Abstract
Despite evidence implicating transcription factors, including STAT3, in oncogenesis, these proteins have been regarded as “undruggable”. We developed a decoy targeting STAT3 and performed a phase 0 trial. Expression levels of STAT3 target genes were decreased in the head and neck cancers following injection with the STAT3 decoy compared with tumors receiving saline control. Decoys have not been amenable to systemic administration due to instability. To overcome this barrier, we linked the oligonucleotide strands using hexa-ethyleneglycol spacers. This cyclic STAT3 decoy bound with high affinity to STAT3 protein, reduced cellular viability, and suppressed STAT3 target gene expression in cancer cells. Intravenous injection of the cyclic STAT3 decoy inhibited xenograft growth and downregulated STAT3 target genes in the tumors. These results provide the first demonstration of a successful strategy to inhibit tumor STAT3 signaling via systemic administration of a selective STAT3 inhibitor, thereby paving the way for broad clinical development.
Keywords: STAT3, decoy oligonucleotide, phase 0, head and neck cancer
INTRODUCTION
Transcription factors are attractive as therapeutic targets due to their critical role in regulating gene expression associated with the development and progression of many diseases, including cancer1. Signal Transducers and Activators of Transcription (STAT) are one such class of transcription factors that regulate various aspects of cell proliferation, survival and differentiation2. Among the seven known members of the mammalian STAT family (STAT 1, 2, 3, 4, 5a, 5b and 6), STAT3 functions as a key mediator of oncogenic signaling3. Constitutive STAT3 activation has been detected in a large number of human cancers, where increased STAT3 signaling is commonly associated with a poor clinical prognosis4–7. In vitro studies have shown that inhibition of STAT3 expression or function attenuates the proliferation and survival of a wide variety of cancer cell lines characterized by overexpression/hyperactivation of STAT3, suggesting an addiction to the oncoprotein8, 9. By contrast, although STAT3 gene inactivation results in embryonic lethality10, many normal adult tissues are unaffected by loss of STAT32, 11, 12. Collectively, these findings point to STAT3 as a highly attractive target in cancer therapy.
Several strategies have been developed to inactivate STAT3, including the use of aptamers and peptidomimetics to target STAT3 protein and antisense oligonucleotides to decrease STAT3 expression. However, to date, challenges in drug delivery have limited the clinical translation of these approaches5–7, 13. Small molecules that reportedly inhibit STAT3 generally function by targeting upstream receptor and non-receptor tyrosine kinases, and therefore lack specificity. In hepatocellular carcinoma, sorafenib, a multikinase inhibitor decreased STAT3 phosphorylation in association with inhibition of phosphatidylinositol-3-kinase (PI3K)/Akt and MEK/ERK pathways14. NSC 74859, a chemical probe inhibitor of STAT3 activity, inhibited tumor development in hepatocellular carcinoma model by blocking STAT3, however it’s application has only been as a preclinical tool15. WP1066, a JAK2 inhibitor demonstrated antitumor activity against renal cell carcinoma in conjunction with decreased STAT3 phosphorylation16. Knocking down STAT3 by an RNAi approach, a preclinical tool, suppressed proliferation in vitro and tumorogenicity in vivo17. Curcumin analogs LLL12 and FLLL32 were evaluated for their ability to inhibit STAT3 activity in vitro and antitumor efficacy in vivo18. In osteosarcoma, LLL12 and FLLL32 inhibited STAT3 activity in vitro and reduced tumor growth. However, there is no evidence of direct binding of LLL12 or FLLL32 to pSTAT3 protein. In an effort to develop a highly specific inhibitor of STAT3, we generated a double-stranded STAT3 oligonucleotide decoy19.
Transcription factor decoys consist of nucleotide sequences derived from conserved genomic regulatory elements that are recognized and bound by the transcription factor in question. Transcription factor decoys elicit their biological effects by competitively inhibiting binding of the transcription factor to corresponding cis elements in genomic DNA, preventing expression of target genes. The STAT3 decoy was derived from the conserved hSIE genomic element found in the c-fos gene promoter, and was comprised of a 15-bp duplex oligonucleotide with free ends and phosphorothioate modifications of the three 5’ and 3’ nucleotides19. This STAT3 decoy demonstrated selective binding for STAT3 protein and inhibited the proliferation and survival of head and neck squamous cell carcinoma (HNSCC) cells in vitro19. Intratumoral administration of STAT3 decoy inhibited the growth of HNSCC xenograft tumors in vivo20. Subsequent investigations by others demonstrated that this STAT3 decoy exhibits anti-tumor activity in a variety of preclinical models including cancers of the lung, breast, skin, and brain21–24. Preclinical studies of the STAT3 decoy in animal models demonstrated that it is well tolerated and lacks toxicity25.
The FDA introduced the concept of phase 0 clinical trials in 2006 to accelerate cancer drug development. One of the objectives of a phase 0 cancer clinical trial is to establish at the very earliest opportunity whether an agent is modulating its target in a tumor, and consequently whether further clinical development is justified. Given the paucity of clinical studies testing the biologic effects of a STAT3-selective inhibitor in humans, we designed a trial with a primary endpoint of target gene modulation in the tumor. To ensure that we could obtain high quality tissue specimens, we elected to directly inject the STAT3 decoy immediately prior to HNSCC tumor resection when the patient was under anesthesia. Although control groups are not typically included in early phase clinical trials, we chose to also enroll subjects exposed to a saline injection, rather than the STAT3 decoy, to serve as controls for the assessment of target gene modulation in the decoy-treated tumors. We received support from the (former) NIH Rapid Access to Interventional Development (RAID) program to manufacture clinical grade material. Due to the relative ease of obtaining biopsies of HNSCC prior to surgical resection, cumulative evidence supporting STAT3 as a therapeutic target in this cancer, and the urgent need for more effective therapies, we performed a phase 0 study to evaluate the biologic effects of the STAT3 decoy in HNSCC patients.
Since the potential for broad clinical application of the STAT3 decoy in its original formulation is limited by its sensitivity to degradation and the necessity for intratumoral administration, we also sought to develop STAT3 decoy modifications that would improve stability and facilitate effective systemic administration. These studies resulted in a chemically modified cyclic STAT3 decoy that demonstrates anti-tumor activity following systemic delivery. This strategy of decoy modification should allow further clinical development and testing of the STAT3 decoy and may have important implications for the generation and therapeutic evaluation of a wide variety of decoys targeting previously considered “undruggable” transcription factors.
RESULTS
Intratumoral administration of a STAT3 decoy oligonucleotide abrogates target gene expression in human HNSCC
STAT3 is a plausible therapeutic target in cancers characterized by STAT3 hyperactivation. To date, no STAT3-selective small molecule has reached clinical testing. We developed a novel strategy to specifically target STAT3 using a decoy olignonucleotide. A phase 0 clinical trial was performed to evaluate the pharmacodynamic effects of this STAT3 decoy, compared with saline control, in patients with HNSCC (ClinicalTrials.gov number, NCT00696176). Patients undergoing surgery for HNSCC were enrolled in this phase 0 clinical trial (Figure 1A). STAT3 decoy dose was escalated in successive cohorts at 3 dose levels from 250 µg to 1 mg per injection (5–6 patients per dose). Patients received a single intratumoral injection of STAT3 decoy or vehicle control (saline). Tumors were biopsied prior to treatment and after completion of surgery. Levels of STAT3 target gene expression were assessed in the tumors pre- and post-treatment. In anticipation of the phase 0 trial, we performed xenograft studies to determine the kinetics of downregulation of target gene expression in the tumors and concluded that decreased protein was observed by 4–6 hours (data not shown). Thirty patients were enrolled (Table 1). No grade 3/4 or dose-limiting toxicities were noted. No toxicities were reported and a maximum tolerated dose (MTD) was not reached. The time between pre- and post-treatment biopsies was similar for the group that received the STAT3 decoy and the group that received saline (4.1± 2.7 versus 4.5±2.1 hours; p = 0.643). There was evidence of decreased expression of STAT3 target genes, including cyclin D1 and Bcl-XL in the post-treatment tumors compared with levels in the pre-treatment biopsies in the group that received STAT3 decoy, compared with expression in the tumors from the group that received saline (Figure 1B–F). There was no evidence of a dose-response on the modulation of target gene expression levels (data not shown). There was no apparent association between baseline levels of total or phosphorylated STAT3 in the tumor and degree of modulation of target gene expression (data not shown). To ensure that global gene expression or RNA stability was not impacted, we performed RT-PCR on a subset of tumors. While RNA expression levels for cyclin D1 and Bcl-XL clearly declined, GAPDH levels were unchanged (data not shown). These findings suggest that intratumoral administration of a transcription factor decoy targeting STAT3 is safe and may decrease target gene expression in HNSCC tumors. Future studies of STAT3 inhibitors in cancer patients are warranted (ClinicalTrials.gov number, NCT00696176).
