/2011/wer8636.pdf). [Google Scholar]
- Xing Y, Liu J, Sakamuri RM, Wang Z, Wen Y, Vissa V, Weng X. VNTR typing studies of Mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr. Rev. 2009;80:261–271. [PubMed] [Google Scholar]
- Yu MW, Yan LB, Shen JP, Sun YM, Zhang GC. Epidemiological analysis on leprosy in China, 2009. Zhonghua Liu Xing Bing Xue Za Zhi. 2010;31:1155–1157. Chinese. [PubMed] [Google Scholar]
- Zhang L, Budiawan T, Matsuoka M. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J. Clin. Microbiol. 2005;43:5221–5229. doi: 10.1128/JCM.43.10.5221-5229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB REFERENCES
(most recently accessed on September 16, 2012)
Leprosy statistics:
- http://www.who.int/lep/situation/new_cases/en/index.html.
- http://www.who.int/wer/2011/wer8636.pdf: Weekly epidemiological record. 2011.Leprosy update, 36, 2011, 86, 389–400.
Maps:
- http://en.wikipedia.org/wiki/File:Sichuan_prfc_map.png.
- http://en.wikipedia.org/wiki/File:Yunnan_prfc_map.png.
- http://en.wikipedia.org/wiki/File:Guizhou_prfc_map.png.
- http://www.china.org.cn/english/features/zhenhe/132334.htm.
- http://en.wikipedia.org/wiki/File:Transasia_trade_routes_1stC_CE_gr2.pngThis map linked to http://en.wikipedia.org/wiki/Silk_Road is entitled ‘The Silk Road in the 1st century’. The file history for the map has the following comment: The map depicts trading routes used around the 1st century CE centred on the Silk Road. The routes remain largely valid for the period 500 BCE to 500 CE. Geographical labels for regions are adapted from the Geography of Ptolemy (c. 150 CE), some trading centre names date from later (c. 400 CE). Relying on Ptolemy's names is wrong but neutral.The map has also been used here: https://wiki.eee.uci.edu/index.php/History_134C_%28Spring_2010%29
Maritime Silk Route:
- http://www.harrassowitz-verlag.de/dzo/artikel/201/003/3771_201.pdf?t=1272014903: Aspects of the Maritime Silk Road: From the Persian Gulf to the East China Sea Edited by Ralph Kauz
- http://www.npr.org/templates/story/story.php?storyId=128113397: The 'Other' Silk Road: China Peers Into Maritime Past by Anthony Kuhn:
- http://english.ningbo.gov.cn/col/col55/index.html: Information on Ningbo port, China
- http://english.ningbo.gov.cn.
- Homepage →Discovering Ningbo→About the City→Overview
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic, clinical and M. leprae genotyping information for the leprosy patients in the study