Background: Polyamines play roles in bacterial cell-to-cell signaling processes.
Results: In Proteus mirabilis, PlaP is important for putrescine uptake, swarming motility, and urothelial cell invasion, and the putrescine transport inhibitor Triamide-44 inhibits these processes.
Conclusion: PlaP is the primary putrescine transporter in P. mirabilis.
Significance: This research suggests that novel drug cocktails that target both microbial putrescine uptake and biosynthesis can be developed.
Keywords: Bacteria, Bacterial Pathogenesis, Drug Design, Microbial Pathogenesis, Microbiology, Polyamines, Transporters
Abstract
Previously, we reported that the speA gene, encoding arginine decarboxylase, is required for swarming in the urinary tract pathogen Proteus mirabilis. In addition, this previous study suggested that putrescine may act as a cell-to-cell signaling molecule (Sturgill, G., and Rather, P. N. (2004) Mol. Microbiol. 51, 437–446). In this new study, PlaP, a putative putrescine importer, was characterized in P. mirabilis. In a wild-type background, a plaP null mutation resulted in a modest swarming defect and slightly decreased levels of intracellular putrescine. In a P. mirabilis speA mutant with greatly reduced levels of intracellular putrescine, plaP was required for the putrescine-dependent rescue of swarming motility. When a speA/plaP double mutant was grown in the presence of extracellular putrescine, the intracellular levels of putrescine were greatly reduced compared with the speA mutant alone, indicating that PlaP functioned as the primary putrescine importer. In urothelial cell invasion assays, a speA mutant exhibited a 50% reduction in invasion when compared with wild type, and this defect could be restored by putrescine in a PlaP-dependent manner. The putrescine analog Triamide-44 partially inhibited the uptake of putrescine by PlaP and decreased both putrescine stimulated swarming and urothelial cell invasion in a speA mutant.
Introduction
Polyamines are aliphatic amines that have two or more amino groups. The basic polyamines are putrescine, spermidine, and spermine, and it has been reported that several plants (1) and bacteria (2) produce a variety of unusual polyamines such as norspermidine. Polyamines are widely distributed from prokaryotic cells (3) to higher plants and animals (4), and polyamines play important roles in a variety of cellular functions, including translation, transcription, and chromatin remodeling.
Recent studies suggest that polyamines play important roles in cell-to-cell signaling in bacteria. In Yersinia pestis, the etiological agent of bubonic and pneumonic plague, SpeA and SpeC, which include the synthetic pathways for putrescine generation, are indispensable in forming normal biofilms. Indeed, the ΔspeA ΔspeC mutant could form biofilms only when the medium was supplemented with putrescine (5). In Vibrio cholerae, a human intestinal pathogen, norspermidine activates biofilm formation in a norspermidine sensor NspS-dependent manner (6) and a mutant in the biosynthetic pathway of norspermidine exhibited severe defects in biofilm formation (7). In Bacillus subtilis, self-produced norspermidine mediates biofilm disassembly by targeting exopolysaccharides (8), and biofilm formation of a speA mutant is dependent on spermidine (9). In Escherichia coli, polyamines are important for both biofilm formation (10) and surface motility (11, 12).
In addition, a growing body of evidence indicates that polyamines are important for bacterial virulence. For example, in Salmonella enterica sv. typhimurium, polyamines are required for virulence (13); in Legionella pneumophila, intracellular growth is enhanced by polyamines derived from host cells (14), and in Streptococcus pneumoniae, polyamine biosynthetic pathways and the spermidine transporter PotABCD are important for infection (15, 16).
In E. coli, putrescine is synthesized from ornithine by ornithine decarboxylase (SpeC and SpeF) or from arginine by the sequential actions of arginine decarboxylase (SpeA) and agmatinase (SpeB) (17). Spermidine can be formed from putrescine by the addition of an aminopropyl group catalyzed by an aminopropyltransferase (SpeE) (18). The source of the propylamine group is decarboxylated S-adenosyl-l-methionine, which is produced by the action of the adenosylmethionine decarboxylase (SpeD) (19). In addition, there are four putrescine transporters, PotE (20), PotFGHI (21), PuuP (22), and PlaP (12), and one spermidine transporter PotABCD (23) in E. coli.
The Gram-negative bacterium Proteus mirabilis is well known for its ability to cause urinary tract infections in humans (24). A prominent feature of P. mirabilis is the ability to carry out a highly coordinated multicellular migration on solid media termed swarming (25–27). Swarming involves a complex repeating cycle of differentiation between two cell types, vegetative and swarmer cells. The vegetative form predominates in liquid and is a typical Gram-negative rod. When vegetative cells are placed on solid surfaces such as agar, the cell differentiates into a swarmer cell after a period of 3–4 h. Swarmer cells express levels of flagellin, encoded by the flaA locus, that are 10-fold higher than vegetative cells (28). The process of swarming requires that swarmer cells align together to form multicellular rafts that translocate across solid surfaces (29). Differentiated swarmer cells are more invasive for urothelial cells than the vegetative cells (30). P. mirabilis also produces several virulence factors that are coordinately regulated with swarmer cell differentiation, such as urease, IgA protease, and hemolysin (31–33).
It has previously been reported that putrescine is required for swarmer cell differentiation in P. mirabilis, possibly by acting as a cell-to-cell signaling molecule (34). When extracellular polyamines are used as signaling molecules, a polyamine sensor or importer is required because the hydrophilic nature of polyamines prevents their transfer across membranes. Consistent with this, we previously reported in E. coli that spermidine and putrescine induce bacterial surface motility on semisolid media in a spermidine importer PotABCD- (11) and putrescine importer PlaP (12)-dependent manner, respectively.
