Background: ThPOK is required for CD4+ T helper cell differentiation and represses CD8 T cell-related genes, including Eomesodermin (Eomes) and IFNγ.
Results: TIP60 stabilizes ThPOK by acetylation and augments ThPOK-mediated transcriptional repression of Eomes.
Conclusion: TIP60 is a positive regulator of ThPOK.
Significance: TIP60 could be a novel target for modulating IFNγ-mediated inflammation against cancer and virus infection.
Keywords: Histone Acetylase, Inflammation, Protein Stability, T Cell, Transcription Regulation, Eomesodermin, TIP60, ThPOK, Acetylation
Abstract
The abundant expression of IFNγ in Th-inducing POK (ThPOK)-deficient CD4+ T cells requires the activation of Eomesodermin (Eomes); however, the underlying mechanism of this phenomenon remains unclear. Here we report that ThPOK binds directly to the promoter region of the Eomes gene to repress its expression in CD4+ T cells. We identified the histone acetyltransferase TIP60 as a co-repressor of ThPOK-target genes, where ectopically expressed TIP60 increased ThPOK protein stability by promoting its acetylation at its Lys360 residue to then augment the transcriptional repression of Eomes. Moreover, knockdown of endogenous TIP60 abolished the stabilization of ThPOK in CD4+ T cells, which led to the transcriptional activation of Eomes and increased production of IFNγ. Our results reveal a novel pathway by which TIP60 and ThPOK synergistically suppresses Eomes function and IFNγ production, which could contribute to the regulation of inflammation.
Introduction
TIP602 (Tat-interactive protein, 60 kDa) was identified as an HIV-1 TAT-interacting protein that augments TAT-mediated transactivation of the HIV-1 promoter (1). Further studies characterized TIP60 as a histone acetyltransferase that acetylates H2A, H3, and H4, but not H2B of the core histones to regulate gene expression (2). However, TIP60 is not only a chromatin modifier that regulates transcription as it also plays a role in DNA repair, cell apoptosis, oncogenesis, and serum deprivation-induced autophagy through exerting its function as a transcriptional co-regulator or through its enzymatic activity on transcription factors such as FOXP3, STAT3, c-Myc, p53, and protein kinases including ATM and UKL1 (3–9).
TIP60 deficiency in mice is lethal, whereas heterozygous deletion of TIP60 leads to haplo-insufficient tumor suppression (10). However, the role of TIP60 in regulating immune responses remains largely uncharacterized. In CD4+ CD25+ regulatory T (Treg) cells, we have shown that TIP60 binds directly to the proline-rich domain of the forkhead family transcription factor FOXP3, which is essential for FOXP3-mediated repression of IL-2 expression (8). In CD4+ T helper 2 (Th2) and Th17 cells, the proinflammatory cytokine IL-9 activates STAT3 to modulate its downstream gene expression (11) and is regulated by TIP60 through its interaction with both IL9Rα and STAT3 (9, 12).
The TCR signal-induced zinc finger and BR-C, Ttk, and Bab (BTB) domain-containing protein family transcription factor Th-inducing POK (ThPOK) is dominantly expressed in CD4+ T cells (13) and is necessary for T helper cell differentiation (14, 15). ThPOK binds to the silencers within the CD4 and ThPOK gene loci and inhibits silencer activity (13, 16). Moreover, a number of studies have shown how ThPOK regulates CD4 versus CD8 T cell lineage commitment by suppressing classical CD8 lineage genes such as CD8, Perforin, Granzyme B, and RUNX3 (14, 17–19). Others have also shown how the function of Eomesodermin (Eomes), a T-box transcriptional activator of IFN-γ, negatively correlates with ThPOK expression (17, 18, 20).
Here we report a previously uncharacterized mechanism by which the gene transcription of Eomes is directly repressed by ThPOK and how TIP60 is a cofactor for ThPOK-mediated repression of Eomes expression. This pathway in turn mitigates the activation of Eomes target genes such as IFNγ in human CD4+ T cells. As ThPOK contains a proline-rich domain, we hypothesized that TIP60 might also bind to its proline-rich domain to mediate T cell lineage differentiation and function and modulate inflammation through regulating the transcriptional induction of Eomes; however, we found that the C-terminal region of ThPOK interacted with TIP60 and is acetylated at the Lys360 residue. Our results thus reveal a direct molecular link between TIP60 function and the modulation of CD4+ T cell-mediated inflammation through cytokine production.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK 293T cells were cultured in DMEM containing 10% FBS and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Jurkat cells were maintained in RPMI 1640 medium containing 10% FBS. Transfection of Jurkat cells with plasmid DNA was performed by electroporation on a Gene Pulser X cell apparatus (Bio-Rad Laboratories). Jurkat cells were activated using soluble antibodies against CD3 (1 μg/ml, Hit3a; Biolegend) and CD28 (2 μg/ml, CD28.2; Biolegend).
Immunoprecipitation and Immunoblotting
Cells were washed with ice-cold PBS and lysed on ice for 30 min in 1× RIPA buffer (50 mm Tris-HCl, pH 7.5, 135 mm NaCl; 1% Nonidet P-40; 0.5% sodium DOC; 1 mm EDTA, 10% glycerol) containing protease inhibitor (1:100, P8340; Sigma-Aldrich), 1 mm NaF, and 1 mm PMSF. Cell lysates were cleared by centrifugation, and supernatants were immunoprecipitated with the appropriate antibodies (Abs) using protein A/G-agarose beads at 4 °C. After washing, 2× sample loading buffer was added to the immunoprecipitates. Samples were then used for immunoblot analysis with the indicated antibodies.
Antibodies and Reagents
The following antibodies were used for flow cytometry analysis: anti-CD4-FITC (RPA-T4; Biolegend), anti-CD8-APC (RPA-T8; BD Biosciences), anti-TCRαβ-PE (IP26; eBioscience), and anti-IFNγ-APC (4S.B3; eBioscience). Fixable viability dye eFluor 780 was purchased from eBioscience. Anti-HA (F-7), anti-ThPOK (A-4), anti-TIP60 (N-17), and goat IgG (sc-2028) were from Santa Cruz Biotechnology. Anti-FLAG (M2), anti-β-actin, and anti-α-tubulin were from Sigma-Aldrich and Tianjin Sungene Biotech (China), respectively. Mouse IgG was from Millipore. Anti-acetyllysine Ab was obtained from Immunechem Pharmaceuticals (Canada). Protein A/G-agarose beads (A10001) were purchased from Abmart (China). Cycloheximide (C7698-5G) and nicotinamide (72340-100G) were purchased from Sigma-Aldrich. EX-527 (S1541) was purchased from Selleck. Human ThPOK was cloned into the pIP-HA2 vector, and pCMV2-FLAG-TIP60 has been described previously (8). Mutagenesis was carried out according to the manufacturer's instructions using the Toyobo mutagenesis kit. ThPOK was cloned into the FUGW plasmid (kindly provided by Lan Ke, Institut Pasteur of Shanghai, Chinese Academy of Sciences).
