Background: Initial activation of the cardiac-specific transcriptional program during cellular differentiation is poorly understood.
Results: Nox4-generated ROS promotes the initial transcriptional activation of GATA-4 via a c-Jun-dependent mechanism in pluripotent progenitor cells.
Conclusion: The redox-dependent activation of a widely expressed transcription factor within pluripotent cells acts to effect cardiac lineage-specific gene transcription.
Significance: Redox-dependent transcriptional regulation mediates the activation of the cardiac-specific cellular differentiation program.
Keywords: AP-1 Transcription Factor, Cardiac Development, Nox, Reactive Oxygen Species (ROS), Redox Regulation, Nox4, Cardiogenesis
Abstract
NADPH oxidase 4 (Nox4) generates reactive oxygen species (ROS) that can modulate cellular phenotype and function in part through the redox modulation of the activity of transcription factors. We demonstrate here the potential of Nox4 to drive cardiomyocyte differentiation in pluripotent embryonal carcinoma cells, and we show that this involves the redox activation of c-Jun. This in turn acts to up-regulate GATA-4 expression, one of the earliest markers of cardiotypic differentiation, through a defined and highly conserved cis-acting motif within the GATA-4 promoter. These data therefore suggest a mechanism whereby ROS act in pluripotential cells in vivo to regulate the initial transcription of critical tissue-restricted determinant(s) of the cardiomyocyte phenotype, including GATA-4. The ROS-dependent activation, mediated by Nox4, of widely expressed redox-regulated transcription factors, such as c-Jun, is fundamental to this process.
Introduction
Understanding the molecular mechanisms that underlie the transition from multipotency to cardiac lineage commitment is crucial to the progression toward both a better understanding of heart development during embryogenesis and potentially to the development of long term regenerative heart therapies. The study in vivo of mammalian cardiomyocyte differentiation is particularly challenging, as the heart develops very early during embryogenesis (reviewed in Ref. 1). However, the establishment of pluripotent cell lines such as embryonic stem (ES)2 and embryonal carcinoma (EC) cells, which can differentiate into derivatives of all three primary germ layers, including cardiomyocytes, has greatly facilitated the analysis of very early cardiogenesis (2, 3). Thus, the differentiation in vitro of ES and EC cells to the cardiac lineage represents highly regulated culture systems in which gene expression and cellular function are modulated in a developmentally controlled manner, equivalent to that found in vivo. We and others have shown that mRNAs encoding early expressed cardiac-restricted transcription factors such as GATA-4, GATA-6, MEF2C, and Nkx2.5 are initially up-regulated in ES and EC cells. These in turn act to promote the full cardiac transcriptional program, including contractile proteins such as α- and β-myosin heavy chain (α- and β-MHC), atrial natriuretic factor, the Na+-Ca2+ exchanger, and phospholamban, etc. (2, 4). In addition, these early expressed transcription factors are known to both cross-regulate each others' expression (5–8) and to potentially autoregulate their own expression (9). However, how these transcription factors themselves are initially activated remains unclear.
There is now increasing evidence to suggest that redox-based mechanisms may be central to the process of early cardiogenesis. Within ES cells that have been induced to differentiate into embryoid bodies (EBs), agents that increase the levels of intracellular ROS, also increase the proportion of beating cardiomyocytes within the EB population (10, 11). By contrast, agents that scavenge or reduce ROS levels (e.g. trolox, pyrrolidine dithiocarbamate, catalase, and N-acetylcysteine) impair cardiomyocyte formation in EBs (12, 13). Exogenous administration of ROS, by incubation of ES cells with hydrogen peroxide (H2O2), either enhances or impairs the cardiomyogenic program, dependent upon the concentration and timing during differentiation, suggesting both a dose dependence and cellular competency in the response to ROS (12, 14, 15).
The precise source(s) of ROS and molecular mechanisms of action underlying the redox regulation of cardiogenesis in ES cells largely remain unclear. However, several recent studies have suggested the potential involvement of NADPH oxidases (Noxs), in particular Nox4, in this process (16). Nox enzymes comprise a family of transmembrane enzymes that catalyze the one-electron reduction of molecular oxygen to superoxide as their primary function (17, 18). Seven family members have been identified in mammalian species (Nox1–5 and DUOX1–2), which are each transcribed from separate genes. Although different isoforms are often co-expressed within individual cell types, they are subject to differential and complex regulation and have been shown to have distinct, nonredundant functions (19). Both Nox2 and Nox4 are co-expressed within cardiotypic cells at different stages of differentiation and development (20, 21), and Nox4 is the more abundant isoform expressed in undifferentiated ES cells and in ES cell-derived or neonatal cardiomyocytes (12). Nox4 has been demonstrated to be constitutively active, and hence its activity is primarily regulated by the level of its expression (22). Another potentially significant difference between Nox4 and other Nox proteins is that the superoxide generated by Nox4 is preferentially further reduced to H2O2, in a process that is apparently mediated, at least in part, by an intrinsic activity within its E-loop (23–25). Thus, Nox4 may elicit signaling pathways distinct from those regulated by other Noxs, including Nox2 within cardiotypic (and other) cells.
