Background: TSC2-null cells that exhibit elevated mTOR activity have up-regulation of ER stress pathways.
Results: mTORC1 activation in TSC2-null Elt3 cells promotes c-MYC translation and c-MYC-dependent ATF4 transcription during bortezomib treatment, which enhances apoptosis.
Conclusion: mTOR can regulate ATF4 and CHOP transcription factor expression independently of unfolded protein accumulation.
Significance: c-MYC level determines whether TSC-null cells die of ER stress.
Keywords: Apoptosis, mTOR, Myc, Tuberous Sclerosis (Tsc), Unfolded Protein Response, ATF4, Bortezomib, Rapamycin
Abstract
Many factors, including duration and intensity of the unfolded protein response (UPR), dictate whether cells will adapt to endoplasmic reticulum stress or undergo apoptosis. In tuberous sclerosis (TSC), elevation of mammalian target of rapamycin complex 1 (mTORC1) activity has been proposed to compound the induction of UPR transcription factors ATF4 and CHOP, suggesting that the UPR could be targeted to eradicate TSC1/2-null cells during patient therapy. Here we report that control of c-MYC translation by mTORC1 plays a key role in determining whether TSC2-null Elt3 rat leiomyoma cells apoptose in response to UPR induction by the proteasome inhibitor bortezomib. Although bortezomib induces eukaryotic initiating factor 2α phosphorylation, mTORC1 activity was also required for downstream induction of the UPR transcription factors ATF4 and CHOP by a mechanism involving increased expression of c-MYC. Although bortezomib-induced c-MYC transcription was resistant to rapamycin treatment, mTORC1 activity was required for efficient c-MYC translation. c-MYC subsequently bound to the ATF4 promoter, suggesting direct involvement of an mTORC1/c-MYC-driven signaling pathway in the activation of the UPR. Consistent with this notion, exogenously expressed c-MYC reversed the ability of rapamycin to prevent bortezomib-induced CHOP and ATF4 expression as well as apoptosis. These findings indicate that the induction of ATF4/CHOP expression occurs via mTORC1 regulation of c-MYC and that this signaling pathway is a major determinant in the ability of bortezomib to induce apoptosis.
Introduction
Tuberous sclerosis complex (TSC)3 is a disease characterized by benign, tuberous growths in multiple organ systems that exhibit high levels of mTORC1 activation (1). Patients with TSC carry an autosomal dominant mutation in their TSC1 or TSC2 tumor suppressor genes, both of which are required to suppress high levels of mTOR complex 1 (mTORC1) activation (1). The TSC1 and TSC2 gene products, designated hamartin and tuberin, control mTORC1 activity via the small GTPase Rheb (2–5). Tuberin acts as a GTPase activating protein (GAP) that switches Rheb from an active GTP-bound state to an inactive GDP-bound form (2–4, 6). Meanwhile, hamartin stabilizes tuberin to prevent its degradation (7). Inactivating mutations in either TSC1 or TSC2, like those found in TSC patients, result in higher levels of GTP-loaded Rheb that leads to activation of mTORC1 and high levels of protein synthesis. In cell culture models of TSC where TSC1 or TSC2 is deleted from the genome, mouse embryonic fibroblasts (MEFs) show increased sensitivity to a class of compounds known to cause stress to the endoplasmic reticulum (ER) (8–10).
Proteins destined for secretion are synthesized at the rough ER and folded within its lumen. Perturbations caused by the accumulation of misfolded proteins, changes in calcium homeostasis, and nutrient or oxygen deprivation can cause stress to the ER and activation of the unfolded protein response (UPR). The UPR involves three transmembrane proteins: inositol-requiring enzyme-1 (IRE1), activating transcription factor-6 (ATF6), and protein kinase-like ER kinase (PERK) (11, 12). Activation of these three branches of the UPR allows the cell to adapt to the unfolded protein stress by arresting global protein synthesis, preferentially translating pro-survival transcription factors, and inducing the expression of proteins that facilitate the folding, processing, and trafficking of secretory proteins. However, if unfolded protein stress is severe or prolonged, the UPR can trigger apoptosis through a mechanism involving heightened PERK-dependent translation of the transcription factors ATF4 and CCAAT/enhancer-binding protein homologous protein (CHOP) (13, 14). PERK is a member of the eIF2α kinase family. By phosphorylating eIF2α at serine 51, PERK causes a global arrest of mRNA translation but enables the preferential translation of specific stress-responsive mRNAs that contain complex 5′-leader sequences that include regulatory upstream open reading frames (15, 16). These mRNAs, including ATF4 and CHOP, are also transcribed more effectively during PERK activation (17, 18). CHOP is critical for UPR-induced death and knock-out MEFs lacking this transcription factor are more resistant to drugs that induce the UPR (17, 19).
In the current study we explored the effects of bortezomib, a chemotherapeutic drug that can cause ER stress on UPR signaling and death of the TSC2-null rat leiomyoma cell line, Elt3. Pretreatment with the mTORC1 inhibitor rapamycin was used to determine the contribution of the TSC2/mTOR pathway to both activation of the UPR and Elt3 cell death. Bortezomib promoted the UPR, specifically the transcription and translation of ATF4 and CHOP, and UPR-induced apoptosis by mechanisms that were suppressed by rapamycin. This suggested that mTORC1 regulates the expression of a transcription factor that is required for induction of these UPR genes. In support of this notion, rapamycin treatment decreased c-MYC protein expression in Elt3 cells and c-MYC bound to the ATF4 promoter. Exogenous expression of c-MYC overcame the suppressive effects of rapamycin on ATF4/CHOP expression and cell death, whereas inhibition of c-MYC suppressed these bortezomib-induced events. These findings demonstrate that activation of an mTORC1/c-MYC pathway is required for bortezomib-induced expression of ATF4 and CHOP to promote UPR-mediated apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture
Elt3 cells were a gift from Cheryl Walker (MD Anderson Cancer Center). All experiments were performed on cells between passages 40 and 50 that were maintained in DF-8 media as described by Walker and Ginsler (20). Cells were plated at 70% confluence. The following day DF-8 media was replaced with serum-free DMEM (Lonza) containing DMSO vehicle control or 50 nm rapamycin (Calbiochem). 24 h later bortezomib (LC Laboratories) was added to each plate to a final concentration of 20 nm. In experiments using c-MYC inhibitor II (EMD Millipore), cells were starved of serum overnight and treated with 5 μm c-MYC inhibitor II 2 h before treatment with 20 nm bortezomib. Experiments also used 10 mm 2-DG and 1 μm thapsigargin when described.
Nuclear Lysates
To increase the detectability of ATF4 and CHOP proteins in immunoblots, nuclear lysates were prepared from cultured cells. For the nuclear preparation, 60-mm plates of Elt3 cells were washed with ice-cold PBS. Cells were then harvested into 1 ml of 10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, and 0.25% IGEPAL. Cells were incubated with rotation at 4 °C for 10 min. Nuclei were then pelleted at 3000 rpm in an accuSpin Micro 17R microcentrifuge (Fisher) at 4 °C for 10 min. The supernatant was discarded, and the pellet was resuspended in high detergent lysis solution (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton-100, and 1% SDS). Nuclear lysates were incubated for 10 min on ice and pelleted for 10 min at 13,300 rpm at 4 °C. Supernatants were transferred to ice-cold tubes and frozen in liquid nitrogen for later analysis.
Western Blot Analyses and Antibodies
Whole cell lysates used in Western blot analyses were prepared using high detergent lysis solution with protease and phosphatase inhibitors. Alternatively, nuclear lysates were prepared for detection of ATF4, ATF6, and CHOP proteins as detailed above. Lysates were normalized by protein concentration using a Bradford assay (Bio-Rad) and analyzed using 10 or 15% SDS-PAGE gels. Proteins separated in these gels were transferred to PVDF-FL membranes (Millipore). Membranes were blocked in 5% nonfat dry milk and probed with primary antibodies. Antibodies specific to cleaved caspase-3 (#9661), Lamin A/C (#4777), and c-MYC (#5606) were obtained from Cell Signaling Technologies. Antibodies specific to ATF4 (sc-200), U1snRNP70 (sc-9571), CHOP (sc-7351), and β-actin (sc-47778) were obtained from Santa Cruz Biotechnology. The ATF6 antibody was as described in Teske et al. (21). Western blots were visualized by x-ray film using SuperSignal West Femto Maximum Sensitivity substrate (Thermo Scientific) or scanned using an Odyssey LiCOR machine.