In E. coli, PlaP is the most recently discovered putrescine importer, and its activity depends on the protonmotive force (12). The Km of PlaP (155 μm) is 40–300 times higher than that of other importers reported previously (12). The low affinity of PlaP for putrescine may allow E. coli to sense the cell density depending on the concentration of extracellular putrescine (12). Orthologs of PlaP are distributed among Enterobacteriaceae such as Yersinia, Serratia, Photorhabdus, Pectobacterium, and Morganella. In P. mirabilis, a putative PlaP ortholog (PMI0843) is 459 amino acids and is predicted to be localized to the cytoplasmic membrane. In this study, we report that PlaP is important for putrescine uptake, swarming motility, and urothelial cell invasion of P. mirabilis. In addition, the inhibition of putrescine uptake using the putrescine transport inhibitor Triamide-44 decreased swarming and urothelial cell invasion.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions
E. coli and P. mirabilis were grown in modified Luria-Bertani (LB) broth (1% (w/v) tryptone, 0.5% yeast extract, 0.5% NaCl) with reciprocal shaking at 250 rpm at 37 °C or on LB plates (containing 1.5% agar) at 37 °C. For construction of P. mirabilis mutants, LSW plates (1% tryptone, 0.5% yeast extract, 0.5% glycerol, 2% agar) were used. For sucrose selection, 10% sucrose was added to LSW plates. For the analysis of intracellular putrescine concentrations, overnight cultures of P. mirabilis strains were inoculated to the same initial cell density (A600 of 0.03) into 2 ml of LB or LB + 0.25% NaCl (1% tryptone, 0.5% yeast extract, 0.25% NaCl) broth in test tubes (13 × 100 mm) and were grown at 37 °C with reciprocal shaking at 250 rpm. When the absorbance (A600) reached 0.2 (∼1.5 h after inoculation), putrescine (Sigma) and/or Triamide-44 (35, 36) were added in the concentrations shown in the figure legends. Cells were then harvested after 40 min (when the concentration of putrescine was 25 μm) or 1 h (when the concentration of putrescine was 50 μm or media used was LB + 0.25% NaCl). For cell invasion assays, overnight cultures of P. mirabilis strains were inoculated to the same initial cell density (A600 of 0.03) into 2 ml of LB broth in test tubes (13 × 100 mm) and were grown at 37 °C with reciprocal shaking at 250 rpm. When the A600 reached 0.2 (∼1.5 h after inoculation), cells were harvested and subjected to the infection assay. Antibiotics were used for selection at concentrations of 25 μg/ml for both chloramphenicol and streptomycin and 100 μg/ml for ampicillin for E. coli. Antibiotic concentrations for the selection of P. mirabilis were 100 μg/ml for chloramphenicol, 35 μg/ml for streptomycin, and 15 μg/ml for tetracycline.
Strain and Plasmid Constructions
For cloning purposes, E. coli strains XL1-Blue and CC118 were used (Table 1). For conjugal matings described previously (37), E. coli strain either SK680 (for introducing ΔplaP) or SK707 (for complementation of plaP+) was used as the donor strains, and either P. mirabilis PM7002 or PM437 was used as the recipients. Exconjugants were selected on tetracycline and streptomycin to select for a Campbell-type insertion into the plaP region. The resulting strains, SK685 and SK732 harboring both the wild-type allele and the mutated plaP alleles, were subjected to sucrose selection to select for the second recombination event, which results in excision of the vector leaving either the wild-type allele or the plaP::Cm mutation. The plaP::Cm mutation in the resulting strains SK694 and SK738 was confirmed by Southern blot analysis and PCR (supplemental Fig. S1). For complementation of the plaP mutant, the wild-type plaP gene was inserted into the chromosome of the plaP mutant via pKNG101 integration using SK707 as the donor strain and either SK694 or SK738 as the recipient strains. The complemented allele of the resulting strains (SK713 and SK739) was confirmed by Southern blotting and PCR (supplemental Fig. S1). To construct pSK+CmR, plasmid pUT::mini-Tn5Cm was digested using EcoRI, yielding a 3-kb CmR fragment that was subcloned into pBluescriptII SK(−) digested with EcoRI. To construct pKNG101-plaP+, the plaP (PMI0843) gene and 500-bp upstream and downstream region of plaP was amplified by PCR using Finnzyme (Thermo scientific) as polymerase, “plaP.up” and “plaP.down” as primers, and P. mirabilis HI4320 genomic DNA as template. The resulting 2,520-bp fragment was digested by SpeI and SmaI, whose sites were present in the PCR primers. The resulting fragment was ligated to pKNG101 digested with SpeI and SmaI. To construct pKNG101-ΔplaP, pKNG101-plaP+ was digested using ScaI and AleI, which removes an internal segment of the plaP gene. The resulting 8.2-kb fragment was ligated to the 3.6-kb fragment containing a chloramphenicol resistance gene derived by PCR using pSK+CmR as a template, the M13For and M13Rev primers, and Finnzyme as polymerase. To construct a plaPp-lacZ transcriptional fusion in pQF50, a 1,030-bp region, including the plaP upstream region and 30 bp of the plaP gene, was amplified by PCR using Finnzyme polymerase, “plaP up 1,000 bp” and “plaP 30-bp down ATG” as primers, and HI4320 genomic DNA. Amplified 1-kbp fragment was digested with HindIII (in plaP 30-bp down ATG) and BamHI (in “plaP up 1000 bp”). The resulting fragment was ligated to pQF50 digested by HindIII and BamHI. To construct pQF50-plaPp-lacZ+ (SmR), pQF50-plaPp-lacZ+ (AmpR) was partially digested using ScaI. The resulting 7.8-kbp fragment (digested only once) was ligated to the 2.1-kbp SmR fragment generated by the digestion of pKNG101 by EcoRV and SmaI.
TABLE 1.