Many of the agents that increase intracellular ROS and promote cardiogenesis in pluripotent cells have also been demonstrated to increase the expression of Nox4 in these cells (10, 26). Furthermore, during cardiomyocyte differentiation in vitro, siRNA-mediated down-regulation of Nox4 reduced the potential of ES cells to express cardiac-specific markers and form-beating EBs. This effect could be reversed by a pulse of exogenous H2O2, administered at a time point coinciding with the onset of expression of the cardiac-restricted transcription factors (12), suggesting the ROS generated by Nox4 play a role in this very early stage of cardiomyocyte differentiation.
ROS act as transducers in intracellular signaling pathways through the reversible oxidation of specific amino acid residues within proteins (27). These post-translational modifications have been shown to act to alter the structure and hence modify the function(s) of many proteins, including protein phosphatases and kinases, ion channels, and transcription factors. Such “redox-sensitive” transcription factors include p53, NF-κB, HIF-1α, AP-1, and Sp1 (28). Their DNA binding activities and/or nuclear translocation mechanisms are directly or indirectly regulated by levels of ROS. They are typically widely expressed within different cell types and provide the cell with a mechanism to effect changes in transcriptional programs in response to cellular redox changes.
In this study, we aimed to elucidate the redox-dependent molecular mechanism(s) underlying the regulation of cardiomyocyte differentiation in pluripotent cells by Nox4. We show that ROS generated by Nox4 during the early stages of cardiomyocyte differentiation activates transcription of GATA-4. We demonstrate that this activation is dependent upon a cis-acting AP-1-binding motif within the GATA-4 promoter and is mediated by the redox-sensitive transcription factor c-Jun. The data presented here therefore suggest that redox regulation of the ubiquitously expressed transcription factor, c-Jun, acts to initiate the cardiac-specific transcriptional program, prior to the expression of cardiac transcription factors.
EXPERIMENTAL PROCEDURES
Cell Culture
P19 CL6 cells were a kind gift from Issei Komuro. Cells were maintained in α-minimum essential media (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 4 mm l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Sigma). To induce cardiotypic differentiation, cells were plated at 3.7 × 105 cells per 60-mm dish in growth media supplemented with 1% DMSO. The medium was renewed every 2 days. In experiments with ROS scavengers or Nox enzyme inhibitors, cells were supplemented with 4 mm sodium 4,5-dihydroxybenzene-1,3-disulfonate (tiron) for 24 h or 10 μm diphenyliodonium (DPI) for 6 h. For the inhibition of c-Jun N-terminal kinase (JNK) activity, JNK inhibitor II (Calbiochem) was used at a final concentration of 20 μm. Neonatal rat cardiomyocyte isolation and culture were performed as described previously (20).
Adenoviral Transduction
Full-length mouse cDNA of Nox4 (a kind gift from Thomas Leto) was cloned into pcDNA3.1 (Invitrogen). Adenoviral expression vectors were generated for Nox4 (AdNox4) and β-galactosidase (AdβGal) using the AdEasy adenoviral vector system (Qbiogene) according to the manufacturer's protocol. Cells were transduced with adenoviral constructs at a multiplicity of infection (m.o.i.) of 20 and incubated overnight prior to use. Adenoviral shRNAs against Nox4 and GFP (kind gifts of R. Davisson) were used as described previously (29) at an m.o.i. of 100.
Western Blotting
Cells were homogenized and lysed in whole cell lysis buffer (25 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EGTA, 5 mm EDTA, 30 mm NaF, 40 mm β-glycerophosphate, 20 mm sodium pyrophosphate, 1 mm sodium orthovanadate, 1 mm PMSF, 0.5% Nonidet P-40, and protease inhibitor mixture (Sigma)). Protein concentrations were determined by Bradford assay to ensure equal protein loading. Lysates were denatured in Laemmli loading buffer. Proteins were separated on 10% SDS-polyacrylamide gels and transferred to a PVDF membrane. Membranes were probed with antibodies against GATA-4 (eBioscience), NF-κB, p53, c-Jun, phospho-c-Jun (Ser-63) (Cell Signaling), and Nox4 (19). Blots were probed for β-actin (Sigma) as a loading control. Protein bands were visualized using Amersham Biosciences ECL Western blotting detection reagent (GE Healthcare) according to the manufacturer's instructions.