Luciferase Reporter Assay
The 1000-bp region upstream of the human Eomes transcriptional starting site (NCBI: human, chromosome 3 NC_000003.11; mouse, chromosome 9 NC_000075.6) was cloned into the pGL3-Basic vector to generate the pGL3-Eomes-Luc reporter construct. Jurkat cells were co-transfected with the reporter plasmid and a Renilla luciferase encoding plasmid as a control, and/or FLAG-TIP60 as indicated. 48 h later, cells were lysed, and luciferase assays were performed using the Dual-luciferase reporter kit (Promega).
Chromatin Immunoprecipitation
Primary human CD4+ T cells were stimulated using anti-CD3/CD28 dynal beads (Invitrogen) at a cell to bead ratio of 1:1 for the indicated time periods. Cells were then cross-linked with formaldehyde, and the chromatin was sonicated into ∼500-bp fragments. The sheared chromatin was immunoprecipitated with anti-ThPOK antibody, and mouse IgG was used as a negative control. The pulled down DNA fragments were subjected to qPCR analysis. Primers that were used are as follows: Probe N forward, 5′-ccagtctaaggagggtgctg-3′ and reverse, 5′-tgaaatgggctttccttttg-3′; Probe B forward, 5′-agtcacaggcgacttgatcc-3′ and reverse, 5′-agatctttgtccccatccac-3′.
Quantitative Real Time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). cDNA was synthesized using a reverse transcriptase kit (TaKaRa, Japan), followed by qRT-PCR analysis (SYBR Green; TaKaRa). The primers that were used are as follows: CD8 forward, 5′-cctgagcaactccatcatgt-3′ and reverse, 5′-gatatcacaggcgaagtccag-3′; CD4 forward, 5′-gctggaatccaacatcaagg-3′ and reverse, 5′-ctgaaaccggtgaggacact-3′; ThPOK forward, 5′-gtctgccacaagatcatcca-3′ and reverse, 5′-tcgtagctgtgcaggaagc-3′; TIP60 forward, 5′-agatcctgagcgtgaaggac-3′ and reverse, 5′-tctctctggagagccaggac-3′; Eomes forward, 5′-tgcaaaaggcttcagagaca-3′; and reverse, 5′-ctctgttggggtgaaaggag-3′; Tbx21 forward, 5′-ccgtgactgcctaccagaat-3′ and reverse, 5′-atctcccccaaggaattgac-3′; IFNγ forward, 5′-aaacgagatgacttcgaaaagc-3′ and reverse, 5′-atattgcaggcaggacaacc-3′; β-actin forward, 5′-ctcttccagccttccttcct-3′ and reverse, 5′-cagggcagtgatctccttct-3′.
T Cell Isolation
Primary human CD4+ and CD8+ T cells from healthy donors were isolated by FACS on a BD FACS ARIA II sorter (BD Biosciences). Primary T cells were expanded using anti-CD3/CD28 dynal beads (Invitrogen) at a cell to bead ratio of 1:1 in X-VIVO-15 medium (Lonza, Switzerland) supplemented with 10% human AB serum, 1% GlutaMax (GIBCO), 1% sodium pyruvate (GIBCO), and 1% Pen/Strep (GIBCO).
Intracellular Staining
In brief, CD4+ T cells were restimulated with phorbol 12-myristate 13-acetate (50 ng/ml), ionomycin (1 μm), and Golgi Stop for 4 h. At the end of stimulation, cells were stained with fixable viability dye eFluor 780 and anti-CD4-FITC, then washed with PBS. Staining of IFNγ was carried out with the IC Fixation Buffer (eBioscience catalog number 00-8222) and Permeabilization Buffer (10×) (eBioscience catalog number 00-8333) according to the manufacturer's instructions.
Lentiviral Constructs and Transduction
The shRNA lentiviral vector pLKO.1 shTIP60, pLKO.1 shThPOK, or pLKO.1 shCK was transfected into HEK 293T cells via calcium phosphate transfection with the lentivirus packing vector Delta 8.9 and VSVG envelope glycoprotein. Viral supernatants were harvested after 48 h. Primary T cells were transduced with virus along with anti-CD3/28 stimuli (1 cell to 1 bead). The following shRNA sequences were used: shCK, 5′-caacaagatgaagagcaccaa-3′; shTIP60, 5′-cctcaatctcatcaactacta-3′; shThPOK, 5′-ccgcctctctctagctcgatt-3′.
RESULTS
Differential Expression of TIP60 in Human CD4+ and CD8+ T Cells
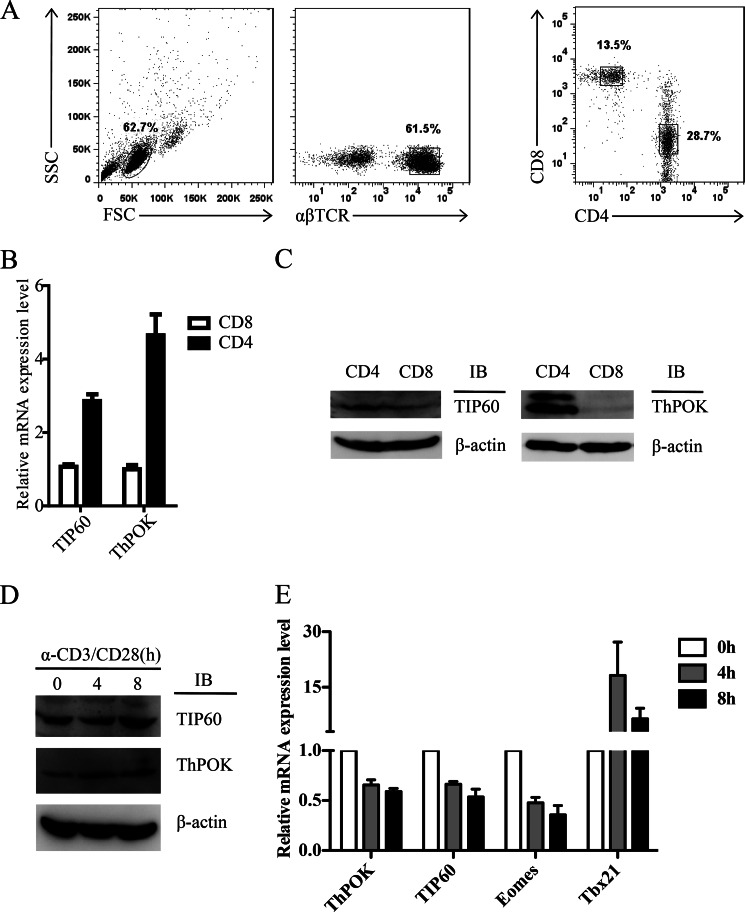
CD4+ and CD8+ T cells were isolated from human peripheral blood mononuclear cells (Fig. 1A) and subsequently examined for TIP60 mRNA and protein expression levels alongside the CD4 T cell lineage-specific transcription factor ThPOK as a control. We found that TIP60 mRNA was highly expressed in CD4+ T cells compared with CD8+ T cells (Fig. 1B). Also, protein expression was found to be consistent with the mRNA levels (Fig. 1C). Therefore, both TIP60 and ThPOK are higher expressed in CD4+ T cells.
FIGURE 1.