Strains, plasmids, and oligonucleotides in this study
| Strain, plasmid, or oligonucleotide | Characteristic or sequence | Source or Ref. |
|---|---|---|
| Strains (E. coli) | ||
| SK680 | SM10 (λpir)/pKNG101-ΔplaP | This study |
| SK707 | SM10 (λpir)/pKNG101-plaP+ | This study |
| SM10 (λpir) | thi-1 thr leu tonA lacY supE recA RP4–2-Tc::Mu KmR λpir | V. de Lorenzo |
| Strains (P. mirabilis) | ||
| HI4320 | Wild type, complete genome sequence was performed | 51, 52 |
| PM7002 | Wild type | ATCC |
| PM437 | speA::mini-Tn5 (KmR) lacZ1 | 34 |
| SK685 | speA::mini-Tn5 (KmR) lacZ1, plaP+ ΔplaP::CmR | This study |
| SK694 | speA::mini-Tn5 (KmR) lacZ1 ΔplaP::CmR | This study |
| SK713 | speA::mini-Tn5 (KmR) lacZ1 ΔplaP::CmR/plaP+ complemented | This study |
| SK732 | plaP+ ΔplaP::CmR | This study |
| SK738 | ΔplaP::CmR | This study |
| SK739 | ΔplaP::CmR/plaP+ complemented | This study |
| SK750 | pQF50- plaPp-lacZ+ (SmR)/PM7002 | This study |
| Plasmids | ||
| pBluescriptII SK(−) | ColE1 replicon lacZα AmpR | Stratagene |
| pKNG101 | R6K replicon mob+ sacB+R+ SmR | 37 |
| pSK+CmR | ColE1 replicon lacZα AmpR CmR | Laboratory stock |
| pKNG101-plaP+ | R6K replicon mob+ sacB+R+ plaP+ SmR | This study |
| pKNG101-ΔplaP | R6K replicon mob+ sacB+R+ ΔplaP::CmR SmR | This study |
| pQF50- plaPp-lacZ+ (AmpR) | pRO1600 replicon plaPp-lacZ+ AmpR | This study |
| pQF50- plaPp-lacZ+ (SmR) | pRO1600 replicon plaPp-lacZ+ SmR | This study |
| pQF50 | pRO1600 replicon lacZ (promoterless) AmpR | 41 |
| pUT::mini-Tn5Cm | R6K replicon mini-Tn5Cm AmpR | 45 |
| Oligonucleotides | ||
| plaP.up | 5′-CCCACTAGTGATTTGTTAATGTGTTAATGA-3′ | |
| plaP.down | 5′-AAACCCGGGTAAAAATAAGGCGAATTACA-3′ | |
| plaP.outside | 5′-CGTGCTGGTGCATCTAATAC-3′ | |
| plaP.cDNA | 5′-GCGACTAACGCAACAACAAA-3′ | |
| plaP RT for | 5′-ATGGGGTTGGCATTTTTACA-3′ | |
| plaP RT rev | 5′-CCAACATGTGGACTCATGGA-3′ | |
| QRT 16S FWD | 5′-GGCTCAGATTGAACGCTGGC-3′ | |
| QRT 16S REV | 5′-CGAAGAGCCCCTGCTTGG-3′ | |
| M13For | 5′-GTAAAACGACGGCCAGT-3′ | |
| M13Rev | 5′-CACACAGGAAACAGCTATGACCAT-3′ | |
| plaP up 1,000 bp | 5′-TTTGGATCCTTCAACCTTAGAAGGCCGTC-3′ | |
| plaP 30-bp down ATG | 5′-CCCAAGCTTTGCCGATAGAGGAGATGACA-3′ | |
Southern Blot Analysis
To confirm the appropriate plaP gene disruption or integration of the wild-type plaP gene for complementation in single copy, chromosomal DNA from the plaP mutants (SK694 and SK738) or the complemented strains (SK713 and SK739) were extracted and separately digested with BamHI, HindIII, PstI, and EcoRI before being transferred to a nitrocellulose membrane and probed with a plaP-specific digoxigenin-labeled probe (supplemental Fig. S1).
Swarm Assays
To examine swarming phenotypes, overnight cultures of P. mirabilis strains were diluted in LB to an A600 of 0.4, and 3 μl of the diluted culture was spotted onto the center of LB or LB + 0.25% NaCl (w/v) plates containing 1.5% agar. After incubation at 37 °C, the picture was taken or the diameter of swarming was measured.
β-Galactosidase Assays
β-Galactosidase assays were carried out as described previously (38). Overnight cultures of P. mirabilis strains were inoculated to the same initial cell density (A600 of 0.03) into 2 ml of LB or LB + 0.25% NaCl broth in test tubes (13 × 100 mm) and were grown at 37 °C with reciprocal shaking at 250 rpm, and cells were harvested at different growth phases. Putrescine (50 μm) or T443 (100 μm) were added to LB broth when the A600 reached 0.2 (∼1.5 h after inoculation), and cells were harvested after 40 min (when T44 was added) or 1 h (when putrescine was added).
Semi-quantitative RT-PCR
The cDNA synthesis reaction on total RNA from PM7002 harvested at different growth phases was performed using iScriptTM cDNA synthesis kit (Bio-Rad) and the gene-specific primer “plaP.cDNA,” following manufacturer's instructions. The expression of plaP gene was monitored by semi-quantitative RT-PCR using the cDNA product as template and primers “plaP RT for” and “plaP RT rev.” As an internal control, expression of the 16 S rRNA gene was also examined from the same RNA samples after random cDNA synthesis using primers “QRT 16S FWD” and “QRT 16 S REV.” The absence of contaminating DNA from both samples was confirmed by the inability to generate PCR products in the absence of cDNA synthesis.
Analysis of Intracellular Putrescine Concentration
The concentration of putrescine in the cells was measured as described previously (39). Briefly, an HPLC system equipped with a cation exchange column, TSKgel PolyaminePak (Toso, Tokyo, Japan), was used for separation, identification, and quantification of putrescine. o-Phthalaldehyde was used as the detection reagent, and fluorescence was detected with a fluorescence detector. Standard putrescine was purchased from Sigma.
Urothelial Cell Invasion Assays
Cell invasion assays were performed as described previously (40), with some modifications. UMUC-3 urothelial cells (ATCC) were cultured in EMEM supplemented with 10% fetal calf serum (FCS) in 24-well plates until reaching confluency. Prior to being infected with bacterial cells, the monolayer was washed twice with serum-free EMEM. Harvested P. mirabilis cells were washed twice with serum-free EMEM and resuspended in serum-free EMEM at an A600 of 0.1. The suspension of P. mirabilis (500 μl) was overlaid on the UMUC-3 urothelial cell monolayer (multiplicity of infection was ∼100 P. mirabilis cells per 1 urothelial cell) and incubated for 1 h. Urothelial cell monolayers were washed twice with 500 μl of serum-free EMEM containing 100 μg/ml gentamicin to remove free bacteria and incubated for 1 h in serum-free EMEM containing 100 μg/ml gentamicin to kill extracellular bacteria. Finally, the monolayers were washed once with serum-free EMEM and lysed with 500 μl of 0.1% Triton X-100. A 1:10 dilution series was made with PBS and plated on LB plates containing 3% agar and incubated at 37 °C for 24 h, and viable cells were determined by colony counts. Triplicate wells were used to determine the total number of bacteria per well (the percentage of bacterial invasion = (bacteria recovered/input bacteria) × 100). Data are expressed as fold increases in invasion over that of PM7002 (wild type) without additives and represent the means ± S.E. of results from three independent experiments.