Quantitative Real-time-PCR Measurements
RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer's protocol. First-strand cDNA synthesis was performed using Moloney MuLV reverse transcriptase (New England Biolabs), and quantitative real-time-PCR detection of transcripts was performed using the Light Cycler 480 (Roche Applied Science) and the Roche Universal Probes Library and β-actin control primers according to the manufacturers' protocols. Primer sequences used for ATF4 were TCAGACACCGGCAAGGAG and GTGGCCAAAAGCTCATCTG, for CHOP were ACCACCACACCTGAAAGCA and AGCTGGACACTGTCTCAAAGG, for NOXA were GCGAAAGAGCACGATGAGA and GATCACACTCGTCCTTCAGGT, and for c-MYC were GCTCCTCGCGTTATTTGAAG and GCATCGTCGTGACTGTCG.
Trypan Blue Cell Viability Assays
After bortezomib treatment, cells were washed with ice-cold PBS and trypsinized for 5 min at 37 °C. The media from the plate, PBS wash, and trypsinized cells were pooled into a 15-ml conical tube, and cells were pelleted for 5 min at 1000 rpm in a Beckman GS-15R centrifuge at 4 °C. The supernatant was discarded, and cells were resuspended in a 1:1 solution of PBS:0.4% trypan blue (Sigma). The number of live and dead cells was counted using a hemocytometer. Clumps of cells where individual cells could not be accurately counted were excluded from these counts.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed on 107 Elt3 cells that had not been treated with any drug, pretreated with 50 nm rapamycin for 24 h, treated for 4 h with 20 nm bortezomib, or treated with both drugs. The SimpleChIP Chromatin IP kit (Cell Signaling #9003) was used according to the manufacturer's specifications. Enzymatic digestion was calibrated to yield genomic DNA fragments between 100 and 500 bp. Fragmented DNA was immunoprecipitated using nonspecific rabbit IgG or a c-MYC-specific rabbit monoclonal antibody. PCR detection of immunoprecipitated DNA fragments was performed using Maxima Hot Start 2× PCR master mix (Fermentas). Primer sequences were designed using the NCBI Primer Blast program to flank and specifically amplify DNA regions containing the E-box sequences at −78 and +632 of the rat ATF4 promoter (mRNA start site defined by comparison with mouse sequence (16)). The primer sequences used to amplify the DNA region +534 to +752 containing the putative E-box at +632 were AAGCTGCTTCCTCCGGGTGG (forward) and GCAACGCTGCTGCTGGGTTTC (reverse) and the DNA region −131 to +137 containing the E-box at −78 of the rat ATF4 promoter CGGGCCAGAGCGTCAATGGG (forward) and CTGCAAAGGCCAACGCTGCC (reverse). Control primers spanning +213 to +446 TGCTTTGCTGTGTTTGGGTG (forward) and CCACGTTCGCAGAATGACAC (reverse) or −486 to −328 TCTGGTGGCTCTTCCCGATA (forward) and GACGGTCAAAGCCAAAGCTG (reverse) failed to differentially amplify chromatin immunoprecipitation (ChIP) DNA from IgG versus anti-MYC immunoprecipitates after bortezomib treatment.4
Cloning and Lentiviral Production
A cDNA fragment encoding the human c-MYC was generated by PCR using primers that included 5′-BamHI and 3′-NotI sites. This fragment was then inserted between the same sites of the pCDH1-CMV-MCS-EF1-Hygro lentiviral expression vector (System Biosciences). Empty vector or the pCDH1-c-MYC plasmid was co-transfected into 293T cells along with pCMV-VSV-G (Addgene #8454), pRSV-REV (Addgene #12253), and pMDLg/pRRE (Addgene #12251) plasmids after calcium phosphate precipitation. Two days post-transfection the viral supernatant was filtered (0.45 μm) and stored at −80 °C until needed.
Luciferase Assays
1.5 kb of the rat ATF4 gene were amplified by PCR from Elt3 cell genomic DNA and cloned as a NheI-XhoI fragment into the pGL3-basic plasmid (Promega). Primer sequences were GCTAGCTCTATATGCCTAAAGCTCTGC and CTCGAGGTTGTAGGGATTTGCTGGTAT. The E-box located between bp −78 and −83 was mutated from CACGTG to CAAATG using the primer overlap method (22). A second potential E-box present at bp +632 to +637 in intron 1 was similarly mutated alone or in tandem with the −78 site. No other E-box sequences were detected within 4 kb of the transcript start site. 0.5 μg of the luciferase reporter and 0.5 μg of c-MYC or empty vector plasmid were transfected into 293T cells. Cells were lysed, and transcription was measured 48 h later according to the Promega luciferase assay system protocol.
Generation of c-MYC and Empty Vector Stable Cell Lines
Elt3 stable cell lines were generated by plating the cells into media containing lentivirus and 5 μg/ml Polybrene. As a control, an additional culture plate was left uninfected. This plate served as a control for complete drug selection. On the day after infection, cells were selected using 300 μg/ml hygromycin B. This selection was continued by refreshing the media and selection drug every 2 days until cells on the uninfected control plate were completely killed.
Statistical Analysis
Bar graphs represent the mean measurement of 3 or 4 experiments with error bars representing the S.D. from this mean. p values were calculated using the t test function in Microsoft Excel.
RESULTS
Bortezomib-induced Cell Death Is Reduced by Rapamycin
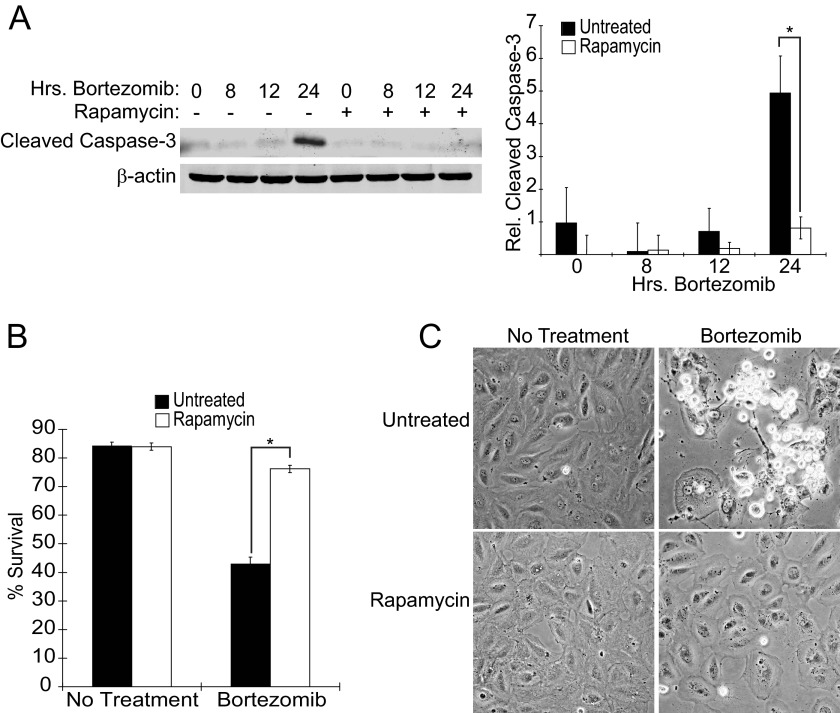
It was previously reported that 24 h pretreatment with the mTORC1 inhibitor rapamycin decreases the ability of UPR inducers such as thapsigargin and tunicamycin to induce the death of TSC2−/− MEFs (9). This suggested that ER stress is only lethal in the presence of high mTORC1 activity and that this might be taken advantage of in the elimination of TSC1/2-null lesions. We began our study by confirming that Elt3 cells, a TSC2-null rat leiomyoma cell line, similarly respond to the clinically approved proteasome inhibitor and UPR inducer, bortezomib. Bortezomib treatment induced apoptosis of Elt3 cells as shown by an increase in cleaved caspase-3 at 24 h (Fig. 1A). However, cells pretreated with 50 nm rapamycin for 24 h showed significantly less caspase activity or overall cell death upon exposure to bortezomib. In support of the notion that caspase-3 cleavage accompanies apoptosis, Elt3 cells at the same 24-h time point showed reduced viability as determined by trypan blue staining; bortezomib lowered the viability of Elt3 cells to 43%, whereas it only modestly decreased the survival of the rapamycin-pretreated cell from 84 to 76% (Fig. 1B). Bortezomib treatment caused cells to round and lose adherence, whereas rapamycin-pretreated cells remained mostly flat and attached (Fig. 1C). Despite the failure of bortezomib to induce robust apoptosis in rapamycin-pretreated cells, it functioned in conjunction with the mTORC1 inhibitor to decrease proliferation as evidenced by a decrease in cell density (Fig. 1C, lower right panel).