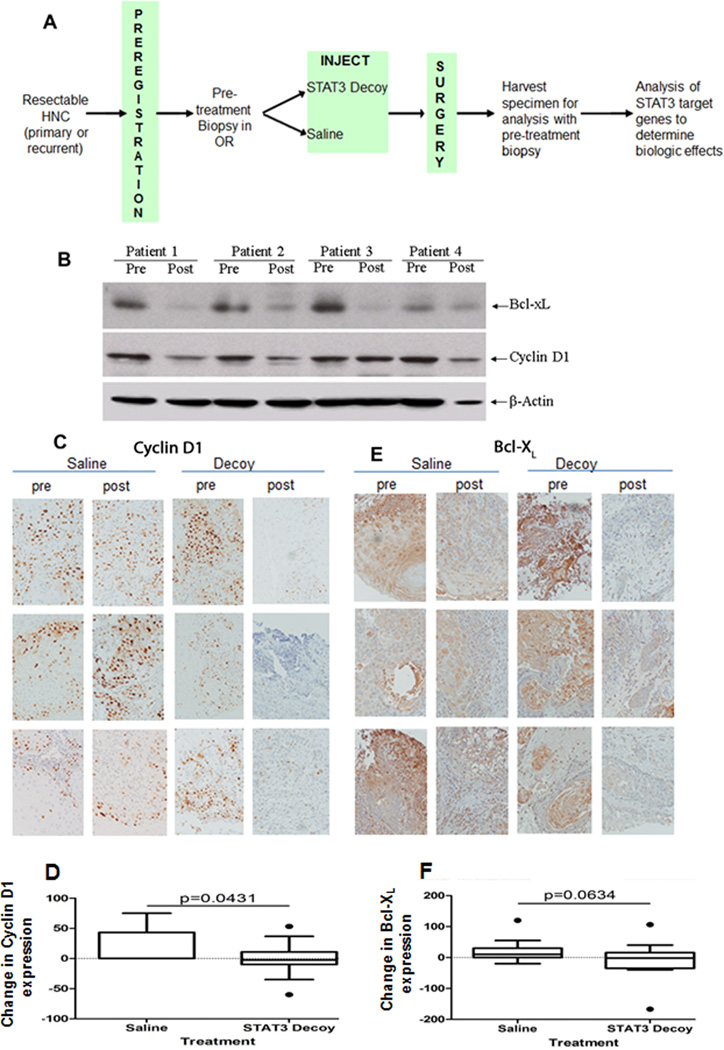
Figure 1.
Intratumoral administration of a STAT3 decoy oligonucleotide abrogates target gene expression in HNSCC patients. (A) Schema of phase 0 trial. HNSCC tumors were biopsied and injected with a single dose of a STAT3 decoy oligonucleotide (or saline) followed by tumor resection and analysis of target gene expression in the paired tumor samples. (B) Downmodulation of STAT3 target genes in representative HNSCC tumors injected with STAT3 decoy as shown by western blot analyses. Whole tumor lysates were prepared from 4 HNSCC tumors pre- and post-injection with the STAT3 decoy enrolled on the first dose tier. Proteins (40 ug) were resolved on a 12.5% SDS/PAGE gel and subjected to immunoblotting with anti-Bcl-XL and cyclin D1 antibody. β-actin was used as a loading control. (C) Representative images of IHC staining for cyclin D1 protein expression in pre- and post-STAT3 decoy oligonucleotide or saline injections from 6 patients. (D) Cumulative quantitative determination of cyclin D1 expression in all 30 HNSCC patient tumors showed a significant decrease in expression in tumors injected with STAT3 decoy oligonucleotide compared to saline injection (p=0.0431). (E) Representative images of IHC staining for Bcl-XL protein expression in pre- and post-STAT3 decoy oligonucleotide or saline injections from 6 HNSCC patients. (F) Cumulative quantitative determination of Bcl-XL expression in all 30 HNSCC patient tumors demonstrated decreased expression in STAT3 decoy injected patient tumors compared to the patient tumors injected with saline (p=0.0634).
Table 1.
Clinical and Pathological features of HNSCC subjects enrolled on phase 0 STAT3 decoy trial
| Saline (n=14) |
Decoy (n=16) |
|
|---|---|---|
| Age (yrs) | 51.4 ± 15.4 | 63.6 ± 15.1 |
| Sex (male:female) | 8:6 | 13:3 |
| Race (% white) | 86% | 88% |
| Site (n) | ||
| Oral cavity | 11 | 9 |
| Pharynx | 0 | 2 |
| Larynx | 3 | 5 |
| Primary:recurrent tumor | 12:2 | 9:7 |
| T stage (n) | ||
| 1 | 6 | 0 |
| 2 | 2 | 6 |
| 3 | 0 | 1 |
| 4 | 4 | 2 |
| N stage (n) | ||
| 0 | 9 | 4 |
| 1 | 1 | 3 |
| 2 | 2 | 2 |
| Prior HNSCC treatment (n) | ||
| Surgery | 0 | 4 |
| Radiation | 2 | 8 |
| Chemotherapy | 1 | 2 |
| Biologic (cetuximab) | 0 | 3 |
| Time between pre-treatment and post-treatment biopsies (hrs) | 4.1 ± 2.7 | 4.5 ± 2.1 |
Intravenous injection of parental STAT3 decoy fails to abrogate xenograft tumor growth
The STAT3 decoy used in the phase 0 trial consists of a 15-mer duplex oligonucleotide with phosphorothioate modifications at the 5’ and 3’ ends to enhance stability as described previously27. To determine whether this parental STAT3 decoy used could be administered systemically and retain anti-tumor effects, mice harboring cancer xenografts were given daily IV injection of the decoy (5 mg/kg). No reduction of tumor growth (Supplementary Figure S1A) or downmodulation of STAT3 target genes in the tumors (Supplementary Figure S1B) was observed, demonstrating that the parental STAT3 decoy requires local/intratumoral delivery to inhibit STAT3 signaling.