RESULTS
PlaP Is Required for the Response to Extracellular Putrescine in P. mirabilis
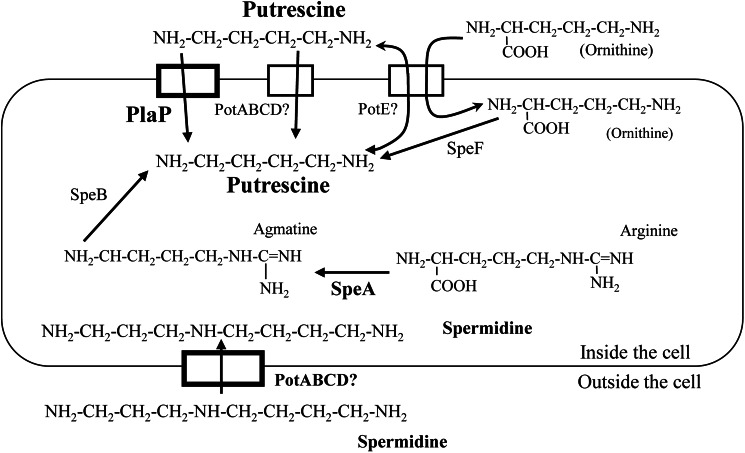
In P. mirabilis, putrescine is likely synthesized from ornithine by ornithine decarboxylase (SpeF) or from arginine by the sequential actions of arginine decarboxylase (SpeA) and agmatinase (SpeB) (Fig. 1) (34). Interestingly, P. mirabilis lacks the spermidine biosynthetic pathway composed of SpeD and SpeE. In addition, genes encoding enzymes forming alternative pathways for spermidine and norspermidine synthesis are not predicted to be encoded in P. mirabilis (7). Also, in contrast with E. coli having numerous putrescine importers, P. mirabilis appears to have only two, PotE and PlaP, and one spermidine transporter, PotABCD, according to bioinformatic analysis (see Fig. 1). Previously, an E. coli mutant deficient in putrescine synthesis (ΔspeAB and ΔspeC) and missing the polyamine importers PotABCD and YdcSTUV was dependent on both putrescine and the putrescine importer PlaP for surface motility on semisolid media (12).
FIGURE 1.
Proposed biosynthetic and transport pathways for polyamines in P. mirabilis. The locations of putative polyamine transporters in the cytoplasmic membrane are shown. Question marks were added to PotABCD and PotE because their roles are unknown.
In P. mirabilis, a putative PlaP homolog (PMI0843) is present, exhibiting 60% amino acid identity to PlaP of E. coli. To investigate the role of the P. mirabilis PlaP homolog, a plaP null mutant was constructed (see under “Experimental Procedures”). The plaP::Cm mutation was first constructed in a P. mirabilis speA::Km mutant deficient in putrescine biosynthesis, as the lower intracellular levels of putrescine in this background would emphasize a role for PlaP in putrescine import. The speA::Km mutant was previously shown to exhibit a severe swarming defect, which could be rescued by the addition of putrescine to agar plates (34). Three strains, PM437 (speA::Km), SK694 (speA::Km, plaP::Cm),and SK713 (speA::Km, plaP::Cm/plaP+complemented), did not swarm on an LB plate without putrescine because they share the speA::Km mutation and are deficient in putrescine synthesis (Fig. 2B) (34). In contrast, PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+complemented) strains swarmed normally on an LB plate supplemented with putrescine (Fig. 2C). However, SK694 (speA::Km, plaP::Cm) did not swarm on the LB agar supplemented with putrescine (Fig. 2C), indicating that plaP is required for the response to extracellular putrescine.
FIGURE 2.

Phenotypes involved in plaP in a putrescine-deficient speA mutant. A–C, PlaP- and putrescine-dependent swarming in a putrescine-deficient speA mutant. A diluted cell suspension (3 μl, A600 = 0.4) of PM437 (speA::Km), SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+complemented) were spotted on a 150-mm plate containing 50 ml of LB agar (B) or LB agar supplemented with 50 μm putrescine (C). Plates were photographed after 9 h of incubation at 37 °C. D, PlaP-dependent putrescine uptake in a putrescine-deficient speA mutant. Intracellular putrescine concentrations were measured in the cells of PM437 (speA::Km); SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+complemented) grown in LB broth (A) supplemented with (gray bars) or without (white bars) 50 μm putrescine. The total cultivation time was ∼2.5 h. The assays were performed three times, and values were expressed as the mean ± S.D.
PlaP Is the Main Putrescine Importer in P. mirabilis
To directly test the role of PlaP in putrescine import, the intracellular putrescine concentrations for PM437 (speA::Km), SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+ complemented) were determined after growth in media supplemented with or without 50 μm putrescine (Fig. 2D). The intracellular concentration of putrescine in PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+complemented) grown in the broth supplemented with putrescine (dark gray bars in Fig. 2D) were 7-fold higher than in media without putrescine (white bars in Fig. 2D). However, this increase in the levels of intracellular putrescine was not observed in the plaP::Cm mutant (SK694) (Fig. 2D).
Role of plaP in a Wild-type Background
To assess the role of PlaP in a wild-type (speA+) genetic background, strains PM7002 (wild type), SK738 (plaP::Cm), and SK739 (plaP::Cm/plaP+complemented) were subjected to swarming assays, and the intracellular levels of putrescine were determined in each strain. Relative to wild-type PM7002, strain SK738 (plaP::Cm) exhibited a swarming defect with a migration distance that was reduced to 60% of either PM7002 or SK739 (plaP::Cm/plaP+complemented) on LB + 0.25% NaCl plates (Fig. 3A). The intracellular concentrations of putrescine concentration in PM7002, SK738 (plaP::Cm), and SK739 (plaP::Cm/plaP+complemented) were not significantly different when P. mirabilis strains were grown in LB + 0.25% NaCl broth (Fig. 3B). When normal LB (containing 0.5% NaCl) was used, neither the swarming defect nor the decrease in intracellular putrescine associated with the deletion of plaP gene was observed.
FIGURE 3.
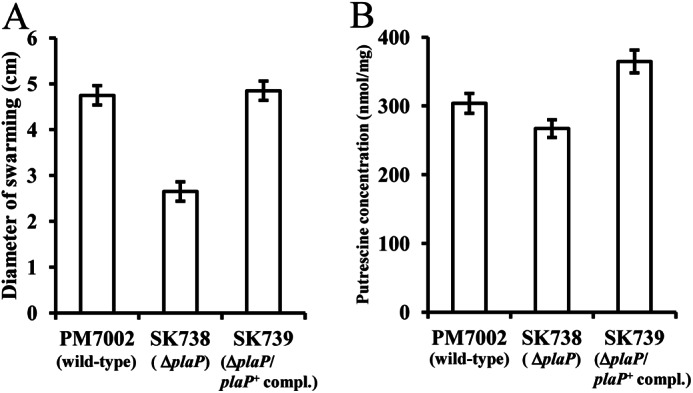
PlaP-dependent swarming and putrescine uptake of wild-type P. mirabilis. A, diluted cell suspension (3 μl, A600 = 0.4) of PM7002 (wild type), SK738 (plaP::Cm), and SK739 (plaP::Cm/plaP+complemented) were spotted on LB + 0.25% NaCl plates. After 15 h of inoculation at 37 °C, the plates were photographed, and the diameter of swarming was measured. B, cells were grown in LB 0.25% NaCl broth for 2.5 h, and the concentration of putrescine in the cell was measured. The assays for swarming and determining the putrescine concentration in cells were performed at least twice, and values are reported as the mean ± S.D.