FIGURE 1.
Elt3 cells undergo rapamycin-sensitive apoptosis when treated with bortezomib. A, Elt3 cells were pretreated with 50 nm rapamycin or vehicle control for 24 h before being exposed to 20 nm bortezomib for an additional 8, 12, or 24 h. Lysates were prepared after these drug treatments and analyzed by Western blot using antibodies selective for cleaved caspase-3 or β-actin. B, trypan blue staining of Elt3 cells exposed to bortezomib for 24 h in the presence or absence of rapamycin pretreatment was used to quantitate cell death. *, p < 0.05. C, shown is phase contrast microscopy of the Elt3 cells treated with rapamycin and bortezomib as indicated.
Early UPR Markers Are Induced by Bortezomib Treatment but Unaffected by Rapamycin
In its earliest stages, ER stress leads to the PERK-dependent phosphorylation of eIF2α at serine 51, cleavage of ATF6 to its active form, and inositol-requiring enzyme-1 (IRE1)-facilitated splicing of X-box protein 1 mRNA, allowing translation of an activated transcription factor (11). Phosphorylation of eIF2α not only attenuates global translation but also promotes the transcription and translation of the pro-apoptotic transcription factors ATF4 and CHOP (11, 12). Both of these transcription factors have been linked to proteasome inhibitor-induced apoptosis, and both factors likely contributed to the cell death that was observed in Fig. 1 (18, 19).
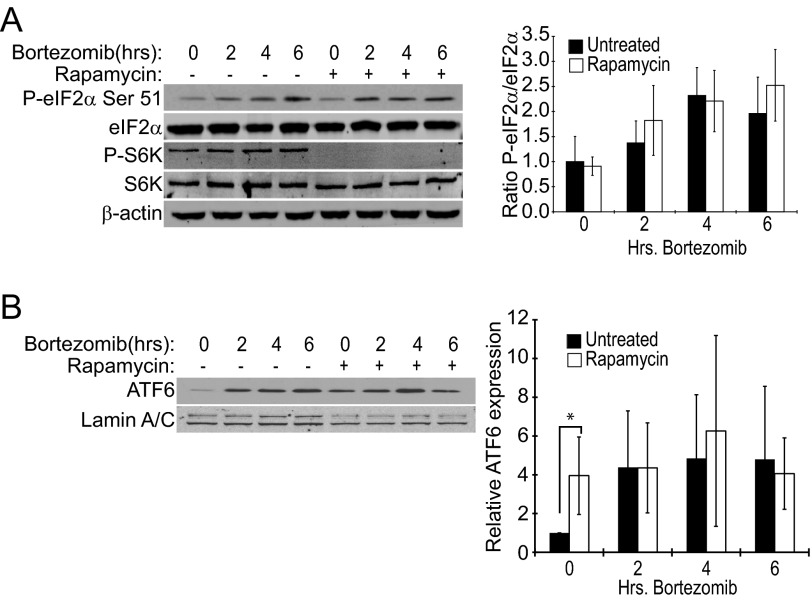
We found that although eIF2α phosphorylation was consistently increased after 4 or 6 h bortezomib treatment (Fig. 2A), rapamycin pretreatment had no effect on eIF2α phosphorylation at any time point tested. Likewise, there was no observable difference in the accumulation of the cleaved (active) fragment of ATF6 in the nucleus (Fig. 2B). Phosphorylation of the mTORC1 substrates, S6 kinase (S6K) (see Fig. 2a) and 4EBP1,4 was completely inhibited by rapamycin in these experiments, indicating that drug treatment had effectively blocked mTORC1. The lack of effect of rapamycin pretreatment on bortezomib-induced eIF2α phosphorylation and activated ATF6 suggested that activation of mTORC1 in these cells does not greatly impact the level of unfolded protein to further exacerbate ER stress and PERK phosphorylation of eIF2α when combined with proteasome inhibition.
FIGURE 2.
Early UPR markers are induced by bortezomib but unaffected by rapamycin treatment of Elt3 cells. Elt3 cells were pretreated with 50 nm rapamycin for 24 h before being exposed to 20 nm bortezomib for the times shown. A, whole cell lysates were prepared, and the indicated proteins were measured by Western blot using specific antibodies. B, nuclear lysates were prepared and probed for the nuclear cleaved fragment of ATF6 using Lamin A/C as a loading control.
ATF4 and CHOP Protein and mRNA Levels Are Induced by Bortezomib in a Rapamycin-dependent Manner
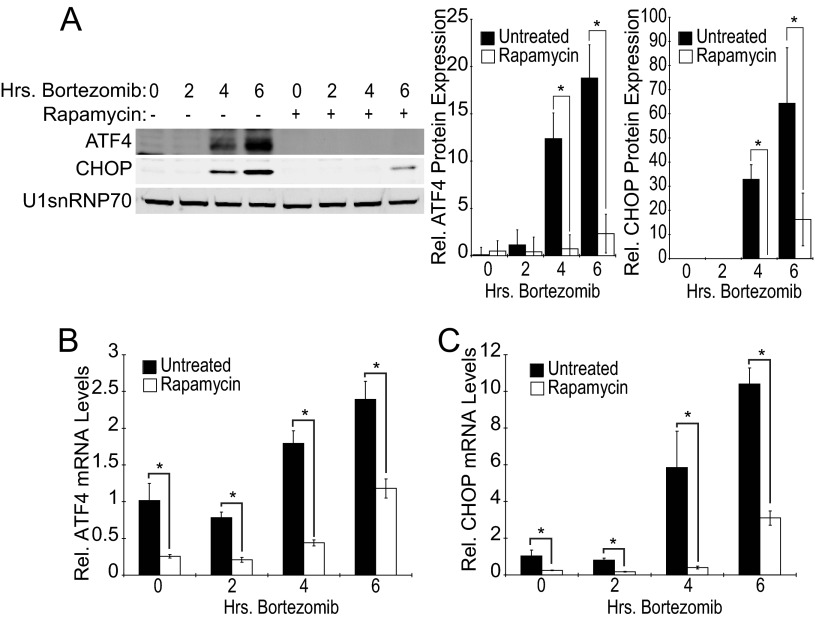
Coincident with elevated eIF2α phosphorylation, the expression of both ATF4 and CHOP was induced after 4- or 6-h bortezomib exposure (Fig. 3A). However, in contrast to eIF2α phosphorylation, rapamycin reduced expression of the ATF4 and CHOP proteins. Further investigation determined that the induction of both ATF4 and CHOP mRNAs by bortezomib was also suppressed by rapamycin pretreatment (Fig. 3, B and C). These data suggest that although proximal events of the UPR are not mTORC1-dependent, the downstream pro-apoptotic signals emanating from the ATF4/CHOP portion of this pathway are inhibited at the level of transcription by rapamycin pretreatment.
FIGURE 3.
Rapamycin prevents induction of downstream UPR markers. Elt3 cells were pretreated with 50 nm rapamycin or vehicle control for 24 h before being exposed to 20 nm bortezomib for an additional 2, 4, or 6 h. A, nuclear lysates were prepared and subjected to SDS-PAGE. ATF4, CHOP, and U1snRNP70 were measured by Western blot. Relative levels of ATF4 and CHOP proteins in each treatment group are presented as histograms on the right side of the panel. B and C, shown are quantitative real-time-PCR measurement of ATF4 and CHOP mRNA levels in Elt3 cells pretreated with rapamycin (24 h) followed by exposure to bortezomib for up to 6 h, as indicated (*, p < 0.05).