Short term TCR stimulation stabilizes ThPOK in human CD4+ T cells. A–C, CD4 SP and CD8 SP T cells were isolated from human peripheral blood mononuclear cells of healthy donors. B, total RNA was extracted, and the mRNA levels of TIP60 and ThPOK were measured by qRT-PCR analysis. Data show the mean of three separate experiments. Error bars indicate S.D. C, cells were lysed with RIPA buffer; TIP60 and ThPOK protein levels were analyzed by Western blotting (IB) using β-actin as a loading control. D and E, resting CD4+ T cells were stimulated with α-CD3 and α-CD28 antibodies for the indicated time periods. D, cells were lysed with RIPA buffer; cell lysates were directly tested by Western blotting with the indicated antibodies. E, total RNA was extracted, and the mRNA levels of ThPOK, TIP60, Eomes, and Tbx21 were measured by qRT-PCR. Data show the mean of at least three independent experiments. Error bars indicate S.D.
Short Term TCR Stimulation Stabilizes ThPOK in CD4+ T Cells
Previous studies indicated that long term TCR stimulation triggers ThPOK transcription in CD4+CD8lo thymocytes (13). However, it remains unclear how short term TCR stimulation regulates ThPOK expression in CD4 single positive (SP) cells. To reveal the role of TCR signaling, resting CD4 SP T cells were stimulated with soluble antibodies against CD3 and CD28 for a short period (within 8 h) prior to the analysis of ThPOK protein and mRNA levels. We observed an increasing level of ThPOK protein upon TCR stimulation, as well as TIP60 (Fig. 1D). However, the transcription of both genes was slightly repressed (Fig. 1E), which indicated that ThPOK and TIP60 were stabilized posttranslationally. Consistent with the previous findings, we identified that stabilized ThPOK correlates with the repression of Eomes gene transcription, but not T-bet (Fig. 1E).
TIP60 Associates with ThPOK to Promote Its Acetylation and Stability
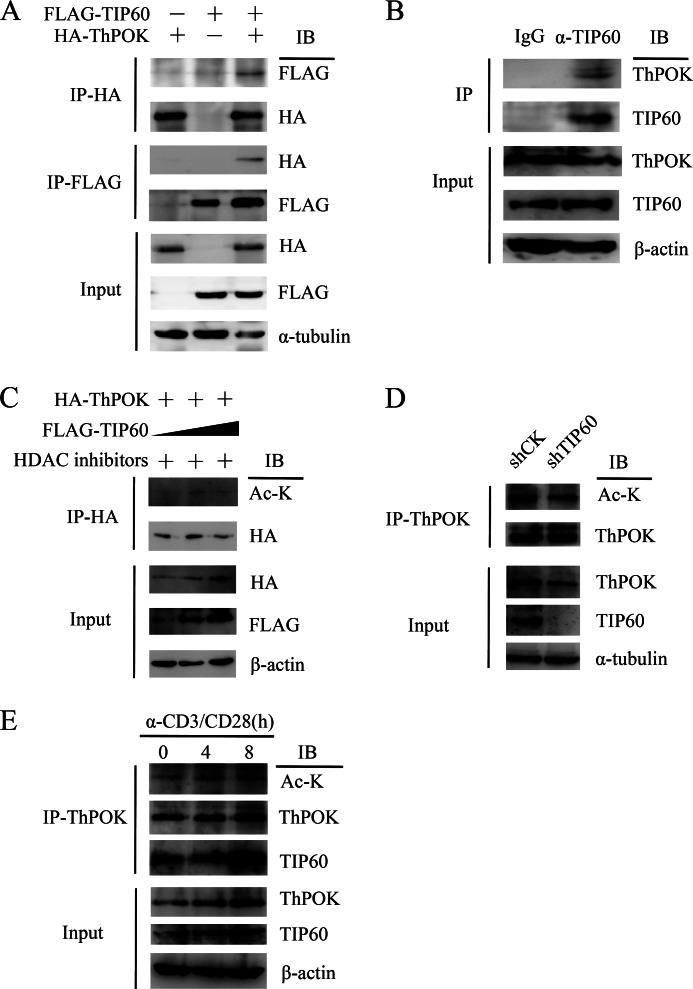
As ThPOK contains a proline-rich domain that could be potentially recognized by TIP60, we tested whether TIP60 and ThPOK could interact with each other by reciprocal immunoprecipitation. HA-tagged ThPOK (HA-ThPOK) and FLAG-tagged TIP60 (FLAG-TIP60) were ectopically expressed in HEK 293T cells, followed by reciprocal immunoprecipitation. We found a positive interaction between TIP60 and ThPOK (Fig. 2A). We then further showed that endogenous TIP60 could co-immunoprecipitate ThPOK in human primary CD4+ T cells (Fig. 2B).
FIGURE 2.
TIP60 interacts with and acetylates ThPOK. A, HEK 293T cells were co-transfected with expression vectors encoding for HA-ThPOK or FLAG-TIP60, as indicated. Cell lysates were immunoprecipitated (IP) with either anti-FLAG or anti-HA antibody and detected by Western blotting (IB). B, nuclear extracts from CD4+ T cells were immunoprecipitated with anti-TIP60 or control IgG antibodies and then analyzed by Western blotting. C, TIP60 acetylates ThPOK. HEK 293T cells were co-transfected with HA-ThPOK and increasing amounts of FLAG-TIP60; 1 mm nicotinamide and 50 μm Ex-527 were added 6 h before cell harvesting. Cell lysates were immunoprecipitated with anti-HA antibody and detected by Western blotting with the indicated antibodies. D, CD4+ T cells were transduced with lentivirus containing shRNA sequences targeting CK (control) or TIP60. Cells were cultured with anti-CD3/CD28 dynal beads and selected with puromycin for 2 days. Selected cells were lysed, and cell lysates were immunoprecipitated with anti-ThPOK antibody and analyzed by Western blotting. E, resting CD4+ T cells were stimulated with α-CD3 and α-CD28 antibodies for the indicated time periods. Cells were lysed with RIPA buffer; cell lysates were immunoprecipitated with anti-ThPOK antibody and analyzed by Western blotting.
TIP60 is a lysine acetyltransferase, which acetylates not only histones but also nuclear transcription factors such as c-Myc, p53, and FOXP3. We were interested in investigating whether TIP60 could similarly modify ThPOK by lysine acetylation. We co-transfected the HA-ThPOK-expressing plasmid with increasing amounts of FLAG-TIP60-expressing plasmid into HEK 293T cells; cells were then treated with protein deacetylation inhibitors. We found that the overexpression of TIP60 promoted ThPOK acetylation (Fig. 2C). To elucidate the role of TIP60 in primary T cells, lentivirus-mediated shRNA knockdown of endogenous TIP60 was performed. Loss of TIP60 significantly reduced the acetylation level of ThPOK (Fig. 2D). Previously, we found that short term TCR stimulation could stabilize ThPOK. Others have also shown that the acetylation of ThPOK increases its stability (21); therefore, we decided to check the acetylation level of ThPOK after TCR stimulation. As expected, short term TCR stimulation promoted the interaction between TIP60 and ThPOK, which most likely increases the acetylation of ThPOK (Fig. 2E).