Regulation of PlaP
To assess the regulation of plaP, a plaP-lacZ transcriptional fusion was constructed in the plasmid pQF50 (41) and introduced into wild-type PM7002. The expression of plaP-lacZ decreased from lag phase to early exponential phase, and then increased in a density-dependent manner with levels of β-galactosidase ∼2–3-fold higher at late exponential phase and stationary phase (Fig. 4). Semi-quantitative RT-PCR using total RNA of PM7002 harvested in the different growth phases showed a similar trend, where plaP expression in stationary phase was greater than in exponential phase (data not shown). The addition of putrescine (50 μm) or decreasing the NaCl concentration had no effect on plaP-lacZ expression (data not shown).
FIGURE 4.
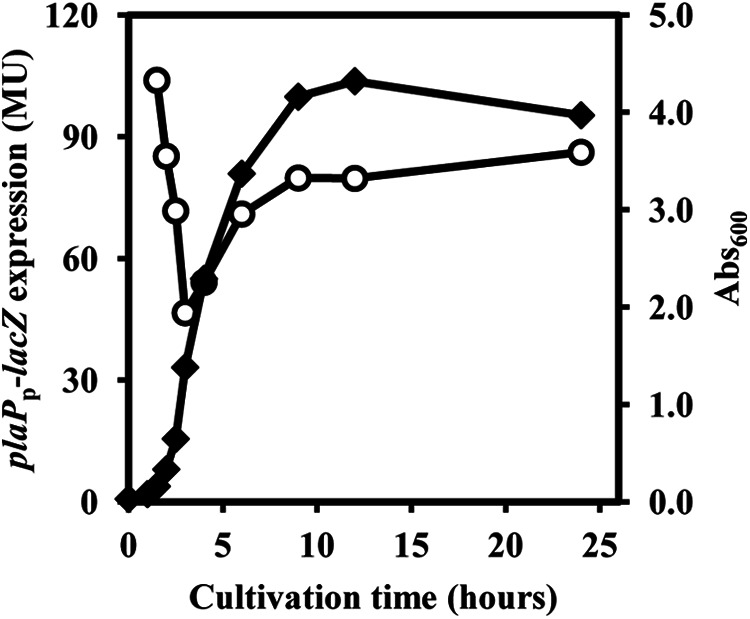
Growth phase regulated expression of plaP. The wild-type P. mirabilis strain SK750 containing the plaP-lacZ fusion was grown in LB broth with 35 μg/ml streptomycin and harvested at various time points during growth. The reported values represent β-galactosidase activity in Miller units (MU). Black diamonds indicate A600 of cells. The assays in one experiment were performed three times, and values are reported as the mean ± S.D. The experiment was performed two additional times, and similar results were obtained.
Putrescine and the Putrescine Importer PlaP Are Important for the Invasion of Urothelial Cells
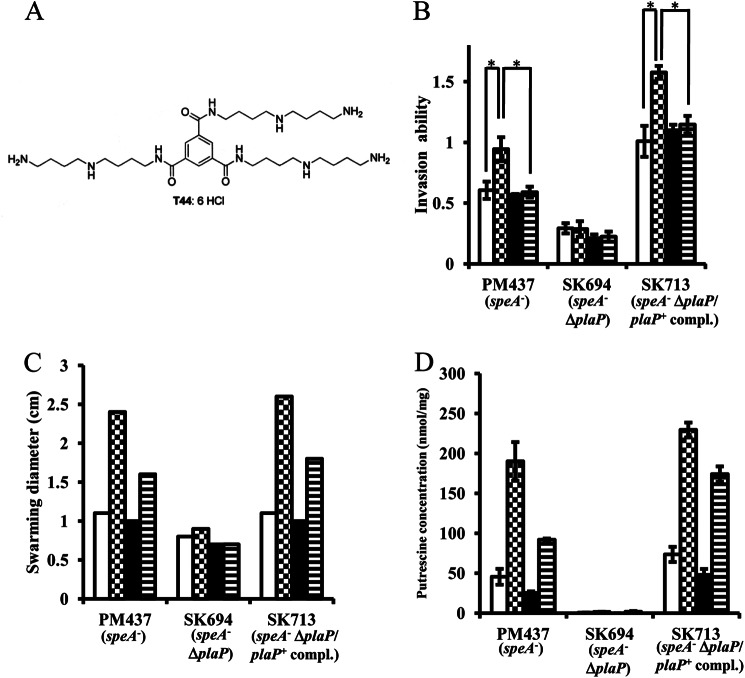
To assess the role of PlaP and putrescine in the invasion of urothelial cells by P. mirabilis, invasion assays were performed using the urothelial cell line UMUC-3. Exogenously added putrescine at the concentration of 25 μm resulted in a 2-fold increase in the invasion ability of P. mirabilis strains PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+complemented) (white and checked bars in Fig. 5B), but putrescine did not increase the invasion ability of SK694 (speA::Km, plaP::Cm) (white and checked bars in Fig. 5B). These results show that extracellular putrescine and its importer PlaP contribute to the ability of a P. mirabilis speA::Km mutant to invade urothelial cells. However, in a speA+ background, the plaP mutation did not effect the invasion of strains grown in either LB or LB + 0.25% NaCl (Fig. 6A).
FIGURE 5.
PlaP-dependent invasion of urothelial cells by P. mirabilis, and inhibition by the putrescine analog T44 on invasion ability, swarming motility, and putrescine uptake of a putrescine-deficient speA mutant. A, chemical structure of T44 is shown. B, strains PM437 (speA::Km), SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+complemented) were tested for the ability to invade UMUC-3 urothelial cells by the gentamicin protection assay. Wells were supplemented with 25 μm putrescine (checked bars), with 100 μm T44 (black bars), and with both 25 μm putrescine and 100 μm T44 (horizontal striped bars) or without additives (white bars). Data are expressed as the fold increase in invasion over that of PM7002 (wild type) without additives. The assays were performed three times, and values were expressed as the mean ± S.D. *, p < 0.01. C, swarming diameter of PM437 (speA::Km), SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+complemented) is shown during growth on an LB plate supplemented with 25 μm putrescine (checked bars), 100 μm T44 (black bars), and with both 25 μm putrescine and 100 μm T44 (horizontal striped bars) or without additives (white bars). D, intracellular putrescine concentrations of PM437 (speA::Km), SK694 (speA::Km, plaP::Cm), and SK713 (speA::Km, plaP::Cm/plaP+complemented) are shown during growth in LB broth supplemented with 25 μm putrescine (checked bars), 100 μm T44 (black bars), and with both 25 μm putrescine and 100 μm T44 (horizontal striped bars) or without additives (white bars). The assays were performed three times, and values were expressed as the mean ± S.D.