Bortezomib-induced Expression of ATF4 and CHOP Requires New mRNA and Protein Synthesis
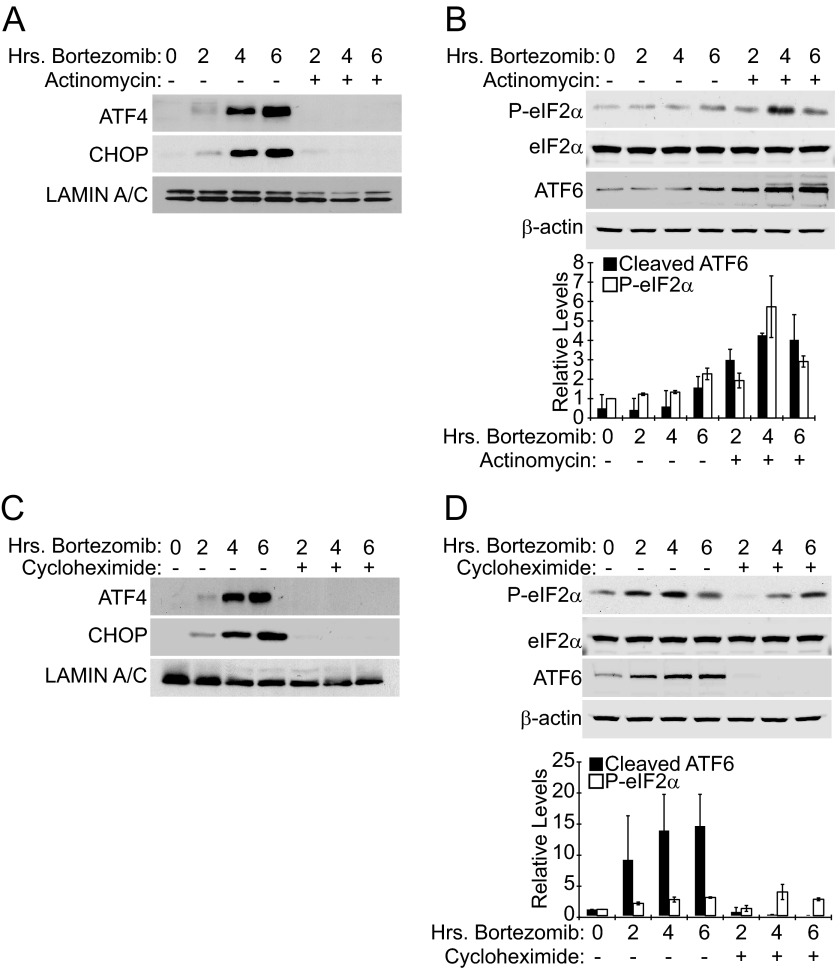
The ability of rapamycin to impede ATF4 and CHOP protein expression upon bortezomib treatment combined with the reduced mRNA expression of these genes after drug pretreatment suggested that rapamycin may deplete cells of a transcription factor that are required for bortezomib to induce ATF4 and CHOP expression. Furthermore, if rapamycin pretreatment decreased the synthesis of these proteins, it could result in reduced accumulation in the presence of proteasome inhibitor. To determine if the increase in ATF4 and CHOP protein levels is the result of stress-induced synthesis (transcription and/or translation) or the result of protein accumulation due to inhibition of the proteasome, we conducted experiments using the RNA polymerase inhibitor actinomycin D and the protein synthesis inhibitor cycloheximide. We found that simultaneously treating Elt3 cells with bortezomib and 5 μg/ml actinomycin blocked ATF4 and CHOP protein expression (Fig. 4A). This result suggests that bortezomib requires new mRNA synthesis to induce expression of either transcription factor. Cycloheximide similarly suppressed bortezomib-induced accumulation of ATF4 and CHOP proteins (Fig. 4C), indicating that new protein synthesis is also required and that the observed increase in protein expression is not merely the result of its accumulation due to proteasome inhibition. As a control, we also measured the phosphorylation of eIF2α and cleavage of ATF6 during actinomycin or cycloheximide treatment time courses. Neither actinomycin nor cycloheximide prevented bortezomib from inducing the phosphorylation of eIF2α (Fig. 4, B and D). We also observe that ATF6 cleavage was somewhat enhanced by actinomycin treatment. Cycloheximide treatment did prevent ATF6 cleavage (Fig. 4D), which is consistent with Teske et al. (21), who reported that ATF6 cleavage requires new protein synthesis and is completely inhibited by cycloheximide. These control experiments confirm that our observed block of ATF4 and CHOP expression during actinomycin and cycloheximide treatment is not merely a result of relieving ER stress by decreasing the load on the ER protein folding machinery.
FIGURE 4.
Elevation of ATF4 and CHOP protein levels by bortezomib requires the synthesis of new mRNA and protein. A, Elt3 cells were treated with 20 nm bortezomib for 2, 4, and 6 h in the presence or absence of 5 μg/ml actinomycin D. ATF4, CHOP, and lamin A/C levels in nuclear lysates were determined by Western blot. B, Elt3 cells were treated as in A. Phosphorylation of eIF2α, total eIF2α, the cleaved amino-terminal fragment of ATF6, and β-actin in cell lysates were determined by Western blot. C, Elt3 cells were treated with 20 nm bortezomib in the presence or absence of 100 μg/ml cycloheximide. Protein levels were measured as in A. D, Elt3 cells were treated as in C. Protein levels were measured as in B.
Transcriptional Expression of c-MYC Is Enhanced in Elt3 Cells during ER Stress Agents
Our experiments suggested that rapamycin inhibits induction of ATF4 and CHOP expression by blocking the synthesis of ATF4 and CHOP mRNA but does not alter upstream PERK phosphorylation of eIF2α. This selective targeting of downstream UPR components could be the result of altered regulation of a transcription factor(s) in response to elevated mTORC1 signaling by exposure to bortezomib or a combination of both stimuli. The oncogenic transcription factor c-MYC is a strong candidate for this link between mTORC1 and bortezomib; c-MYC is translationally up-regulated by mTORC1 activation and down-regulated by rapamycin treatment (23, 24). Additionally, c-MYC is a short-lived protein whose expression is increased rapidly during proteasome inhibition, and c-MYC has been shown to contribute to bortezomib-induced death of several tumor cell lines (25–27). Finally, the ATF4 gene promoter contains E-box consensus binding sites for basic helix-loop-helix transcription factors such as MYC (see Fig. 6A).
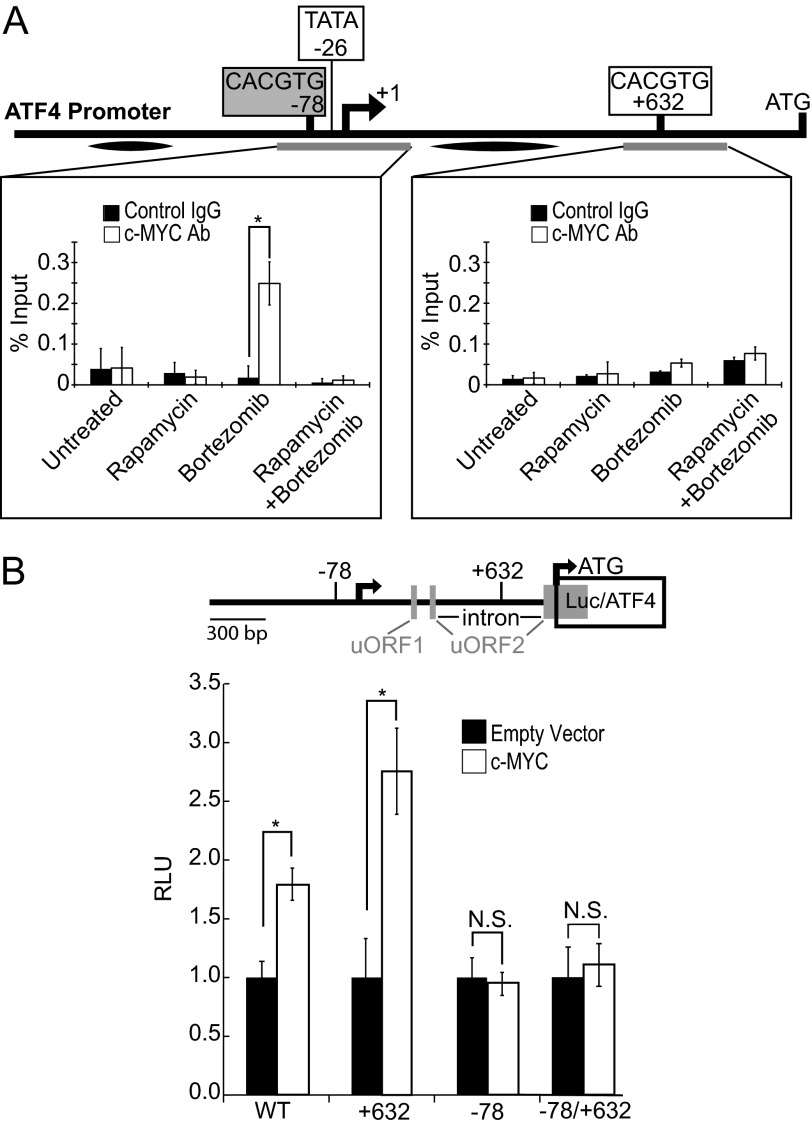
FIGURE 6.