Design of modified STAT3 decoys
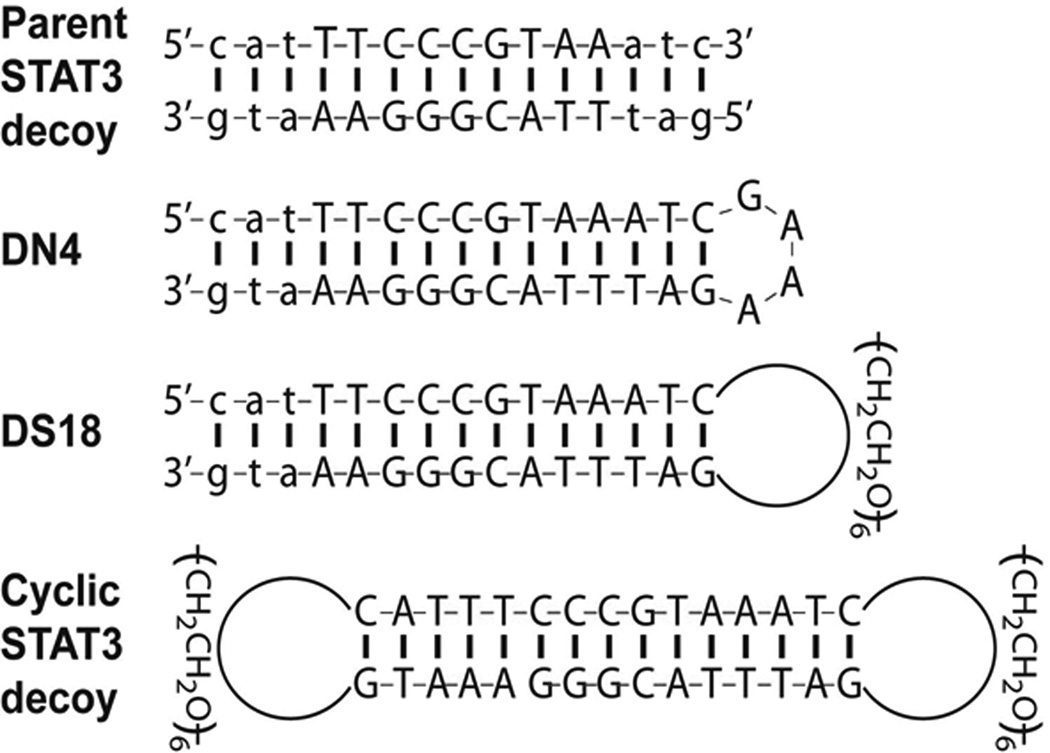
A plausible explanation for the lack of anti-tumor activity of the systemically administered parental STAT3 decoy is the vulnerability of this reagent to degradation and/or thermal denaturation in vivo, due to the presence of free ends. Modifications of the parental STAT3 decoy were undertaken in an effort to improve serum half-life and thermal stability, and thereby facilitate systemic delivery. Since nucleolytic degradation predominantly occurs at the 3’ end of single-stranded DNA or frayed ends of duplexes, we predicted that linkage of the two strands, as well as complete circularization, would improve stability in serum, while also enhancing thermal stability, ensuring that the decoy remains in annealed duplex form. Three distinct, unimolecular, derivatives of the parental STAT3 decoy were generated and evaluated (Figure 2). Figure S2 (Supplementary Figure S2) illustrates the chemistry used to generate the modified decoys. The DN4 decoy contains a single, 4-nucleotide loop (GAAA) linking the 3’ end of the sense strand to the 5’ end of the antisense strand. In the DS18 decoy, this loop is replaced by a single hexa-ethyleneglycol linkage. The cyclic STAT3 decoy utilizes hexa-ethyleneglycol linkages at both ends to generate a completely cyclical structure with no free ends.
Figure 2.
Structure of STAT3 decoys. The parental STAT3 decoy was modified by a 4-nucleotide hairpin (DN4), or a hexa-ethyleneglycol linkage (DS18) to generate unimolecular structures. Two hexa-ethyleneglycol linkages were used to generate the completely circularized cyclic STAT3 decoy.
Modified STAT3 decoys exhibit longer half-lives in serum
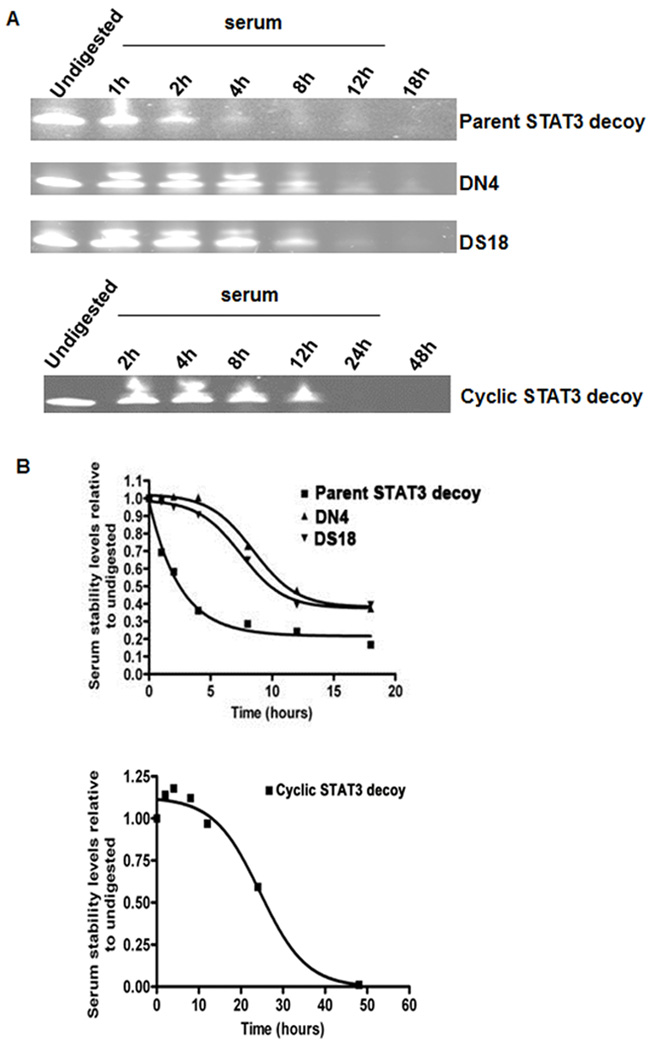
Following incubation in mouse serum for varying lengths of time, approximations of decoy half-life of the parental and modified STAT3 decoys were determined (Figure 3A–B). Consistent with its lack of anti-tumor activity when administered systemically, the parental STAT3 decoy exhibited a relatively short serum half-life of approximately 1.5 hours. By contrast, each of the modified decoys exhibited substantially longer half-lives. The half-life of DN4 was approximately 4 hours, while that of DS18 was about 3.5 hours. The most stable derivative was the cyclic decoy, which was detected up to 12 hours in serum. The markedly improved stability of the cyclic STAT3 decoy indicated that removal of all free ends, via circularization, was important for enhancing resistance to degradation.
Figure 3.
The modified STAT3 decoys exhibit enhanced stabilities in mouse serum. (A) Parental STAT3 decoy, DN4, DS18, and cyclic STAT3 decoy were incubated for varying lengths of time in mouse serum, electrophoresed on 15% TBE + 7 M Urea gels (polyacrylamide), and stained with SYBR-Gold, as described in Materials and Methods. Undigested indicates decoy in the absence of serum. (B) Densitometric analyses were performed on the parental STAT3 decoy, DN4, DS18, and cyclic STAT3 decoy samples shown in Panel A, and results were expressed relative to the corresponding undigested forms.
Since the decoy acts to mimic double-stranded STAT3 response elements in target genes, thermal denaturation temperatures above 37°C will be important for effective systemic administration. UV-denaturation determinations (Supplementary Figure S3) revealed a melting temperature of only 30°C for the parental STAT3 decoy. However, generation of unimolecular decoy forms resulted in enhanced thermal stability, with the DN4 and DS18 STAT3 decoys yielding melting temperatures of 57°C and 54°C, respectively. Moreover, complete circularization resulted in dramatic resistance to thermal denaturation, with cyclic STAT3 decoy demonstrating a melting temperature of 80°C, well above body temperature.