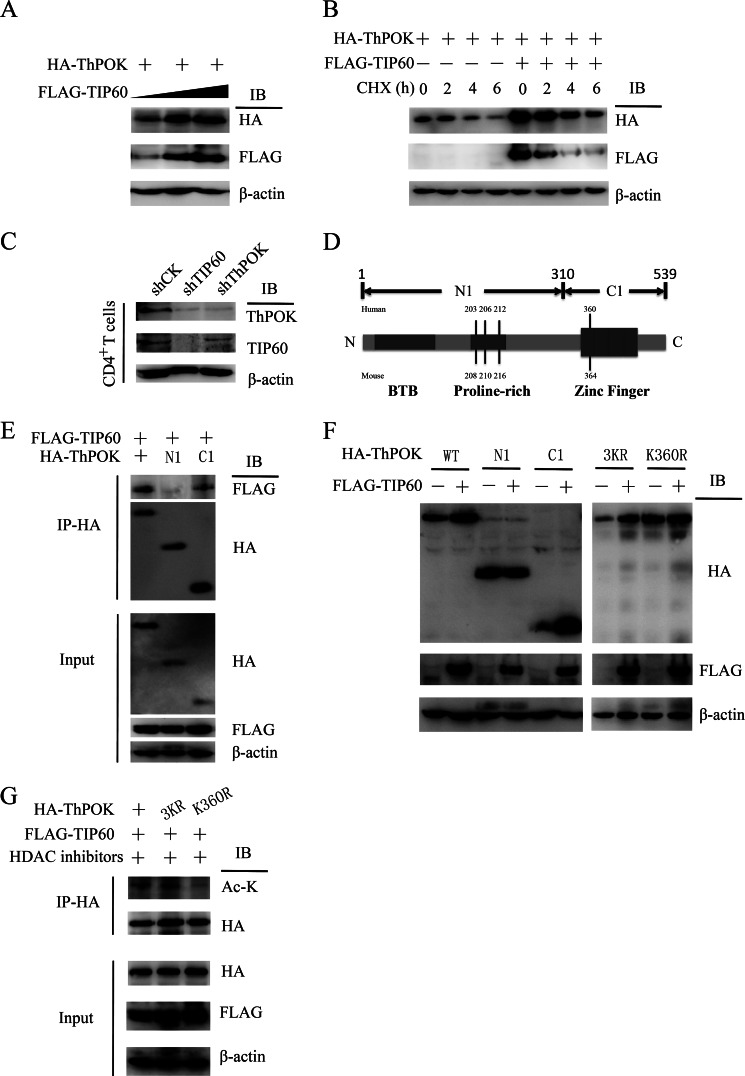
We then further tested the relationship between acetylation and protein stability. We observed that the increase in ThPOK protein level positively correlated with TIP60 level, which indicated that the overexpression of TIP60 facilitated the stabilization of ThPOK (Fig. 3A). To fully confirm this result, we treated the cells with protein synthesis inhibitor cycloheximide and examined ThPOK protein stability. HA-ThPOK was co-transfected with or without FLAG-TIP60, and cells were then treated with cycloheximide for the indicated time periods. Here, we confirmed that ThPOK stability could be positively regulated by TIP60 (Fig. 3B). To further examine TIP60-mediated stabilization of ThPOK under more physiological conditions, knockdown of endogenous TIP60 was performed in primary human CD4+ T cells. The protein level of ThPOK decreased noticeably in TIP60-silenced CD4+ T cells. Meanwhile, CD4+ T cells transduced with shRNA constructs targeting ThPOK were tested as a control (Fig. 3C). We then mapped by co-IP for the regions of ThPOK that associates with TIP60 to the C-terminal domain (residues 310–539), but not the N-terminal subdomain, which contains the proline-rich region (Fig. 3, D and E). These results indicated that the acetylated lysines were likely at the C-terminal region. Furthermore, TIP60 specifically stabilized the C-terminal region (C1) of ThPOK but not the N-terminal region (N1) (Fig. 3F). TIP60 preferentially acetylates the lysine of G(X)GK motif of histones (22). When we screened all of the residues of ThPOK, we identified that lysine 360 (GAGK motif) was contained in the C-terminal region, and thus a potential acetylation site (Fig. 3D). We found that a point mutation of lysine 360 into arginine abrogated TIP60-mediated stabilization, whereas combinatorial mutation of Lys203, Lys206, and Lys212 (lysines located in the proline-rich domain) could not (Fig. 3F). Further experiments showed that the ThPOK-K360R mutant abolished TIP60-mediated acetylation, which indicates that lysine 360 is the target of TIP60 (Fig. 3G).
FIGURE 3.
TIP60-mediated acetylation of lysine 360 stabilizes ThPOK. A, HA-ThPOK was co-transfected with increasing amounts of FLAG-TIP60 into HEK 293T cells; cell lysates were analyzed by Western blotting (IB). B, HA-ThPOK was expressed in HEK 293T cells in the presence or absence of FLAG-TIP60. Cells were treated with 20 μg/ml cycloheximide (CHX) for the indicated periods. Levels of ThPOK were determined by Western blotting. C, CD4+ T cells were transduced with lentivirus containing shRNA sequences targeting CK (control), TIP60, or ThPOK. Cells were cultured with anti-CD3/CD28 dynal beads and selected with puromycin for 5 days. Selected cells were lysed for the detection of ThPOK and TIP60 levels by Western blotting. D, domain structure of ThPOK protein. The potential acetylated lysines by TIP60 were labeled as indicated. E, FLAG-TIP60 was expressed in HEK 293T cells with HA-tagged ThPOK or its deletion variants. Cell lysates were immunoprecipitated (IP) with anti-HA antibody and detected by WB with the indicated antibodies. F, FLAG-TIP60 was expressed in HEK 293T cells with HA-tagged ThPOK, its lysine mutants, or deletion variants. Cell lysates were analyzed by Western blotting. G, HEK 293T cells were transfected with HA-tagged ThPOK, its lysine mutants, or FLAG-TIP60 as indicated. 1 mm nicotinamide and 50 μm Ex-527 were added 6 h before cell harvesting. Cell lysates were immunoprecipitated with anti-HA antibody and detected by Western blotting with the indicated antibodies.
TIP60 Augments ThPOK-mediated Repression of Eomes in CD4+ T Cells
Because CD4+ T helper cells become lineage-committed upon activation to mediate inflammatory responses, our data suggested that TIP60 could regulate inflammatory cytokine expression through modulating the activity of ThPOK. Previous studies have revealed how Eomes activity is up-regulated in the absence of ThPOK or in the presence of insufficient ThPOK expression (17, 18). Overexpression of ThPOK in CD8+ T cells specifically represses Eomes but not T-bet (20). Thus, we intended to clarify whether ThPOK could bind to the Eomes gene promoter to repress its transcription in CD4+ T cells.