RNA Extraction, cDNA Synthesis, and Quantitative Real Time PCR
Total RNA was isolated from tissue or cells using the SV Total RNA Isolation System (Promega) according to the manufacturer's recommendations. cDNA was generated by random-primed reverse transcription of 1 μg of total RNA with Moloney murine leukemia virus RT (Promega) according to the manufacturer's protocol. Relative gene expression was quantified using SYBR Green and the comparative Ct analysis method, on a Mastercycler® ep realplex PCR system (Eppendorf). In all cases control reactions, in which reverse transcriptase was omitted, were performed. Cytoskeletal β-actin levels were used for normalization. Forward and reverse primers used to detect transcripts were as follows (all are 5′ to 3′): β-actin F, CTGTCGAGTCGCGTCCACCC, and R, ATGCCGGAGCCGTTGTCGAC; GATA-4 F, GGGCCAACCCTGGAAGAC, and R, GCCCCACAATTGACACACTCT; Nox4 F, GTCAAACAGATGGGATTCAGAA, and R, CCGAGGACGCCCAATCCGAGGACGCCCAAT; Nkx 2.5 F, CCGATCCATCCCACTTTATTGA, and R, CCTAGTGTGGAATCCGTCGAA; and MEF2C F, TGATCAGCAGGCAAAGATTG, and R, GGATGGTAACTGGCATCTCAA. Standard GoTaqDNA polymerase (Promega) was used for semi-quantitative RT-PCR. Primers used to determine α-MHC and alternative GATA-4 transcript expression were as follows (all 5′ to 3′): α-MHC F, TCATTCCCAACGAGCGAAA, and R, GCCGGAAGTCCCCATAGAGA; E1a-containing transcript, TTTCTGGGAAACTGGAGCTG; E1b-containing transcript, GCTGATCTGTCCCTGAAAGC; E1b′-containing transcript, GTTCACTCTGGAAGCGTTGG; and the common reverse primer located in the shared exon 2 region, CTGGTCTCGAACACCCTGAG.
Construction of Mouse GATA-4 Luciferase Reporter Plasmid
The GATA-4 luciferase reporter plasmid was constructed by inserting a 1407-bp PCR-amplified fragment of the GATA-4 promoter (−1286 bp to +121 bp relative to the cap site) between the KpnI and XhoI sites of pGL4.22(luc2CP/Puro) luciferase vector (Promega). The mouse GATA-4 promoter fragment was generated with the primer pair 5′-GGAATGGTACCAGTCCTTCCTCTGGGATTTT-3′ (incorporating a 5′ KpnI restriction site) and 5′-GGAATCTCGAGCTGGCCATCTCCAGTTTCCC-3′ (incorporating a 3′ XhoI restriction site), using Herculase (Stratagene) and the bacterial artificial chromosome RP23-249C3 clone (BACPAC Resources Center, CHORI), which includes the mouse GATA-4 genomic locus as template. Site-specific mutations were introduced into sequence motifs in the luciferase plasmid using a modified version of a previously described technique (30). Primers used were as follows: forward, 5′-CCCGGAGACCTGGTGCCCCACCGGCGAC-3′, and reverse, 5′-GCACCAGGTCTCCGGGATGCCTTGCTAG-3′. The fidelity of all plasmid constructs was confirmed by sequencing.
Luciferase Reporter Assays
P19 CL6 cells were transfected using Lipofectamine 2000TM reagent (Invitrogen) according to the manufacturer's protocol, with firefly luciferase reporter plasmids containing the proximal GATA-4 promoter region or an AP-1-responsive response element or appropriate promoterless control vectors (Promega). A Renilla luciferase-linked reporter construct (pSV40-pRL; Promega) was co-transfected as control in all cases. After transfection, cells were incubated overnight in normal growth media. Promoter activity was assessed using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's protocol. Luminescence readings were measured, and the data are represented as a ratio of firefly; Renilla luminescence was expressed in relative luminescence units (RLU) or fold induction as indicated. Biological triplicate assays were performed in all cases.