c-MYC binds to and stimulates the ATF4 promoter. A, Elt3 cells were treated with 50 nm rapamycin for 24 h and 20 nm bortezomib for 4 h where indicated. Cells were then fixed and analyzed by ChIP for c-MYC binding to the E-boxes at positions −78 and +632 of the rat ATF4 promoter. Graphs represent the mean ± S.D. of three independent ChIP experiments. Gray bars below the gene indicate the regions amplified. Black ovals indicate additional control sequences found not to be amplified following c-MYC immunoprecipitation.4 Ab, antibody. B, 1.5 kb of the ATF4 gene (5′ of rat ATF4 coding region) were cloned upstream of the luciferase gene. The E-boxes located at −78 and +632 were mutated singly or together to prevent c-MYC binding and activation of luciferase transcription. These reporters were then transfected into 293T cells with empty vector or c-MYC, and luciferase activity was determined 48 h later. *, p < 0.05. RLU, relative light units.
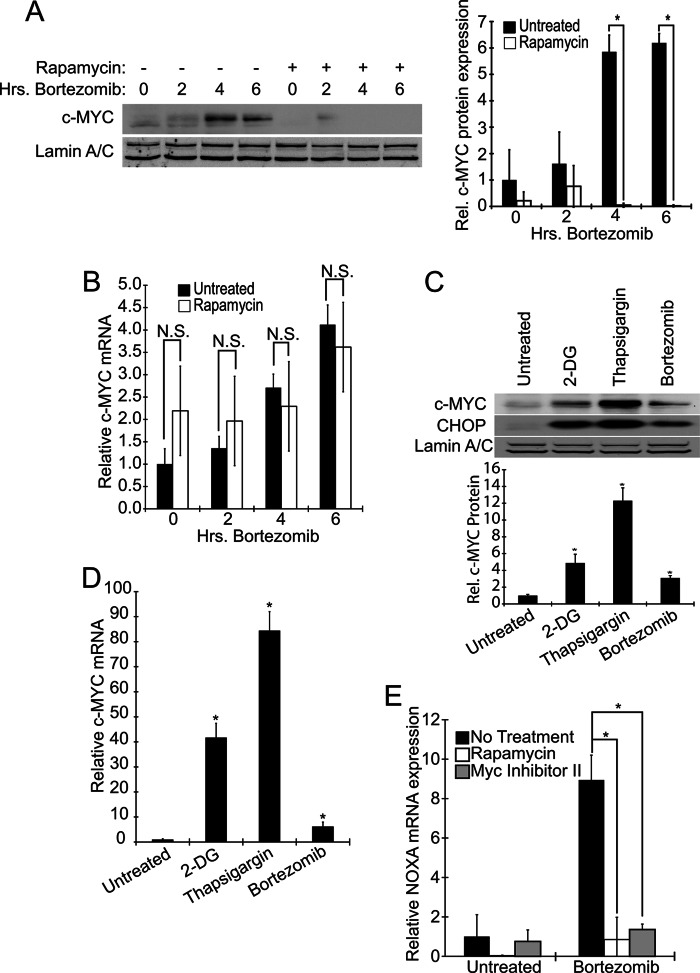
We found that treating Elt3 cells with bortezomib for 2, 4, or 6 h increased c-MYC protein expression (Fig. 5A). Consistent with its potential role as a mediator of mTORC1-induced ATF4 and CHOP expression, c-MYC induction by bortezomib was dampened by pretreatment with rapamycin. Interestingly, although we additionally observed an increase in the levels of c-MYC mRNA in response to bortezomib treatment, this up-regulation was insensitive to rapamycin pretreatment (Fig. 5B). This is consistent with rapamycin specifically inhibiting only the translation of c-MYC into protein. Two other ER stress-inducing drugs, 2-deoxyglucose and thapsigargin, also increased the expression of both c-MYC mRNA and protein (Fig. 5, C and D). How these stresses increase c-MYC transcription is not clear but is likely associated with induction of cell death.
FIGURE 5.
Bortezomib and other ER stressors induce expression and activity of c-MYC in a rapamycin-sensitive manner. Elt3 cells were pretreated with 50 nm rapamycin or vehicle control for 24 h before exposure to 20 nm bortezomib for 2, 4, or 6 h, as indicated. A, lysates were then prepared, and the levels of c-MYC and lamin A/C were measured by Western blot. B, levels of c-MYC mRNA were measured under the same conditions as A. C and D, 10 mm 2-deoxyglucose (2-DG), 1 μm thapsigargin, or 20 nm bortezomib were separately used to induce ER stress for 6 h. c-MYC and CHOP protein and c-MYC mRNA levels were then measured. N.S., not significant. E, basal- and bortezomib (50 nm, 6 h)-induced NOXA mRNA levels were measured after pretreatment with rapamycin (24 h) or MYC inhibitor II (2 h). *, p < 0.05.
To examine the effect of rapamycin on c-MYC activity, we measured mRNA levels of the stress-induced c-MYC transcriptional target NOXA. In these studies, we also evaluated the effectiveness of the small molecule, c-MYC inhibitor II, which was originally reported to inhibit c-MYC activity in Rat1a fibroblasts (28). We found that both rapamycin and the c-MYC inhibitor block the induction of NOXA after bortezomib treatment. These findings collectively suggest that ER stress, caused by bortezomib or other ER stress-inducing agents, promoted the transcription of c-MYC, resulting in up-regulation of c-MYC levels/activity. Rapamycin is able to block the translation of c-MYC under the conditions we studied, thereby inhibiting transcription of the c-MYC target, NOXA (Fig. 5E).
Rapamycin Inhibits Bortezomib-induced c-MYC Expression and Binding to the ATF4 Gene Promoter
To determine if c-MYC plays a direct role in the transcription of ATF4, we performed ChIP assays on the ATF4 promoter. ChIP revealed binding of c-MYC to a genomic DNA region containing the canonical E-box (CACGTG) at −78 of the ATF4 promoter (Fig. 6A) after 4 h of bortezomib treatment. This binding was reduced to basal levels if cells were pretreated with rapamycin. However, c-Myc failed to bind a potential E-box at +632 or two additional regions of the ATF4 promoter or 5′UTR (Fig. 6A).4 We also found that mutating the E-box at −78 but not a similar sequence at +632 of the ATF4 gene blocked the induction of luciferase during c-MYC overexpression in 293T cells (Fig. 6B). It is not clear why mutation of the +632 CACGTG sequence (located in an intron within a 5′ upstream ORF) positively impacted gene expression. Joint mutation of both putative E-box sites in the same reporter construct resulted in a lack of c-MYC responsiveness. These results indicate that c-MYC binds the ATF4 promoter during bortezomib treatment and may play a role in its induced transcriptional expression. Bortezomib treatment also up-regulates c-MYC expression in this cell line in a rapamycin-sensitive manner. These findings support the idea that c-MYC is a transcription factor that is induced by mTORC1 during bortezomib treatment, leading to a direct activation of transcription of the ATF4 gene followed by enhanced expression of ATF4 target genes, such as CHOP.