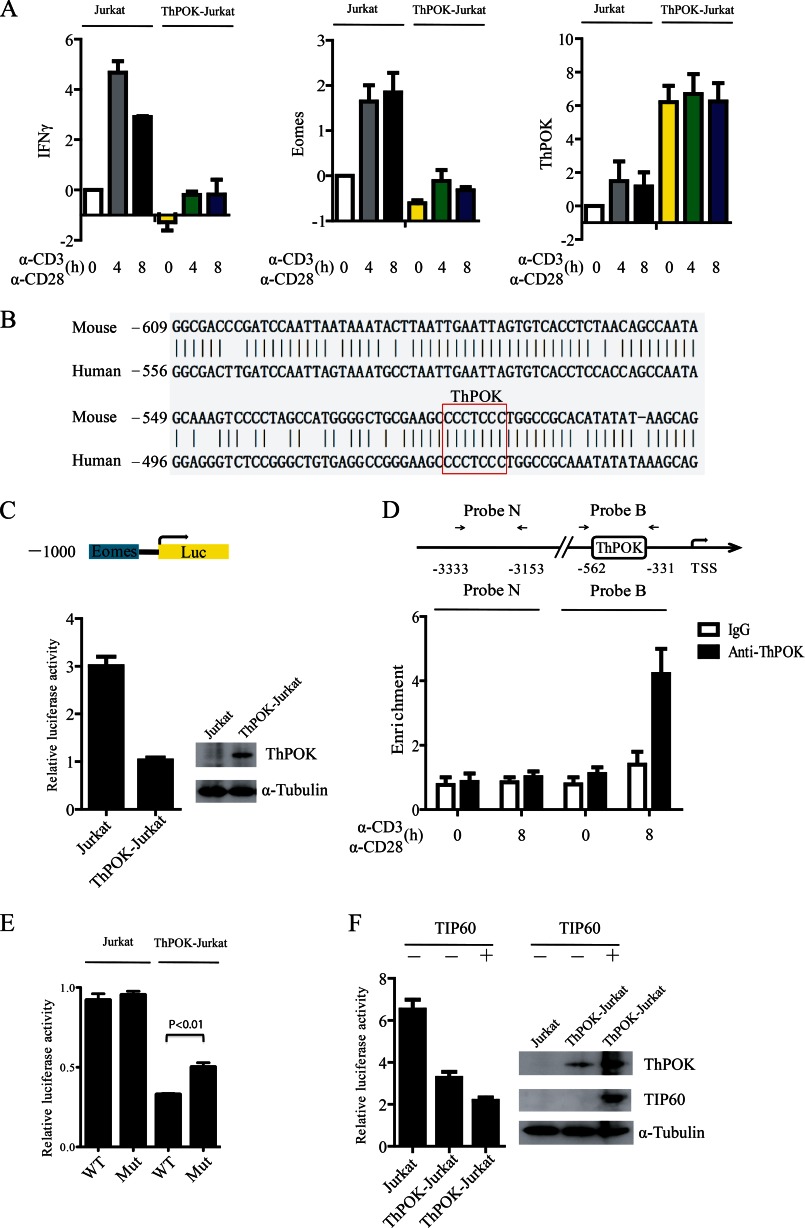
Jurkat and ThPOK-overexpressed Jurkat (ThPOK-Jurkat) cells were stimulated with soluble α-CD3/CD28 antibodies for the indicated time periods (Fig. 4A). We then examined the mRNA levels of Eomes and IFNγ. Both genes were down-regulated in ThPOK-Jurkat cells (Fig. 4A). We aligned the 1-kb region (−1,000 to 0) of the Eomes promoter from mouse and human. The alignment result showed that there was a consensus ThPOK binding sequence CCCTCCC (19, 23, 24), which suggested that Eomes is potentially a direct transcriptional target of ThPOK (Fig. 4B). We next subcloned the 1-kb region of the human Eomes promoter into the pGL3-Basic Luciferase vector and transfected the Eomes luciferase (Eomes-Luc) reporter plasmid into Jurkat or ThPOK-Jurkat cells. We found that the Eomes-Luc reporter activity was significantly repressed in ThPOK-Jurkat cells (Fig. 4C). To further confirm the involvement of ThPOK in Eomes gene transcription we analyzed whether ThPOK could bind to the Eomes promoter in primary CD4+ T cells using a ChIP assay. Human CD4+ T cells were stimulated with anti-CD3/CD28 dynal beads for the indicated periods, and the binding of ThPOK to the promoter (Probe B region with a consensus ThPOK binding site) was notably higher after anti-CD3/CD28 stimulation (Fig. 4D). Furthermore, mutation of the ThPOK binding site could reverse ThPOK-mediated repression of Eomes-Luc reporter activity (Fig. 4E), but not completely, perhaps due to other less consensus binding sites that may be responsible for ThPOK binding and activity at the Eomes promoter.
FIGURE 4.
Overexpression of TIP60 augments ThPOK-mediated repression of Eomes. A, a Jurkat cell line stably expressing ThPOK (ThPOK-Jurkat) was established. Jurkat and ThPOK-Jurkat cells were stimulated with α-CD3 and α-CD28 antibodies for the indicated time periods. Total RNA was extracted, and the mRNA levels of Eomes, IFNγ, and ThPOK were measured by qRT-PCR. Data show the mean of three separate experiments. Error bars indicate S.D. B, The 1-kb region (−1,000 to 0) of the Eomes promoter from mouse and human were aligned online using NCBI Blast (bl2seq). One consensus ThPOK binding site CCCTCCC was found on the Eomes promoter from both mouse and human. C, reporter transfection assay of the repressive activity of Eomes promoter by ThPOK in Jurkat and ThPOK-Jurkat cells. Cells were transfected with the pGL3-Eomes-Luc reporter which carries a 1,000-bp region of the Eomes promoter. 48 h later, luciferase activity was analyzed. D, CD4+ T cells were stimulated with anti-CD3/CD28 dynal beads for the indicated time periods. The binding of ThPOK to the Eomes promoter was determined by a ChIP assay with probe B, and probe N was used as a control. E, Jurkat or ThPOK-Jurkat cells were transfected with either pGL3-Eomes-Luc reporter or mutated reporter (CCCTCCC to CATTCCC) as indicated. The luciferase activity was analyzed 48 h later. Data show the mean of three separate experiments. Error bars indicate S.D. F, the pGL3-Eomes-Luc reporter was co-transfected with or without FLAG-TIP60 into Jurkat or ThPOK-Jurkat cells as indicated. The luciferase activity in the transfected cells was then determined. Error bars of C, D, and F show the S.D. from the mean in one experiment representative of three independent experiments.
As we found that TIP60 could stabilize ThPOK in CD4+ T cells we decided to test whether TIP60 could promote ThPOK-mediated repression of Eomes expression. Jurkat and ThPOK-Jurkat cells were transfected with the Eomes luciferase reporter and FLAG-TIP60-encoding plasmids as indicated. We found that TIP60 stabilized ThPOK and increased the repression of the Eomes promoter as indicated by the decrease in luciferase activity (Fig. 4F).