Modified STAT3 decoys bind avidly to pSTAT3 protein
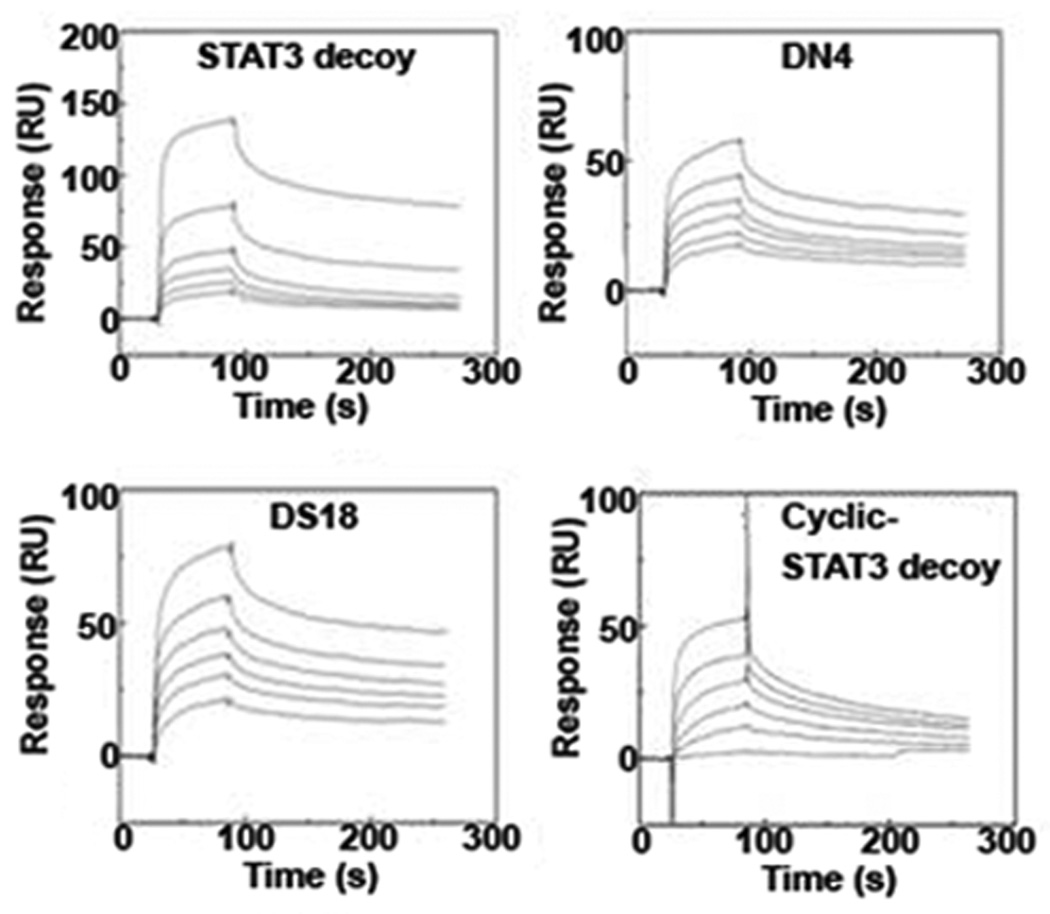
We next determined whether any of the chemical modifications of the parental STAT3 decoy interfered with binding to STAT3 protein. Binding assays were performed using recombinant, tyrosine(Y705)-phosphorylated STAT3 protein (pSTAT3), representing the activated form of the transcription factor28, 29. Parental or modified STAT3 decoys were first incubated with the pSTAT3 protein, followed by nondenaturing polyacrylamide gel electrophoresis and SYBR-Gold staining of the nucleic acid decoys (Supplementary Figure S4). Results from these experiments suggested that the DN4, DS18, and cyclic STAT3 decoys exhibited similar binding as the parental STAT3 decoy to pSTAT3 protein. These findings were subsequently confirmed by surface plasmon resonance (SPR) measurements, which additionally provided quantitative binding interaction parameters (Figure 4). SPR analyses permitted derivation of the rates and affinities of association and dissociation between the decoys in solution and immobilized pSTAT3 protein, as monitored in real-time. In general, the chemical modifications introduced in the DN4, DS18, and cyclic STAT3 decoys did not significantly perturb the kinetics of complex formation, with the ka and kd of the DN4, DS18, and cyclic decoys to immobilized pSTAT3 remaining largely unchanged compared to the parental STAT3 decoy (Figure 4). To quantitatively evaluate the strength of interactions between the four STAT3 decoys and the pSTAT3 protein, their equilibrium dissociation constants (KD) were determined by fitting the SPR data according to a 1:1 Langmuir binding model. The KD values were calculated as a function of their rates of dissociation relative to association, according to the following equation KD=1/KA=kd/ka (KA is the equilibrium association constant). The immobilized pSTAT3 protein bound the parental and modified decoys (DN4, DS18, and cyclic) with similar nanomolar affinities (Supplementary Table S1). Thus, the chemical modifications introduced in the parental decoy, resulting in the enhanced serum half-lives and thermal stabilities of the DN4, DS18, and cyclic STAT3 decoys, did not adversely affect their binding to pSTAT3 protein.
Figure 4.
Modified STAT3 decoys bind to phosphorylated STAT3 protein with similar affinity as the parental STAT3 decoy. Quantitative assessment of the binding of parental STAT3 decoy, DN4, DS18 and cyclic STAT3 decoy to recombinant pSTAT3 protein by Surface Plasmon Resonance. Binding to pSTAT3 protein immobilized on a carboxymethylated dextran matrix (CM5) chip was determined at six different concentrations of the parental, DN4, DS18, and cyclic STAT3 decoys (0.313 µM, 0.625 µM, 1.25 µM, 2.5 µM, 5.0 µM, 10.0 µM).
Modified STAT3 decoys inhibit in vitro viability and expression of STAT3 target genes in cancer cell lines
To determine whether chemical modifications in the STAT3 decoy resulted in altered in vitro activities, HNSCC cells (UM-SCC1, UM-22B) and bladder cancer cells (T24) were treated with varying concentrations of parental STAT3 decoy, DN4, DS18, or cyclic STAT3 decoy to determine EC50 values (Supplementary Table S2). Corresponding mutant control decoys that differed from the parental or modified decoys at a single base-pair (as described in Materials and Methods) were also evaluated. In all three cell lines tested, the parental and modified STAT3 decoys exhibited EC50 values in the low nanomolar range (below 100 nM) at the end of 24h, 48h and 72h. By contrast, none of the mutant control decoys demonstrated nanomolar activity.
Transcription factor decoys act by interfering with the transcription of target genes. To determine the impact of the modified STAT3 decoys on expression of key STAT3 target genes, UM-SCC1 (Supplementary Figure S5A), UM-22B (Supplementary Figure S5B) and T24 cells (Supplementary Figure S5C), were treated with IC50 concentrations of DN4, DS18, cyclic STAT3 decoy, or corresponding mutant control decoys. Following incubation, immunoblotting was used to assess Bcl-XL and cyclin D1 expression levels. Treatment with DN4, DS18, and cyclic STAT3 decoy led to downregulation of both Bcl-XL and cyclin D1, when compared with treatment with vehicle alone, or treatment with the corresponding mutant control decoy. Thus, in cancer cell lines, the modified DN4, DS18, and cyclic STAT3 decoys retained the ability to decrease the expression of STAT3 target genes.
Cyclic STAT3 decoy does not inhibit cell viability or STAT3 target gene expression in STAT3 null cells but potently reduces cell viability and downmodulates STAT3 target genes in cells expressing wild-type STAT3
In order to determine the specificity of the cyclic STAT3 decoy, A4 colon cancer cells expressing human wild-type STAT3 (A4 STAT3 wild-type) or isogenic cells engineered to serve as STAT3 null cells (A4 STAT3 null cells)30 were used to determine the effect of the parental or modified decoys. The A4 STAT3 null cells when treated with the parental or cyclic STAT3 decoy did not show downmodulation of STAT3 target genes (Supplementary Figure S6A) or inhibition of growth. In contrast, the isogenic cells that retain STAT3 expression, were potently growth inhibited by treatment with the parental or cyclic STAT3 decoy (Supplementary Figure S6B) in association with downregulation of STAT3 target gene expression (Supplementary Figure S6C). These results suggest that STAT3 is the selective target of the STAT3 decoys and indicate that tumors that do not express STAT3 are unlikely to be responsive to treatment with the STAT3 decoy.