Chromatin Immunoprecipitation (ChIP) Assays
P19 CL6 cells were induced to differentiate in the presence of 1% DMSO for 4 days, and subsequently ChIP assays were performed on these cells using the Magna-ChIP chromatin immunoprecipitation kit (Millipore), essentially according to the manufacturer's instructions. Immunoprecipitations were performed using 5 μg of a rabbit monoclonal c-Jun antibody (Cell Signaling). Negative controls with normal IgG, p53 antibody (Cell Signaling), and no antibody were also performed. Primers used to amplify the GATA-4 promoter were as follows: forward, 5′-CTTGCGTCTCTAGGGACTGA-3′, and reverse, 5′-AGAAGGTGACCTCGCACACT-3′. PCR conditions were as follows: 94 °C for 3 min, then 32 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for a 2-min final extension. Samples of chromatin before immunoprecipitation were analyzed and used as an input control.
Nuclear Extract Preparation and AP-1 DNA Binding Activity
Nuclear extracts were prepared from freshly harvested undifferentiated P19 CL6 cells and cells at day 4 of differentiation or from NRC cells transduced with an adenoviral vector encoding either Nox4 or β-galactosidase as control as described previously (31). AP-1 DNA binding activity in NRCs was assessed using a TransAMTM transcription factor ELISA kit (Active Motif) according to the manufacturer's recommendations. c-Jun DNA binding activity is expressed as A450 nm ± S.E.
FOX Assay for the Measurement of Hydrogen Peroxide
H2O2 production from differentiating P19 CL6 cells was measured from cell culture supernatants of P19 CL6 cells after 4 days of differentiation or control cells. H2O2 induces oxidation of the ferrous ion (Fe2+) to a ferric ion (Fe3+) that reacts with xylenol orange and results in the development of a blue-purple complex that can be assayed by reading its absorbance at 560 nm. Briefly, 100 μl of supernatant from cells was mixed with 900 μl of FOX reagent (100 μm xylenol orange, 250 μm ammonium ferrous sulfate, 25 mm H2SO4, 4 mm butylated hydroxytoluene, dissolved in 90% methanol) and incubated at RT for 30 min. Samples were then centrifuged at 12,000 × g for 5 min to remove any flocculated material. The absorbance of the supernatant is read at 560 nm, and H2O2 concentrations were determined from a H2O2 standard curve. Catalase-inhibitable signal, i.e. H2O2-specific, portion of the signal is determined by incubating replicate samples with 100 units/ml of catalase for 5 min prior to the addition of the FOX reagent.
Bio-oligo Pulldown Assay
Nuclear extracts were prepared from differentiating P19 CL6 cells at day 4 as described above. Biotinylated promoter fragments were generated by PCR. Forward and reverse primers used to amplify mouse GATA-4 promoter fragments were as follows (all 5′ to 3′): 1.4-kb fragment F, biotin-AGTCCTTCCTCTGGGATTTT, and R, CTGGCCATCTCCAGTTTCCC; AP-1-binding sites (see Fig. 3A–C, respectively), F, biotin-GCCTCCTGACCCTCTCCCGA, and R, GGGCTCATGGTCGCCGGT; F, biotin-CGGCGACCATGAGCCCAG, and R, CTTTCCTCCCTCCTCTCAGT; F, biotin-AGGGGACAAGCCGGAGGCC, and R, CTGGCCATCTCCAGTTTCCC. The 21-bp biotinylated oligonucleotides and unmodified complementary oligonucleotides encoding WT or mutated AP-1-binding sites were obtained from Sigma and annealed into duplexes for use. Oligonucleotides used were as follows: wild type F, biotin-ATCCCGGGTGACCCAGTGCCC, and R, GGGCACTGGGTCACCCGGGAT; mutated, biotin-ATCCCGGGTGACCTGGTGCCC, and R, GGGCACCAGGTCACCCGGGAT. The bio-oligonucleotide pulldown assay was performed as described previously (32). Briefly, biotinylated double-stranded DNA (dsDNA) was generated, purified, and bound to streptavidin-coated magnetic beads (Dynabeads, Invitrogen) as per the manufacturer's protocol. 20 μg of nuclear extract was preincubated with 5 μg of poly(dI-dC) (Sigma) for 15 min at room temperature. dsDNA-coated magnetic beads were added, and the reaction was incubated for 30 min at room temperature. Protein-DNA-bead complexes were separated using a magnet and washed three times with 1× binding buffer (20 mm HEPES-KOH, pH 7.9, 25 mm KCl, 0.1 mm EDTA, 2 mm spermidine, 2 mm MgCl2, 0.025% Nonidet P-40). Bound proteins were eluted from the beads by the addition of NaCl to a final concentration of 500 mm and analyzed by Western blot. Bound dsDNA was released from the beads by incubating the beads in 50% formamide at 70 °C for 5 min.
FIGURE 3.