TIP60 Positively Regulates ThPOK-mediated Repression of Eomes in CD4+ T Cells
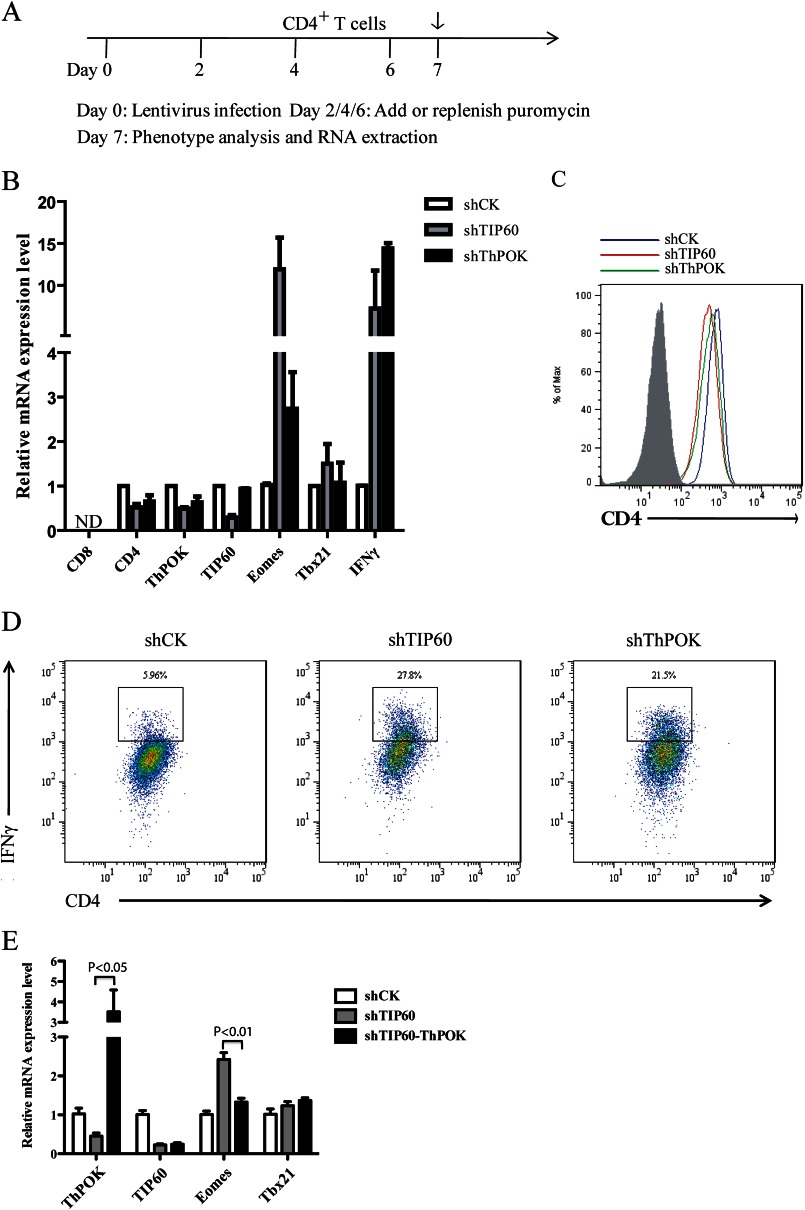
ThPOK was reported to bind to the silencers of the CD4 and ThPOK gene loci to abolish their activity (16). We hypothesized that TIP60 could positively regulate ThPOK-mediated transcriptional regulation of CD4, ThPOK, and Eomes. To test this, primary CD4+ T cells were transduced with shRNA constructs targeting CK (control), TIP60, or ThPOK and then subjected to analysis by qRT-PCR or flow cytometry (Fig. 5A). Total RNA was then extracted, and the mRNA levels of CD8, CD4, ThPOK, TIP60, Eomes, Tbx21, and IFNγ were determined by qRT-PCR. No CD8 transcripts were detected, which indicated that the knockdown of either TIP60 or ThPOK did not induce CD8 expression in CD4+ CD8− T cells (Fig. 5B). Both CD4 and ThPOK were down-regulated upon knockdown of TIP60 or ThPOK, whereas Eomes and IFNγ were both noticeably up-regulated (Fig. 5B). However, the expression of Tbx21, another T-box transcriptional activator of IFN-γ, was not dramatically affected (Fig. 5B). The protein levels of CD4 and IFNγ were further tested by flow cytometry. Knockdown of either TIP60 or ThPOK in CD4+ CD8− T cells slightly decreased CD4 protein level, which was consistent with the previous mRNA expression result (Fig. 5C). Meanwhile, we observed a higher percentage of CD4+ IFNγ+ T cells in cells depleted of TIP60 or ThPOK (Fig. 5D). To test whether the up-regulation of Eomes mRNA in TIP60-depleted cells was ThPOK-dependent, we overexpressed ThPOK in TIP60-depleted cells. The overexpression of ThPOK significantly promoted the repression of Eomes (Fig. 5E), which indicated that the loss of ThPOK activity in TIP60-depleted CD4+ T cells was responsible for the increase in transcription of Eomes. Our data suggest that TIP60 and ThPOK could significantly repress Eomes expression and play a potential role in modulating inflammation (Fig. 6).
FIGURE 5.
TIP60 positively regulates ThPOK-mediated repression of Eomes in CD4+ T cells. A, timeline showing the knockdown of TIP60 and ThPOK in CD4+ T cells. On day 0, CD4+ T cells were transduced with lentivirus. 48 h later, cultures were treated with puromycin and replenished every other day. On day 7, selected CD4+ T cells were subjected to analysis by FACS or qRT-PCR. B, CD4+ T cells transduced with lentivirus-containing shRNA constructs targeting CK (control), TIP60, or ThPOK. Cells were cultured with anti-CD3/CD28 dynal beads and selected with puromycin for 5 days prior to RNA extraction. Total RNA was extracted, and mRNA levels of CD8, CD4, TIP60, ThPOK, Eomes, Tbx21, and IFNγ were determined by qRT-PCR. Data show the mean of three separate experiments. Error bars indicate S.D. C, expression of CD4 in selected CD4+ T cells described in B. Surface CD4 level was analyzed by flow cytometry. The shaded area represents the isotype control. D, IFNγ production by CD4+ T cells described in B. IFNγ level was observed after restimulation of cells with phorbol 12-myristate 13-acetate, ionomycin, and Golgi Stop for 4 h. E, CD4+ T cells transduced with lentivirus-containing shRNA constructs targeting CK (shCK) or TIP60 (shTIP60) as described in A. On day 4, shTIP60-transduced CD4+ T cells were also transduced with lentivirus-encoding ThPOK protein and GFP under the internal ribosome entry site. These cells (shTIP60-ThPOK) were cultured with anti-CD3/CD28 dynal beads and selected with puromycin for another 3 days. Both GFP+ and GFP− cells were selected by FACS prior to RNA extraction. Total RNA was extracted and mRNA levels of ThPOK, TIP60, Eomes, and Tbx21 were determined by qRT-PCR. Data show the mean of three separate experiments. Error bars indicate S.D.