Overexpression of c-MYC Rescues Rapamycin-mediated Suppression of ATF4 and CHOP Expression in Elt3 Cells Treated with Bortezomib
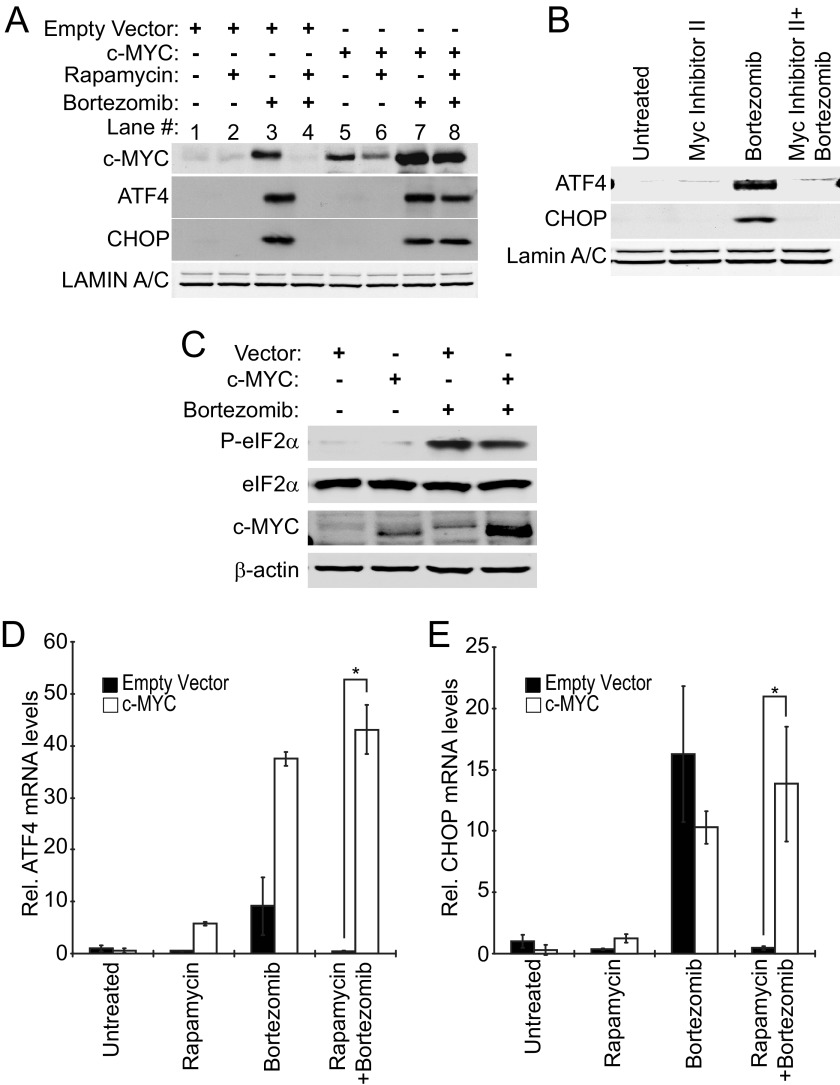
Rapamycin is suggested to prevent the induction of ATF4 and CHOP expression and the accompanying apoptosis in response to bortezomib treatment by decreasing the expression levels of c-MYC. To test this model, we used a lentiviral expression system to restore the c-MYC protein expression that had been lost as a result of rapamycin treatment. Elt3 cells were transduced with lentiviruses expressing c-MYC or control empty vector. Cells transduced with empty vector showed a similar lowering of bortezomib-induced c-MYC, ATF4, and CHOP expression upon rapamycin pretreatment (Fig. 7A, compare lanes 3 and 4) as had been observed in uninfected cells (Figs. 3A and 5A). Meanwhile, c-MYC-transduced cells showed high c-MYC expression basally after 6 h of bortezomib treatment and even after pretreatment with rapamycin (Fig. 7A, lanes 5–8). Additionally, cells expressing exogenous c-MYC showed induction of ATF4 and CHOP when treated with bortezomib alone or after rapamycin pretreatment (Fig. 7A, lanes 7 and 8). The expression of exogenous c-MYC was also able to rescue the expression of ATF4 and CHOP at the mRNA level after bortezomib treatment (Fig. 7, D and E). Furthermore, treatment of cells with MYC inhibitor II, which was shown to block c-MYC induction of NOXA in Fig. 5E, blocked bortezomib-induced ATF4 and CHOP expression (Fig. 7B).
FIGURE 7.
Overexpression of c-MYC rescues rapamycin-mediated suppression of ATF4 and CHOP expression in cells exposed to bortezomib. Lentivirus was used to stably express c-MYC in Elt3 cells. A, vector- and c-MYC-expressing cells were then pretreated with vehicle control or 50 nm rapamycin (24 h) before treatment with bortezomib for 6 h. Nuclear lysates were prepared, and levels of ATF4, CHOP, and Lamin A/C proteins were measured by Western blot. B, Elt3 cells were pretreated with Myc inhibitor II for 2 h before treatment with bortezomib for an additional 6 h. Western blots were executed as in A. C, Elt3 cells overexpressing c-MYC were treated with bortezomib for 6 h. Phosphorylation of eIF2α, total eIF2α, c-MYC, and β-actin were measured by Western blot. D and E, under the same conditions, ATF4 and CHOP mRNAs were measured by quantitative real-time-PCR. *, p < 0.05.
This effect of c-MYC did not appear to be the result of increased ER stress caused by c-MYC overexpression as exogenous c-MYC-expressing cells have similar levels of eIF2α phosphorylation in the absence or presence of bortezomib compared with empty vector cells (Fig. 7C). Together, these results suggest that the ability of rapamycin to suppress ATF4 and CHOP expression is dependent on its down-regulation of c-MYC. These findings also support the notion that c-MYC plays a central role in the induction of the ATF4 and CHOP mRNAs during bortezomib treatment (Fig. 7A).
c-MYC Overexpression Rescues Rapamycin-mediated Suppression of Bortezomib-induced elt3 Cell Apoptosis
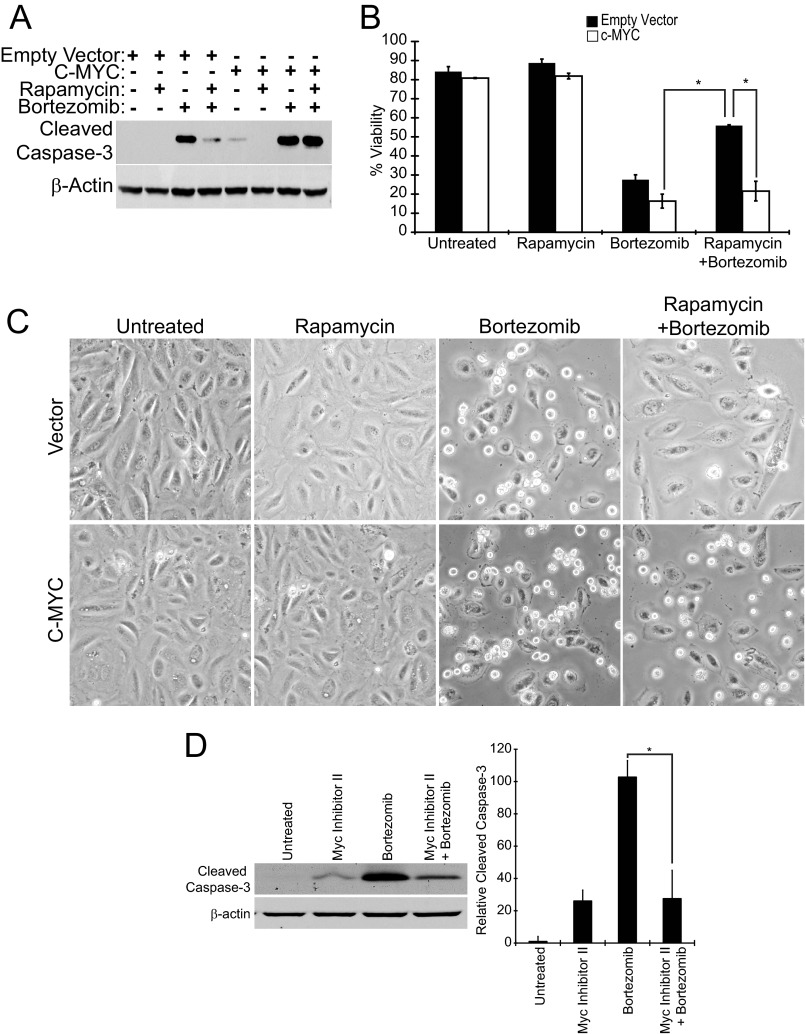
Next we used the lentiviral c-MYC overexpression system to test if c-MYC rescue of ATF4 and CHOP expression in the presence of rapamycin is sufficient to restore bortezomib-induced apoptosis. Elt3 cells transduced with empty vector virus showed caspase-3 cleavage after 24 h of exposure to bortezomib that was dramatically reduced by rapamycin pretreatment (Fig. 8A), which was similar to that observed in Fig. 1. Importantly, cells transduced with c-MYC-expressing virus showed caspase-3 cleavage after 24 h of bortezomib treatment with or without rapamycin pretreatment, indicating that restoring c-MYC expression is sufficient to restore bortezomib-induced apoptosis (Fig. 8A). A similar outcome was seen on cell viability as measured by trypan blue staining; cells transduced with only empty vector showed a 57% decrease in cell viability after bortezomib treatment that was significantly inhibited by rapamycin pretreatment (Fig. 8B). By comparison, c-MYC transduced cells showed a similar decrease in cell viability that could not be rescued by pretreating cells with the mTORC1 inhibitor (Fig. 8B). This decrease in cell viability was also readily apparent by microscopy where c-MYC-overexpressing cells showed a rounded non-adherent morphology after bortezomib treatment with or without rapamycin (Fig. 8C). In contrast, the vector-transduced cells remained flat and adherent when pretreated with rapamycin before exposure to bortezomib.