Functional analysis of the 5′-upstream sequence of the GATA-4 gene. A, structural organization of the mouse GATA-4 gene showing alternative first exons E1a, E1b, and E1b′ (not to scale). B, semi-quantitative RT-PCR analysis of alternative GATA-4 first exon usage in mRNA isolated from undifferentiated P19 CL6 cells (see “Experimental Procedures”). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is included as a positive control. C, semi-quantitative RT-PCR analysis of alternative GATA-4 first exon expression in mRNA isolated from undifferentiated P19 CL6 cells (day 0) and cells at day 4 of differentiation. GAPDH is again included as a loading control. D, luciferase reporter activity resulting from undifferentiated P19CL6 cells (day 0; open bars) or cells at day 4 of differentiation (day 4; black bars), cultured in the presence of vehicle control, or 4 mm tiron or 10 μm DPI as indicated. Cells were transfected with a GATA-4 promoter construct (GATA-4 prom) or the promoterless control vector (empty control), as indicated, for 24 h prior to the measurement of luciferase activity. E, luciferase activity resulting from undifferentiated P19 CL6 cells transduced with either AdNox4 (black bars) or AdβGal (open bars) as control, subsequently transfected with a GATA-4 reporter plasmid or control promoterless vector as indicated, and incubated for 24 h under normal growth conditions or in the presence of tiron (4 mm) for 6 h prior to luciferase activity measurement as indicated. In all cases, luciferase activity, normalized to that of the co-transfected SV40-Renilla luciferase control vector, is expressed as RLU. All data are presented as mean ± S.E. *, p < 0.05; n.s., not significant.
siRNA-mediated Knockdown of c-Jun and p22Phox
siRNA-mediated knockdown of c-Jun or p22Phox in P19 CL6 cells was performed by transfecting siRNAs to c-Jun (Ambion; identification number S68564), p22Phox (Ambion; identification number S201230), or scrambled control (Ambion) using HiPerFect transfection reagent (Qiagen) according to the manufacturer's protocol. The extent of protein knockdown was assessed by Western blot using either an anti-c-Jun antibody (Cell Signaling) or an anti-cytochrome b245 light chain antibody (p22Phox; Abcam).
Statistical Analyses
Data are expressed as mean ± S.E. Comparisons between sample groups or measurements at different time points were made by unpaired Student's t test.
RESULTS
Efficient Cardiotypic Differentiation in P19 CL6 Cells Is Dependent upon Nox4
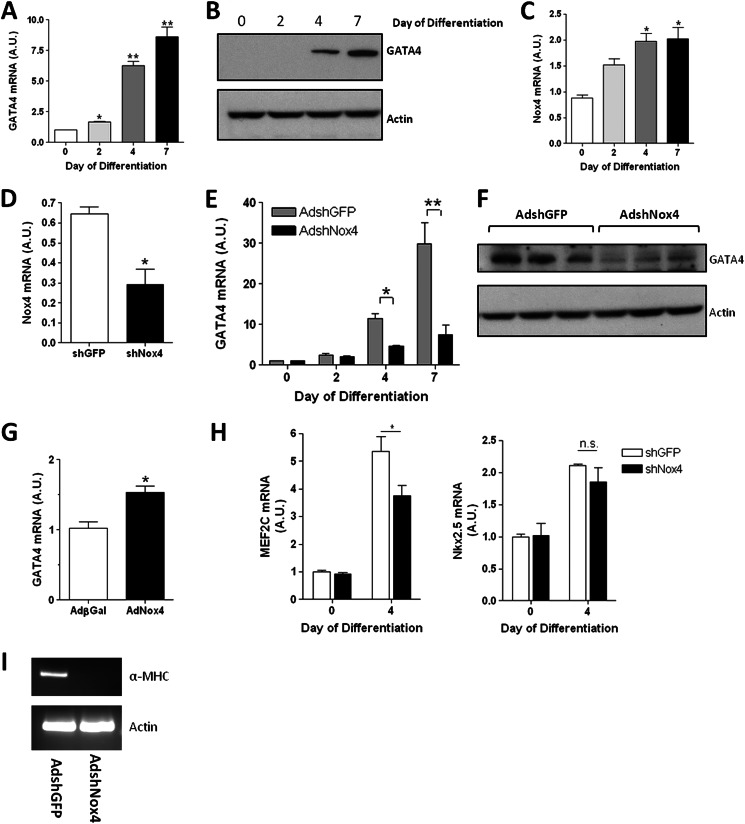
Previously, it had been reported that endogenous Nox4 mRNA expression levels and ROS production increased upon the induction of the cardiogenic differentiation program in EBs in vitro (12) and that ablation of Nox4 was sufficient to impair this differentiation to the cardiac phenotype. We initially sought to determine whether this was similarly true in the differentiation of the EC cell line P19 CL6 cells to the cardiac lineage. P19 CL6 cells were induced to differentiate by culturing the cells in media containing 1% DMSO for the indicated number of days. GATA-4, together with GATA-6, was the earliest cardiac marker in this system to be up-regulated (see Refs. 4, 33 and data not shown). Thus a small (but significant) increase in GATA-4 mRNA expression was apparent by day 2 of differentiation, whereas a highly significant increase in both GATA-4 mRNA and protein levels was detected by day 4 that increased further by day 7 (Fig. 1, A and B). Correspondingly, Nox4 mRNA levels in differentiating P19 CL6 cells also demonstrated a significant increase (∼2-fold), in parallel with the activation of GATA-4 expression, although the magnitude of the increase was less (Fig. 1C). However, although GATA-4 mRNA levels continued to increase after day 4, Nox4 mRNA levels did not change significantly after this point (Fig. 1C and data not shown).