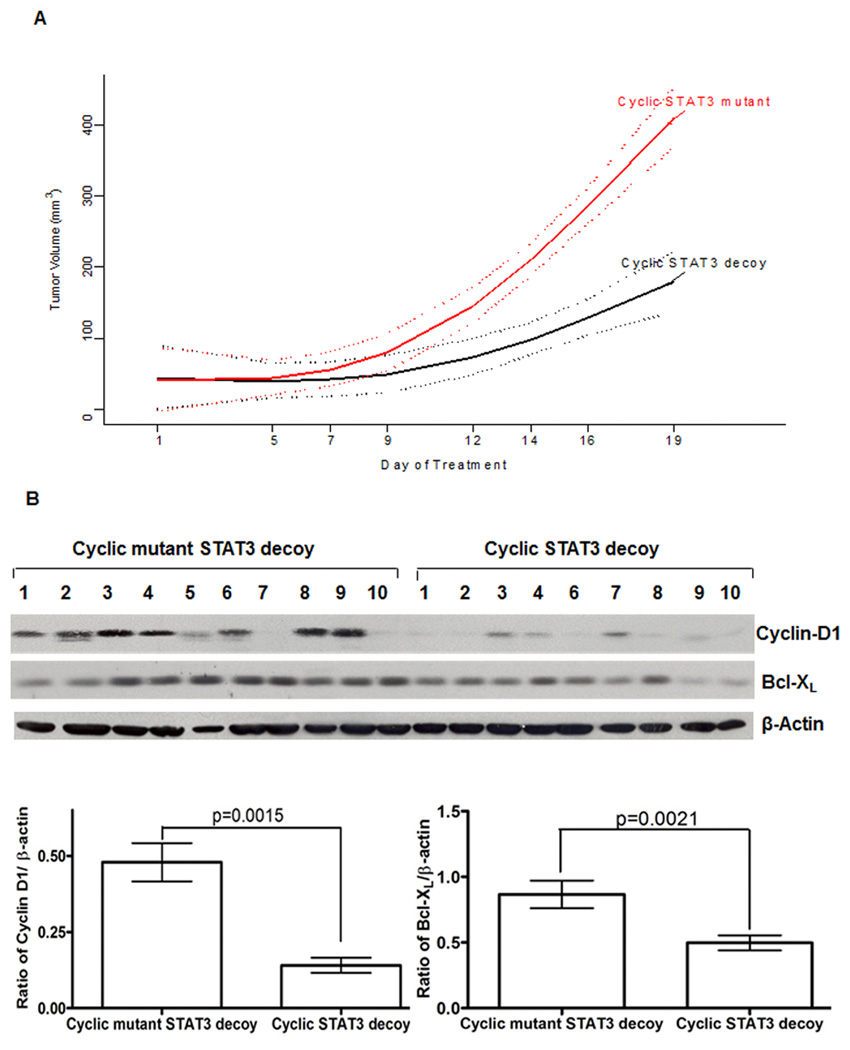
Systemic administration of cyclic STAT3 decoy inhibits tumor growth and expression of STAT3 target genes in vivo
Our in vitro studies revealed that the modified, unimolecular DN4, DS18, and cyclic STAT3 decoys demonstrated enhanced serum half-lives and thermal stabilities, while retaining biological and biochemical activities. Based on these results, we sought to determine whether systemic IV administration of the modified decoys would exert effects on xenograft tumors. To evaluate the anti-tumor effects of systemic administration of the cyclic STAT3 decoy, mice bearing established HNSCC xenografts (10 mice/group) were given daily intravenous injections (tail vein) of the cyclic decoy or the corresponding cyclic mutant control decoy (5 mg/kg/day), and tumor growth was monitored for 19 days. Tumors treated with the cyclic STAT3 decoy exhibited significant growth inhibition relative to tumors treated with cyclic mutant control decoy (p<0.0001, Figure 5A). Moreover, 2 of 10 tumors treated with cyclic STAT3 decoy experienced complete tumor regression. To ascertain the impact of the systemically administered cyclic STAT3 decoy on the expression of STAT3 target genes, tumors were harvested after 19 days of treatment and the levels of cyclin D1 and Bcl-XL in the tumors were determined. Relative to treatment with cyclic mutant control decoy, systemic administration of cyclic STAT3 decoy resulted in a significant decrease in cyclin D1/β-actin ratio (p=0.0015) and Bcl-XL/β-actin ratio (p=0.0021) (Figure 5B). Cyclic STAT3 decoy treatment did not alter the expression of total or phosphorylated STAT3 in the tumors (Supplemental Figure S7). These results indicate that the cyclic STAT3 decoy exerts its anti-tumor effects by impacting STAT3 activity, and not by inhibiting the expression of either total or phosphorylated STAT3.
Figure 5.
Systemic delivery of cyclic STAT3 decoy suppresses HNSCC xenograft tumor growth and expression of STAT3 target genes in the tumors. (A) Mean tumor volume by day of treatment and treatment groups. UM-SCC1 cells (3×106 cells) were inoculated subcutaneously in the right flank of athymic nude mice. Following the development of palpable tumors, mice were randomized, then given daily IV injections of cyclic STAT3 decoy or the corresponding cyclic mutant STAT3 decoy as a control (5 mg/kg/day; 10 mice/group). Tumor volume measurements were obtained 3 times/week and measured to day 19. A linear model was fit to daily tumor volumes with a non - linear day effect described by a 3 knot restricted cubic spline. The test of interaction between day and group was significant (p < 0.0001) indicating differential growth curves with decreased tumor growth for STAT3 decoy compared to mutant control. Solid lines are predicted means according to the linear model; dotted lines indicated 95% confidence bounds for daily mean values. (B) At the end of 19 days of treatment, tumors were harvested, and whole cell lysates were prepared and subjected to immunoblotting for cyclin D1 and Bcl-XL. β-actin was used to assess protein loading. The bar graph is a quantitative representation of the cyclin D1/β-actin (p=0.0015) and Bcl-XL/β-actin (p=0.0021) ratios in tumors from mice treated systemically with cyclic STAT3 decoy versus the cyclic mutant control decoy.
DISCUSSION
To date, there have been only a handful of phase 0 trials reported in cancer patients and none to date have targeted a transcription factor, included a control group, or used an intratumoral route of administration. We selected the phase 0 trial model for this first-in-human study of a STAT3 selective inhibitor in cancer patients to determine if the STAT3 decoy warranted further clinical development. The intraoperative setting used in our trial allowed for the collection of tumor tissue before and after administration of the STAT3 decoy. The inclusion of a control group enabled us to determine the specificity of the STAT3 decoy on target gene expression in human HNSCC tumors. Our results demonstrate significant pharmacodynamic activity of the intratumoral STAT3 decoy relative to saline control. Phase 0 trials have also proven useful in guiding subsequent studies as seen in the development of the poly (ADP-ribose) polymerase (PARP) inhibitor ABT-888 where the phase 0 results demonstrated significant inhibition of poly (ADP-ribose) levels in tumor biopsies and peripheral blood mononuclear cells at specific dose levels31.
STAT3 is hyperactivated in the majority of human cancers, where preclinical evidence supports STAT3 as a therapeutic target. Agents amenable to clinical administration that inhibit STAT3 generally lack specificity, target upstream receptor or non-receptor kinases, or represent natural products, which have a plethora of molecular targets13, 32. Oligonucleotide (ODN) decoys selectively inhibit the action of transcription factors but have been limited by their bioavailability due to rapid degradation. There has only been a single decoy oligonucleotide tested clinically in the setting of vascular disease, and none to date for cancer therapy33. The results of our phase 0 trial demonstrated downregulation of STAT3 target gene expression in the HNSCC tumor after a single intratumoral inoculation. This is especially compelling since expression level changes were compared to levels in paired biopsies from a control group that received an injection of vehicle (saline) alone into their tumor. Furthermore, most preclinical studies have examined the effects of decoys on target gene expression at far later time points (e.g. days) compared to the 2–6 hour exposure in our intraoperative phase 0 trial. Cancer is largely a systemic disease and therapeutic benefits are likely limited for an agent that requires intratumoral injection. These cumulative findings underscore the need for decoy formulations that are amenable to repeat, systemic administration.