FIGURE 6.
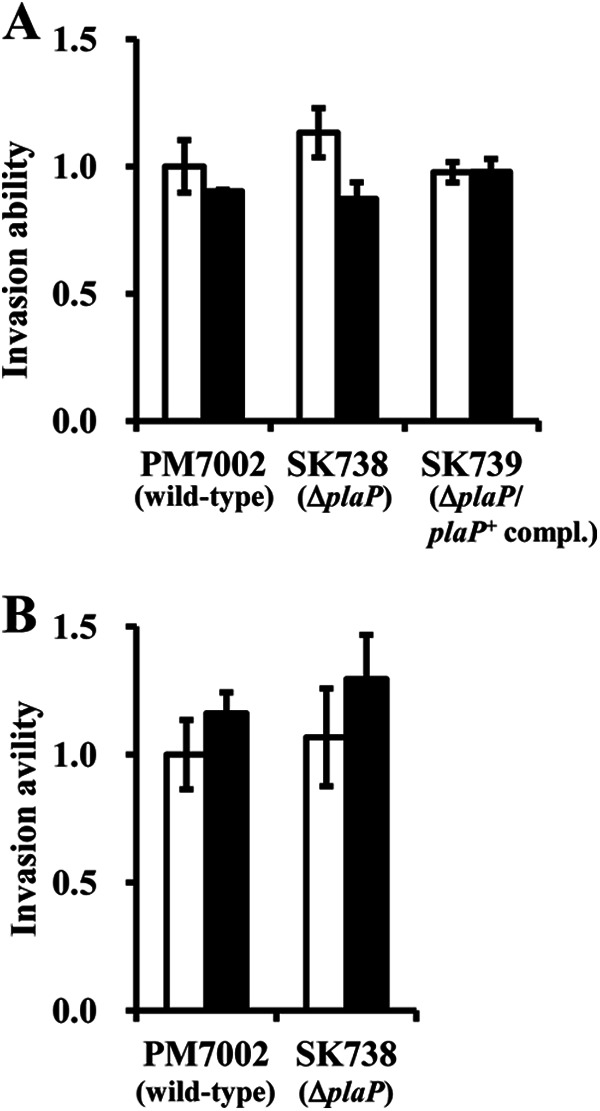
Effect of ΔplaP in speA+ background, NaCl concentration, and DFMO on invasion ability. A, strains PM7002 (wild-type), SK738 (plaP::Cm), and SK739 (plaP::Cm/plaP+complemented) were grown in LB (white bars) or LB + 0.25% NaCl (black bars) and tested for the ability to invade UMUC-3 urothelial cells by the gentamicin protection assay. Data are expressed as the fold increase in invasion over that of PM7002 (wild type) grown in LB. B, strains PM7002 (wild-type) and SK738 (plaP::Cm) were grown in LB (white bars) or LB supplemented with 1 mm DFMO (black bars) and tested for the ability to invade UMUC-3 urothelial cells by the gentamicin protection assay. The assays were performed three times, and values were expressed as the mean ± S.D.
Putrescine Analog Triamide-44 Reduces Putrescine Uptake and Associated Phenotypes
T44 is a putrescine analog designed for selective inhibition of putrescine transport (Fig. 5A) (35, 36). On LB plates supplemented with 100 μm T44 and 25 μm putrescine, the swarming diameter (horizontal striped bars in Fig. 5C) was reduced 30% compared with that on LB plate supplemented with 25 μm of putrescine alone (checked bars in Fig. 5C) in PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+complemented). On LB plates without putrescine, 100 μm T44 slightly reduced swarming ability (white bars versus black bars in Fig. 5C). Similarly, the intracellular putrescine concentration of cells grown with 100 μm T44 and 25 μm putrescine was reduced by 50% in PM437 (speA::Km) and 25% in SK713 (speA::Km, plaP::Cm/plaP+complemented) when compared with that of 25 μm putrescine alone (checked versus horizontal striped bars in Fig. 5D). On LB plates without putrescine, 100 μm T44 reduced putrescine concentration in the cells to some extent (white bar versus black bar in Fig. 5C). β-Galactosidase assays using SK750 (pQF50-plaPp-lacZ/PM7002) showed that T44 supplemented to the media did not have any influence on the expression of plaPp-lacZ (data not shown). T44 added to the LB broth at the concentration of 100 μm reduced growth of P. mirabilis strains slightly (around 10%, data not shown). These results show that T44 inhibits the uptake of putrescine mediated by PlaP, and this results in reduced swarming motility of a speA mutant.
The ability of T44 to reduce the putrescine-stimulated urothelial cell invasion in the speA::Km mutant was also examined. The invasion ability of the P. mirabilis strains PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+ complemented) was stimulated by the addition of putrescine (25 μm) (white versus checked bars in Fig. 5B). In contrast, when PlaP inhibitor T44 (100 μm) and putrescine (25 μm) were both added to the wells, the invasion ability of these P. mirabilis strains was suppressed (checked versus horizontal striped bars in Fig. 5B) to the level of control without supplementation (white bars in Fig. 5B). When T44 alone (100 μm) was added to the wells (black bars in Fig. 5B), the invasion ability of PM437 (speA::Km) and SK713 (speA::Km, plaP::Cm/plaP+ complemented) was not affected. The invasion ability of SK694 (speA::Km, plaP::Cm) was not altered by T44 or putrescine. These results show that T44 inhibited putrescine import by PlaP, and this resulted in the reduction of invasion ability in putrescine-deficient P. mirabilis.
DISCUSSION
In this report, PlaP in P. mirabilis was shown to be important for swarming motility and to act as a putrescine importer (Fig. 2). These results are similar to those found in E. coli, where PlaP encodes a putrescine importer that is important for surface motility (12). Putrescine can potentially act as a signaling molecule to regulate swarmer cell differentiation in P. mirabilis (34). In addition, studies in other bacteria suggest that polyamines can act as signaling molecules for bacterial communication (5–12). It should be noted that these previous studies were performed using polyamine-deficient strains, and former results could stem from severe polyamine deficiency itself (5, 7, 9–12, 34). In this report, however, the plaP mutation also resulted in a statistically significant reduction in swarming in a wild-type background (Fig. 3A), indicating that response to extracellular putrescine is required for normal swarming. This further supports the possibility that P. mirabilis uses putrescine as a cell-cell signaling molecule.