FIGURE 1.
Efficient cardiotypic differentiation in P19 CL6 cells is dependent upon Nox4. A, Q-PCR analysis of GATA-4 mRNA in differentiating P19 CL6 cells. B, GATA-4 protein levels in differentiating P19 CL6 cells. β-Actin is included as a loading control. C, Q-PCR analysis of Nox4 mRNA in P19 CL6 cells over the indicated differentiation time course. D, Q-PCR analysis of Nox4 mRNA levels in undifferentiated P19 CL6 transduced for 72 h with either AdshRNANox4 (black bar) or control AdshGFP (open bar), at an m.o.i. of 100. E, Q-PCR analysis of GATA-4 mRNA levels in P19 CL6 cells during the indicated differentiation time course. Cells were transduced with either AdshRNANox4 (black bars) or control AdshGFP (gray bars) at an m.o.i. of 100 and allowed to differentiate for up to 7 days as indicated. F, GATA-4 proteins levels at day 4 of differentiation after transduction with either AdshGFP or AdshNox4. G, Q-PCR analysis of GATA-4 mRNA in undifferentiated P19 CL6 cells transduced with either AdβGal or AdNox4 for 24 h. H, Q-PCR analysis of MEF2C and Nkx 2.5 mRNA in P19 CL6 cells after 4 days of differentiation. Cells were transduced with for 72 h with either AdshRNANox4 (black bar) or control AdshGFP (open bar), at an m.o.i. of 100. I, semi-quantitative RT-PCR analysis of α-MHC mRNA levels in equivalent samples to those assessed in E, at day 7 of differentiation. All Q-PCR values are shown normalized to β-actin and are plotted in arbitrary units (A.U.) relative to appropriate control. **, p < 0.01; *, p < 0.05.
To determine whether Nox4 activity was a requirement for cardiotypic differentiation in the P19 CL6 system, we knocked down the expression of endogenous Nox4 using an adenovirus-mediated delivery of shRNA. Undifferentiated P19 CL6 cells were transduced with adenoviral shRNA directed against either Nox4 (AdshNox4) or GFP (AdshGFP) as a control. As expected, shRNA knockdown decreased Nox4 mRNA expression levels by ∼55% (Fig. 1D), and this depletion of Nox4 was alone sufficient to significantly inhibit the time-dependent up-regulation of GATA-4 mRNA and protein expression when compared with controls (Fig. 1, E and F). Conversely, in undifferentiated P19 CL6 cells, ectopic (adenovirus-mediated) expression of Nox4 induced a significant induction of GATA-4 mRNA (Fig. 1G). To address the specificity of this inhibition, we additionally assessed the effect of Nox4 depletion upon the expression of other cardiac-restricted transcription factors. Thus at day 4 of induction, we observed no change in the mRNA levels of either GATA-6 or Nkx2.5, upon Nox4 depletion, although a significant reduction of MEF2C mRNA was also observed (see under “Discussion,” Fig. 1H, and data not shown). Furthermore, Nox4 knockdown resulted in complete ablation of the onset of transcriptional expression of the cardiac-specific contractile protein, α-MHC, at day 7 (Fig. 1I). Thus, Nox4-generated ROS is required for the efficient up-regulation of GATA-4 during early cardiomyogenesis in P19 CL6 cells and is sufficient to activate GATA-4 expression in the undifferentiated cells.