Transcription factors decoys are generally comprised of double-stranded oligonucleotides with a high affinity for a transcription factor and compete for binding to the protein with specific cis elements present in the promoter region of target genes. While phosphorothioate modifications represent the most common strategy to improve stability, we and others have found that decoys with a fully modified phosphorothioate backbone have reduced affinity for the specific DNA binding site and hence, reduced efficacy27, 34. Oligonucleotides modified with only terminal phosphorothioate linkages exhibit increased resistance to exonucleases but retain susceptibility to endonuclease activity35, 36. The unmodified parent STAT3 decoy with terminal phosphorothioate modifications demonstrated high affinity as well as efficacy both in vitro27 and when administered intratumorally20, but failed to demonstrate anti-tumor efficacy when injected intravenously, indicating degradation of the STAT3 decoy by serum nucleases as a significant limitation to systemic delivery.
To date, chemical modifications of decoy oligonucleotides to improve serum stability have been associated with reduced biologic efficacy and diminished binding to target proteins37. Several strategies have been adopted to structurally modify transcription factor decoys in attempts to overcome some of the limitations associated with phosphorothioation. Transcription factor decoys modified with peptide nucleic acids (PNAs) have shown increased serum stability but often at the cost of binding specificity and affinity to target proteins37, 38. Oligonucleotides have also been modified with locked nucleic acids (LNA), a nucleic acid analog to improve resistance to nuclease degradation. However, substitution of nucleotides with LNA close to the transcription factor binding region induces conformational changes of adjacent nucleotides that can interfere with binding affinity37. Crinelli et al, reported that substituting nucleotides in an NF-κB decoy with LNA at different positions increased the half-life of the decoys in serum to 40–48 hours, but led to failure of the LNA-modified decoys to bind to NF-κB protein37. Osako et al, modified an NF-κB decoy into a circular oligonucleotide (RODN) and compared it to a phosphorothioate modified (PODN) and unmodified (NODN) NF-kB decoys39. Although RODN and PODN had serum stabilities of 6 h and 24 h, respectively, compared to less than an hour for NODN, binding assays showed that PODN had very low affinity for NF-κB protein. Another transcription factor decoy targeting activator protein-1 (AP-1) was modified to form a dumbbell-like structure (CD AP-1)40. The activity of CD AP-1 has been studied in vitro, however, serum stability data pertaining to the resistance of CD AP-1 to nuclease degradation has not been reported.
Our results suggest that altering the STAT3 decoy to create a unimolecular structure by a hairpin loop containing 4 single-stranded nucleotides (DN4) or with a hexa-ethyleneglycol spacer (DS18), or by complete cyclization results in a more stable therapeutic compound by making it more resistant to serum nucleases, while retaining potency and target specificity, in contrast to the parent decoy which is highly susceptible to degradation and thermal denaturation. Our modified STAT3 decoys inhibited cell proliferation and reduced the expression of STAT3 target genes in vitro and in vivo, and inhibited the growth of xenograft tumors.
Cumulative evidence has implicated a variety of transcription factors, including STAT3, in the development and/or maintenance of an oncogenic phenotype. To date, however, transcription factors have generally been regarded as “undruggable” target molecules. This is the first report of administration of a STAT3-selective molecular targeting agent to cancer patients resulting in a pharmacodynamic signature of biologic activity. Cyclization of this decoy allowed for successful systemic administration and suggests that clinical development of the STAT3 decoy, and other transcription factor decoys may yield effective therapeutic agents.
METHODS
Production of clinical grade STAT3 decoy
The STAT3 decoy (NSC-741763), which is an annealed, double-stranded oligonucleotide that is fully complementary and partially phosphorothioated, with the sense sequence being 5’ C*A*T*TTCCCGTTA*A*T*C 3’ and the antisense sequence 5’ G*A*T*TTACGGGAA*A*T*G 3’ (“*” denotes phosphorothioated sites), was manufactured at NCI-Frederick, Biopharmaceutical Developmental Program (Frederick, MD). The STAT3 decoy was formulated in phosphate buffered saline (PBS, purchased from Amresco Solon, OH) at 3.5 mg/ml stock concentration. The National Cancer Institute Rapid Access to Intervention Development (RAID) program assisted in the manufacture of the decoy compound under GMP conditions and oversaw the preclinical toxicology studies25. The potency of the clinical grade decoy was confirmed by assessing its effects on STAT3 promoter activity and target gene expression as described previously27.
Phase 0 clinical trial
Patients ≥18 years with primary or recurrent head and neck squamous cell carcinoma that was histologically confirmed and amenable to surgical resection were eligible to enroll in this study. Other eligibility criteria included: Eastern Cooperative Oncology Group (ECOG) scale 0–2, hemoglobin ≥10 G/dl, ANC ≥1500/cc, platelets ≥100,000/cc, creatinine ≤1.5 times upper limit of normal, bilirubin ≤1.5 times upper limit of normal, SGOT ≤2.5 upper limit of normal, and corrected serum calcium ≤10.5mg/dl. Patients treated previously with radiation, chemotherapy, or targeted agents were permitted. Ineligible patients included those who were pregnant, had tumors too small to reserve a portion for research purposes, or had received neoadjuvant radiotherapy and/or chemotherapy within a four-week period prior to enrollment. The study was approved by the University of Pittsburgh Institutional Review Board (IRB# 08020216, UPCI# 07-022) and registered on clinicaltrials.gov (NCT00696176). Written informed consent was obtained from patients. Patients were enrolled sequentially on 1 of 3 dose tiers (250 mg decoy in 250 microliters of saline, 500 mg in 500 microliters, 1,000 mg in 1 ml), which were based on extrapolation of the relative size of a xenograft in a mouse to the average volume of a human head and neck tumor. After the administration of general anesthesia for the tumor resection, the pre-treatment tumor biopsy was obtained. Next, the STAT3 decoy was administered by direct inoculation of the tumor with a 25-gauge needle. After resection of the tumor, a post-treatment biopsy was obtained from that area of the tumor that had been injected with the decoy. Control patients, who were injected with saline rather than the decoy, were included to determine the specificity of the STAT3 decoy and to distinguish between the effects of the decoy from the effects of surgery. Patients were monitored for adverse events using the NCI Common Terminology Criteria for Adverse Events version 3.0 criteria. Patients were followed for survival until 2 years after the date of surgery.
Tissue acquisition, processing, TMA construction and immunohistochemistry
Tumors were biopsied before and after administration of the STAT3 decoy (or saline) intraoperatively. The injection site was marked to assure that the post-treatment sample represented tissue that was exposed to the decoy. Tissue was processed primarily for the construction of a tissue microarray (TMA) to allow for assessment of protein expression across the entire cohort. When possible, fresh frozen material was processed for western blotting as described previously26. A tissue microarray (TMA) of the paired specimens from decoy- and saline-treated HNSCC tumors was constructed as described previously41. Tissues were snap frozen in a cryobath before being placed in a freezer at −80°C. Sections were cut out at 5 microns and mounted on Superfrost Plus slides, dried overnight at RT, and then 60°C for an hour. The sections were deparaffinized and hydrated before heat-induced epitope retrieval by a retrieval buffer. Blocking was performed with Invitrogen CAS block. Slides were washed with antibodies and Dako Substrate Chromagen and incubated with TBS. Slides were then counterstained with Harris Hematoxylin, dehydrated, and cleared. The TMA was stained with STAT3 (Cat# 9132, Cell Signaling Technology), p-STAT3 (Cat#9145, Cell Signaling Technology), Cyclin D1 (Clone SP4,Cat# M3635, Dako North America, Inc) and Bcl-XL (Cat# 2762, Cell Signaling Technology). All assessments of levels of STAT3 activation or target gene expression were determined by laboratory personnel blinded to patient identity, the treatment group (STAT3 decoy versus saline), and to the time of sampling (pre- vs post-injection). Assays were performed only after the specimens from the entire cohort were collected to minimize assay variability. The immunostaining was interpreted as the intensity of staining (0 to 3 scale) and the percentage of positively staining cells in the tissue section (0 to 100%) reported as intensity multiplied by expression.