In this study, the role of PlaP in putrescine uptake and the resulting defects in swarming and invasion were more pronounced in a speA mutant due to the very low levels of putrescine present in this strain (Fig. 2D). There are two predicted pathways for putrescine synthesis in P. mirabilis, the SpeA/B-dependent pathway that utilizes arginine and the ornithine decarboxylase-dependent pathway that utilizes ornithine (Fig. 1). E. coli has two genes encoding ornithine decarboxylase (speC and speF). Among them, it is thought that speF is inactive when bacteria grow at near-neutral pH (42) and ΔspeAB ΔspeC E. coli strains completely lose putrescine inside the cell (12). Additionally, in E. coli, speF forms an operon with potE, encoding a putrescine-ornithine antiporter. In P. mirabilis, the gene encoding ornithine decarboxylase is also located next to potE, suggesting this is not speC but speF. The results obtained in this study suggest that the contribution of SpeF to putrescine biosynthesis is insignificant when compared with SpeA because the putrescine concentration in PM437 speA::Km is decreased by 90% compared with PM7002 wild ype (compare white bar of PM437 in Fig. 2C with white bar of PM7002 in Fig. 3B). In addition, it was previously reported that a speF mutant does not exhibit a swarming defect, and a speB/speF double mutant is further reduced for swarming (43). These results suggest that putrescine is predominantly made from arginine in P. mirabilis. Additional studies using difluoromethylornithine (DFMO), a selective ornithine decarboxylase inhibitor, support this hypothesis. In short, DFMO at 1 mm had no effect on growth or invasion ability (Fig. 6B). In addition to lacking orthologs of genes involved in putrescine import (20, 22, 44) and the spermidine synthetic pathway (19), which was discovered in E. coli, P. mirabilis lacks degradation pathways (39, 46–50) for putrescine. Therefore, it is thought that P. mirabilis has a simple system for the transport and metabolism of putrescine, i.e. a functional synthetic pathway (SpeA and SpeB), a putative exporter (PotE), and an importer (PlaP). This would facilitate P. mirabilis in using putrescine as a signaling molecule because the putrescine concentration inside the cell could easily change in response to the concentration of extracellular putrescine.
In P. mirabilis, the ability to invade human urothelial cells is coupled to motility and swarmer cell differentiation (30). The polyamine analog T44 partially inhibited the uptake of putrescine mediated by PlaP (Fig. 5D), and reduced the ability of putrescine to rescue swarming in the speA mutant (Fig. 5C). Furthermore, the ability of the P. mirabilis speA mutant to invade human urothelial cells was stimulated by putrescine imported by PlaP and decreased by putrescine transport inhibitor T44 (Fig. 5B). However, deletion of plaP did not have an effect on the invasion activity of speA+ strains (Fig. 6A), and DFMO, a selective inhibitor for ornithine decarboxylase encoded by speF, did not have an effect on either PM7002 (wild-type) or SK738 (ΔplaP) (Fig. 6B). These results suggest that concurrent use of putrescine transport inhibitor and future inhibitors, which target the arginine to putrescine pathway (SpeA and/or SpeB), may be very effective agents against P. mirabilis.
Supplementary Material

This article contains supplemental Fig. S1.
- T44
- Triamide-44
- EMEM
- Eagle's minimal essential medium
- DFMO
- difluoromethylornithine.
REFERENCES
- 1. Kuehn G. D., Rodriguez-Garay B., Bagga S., Phillips G. C. (1990) Novel occurrence of uncommon polyamines in higher plants. Plant Physiol. 94, 855–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Terui Y., Ohnuma M., Hiraga K., Kawashima E., Oshima T. (2005) Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile, Thermus thermophilus. Biochem. J. 388, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tabor C. W., Tabor H. (1985) Polyamines in microorganisms. Microbiol. Rev. 49, 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pegg A. E. (1986) Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 234, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel C. N., Wortham B. W., Lines J. L., Fetherston J. D., Perry R. D., Oliveira M. A. (2006) Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188, 2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karatan E., Duncan T. R., Watnick P. I. (2005) NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187, 7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J., Sperandio V., Frantz D. E., Longgood J., Camilli A., Phillips M. A., Michael A. J. (2009) An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J. Biol. Chem. 284, 9899–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolodkin-Gal I., Cao S., Chai L., Böttcher T., Kolter R., Clardy J., Losick R. (2012) A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149, 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Burrell M., Hanfrey C. C., Murray E. J., Stanley-Wall N. R., Michael A. J. (2010) Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J. Biol. Chem. 285, 39224–39238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakamoto A., Terui Y., Yamamoto T., Kasahara T., Nakamura M., Tomitori H., Yamamoto K., Ishihama A., Michael A. J., Igarashi K., Kashiwagi K. (2012) Enhanced biofilm formation and/or cell viability by polyamines through stimulation of response regulators UvrY and CpxR in the two-component signal transducing systems and ribosome recycling factor. Int. J. Biochem. Cell Biol. 44, 1877–1886 [DOI] [PubMed] [Google Scholar]
- 11. Kurihara S., Suzuki H., Tsuboi Y., Benno Y. (2009) Dependence of swarming in Escherichia coli K-12 on spermidine and the spermidine importer. FEMS Microbiol. Lett. 294, 97–101 [DOI] [PubMed] [Google Scholar]
- 12. Kurihara S., Suzuki H., Oshida M., Benno Y. (2011) A novel putrescine importer required for type 1 pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J. Biol. Chem. 286, 10185–10192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jelsbak L., Thomsen L. E., Wallrodt I., Jensen P. R., Olsen J. E. (2012) Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One 7, e36149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nasrallah G. K., Riveroll A. L., Chong A., Murray L. E., Lewis P. J., Garduño R. A. (2011) Legionella pneumophila requires polyamines for optimal intracellular growth. J. Bacteriol. 193, 4346–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah P., Nanduri B., Swiatlo E., Ma Y., Pendarvis K. (2011) Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology 157, 504–515 [DOI] [PubMed] [Google Scholar]
- 16. Ware D., Jiang Y., Lin W., Swiatlo E. (2006) Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infect. Immun. 74, 352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boyle S. M., Markham G. D., Hafner E. W., Wright J. M., Tabor H., Tabor C. W. (1984) Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (SpeA, SpeB, SpeC, and MetK). Gene 30, 129–136 [DOI] [PubMed] [Google Scholar]