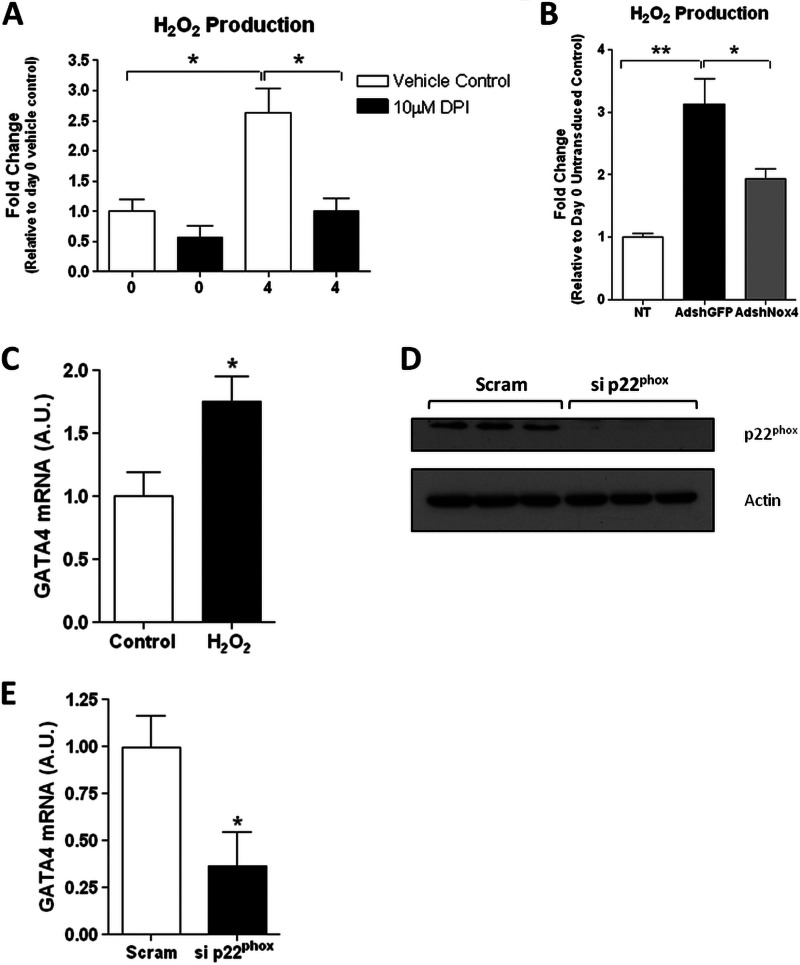
Activation of GATA-4 Transcription Is Dependent upon H2O2 Production and Requires p22Phox
Nox4, in association with the subunit p22Phox alone, is constitutively active (25). Therefore, given sufficient expression of p22Phox, the activity of Nox4 is primarily regulated by the level of its expression (22). Furthermore, it has been demonstrated that Nox4 primarily generates H2O2 as a product, rather than superoxide (O2˙̄). Thus, we assayed extracellular H2O2 production during the differentiation process as a measure of Nox4 activity. A significant increase in H2O2, which was ablated in the presence of the NADPH oxidase inhibitor DPI, was associated with differentiation at day 4 (Fig. 2A), consistent with the observed increase in Nox4 expression at this point (Fig. 1C). Furthermore, specific ablation of Nox4 by siRNA (Fig. 1D) similarly resulted in a significant inhibition of differentiation-associated ROS production (Fig. 2B).
FIGURE 2.
Activation of GATA-4 transcription is dependent upon H2O2 production and requires p22Phox. A, extracellular H2O2 production in cell culture media from undifferentiated (day 0) and differentiated (day 4) P19CL6 cells treated with either 10 μm DPI or 0.1% DMSO (vehicle control) for 6 h prior to assay. B, H2O2 production in cell culture media from undifferentiated P19 CL6 cells (NT (not transduced); open bar) or from cells at day 4 of differentiation transduced with either AdshGFP (black bar) or AdshRNANox4 (gray bar). C, Q-PCR analysis of GATA-4 mRNA levels in P19 CL6 cells after H2O2 treatment. Cells were treated with 10 nm H2O2 for 24 h and cultured for a further 3 days before analysis. D, Western blot analysis of p22Phox protein levels in undifferentiated P19 CL6 cells, 72 h after transfection with control scrambled siRNA (Scram) or siRNA directed against p22Phox (si p22phox). β-Actin is included as a loading control. E, Q-PCR analysis of GATA-4 mRNA levels in P19 CL6 cells transfected with control scrambled siRNA (Scram; black bar) or siRNA directed against p22Phox (si p22phox) for 24 h and subsequently induced to differentiate for 4 days as indicated.
To determine whether these ROS are sufficient to activate GATA-4 transcription and to demonstrate the functional requirement for H2O2, specifically, in this process, undifferentiated p19 CL6 cells were treated with H2O2. This treatment was sufficient to significantly increase GATA-4 transcription (Fig. 2C). The requirement for p22Phox in the activation of GATA-4 was also demonstrated as specific depletion of p22Phox by siRNA also acted to significantly reduce GATA-4 transcription at day 4 of differentiation (Fig. 2, D and E). These data are therefore consistent with H2O2 generated by Nox4 functioning to increase the transcriptional expression of GATA-4 in the P19 CL6 cells.