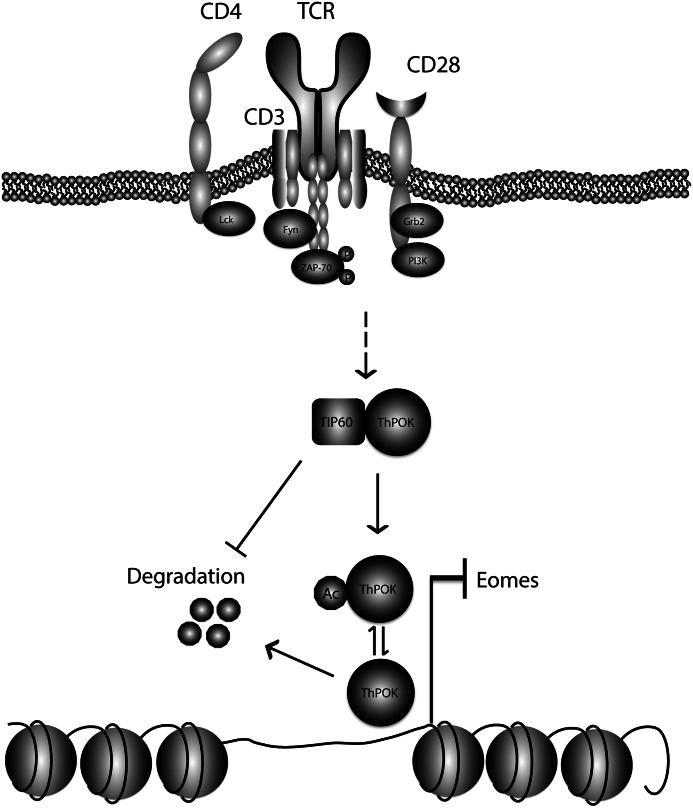
FIGURE 6.
Working model describing the role of TIP60 for ThPOK-mediated repression of Eomes. In human CD4+ T cells, short term TCR stimulation promotes TIP60 association with and acetylation of ThPOK to prevent its degradation. TIP60-mediated stabilization of ThPOK augments the transcriptional repression of Eomes, a T-box transcriptional activator of IFNγ, which leads to the reduced production of IFNγ. Thus, TIP60 and ThPOK may contribute to the dampening of Th1-type inflammation.
DISCUSSION
Eomes was originally characterized as an inflammation-inducing gene in CD8+ T cells and Natural Killer cells, indispensible for IFNγ production during host defense responses against intracellular bacteria and virus infections (25–28). Recently, more attention has been paid to the function of Eomes in CD4+ T helper cells; Eomes is required for Th1 cell differentiation and is generally repressed in other T helper cell lineages. For example, Eomes directs naïve CD4+ T cell polarization toward the Th1 lineage versus Th17, where TGF-β (required for Th17 skewing) suppresses Eomes expression via the c-Jun N-terminal kinase (JNK)-c-Jun signaling pathway (29–31). Eomes is also suppressed in effector Th2 cells; GATA3-deficient naïve CD4+ T cells produce a significant amount of IFNγ during Th2 differentiation, which is dependent on Eomes but not T-bet (32). GATA3 was reported to induce ThPOK expression (33) and may suppress Eomes in a ThPOK-dependent manner. Moreover, in memory Th2 cells, Eomes is highly expressed and interacts with GATA3 to prevent its binding to the IL-5 promoter (34). Here, we have identified another novel molecular pathway which could explain the molecular basis by which Eomes is repressed in CD4+ T helper cells in a TIP60- and ThPOK-dependent manner.
As we postulated, TIP60 was found to augment ThPOK-mediated repression of Eomes through stabilizing ThPOK; we speculate that TIP60 may also recruit histone deacetylases to ThPOK transcriptional complexes to epigenetically silence Eomes by the deacetylation of its promoter, such as HDAC7/9 and SIRT1, which have been reported to associate with TIP60 (8, 9, 35). In addition, ThPOK can recruit HDAC4/5 to the CD8 gene loci to impair CD8 transcription (19). However, it needs to be further substantiated as to whether these histone deacetylases could be recruited to the promoter of Eomes in CD4+ T cells.
The incontrollable overproduction of IFNγ by CD4+ T helper cells may coincide with the development of autoimmune disease (36, 37). CD4+ CD25+ CD127low FOXP3+ regulatory T cells are essential for peripheral tolerance and the prevention of autoimmune diseases (38). Strong evidence shows that autoreactive Treg cells can suppress adaptive immune responses and dampen inflammation (39). Therefore, TIP60, through positive regulation of its function, may be a potential anti-inflammation target because it inhibits IFNγ production in CD4+ T helper cells and also strengthens the suppressive function of Treg cells by promoting FOXP3-mediated transcriptional repression as we had previously identified (8). Although TIP60 has been shown to be involved in DNA repair, apoptosis, and Treg function, here we give a new insight into the molecular mechanism by which TIP60 suppresses Eomes and attenuates the production of the inflammatory cytokine IFNγ in CD4+ T cells. Thus, our work may provide a novel strategy for therapeutic studies toward the treatment of Th1-related diseases such as cancer, autoimmunity, and infectious diseases (36, 40).
This work was supported, in whole or in part, by National Institutes of Health-National Science Foundation of China Collaborative Grant 81161120417. This work was also supported by National Science Foundation of China Grants 30972702, 31170825, 81270083, 31150110337, 31200646, 31200647, and 81271835; Shanghai Rising Star Program 10QA1407900; the Novo Nordisk-Chinese Academy of Sciences Foundation; the Sanofi-Aventis-Shanghai Institutes for Biological Sciences scholarship program, and the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Grants 2010KIP205 and 2012KIP204.
- TIP60
- Tat-interactive protein, 60 kDa
- Eomes
- Eomesodermin
- ThPOK
- Th-inducing POK
- Luc
- luciferase
- qRT-PCR
- quantitative RT-PCR
- RIPA
- radioimmuneprecipitation assay
- SP
- single positive
- TCR
- T cell receptor
- Treg
- regulatory T.