Cell line validation
Cell lines were validated using the AmpFlSTR® Profiler Plus™ kit from PE Biosystems (Foster City, CA) according to the manufacturer's instructions. T24 bladder cancer cell line was obtained from American type culture collection (ATCC). UM-SCC1 and UM-22B were a kind gift from Dr. Thomas E. Carey (University of Michigan).
Generation of modified STAT3 decoys
Our initial design was to convert the bimolecular parental decoy19 into a unimolecular system by bridging the sense and antisense strand through a 4-base linker (DN4) or by a hexa-ethyleneglycol linkage (DS18). The parental STAT3 decoy was also circularized using two hexa-ethyleneglycol linkers attached to the sense and the antisense strands followed by enzymatic ligation of the 3' and 5' ends of the oligonucleotides. The mutant controls differed by one nucleotide at position 9 (G to T). We have previously shown that mutation of this nucleotide position abrogates decoy binding to STAT3 protein19. The single-stranded sense and antisense oligonucleotides of the STAT3 decoy/STAT3 mutant, DN4/MN4, and DS18/MS18 were obtained from Integrated DNA Technologies (Coralville, IA). The cyclic STAT3 decoy/cyclic STAT3 mutant were obtained from Oligo etc (Wilsonville, OR).
Serum stability assays
For the serum stability assay, the parental STAT3 decoy, DN4, DS18 and cyclic STAT3 decoy were incubated in 20 µl mouse serum isolate at a final concentration of 0.05 µg/µl according to standard protocol42. Following separation, the gels were stained with SYBR-Gold and imaged using the Gel Logic 2200 imaging system.
Thermal denaturation assay
Thermal denaturation studies were performed using a Varian Cary 300 Bio spectrophotometer equipped with a thermoelectrically controlled multicell holder, using 1.5 µM strand concentration each in 10 mM Tris and 1 mM EDTA, pH 8.0. Thermal denaturation of STAT3 decoy, DN4, DS18 and cyclic STAT3 decoy were monitored at 260 nm. Both the heating and cooling runs were performed at the rate of 1 °C/min. Melting transitions (Tm values) were determined by taking the first derivatives of the UV-melting curves.
STAT3 binding assays
For in vitro binding assays, parental and modified STAT3 decoys were incubated with 1 µg recombinant, tyrosine (Y705)-phosphorylated STAT3 for 30 minutes at room temperature. Complexes were electrophoresed on a nondenaturing 15% polyacrylamide-TBE gel, followed by visualization of the nucleic acids by staining wit SYBR-Gold (Molecular Probes, Carlsbad, CA).
Quantitative determination of the binding affinities of parental and modified decoys for pSTAT3 protein was accomplished by Surface Plasmon Resonance analyses, using a BIAcore 3000 instrument (GE Healthcare, Piscataway, NJ) following standard protocols43, 44. Unreacted sites on the chip surface were blocked using 1.0 M ethanolamine-HCl (pH 8.5). Binding of parental STAT3 decoy, DN4, DS18 and cyclic STAT3 decoy to pSTAT3 protein were determined at several concentrations of analyte solutions, at a flow rate of 30 µL/min in a running buffer (20 mM HEPES, pH 7.0, 200 mM NaCl, 10 mM MgCl2).
Dose response to determine EC50 value
UM-SCC1, UM-22B and T24 cells (30,000 cells/well) were seeded in 24-well plates in DMEM containing FBS. After 24h, cells were transfected with varying concentrations of STAT3 decoy, DN4, DS18 and cyclic STAT3 decoy. After 4h, the transfection media was replaced with DMEM containing 10% serum. At the end of 24, 48 and 72h, MTT assays were performed to determine the percent cell viability.
Immunoblotting
Immunoblotting was performed as previously described20. Antibodies used for immunoblotting included: rabbit anti-human cyclin D1 polyclonal antibody, mouse anti-human Bcl-XL monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-mouse pSTAT3 monoclonal antibody, rabbit anti-mouse STAT3 polyclonal antibody (Cell signaling, Danvers, MA), Blots were developed using the enhanced chemiluminescence (ECL) detection system (Amersham Life Sciences Inc., Arlington Heights, IL). The membranes were stripped and then probed with rabbit anti-human β-tubulin polyclonal antibody (Abcam Inc, Cambridge, MA). Densitometric analyses were performed using Image J software.
Systemic administration of parental STAT3 decoy in vivo
Female athymic nude mice nu/nu (4–6 weeks old; 20 g; Harlan Sprague–Dawley) with T24 xenograft tumors were treated with intravenous injection of STAT3 decoy or saline (vehicle) (100 µg) on a daily basis. Tumor volumes were measured three times a week. On day 18, the tumors were harvested and immunoblotting of the tumor tissues was performed to detect Bcl-XL and cyclin D1. Detection of β-tubulin was used to assess protein loading. Animal care was in strict compliance with institutional guidelines established by the University of Pittsburgh, the Guide for the Care and Use of Laboratory Animals (National Academy of Sciences, 1996), and the Association for Assessment and Accreditation of Laboratory Animal Care International.
Systemic delivery of cyclic STAT3 decoy in vivo
Female athymic nude mice nu/nu (4–6 weeks old; 20 g; Harlan Sprague-Dawley) with UM-SCC1 tumors were treated daily with intravenous injections of cyclic STAT3 decoy or cyclic mutant STAT3 decoy (100 µg). Palpable tumors were detected by day 3 and there was 100% tumor take for the cell lines used. During the treatment period, tumors were measured three times a week for 19 days. The size of the control tumors (treated with the mutant cyclic decoy) reached the maximum allowable tumor volume by day 19 so we elected to stop the experiment at that time point. At the end of the treatment period, the tumors were harvested and subjected to immunoblot analyses.
Statistical analyses
The goal of the clinical trial was to monitor toxicity and to obtain preliminary estimates of biologic efficacy of the STAT3 decoy. As a phase 0 trial, no hypotheses regarding therapeutic efficacy, biological activity, or optimum dose level were specified. A minimum of 5 patients were accrued to each dose level to provide a one tailed signed rank test to reject the null hypothesis of no change at .α = .03125. In vivo tumor volumes for the systemic delivery of cyclic STAT3 decoy experiment have been estimated with linear regression using a smoothing spline to capture non-linear tumor volume growth profiles. The spline fit is a flexible model that summarizes the overall trend and unlike a simple mean plot, borrows information from all data points. Comparison of treatment groups was based on a test of interaction between measurement day and treatment group. Last-day tumor volumes were also tested with a Wilcoxon test. In vitro and in vivo comparisons of Bcl-XL, cyclin D1, VEGF and Mcl-1 expression levels were conducted with the Jonckheere-Terpstra test for detecting a trend with increasing dose and with the Wilcoxon test for comparing two groups including comparing placebo to all decoy dose levels. All non-parametric tests were exact and two-tailed.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This is the first study of a STAT3 selective inhibitor in humans and the first evidence that a transcription factor decoy can be modified to enable systemic delivery. These findings have therapeutic implications beyond STAT3 to other "undruggable" targets in human cancers.