FIGURE 8.
Overexpression of c-MYC restores bortezomib-induced apoptosis. Elt3 cells overexpressing c-MYC or containing empty vector were incubated for 24 h with 20 nm bortezomib with or without an additional 24-h rapamycin pretreatment. A, cleaved caspase-3 and β-actin were detected in cell lysates by Western blotting. B, trypan blue staining was carried out to measure the survival of similarly treated Elt3 cells. C, photographs of the two cell lines after 24 h bortezomib exposure in the presence or absence of rapamycin pretreatment are shown. D. Elt3 cells were pretreated with MYC Inhibitor II for 2 h before the addition of bortezomib for a further 24 h. Cleaved caspase-3 and β-actin were detected in cell lysates by Western blotting. Levels of cleavage caspase-3 from three independent experiments are shown in the histogram to the right of these Western blots. *, p < 0.05.
As anticipated, blocking the activity of c-MYC using c-MYC inhibitor II resulted in reduced ATF4 and CHOP expression after bortezomib treatment (Fig. 7B). Caspase-3 activation was also reduced after 24 h of bortezomib treatment for those cells treated with the c-MYC inhibitor II (Fig. 8D). These results indicate that in this cell line c-MYC expression contributes to the induction of ATF4 and CHOP and consequent cell death during bortezomib treatment.
DISCUSSION
This study demonstrates the feasibility of using a clinically approved drug to induce the death of cells that have elevated mTORC1 activity due to the loss of TSC2. It also demonstrates for the first time that mTORC1 regulates the UPR at the level of ATF4 and CHOP transcription factors by promoting increased expression of these genes. This is achieved at least in part by the translation of c-MYC that can activate the transcription of ATF4. In concert with bortezomib treatment, which elevates ER stress and induces the expression of c-MYC, high mTORC1 activity contributes to cell death in a manner that can be prevented by rapamycin pretreatment. These data not only suggest a means of eradicating cells exhibiting high mTOR activity by use of a proteasome inhibitor but may also help explain why myeloma cells with elevated c-MYC levels are more sensitive to the proteasome inhibitor, bortezomib/Velcade (27).
The above findings were made possible by a key signaling difference between the Elt3 leiomyoma cells used here and mouse embryo fibroblasts studied in previous reports (8, 9). Pretreatment of Elt3 cells with rapamycin did not significantly impact the bortezomib-induced phosphorylation of eIF2α; however, this rapamycin treatment blocked the induction of downstream ATF4 and CHOP transcription factors. Three previous studies documented increased ER stress-induced apoptosis after the knock-out of TSC1 or -2 (8–10). These studies showed that the induction of these ER stress markers can be decreased with rapamycin treatment. Although only Ozcan et al. (9) showed that rapamycin reduces this ER stress-induced apoptosis, the other studies reported no effect of the compound on cell death (8, 10). Our current findings not only support several aspects of the study by Ozcan et al. (9) but contribute additional mechanistic insight into how mTORC1 contributes to ER stress and cell death. The two studies that used MEFs reported increased PERK and eIF2α phosphorylation upon TSC1 or -2 loss (8, 9), whereas a study of TSC-null rodent neurons primarily focused on the induction of CHOP expression and did not measure eIF2α phosphorylation (10). Because eIF2α phosphorylation was not affected by rapamycin treatment in the current study, it allowed us to unmask a layer of UPR regulation by mTORC1 distal to eIF2α phosphorylation. At this time we have not established why Elt3 cells have an attenuated unfolded protein response compared with MEFs. However, this is likely related to genetic differences between cell lines. For example, Elt3 cells retain wild type p53, whereas the TSC-null MEFs required deletion of p53 for survival (29). Alternatively, MEFs may have lower basal MYC expression. It should also be pointed out that the two MEF studies (8, 9) conflicted in whether TSC loss impacts downstream ATF4 and CHOP activation. Thus there are likely to be mechanistic differences based on both cell type and metabolic state.
Consistent with the failure of rapamycin to impact eIF2α phosphorylation, this drug also did not affect the cleavage of ATF6, suggesting that mTORC1 is not promoting a UPR through increased global translation. Although rapamycin has been documented to more effectively suppress mTORC1-mediated phosphorylation of S6 kinase than 4E-BP (30, 31), which might limit impact of the drug on global translation initiation, we found that our rapamycin pretreatment effectively suppressed both Elt3 cell S6 kinase and 4EBP phosphorylation.4 Bortezomib-induced transcription and translation of ATF4 and CHOP were both suppressed by rapamycin pretreatment and could also be blocked by actinomycin D. These data indicate that although proximal events of the UPR are not mTORC1-dependent, the downstream pro-apoptotic signals emanating from the ATF4/CHOP portion of this pathway are inhibited at the level of transcription by rapamycin pretreatment. Therefore, we speculated that rapamycin may deplete cells of a transcription factor that is required for bortezomib to induce ATF4 and CHOP expression.
Several documented stresses, such as UV irradiation, brain ischemia, or LPS stimulation of macrophages, trigger robust eIF2α phosphorylation yet fail to induce ATF4 and CHOP expression (17, 32, 33). For example, in the case of UV irradiation, ATF4 transcription is repressed, resulting in little ATF4 mRNA being available for preferential translation after eIF2α phosphorylation (17). Although it was proposed in this report that UV causes this reduction of ATF4 mRNA by inducing the expression of a repressor protein, our study showed that rapamycin suppresses the transcription of ATF4 via lowered levels of an activating transcription factor, c-MYC.
The selective targeting of downstream UPR components by rapamycin suggested the expression and/or activation of a transcription factor(s) in response to elevated mTORC1 signaling in TSC2-null Elt3 cells. c-MYC was a logical target given its action as an E-box transcription factor that is translationally controlled by mTORC1. E-box transcription factors were also identified by microarray studies to be more active in TSC1 and TSC2 knock-out MEFs and to be inhibited by rapamycin (34). Although c-MYC was recently reported to accumulate at the regulatory sequences of already active genes to boost their transcription (35, 36), the association of c-MYC with an E-box at −78 of the ATF4 promoter and the requirement of the −78 site for c-MYC stimulation of this gene are more consistent with c-Myc acting in a more traditional trans-acting manner. Despite the poor conservation of the ATF4 5′ promoter region between rats, mice, and humans, all three species have at least two c-MYC binding sites in either their promoter region or first exon. Furthermore, two laboratories have observed c-MYC association with the human ATF4 gene in ChIP on ChIP experiments (37, 38). These data suggest that c-MYC plays a role in controlling ATF4 expression in Elt3 rat leiomyoma cells that is likely conserved in humans.