REFERENCES
- 1. Kamine J., Elangovan B., Subramanian T., Coleman D., Chinnadurai G. (1996) Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology 216, 357–366 [DOI] [PubMed] [Google Scholar]
- 2. Yamamoto T., Horikoshi M. (1997) Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 272, 30595–30598 [DOI] [PubMed] [Google Scholar]
- 3. Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin S. Y., Li T. Y., Liu Q., Zhang C., Li X., Chen Y., Zhang S. M., Lian G., Liu Q., Ruan K., Wang Z., Zhang C. S., Chien K. Y., Wu J., Li Q., Han J., Lin S. C. (2012) GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481 [DOI] [PubMed] [Google Scholar]
- 5. Ikura T., Ogryzko V. V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 [DOI] [PubMed] [Google Scholar]
- 6. Patel J. H., Du Y., Ard P. G., Phillips C., Carella B., Chen C. J., Rakowski C., Chatterjee C., Lieberman P. M., Lane W. S., Blobel G. A., McMahon S. B. (2004) The c-Myc oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24, 10826–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang Y., Luo J., Zhang W., Gu W. (2006) Tip60-dependent acetylation of p53 modulates the decision between cell cycle arrest and apoptosis. Mol. Cell 24, 827–839 [DOI] [PubMed] [Google Scholar]
- 8. Li B., Samanta A., Song X., Iacono K. T., Bembas K., Tao R., Basu S., Riley J. L., Hancock W. W., Shen Y., Saouaf S. J., Greene M. I. (2007) FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. U.S.A. 104, 4571–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao H., Chung J., Kao H. Y., Yang Y. C. (2003) Tip60 is a co-repressor for STAT3. J. Biol. Chem. 278, 11197–11204 [DOI] [PubMed] [Google Scholar]
- 10. Gorrini C., Squatrito M., Luise C., Syed N., Perna D., Wark L., Martinato F., Sardella D., Verrecchia A., Bennett S., Confalonieri S., Cesaroni M., Marchesi F., Gasco M., Scanziani E., Capra M., Mai S., Nuciforo P., Crook T., Lough J., Amati B. (2007) Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448, 1063–1067 [DOI] [PubMed] [Google Scholar]
- 11. Goswami R., Kaplan M. H. (2011) A brief history of IL-9. J. Immunol. 186, 3283–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sliva D., Zhu Y. X., Tsai S., Kamine J., Yang Y. C. (1999) Tip60 interacts with human interleukin-9 receptor α-chain. Biochem. Biophys. Res. Commun. 263, 149–155 [DOI] [PubMed] [Google Scholar]
- 13. He X., Park K., Wang H., He X., Zhang Y., Hua X., Li Y., Kappes D. J. (2008) CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 [DOI] [PubMed] [Google Scholar]
- 14. He X., He X., Dave V. P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B. A., Kappes D. J. (2005) The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 [DOI] [PubMed] [Google Scholar]
- 15. Carpenter A. C., Grainger J. R., Xiong Y., Kanno Y., Chu H. H., Wang L., Naik S., dos Santos L., Wei L., Jenkins M. K., O'Shea J. J., Belkaid Y., Bosselut R. (2012) The transcription factors Thpok and LRF are necessary and partly redundant for T helper cell differentiation. Immunity 37, 622–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muroi S., Naoe Y., Miyamoto C., Akiyama K., Ikawa T., Masuda K., Kawamoto H., Taniuchi I. (2008) Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 9, 1113–1121 [DOI] [PubMed] [Google Scholar]
- 17. Wang L., Wildt K. F., Castro E., Xiong Y., Feigenbaum L., Tessarollo L., Bosselut R. (2008) The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egawa T., Littman D. R. (2008) ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9, 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rui J., Liu H., Zhu X., Cui Y., Liu X. (2012) Epigenetic silencing of CD8 genes by ThPOK-mediated deacetylation during CD4 T cell differentiation. J. Immunol. 189, 1380–1390 [DOI] [PubMed] [Google Scholar]
- 20. Jenkinson S. R., Intlekofer A. M., Sun G., Feigenbaum L., Reiner S. L., Bosselut R. (2007) Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J. Exp. Med. 204, 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang M., Zhang J., Rui J., Liu X. (2010) p300-mediated acetylation stabilizes the Th-inducing POK factor. J. Immunol. 185, 3960–3969 [DOI] [PubMed] [Google Scholar]
- 22. Kimura A., Horikoshi M. (1998) Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells 3, 789–800 [DOI] [PubMed] [Google Scholar]
- 23. Galéra P., Musso M., Ducy P., Karsenty G. (1994) c-Krox, a transcriptional regulator of type I collagen gene expression, is preferentially expressed in skin. Proc. Natl. Acad. Sci. U.S.A. 91, 9372–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galéra P., Park R. W., Ducy P., Mattéi M. G., Karsenty G. (1996) c-Krox binds to several sites in the promoter of both mouse type I collagen genes: structure/function study and developmental expression analysis. J. Biol. Chem. 271, 21331–21339 [DOI] [PubMed] [Google Scholar]
- 25. Pearce E. L., Mullen A. C., Martins G. A., Krawczyk C. M., Hutchins A. S., Zediak V. P., Banica M., DiCioccio C. B., Gross D. A., Mao C. A., Shen H., Cereb N., Yang S. Y., Lindsten T., Rossant J., Hunter C. A., Reiner S. L. (2003) Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science 302, 1041–1043 [DOI] [PubMed] [Google Scholar]
- 26. Intlekofer A. M., Takemoto N., Wherry E. J., Longworth S. A., Northrup J. T., Palanivel V. R., Mullen A. C., Gasink C. R., Kaech S. M., Miller J. D., Gapin L., Ryan K., Russ A. P., Lindsten T., Orange J. S., Goldrath A. W., Ahmed R., Reiner S. L. (2005) Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 [DOI] [PubMed] [Google Scholar]
- 27. Takemoto N., Intlekofer A. M., Northrup J. T., Wherry E. J., Reiner S. L. (2006) Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 177, 7515–7519 [DOI] [PubMed] [Google Scholar]
- 28. Gordon S. M., Chaix J., Rupp L. J., Wu J., Madera S., Sun J. C., Lindsten T., Reiner S. L. (2012) The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y., Xu J., Niu Y., Bromberg J. S., Ding Y. (2008) T-bet and eomesodermin play critical roles in directing T cell differentiation to Th1 versus Th17. J. Immunol. 181, 8700–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ichiyama K., Sekiya T., Inoue N., Tamiya T., Kashiwagi I., Kimura A., Morita R., Muto G., Shichita T., Takahashi R., Yoshimura A. (2011) Transcription factor Smad-independent T helper 17 cell induction by transforming growth factor-β is mediated by suppression of eomesodermin. Immunity 34, 741–754 [DOI] [PubMed] [Google Scholar]
- 31. Narayanan S., Silva R., Peruzzi G., Alvarez Y., Simhadri V. R., Debell K., Coligan J. E., Borrego F. (2010) Human Th1 cells that express CD300a are polyfunctional and after stimulation up-regulate the T-box transcription factor eomesodermin. PloS One 5, e10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yagi R., Junttila I. S., Wei G., Urban J. F., Jr., Zhao K., Paul W. E., Zhu J. (2010) The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 32, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L., Carr T., Xiong Y., Wildt K. F., Zhu J., Feigenbaum L., Bendelac A., Bosselut R. (2010) The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur. J. Immunol. 40, 2385–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Endo Y., Iwamura C., Kuwahara M., Suzuki A., Sugaya K., Tumes D. J., Tokoyoda K., Hosokawa H., Yamashita M., Nakayama T. (2011) Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity 35, 733–745 [DOI] [PubMed] [Google Scholar]
- 35. Yamagata K., Kitabayashi I. (2009) Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem. Biophys. Res. Commun. 390, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J. (2007) Yin and yang interplay of IFN-γ in inflammation and autoimmune disease. J. Clin. Invest. 117, 871–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen J., Liu X. (2009) The role of interferon γ in regulation of CD4+ T-cells and its clinical implications. Cell. Immunol. 254, 85–90 [DOI] [PubMed] [Google Scholar]
- 38. Vignali D. A., Collison L. W., Workman C. J. (2008) How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. von Herrath M. G., Harrison L. C. (2003) Antigen-induced regulatory T cells in autoimmunity. Nat. Rev. Immunol. 3, 223–232 [DOI] [PubMed] [Google Scholar]
- 40. Berry M. P., Graham C. M., McNab F. W., Xu Z., Bloch S. A., Oni T., Wilkinson K. A., Banchereau R., Skinner J., Wilkinson R. J., Quinn C., Blankenship D., Dhawan R., Cush J. J., Mejias A., Ramilo O., Kon O. M., Pascual V., Banchereau J., Chaussabel D., O'Garra A. (2010) An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]