Acknowledgments
Financial Support, including source and number of grants for each author: P50 CA097190, American Cancer Society and the PNC Foundation (to JRG); R01 GM082251 (to JIY); NIGMS R01 GM068566 (to XC) and R01 CA137260 (to DEJ)
Disclosure of COI: Research support from Bristol-Myers Squibb (to JRG)
References
- 1.Libermann TA, Zerbini LF. Targeting transcription factors for cancer gene therapy. Current gene therapy. 2006;6:17–33. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nature reviews. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE. Validating Stat3 in cancer therapy. Nature medicine. 2005;11:595–596. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- 4.Takemoto S, Ushijima K, Kawano K, Yamaguchi T, Terada A, Fujiyoshi N, et al. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. British journal of cancer. 2009;101:967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert opinion on investigational drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao SP, Bromberg JF. Touched and moved by STAT3. Sci STKE. 2006;2006:pe30. doi: 10.1126/stke.3432006pe30. [DOI] [PubMed] [Google Scholar]
- 7.Chen CL, Cen L, Kohout J, Hutzen B, Chan C, Hsieh FC, et al. Signal transducer and activator of transcription 3 activation is associated with bladder cancer cell growth and survival. Molecular cancer. 2008;7:78. doi: 10.1186/1476-4598-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masuda M, Wakasaki T, Suzui M, Toh S, Joe AK, Weinstein IB. Stat3 orchestrates tumor development and progression: the Achilles' heel of head and neck cancers? Current cancer drug targets. 10:117–126. doi: 10.2174/156800910790980197. [DOI] [PubMed] [Google Scholar]
- 9.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer letters. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlessinger K, Levy DE. Malignant transformation but not normal cell growth depends on signal transducer and activator of transcription 3. Cancer research. 2005;65:5828–5834. doi: 10.1158/0008-5472.CAN-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. The EMBO journal. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–241. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- 14.Gu FM, Li QL, Gao Q, Jiang JH, Huang XY, Pan JF, et al. Sorafenib inhibits growth and metastasis of hepatocellular carcinoma by blocking STAT3. World J Gastroenterol. 17:3922–3932. doi: 10.3748/wjg.v17.i34.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu WY, Li J, Wu ZS, Zhang CL, Meng XL. STAT3 activation in monocytes accelerates liver cancer progression. BMC cancer. 11:506. doi: 10.1186/1471-2407-11-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiguchi A, Asano T, Kuroda K, Sato A, Asakuma J, Ito K, et al. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. British journal of cancer. 102:1592–1599. doi: 10.1038/sj.bjc.6605691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Yang G, Jiang T, Cao J, Huang KJ, Qiu ZJ. Down-regulation of STAT3 expression by vector-based small interfering RNA inhibits pancreatic cancer growth. World J Gastroenterol. 17:2992–3001. doi: 10.3748/wjg.v17.i25.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onimoe GI, Liu A, Lin L, Wei CC, Schwartz EB, Bhasin D, et al. Small molecules, LLL12 and FLLL32, inhibit STAT3 and exhibit potent growth suppressive activity in osteosarcoma cells and tumor growth in mice. Investigational new drugs. doi: 10.1007/s10637-011-9645-1. [DOI] [PubMed] [Google Scholar]
- 19.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–979. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Zhang J, Wang L, Wei H, Tian Z. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 23.Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. The Journal of clinical investigation. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Li R, Li G. Inhibitory effects of decoy-ODN targeting activated STAT3 on human glioma growth in vivo. In vivo (Athens, Greece) 2009;23:237–243. [PubMed] [Google Scholar]
- 25.Sen M, Tosca PJ, Zwayer C, Ryan MJ, Johnson JD, Knostman KA, et al. Lack of toxicity of a STAT3 decoy oligonucleotide. Cancer chemotherapy and pharmacology. 2009;63:983–995. doi: 10.1007/s00280-008-0823-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai SY, Koppikar P, Thomas SM, Childs EE, Egloff AM, Seethala RR, et al. Intratumoral epidermal growth factor receptor antisense DNA therapy in head and neck cancer: first human application and potential antitumor mechanisms. J Clin Oncol. 2009;27:1235–1242. doi: 10.1200/JCO.2008.17.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. 2003:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker S, Corthals GL, Aebersold R, Groner B, Muller CW. Expression of a tyrosine phosphorylated, DNA binding Stat3beta dimer in bacteria. FEBS letters. 1998;441:141–147. doi: 10.1016/s0014-5793(98)01543-9. [DOI] [PubMed] [Google Scholar]
- 29.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proceedings of the National Academy of Sciences of the United States of America. 107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bill MA, Fuchs JR, Li C, Yui J, Bakan C, Benson DM, Jr, et al. The small molecule curcumin analog FLLL32 induces apoptosis in melanoma cells via STAT3 inhibition and retains the cellular response to cytokines with anti-tumor activity. Molecular cancer. 9:165. doi: 10.1186/1476-4598-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann MJ, Whittemore AD, Donaldson MC, Belkin M, Conte MS, Polak JF, et al. Ex-vivo gene therapy of human vascular bypass grafts with E2F decoy: the PREVENT single-centre, randomised, controlled trial. Lancet. 1999;354:1493–1498. doi: 10.1016/S0140-6736(99)09405-2. [DOI] [PubMed] [Google Scholar]
- 34.Morishita R, Higaki J, Tomita N, Ogihara T. Application of transcription factor "decoy" strategy as means of gene therapy and study of gene expression in cardiovascular disease. Circulation research. 1998;82:1023–1028. doi: 10.1161/01.res.82.10.1023. [DOI] [PubMed] [Google Scholar]
- 35.Sands H, Gorey-Feret LJ, Ho SP, Bao Y, Cocuzza AJ, Chidester D, et al. Biodistribution and metabolism of internally 3H-labeled oligonucleotides. II. 3',5'-blocked oligonucleotides. Molecular pharmacology. 1995;47:636–646. [PubMed] [Google Scholar]
- 36.Uhlmann E, Peyman A, Ryte A, Schmidt A, Buddecke E. Use of minimally modified antisense oligonucleotides for specific inhibition of gene expression. Methods in enzymology. 2000;313:268–284. doi: 10.1016/s0076-6879(00)13017-4. [DOI] [PubMed] [Google Scholar]
- 37.Crinelli R, Bianchi M, Gentilini L, Palma L, Sorensen MD, Bryld T, et al. Transcription factor decoy oligonucleotides modified with locked nucleic acids: an in vitro study to reconcile biostability with binding affinity. Nucleic acids research. 2004;32:1874–1885. doi: 10.1093/nar/gkh503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomita N, Kashihara N, Morishita R. Transcription factor decoy oligonucleotide-based therapeutic strategy for renal disease. Clinical and experimental nephrology. 2007;11:7–17. doi: 10.1007/s10157-007-0459-6. [DOI] [PubMed] [Google Scholar]
- 39.Osako MK, Tomita N, Nakagami H, Kunugiza Y, Yoshino M, Yuyama K, et al. Increase in nuclease resistance and incorporation of NF-kappaB decoy oligodeoxynucleotides by modification of the 3'-terminus. The journal of gene medicine. 2007;9:812–819. doi: 10.1002/jgm.1077. [DOI] [PubMed] [Google Scholar]
- 40.Ahn JD, Morishita R, Kaneda Y, Kim HJ, Kim YD, Lee HJ, et al. Transcription factor decoy for AP-1 reduces mesangial cell proliferation and extracellular matrix production in vitro and in vivo. Gene therapy. 2004;11:916–923. doi: 10.1038/sj.gt.3302236. [DOI] [PubMed] [Google Scholar]
- 41.Oakley GJ, Fuhrer K, Seethala RR. Brachyury, SOX-9, and podoplanin, new markers in the skull base chordoma vs chondrosarcoma differential: a tissue microarray-based comparative analysis. Mod Pathol. 2008;21:1461–1469. doi: 10.1038/modpathol.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy M, Jason-Moller L, Bruno J. Using Biacore to measure the binding kinetics of an antibody-antigen interaction. In: Coligan John E, et al., editors. Current protocols in protein science. Unit 19. Chapter 19. 2006. p. 4. editorial board. [DOI] [PubMed] [Google Scholar]
- 44.Jason-Moller L, Murphy M, Bruno J. Overview of Biacore systems and their applications. In: Coligan John E, et al., editors. Current protocols in protein science. Unit 19. Chapter 19. 2006. p. 3. editorial board. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.