Although we observed an up-regulation of c-MYC transcript in response to bortezomib treatment, this induction was insensitive to rapamycin pretreatment, consistent with rapamycin specifically inhibiting only the translation of c-MYC into protein. This induction of c-MYC was not limited to bortezomib as several ER stressors (2-deoxyglucose, thapsigargin) induced its transcription/translation. This was consistent with a previous demonstration of thapsigargin and dithiothreitol increasing c-MYC mRNA expression in MEF cells (39). These findings collectively suggest that ER stress, caused by bortezomib or other ER stressing-inducing agents, promotes the transcription of c-MYC, resulting in up-regulation of c-MYC levels/activity. Elevation of exogenous MYC levels in the presence of bortezomib also affected the impact of the proteasome inhibitor on MYC protein levels. Rescue with exogenous MYC suggests that the ability of rapamycin to suppress ATF4 and CHOP expression is dependent on its down-regulation of c-MYC. These findings also support the notion that c-MYC plays a central role in the induction of the ATF4 and CHOP mRNAs during bortezomib treatment. This was also reflected in exogenous c-MYC negating the ability of rapamycin to suppress apoptosis and the ability of MYC inhibitor II to block bortezomib-induced ATF4 and CHOP expression and caspase-3 cleavage. In terms of clinical implications, our studies demonstrate that rapamycin may not be an appropriate drug to combine with bortezomib or other compounds that rely on ATF4- and CHOP-induced cell death.
Acknowledgments
We thank Drs. Cheryl Walker and Clark Wells for Elt3 cells and c-MYC cDNA, respectively, plus Thomas Baird and Michael Fusakio for quantitative PCR assistance.
This work was supported by the United States Department of Defense Award W81XWH1110355 and The LAM foundation (to L. A. Q.).
J. T. Babcock, and L. A. Quilliam, unpublished observations.
- TSC
- tuberous sclerosis complex
- ATF4 or 6
- activating transcription factor 4 or 6
- CHOP
- CCAAT/enhancer-binding protein homologous protein
- ER
- endoplasmic reticulum
- mTORC1
- mammalian target of rapamycin complex 1
- PERK
- protein kinase-like endoplasmic reticulum kinase
- UPR
- unfolded protein response
- MEF
- mouse embryonic fibroblast
- ATF6
- activating transcription factor-6.
REFERENCES
- 1. Crino P. B., Nathanson K. L., Henske E. P. (2006) The tuberous sclerosis complex. New Eng. J. Med. 355, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 2. Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A. (2003) Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278, 32493–32496 [DOI] [PubMed] [Google Scholar]
- 3. Inoki K., Li Y., Xu T., Guan K. L. (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. (2003) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581 [DOI] [PubMed] [Google Scholar]
- 5. Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C., Blenis J. (2002) Tuberous sclerosis complex-1 and 2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 7. Benvenuto G., Li S., Brown S. J., Braverman R., Vass W. C., Cheadle J. P., Halley D. J., Sampson J. R., Wienecke R., DeClue J. E. (2000) The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene 19, 6306–6316 [DOI] [PubMed] [Google Scholar]
- 8. Kang Y. J., Lu M. K., Guan K. L. (2011) The TSC1 and TSC2 tumor suppressors are required for proper ER stress response and protect cells from ER stress-induced apoptosis. Cell Death Differ. 18, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozcan U., Ozcan L., Yilmaz E., Düvel K., Sahin M., Manning B. D., Hotamisligil G. S. (2008) Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol. Cell 29, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Nardo A., Kramvis I., Cho N., Sadowski A., Meikle L., Kwiatkowski D. J., Sahin M. (2009) Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 29, 5926–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marciniak S. J., Ron D. (2006) Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 12. Schröder M., Kaufman R. J. (2005) The mammalian unfolded protein response. Ann. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 13. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 14. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 15. Palam L. R., Baird T. D., Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dey S., Baird T. D., Zhou D., Palam L. R., Spandau D. F., Wek R. C. (2010) Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33165–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Armstrong J. L., Flockhart R., Veal G. J., Lovat P. E., Redfern C. P. (2010) Regulation of endoplasmic reticulum stress-induced cell death by ATF4 in neuroectodermal tumor cells. J. Biol. Chem. 285, 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang H. Y., Wek R. C. (2005) Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 280, 14189–14202 [DOI] [PubMed] [Google Scholar]
- 20. Walker C., Ginsler J. (1992) Development of a quantitative in vitro transformation assay for kidney epithelial cells. Carcinogenesis 13, 25–32 [DOI] [PubMed] [Google Scholar]
- 21. Teske B. F., Wek S. A., Bunpo P., Cundiff J. K., McClintick J. N., Anthony T. G., Wek R. C. (2011) The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell 22, 4390–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Higuchi R., Krummel B., Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments. Study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Düvel K., Yecies J. L., Menon S., Raman P., Lipovsky A. I., Souza A. L., Triantafellow E., Ma Q., Gorski R., Cleaver S., Vander Heiden M. G., MacKeigan J. P., Finan P. M., Clish C. B., Murphy L. O., Manning B. D. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molec. Cell 39, 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hosoi H., Dilling M. B., Liu L. N., Danks M. K., Shikata T., Sekulic A., Abraham R. T., Lawrence J. C., Jr., Houghton P. J. (1998) Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol. Pharmacol. 54, 815–824 [DOI] [PubMed] [Google Scholar]
- 25. Chen S., Blank J. L., Peters T., Liu X. J., Rappoli D. M., Pickard M. D., Menon S., Yu J., Driscoll D. L., Lingaraj T., Burkhardt A. L., Chen W., Garcia K., Sappal D. S., Gray J., Hales P., Leroy P. J., Ringeling J., Rabino C., Spelman J. J., Morgenstern J. P., Lightcap E. S. (2010) Genome-wide siRNA screen for modulators of cell death induced by proteasome inhibitor bortezomib. Cancer Res. 70, 4318–4326 [DOI] [PubMed] [Google Scholar]
- 26. Nikiforov M. A., Riblett M., Tang W. H., Gratchouck V., Zhuang D., Fernandez Y., Verhaegen M., Varambally S., Chinnaiyan A. M., Jakubowiak A. J., Soengas M. S. (2007) Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc. Natl. Acad. Sci., U.S.A. 104, 19488–19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nawrocki S. T., Carew J. S., Maclean K. H., Courage J. F., Huang P., Houghton J. A., Cleveland J. L., Giles F. J., McConkey D. J. (2008) Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood 112, 2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin X., Giap C., Lazo J. S., Prochownik E. V. (2003) Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22, 6151–6159 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Cicchetti G., Onda H., Koon H. B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C. L., Kwiatkowski D. J. (2003) Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through down-regulation of PDGFR. J. Clin. Invest. 112, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choo A. Y., Yoon S. O., Kim S. G., Roux P. P., Blenis J. (2008) Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl. Acad. Sci, U.S.A. 105, 17414–17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woo C. W., Cui D., Arellano J., Dorweiler B., Harding H., Fitzgerald K. A., Ron D., Tabas I. (2009) Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat. Cell Biol. 11, 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar R., Krause G. S., Yoshida H., Mori K., DeGracia D. J. (2003) Dysfunction of the unfolded protein response during global brain ischemia and reperfusion. J. Cereb. Blood Flow Metab. 23, 462–471 [DOI] [PubMed] [Google Scholar]
- 34. Yecies J. L., Manning B. D. (2011) Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 71, 2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin C. Y., Lovén J., Rahl P. B., Paranal R. M., Burge C. B., Bradner J. E., Lee T. I., Young R. A. (2012) Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell 151, 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D. R., Tessarollo L., Casellas R., Zhao K., Levens D. (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z., Van Calcar S., Qu C., Cavenee W. K., Zhang M. Q., Ren B. (2003) A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci., U.S.A. 100, 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zeller K. I., Zhao X., Lee C. W., Chiu K. P., Yao F., Yustein J. T., Ooi H. S., Orlov Y. L., Shahab A., Yong H. C., Fu Y., Weng Z., Kuznetsov V. A., Sung W. K., Ruan Y., Dang C. V., Wei C. L. (2006) Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci., U.S.A. 103, 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liang S. H., Zhang W., McGrath B. C., Zhang P., Cavener D. R. (2006) PERK (eIF2α kinase) is required to activate the stress-activated MAPKs and induce the expression of immediate-early genes upon disruption of ER calcium homoeostasis. Biochem. J. 393, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]