- 18. Tabor C. W., Tabor H., Xie Q. W. (1986) Spermidine synthase of Escherichia coli localization of the speE gene. Proc. Natl. Acad. Sci. U.S.A. 83, 6040–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tabor C. W., Tabor H. (1987) The speEspeD operon of Escherichia coli. Formation and processing of a proenzyme form of S-adenosylmethionine decarboxylase. J. Biol. Chem. 262, 16037–16040 [PubMed] [Google Scholar]
- 20. Kashiwagi K., Miyamoto S., Suzuki F., Kobayashi H., Igarashi K. (1992) Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pistocchi R., Kashiwagi K., Miyamoto S., Nukui E., Sadakata Y., Kobayashi H., Igarashi K. (1993) Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J. Biol. Chem. 268, 146–152 [PubMed] [Google Scholar]
- 22. Kurihara S., Tsuboi Y., Oda S., Kim H. G., Kumagai H., Suzuki H. (2009) The putrescine importer PuuP of Escherichia coli K-12. J. Bacteriol. 191, 2776–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Furuchi T., Kashiwagi K., Kobayashi H., Igarashi K. (1991) Characteristics of the gene for a spermidine and putrescine transport system that maps at 15 min on the Escherichia coli chromosome. J. Biol. Chem. 266, 20928–20933 [PubMed] [Google Scholar]
- 24. Mobley H. L., Belas R. (1995) Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 3, 280–284 [DOI] [PubMed] [Google Scholar]
- 25. Harshey R. M. (2003) Bacterial motility on a surface: Many ways to a common goal. Annu. Rev. Microbiol. 57, 249–273 [DOI] [PubMed] [Google Scholar]
- 26. Morgenstein R. M., Szostek B., Rather P. N. (2010) Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis. FEMS Microbiol. Rev. 34, 753–763 [DOI] [PubMed] [Google Scholar]
- 27. Rather P. N. (2005) Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 28. Belas R. (1994) Expression of multiple flagellin-encoding genes of Proteus mirabilis. J. Bacteriol. 176, 7169–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones B. V., Young R., Mahenthiralingam E., Stickler D. J. (2004) Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect. Immun. 72, 3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allison C., Emödy L., Coleman N., Hughes C. (1994) The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J. Infect. Dis. 169, 1155–1158 [DOI] [PubMed] [Google Scholar]
- 31. Allison C., Lai H. C., Hughes C. (1992) Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6, 1583–1591 [DOI] [PubMed] [Google Scholar]
- 32. Walker K. E., Moghaddame-Jafari S., Lockatell C. V., Johnson D., Belas R. (1999) ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32, 825–836 [DOI] [PubMed] [Google Scholar]
- 33. Fraser G. M., Claret L., Furness R., Gupta S., Hughes C. (2002) Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sturgill G., Rather P. N. (2004) Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51, 437–446 [DOI] [PubMed] [Google Scholar]
- 35. Muth A. F. (2012) Design, Synthesis, and Biological Evaluation of Novel Polyamine Transport System Probes and Their Application to Human Cancers, Ph.D. thesis, University of Central Florida, Orlando, FL [Google Scholar]
- 36. Kaur N., Delcros J. G., Imran J., Khaled A., Chehtane M., Tschammer N., Martin B., Phanstiel O., 4th (2008) A comparison of chlorambucil- and xylene-containing polyamines leads to improved ligands for accessing the polyamine transport system. J. Med. Chem. 51, 1393–1401 [DOI] [PubMed] [Google Scholar]
- 37. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 38. Miller J. H. (1972) Experiments in Molecular Genetics. pp. 1.1–2.110, Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY [Google Scholar]
- 39. Kurihara S., Oda S., Tsuboi Y., Kim H. G., Oshida M., Kumagai H., Suzuki H. (2008) γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 283, 19981–19990 [DOI] [PubMed] [Google Scholar]
- 40. Alamuri P., Löwer M., Hiss J. A., Himpsl S. D., Schneider G., Mobley H. L. (2010) Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect. Immun. 78, 4882–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farinha M. A., Kropinski A. M. (1990) Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172, 3496–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Applebaum D., Sabo D. L., Fischer E. H., Morris D. R. (1975) Biodegradative ornithine decarboxylase of Escherichia coli. Purification, properties, and pyridoxal 5′-phosphate-binding site. Biochemistry 14, 3675–3681 [DOI] [PubMed] [Google Scholar]
- 43. Armbruster C. E., Hodges S. A., Mobley H. L. (2013) Initiation of swarming by Proteus mirabilis occurs in response to specific cues present in urine and requires excess l-glutamine. J. Bacteriol. 195, 1305–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kashiwagi K., Hosokawa N., Furuchi T., Kobayashi H., Sasakawa C., Yoshikawa M., Igarashi K. (1990) Isolation of polyamine transport-deficient mutants of Escherichia coli and cloning of the genes for polyamine transport proteins. J. Biol. Chem. 265, 20893–20897 [PubMed] [Google Scholar]
- 45. de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. (1990) Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J. Bacteriol. 172, 6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kurihara S., Oda S., Kumagai H., Suzuki H. (2006) γ-Glutamyl-γ-aminobutyrate hydrolase in the putrescine utilization pathway of Escherichia coli K-12. FEMS Microbiol. Lett. 256, 318–323 [DOI] [PubMed] [Google Scholar]
- 47. Kurihara S., Oda S., Kato K., Kim H. G., Koyanagi T., Kumagai H., Suzuki H. (2005) A novel putrescine utilization pathway involves γ-glutamylated intermediates of Escherichia coli K-12. J. Biol. Chem. 280, 4602–4608 [DOI] [PubMed] [Google Scholar]
- 48. Kurihara S., Kato K., Asada K., Kumagai H., Suzuki H. (2010) A putrescine-inducible pathway comprising PuuE-YneI in which γ-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192, 4582–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samsonova N. N., Smirnov S. V., Novikova A. E., Ptitsyn L. R. (2005) Identification of Escherichia coli K12 YdcW protein as a γ-aminobutyraldehyde dehydrogenase. FEBS Lett. 579, 4107–4112 [DOI] [PubMed] [Google Scholar]
- 50. Samsonova N. N., Smirnov S. V., Altman I. B., Ptitsyn L. R. (2003) Molecular cloning and characterization of Escherichia coli K12 ygjG gene. BMC Microbiol. 3, 2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mobley H. L., Warren J. W. (1987) Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 25, 2216–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pearson M. M., Sebaihia M., Churcher C., Quail M. A., Seshasayee A. S., Luscombe N. M., Abdellah Z., Arrosmith C., Atkin B., Chillingworth T., Hauser H., Jagels K., Moule S., Mungall K., Norbertczak H., Rabbinowitsch E., Walker D., Whithead S., Thomson N. R., Rather P. N., Parkhill J., Mobley H. L. (2008) Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190, 4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.