1.4-kb Proximal GATA-4 Promoter Fragment Is Sufficient to Mediate Increased Transcriptional Expression during Early Cardiogenesis
From these data, ROS generated by Nox4 are seen to effect an up-regulation in the transcription of GATA-4 at a time point that precedes the onset of expression of other cardiogenic transcription factors. We reasoned that a possible molecular mechanism underlying this increase in GATA-4 transcription might involve the Nox4-mediated activation of a redox-sensitive transcription factor(s) present in the undifferentiated P19 CL6 (precursor) cells.
To investigate this further, we sought to identify the cis-acting sequences of the GATA-4 promoter that transduce the redox-mediated up-regulation of transcription during the early differentiation process. Three distinct transcripts of GATA-4 have been described in the mouse (34). GATA-4 transcript variants use alternative noncoding first exons, exons E1a, E1b, and E1b′ (34), whereas the remaining exon usage is common to all three transcripts (Fig. 3A). GATA-4 is expressed in several mesoderm- and endoderm-derived tissues, and the use of different first exons is thought to determine tissue-specific expression (35). To establish which GATA-4 transcript(s) was up-regulated during early P19 CL6 differentiation, we compared the expression of these three distinct transcripts by PCR analysis. We utilized a reverse primer situated in exon 2, which is common to all transcripts, and used primers specific to each alternative first exon to distinguish between individual transcripts.
In undifferentiated P19 CL6 cells, two of these transcripts, E1a- and E1b-containing, were expressed (Fig. 3B), with E1a-containing transcripts being the more abundant. Expression of E1b′ could not be detected (Fig. 3B). Semi-quantitative RT-PCR further demonstrated that the expression of both the E1a-containing and E1b-containing transcripts was increased by the induction of differentiation (Fig. 3C). However, the E1a-containing transcript was apparently both more highly expressed and more strongly up-regulated in the differentiated cells. E1b′-containing transcripts were again undetectable in the differentiated P19 CL6 cells at this time point. We therefore aimed to investigate the ROS-dependent regulation of GATA-4 through cis-acting sequences within the E1a promoter.
Previously, a genomic region (∼1.3 kb) comprising mouse GATA-4 promoter sequences immediately proximal to the E1a CAP site was cloned upstream of a luciferase reporter gene. This was shown to exhibit significantly increased activity within P19 CL6 cells after 8 days of DMSO-mediated cardiogenic induction, compared with undifferentiated cells (36). By this time point, the expression of cardiac-specific contractile proteins such as α-MHC (characteristic of late cardiomyocyte differentiation) is readily detectable (4, 36). To determine whether these sequences could effect the increase in GATA-4 expression in P19 CL6 cells during early cardiogenesis (i.e. before day 8), we generated a similar reporter construct to that described previously (36). Thus, a fragment comprising the equivalent 1.3-kb upstream promoter region, in addition to the whole of exon E1a, was inserted upstream of a luciferase reporter gene. Exon 1a was included as we have shown previously that the first noncoding exon of GATA genes can be functionally important in the regulation of transcription (37).
We examined the expression of the luciferase reporter gene during a time course of early cardiotypic differentiation. As shown in Fig. 3D, we found a significant increase in luciferase activity in cells by day 4 of differentiation, compared within the undifferentiated cells. We further demonstrated that the activity of this region of the promoter was subject to ROS-mediated regulation. Thus, differentiating cells incubated with either the broad spectrum free radical scavenger sodium 4,5-dihydroxybenzene-1,3-disulfonate (tiron) or the Nox enzyme inhibitor DPI showed a reduction in reporter activity at day 4. Differentiation did not affect the expression of an empty vector control (i.e. the reporter vector without the GATA-4 promoter sequence inserted) showing the specificity of the inserted GATA-4 promoter sequences to differentiation-induced regulation.
Similarly, ectopic expression of Nox4 in undifferentiated P19 CL6 cells was sufficient to increase significantly the expression of the luciferase reporter, while not affecting the expression of an empty vector control (Fig. 3E). Once again, this increase was ablated by the addition of the ROS scavenger tiron. Therefore, the transcriptional activation of GATA-4 during cardiomyocyte differentiation may be in part mediated by redox-regulated transcription factor(s) binding to this proximal promoter region. In addition, Nox4 is the apparent source of ROS effecting the redox state changes.
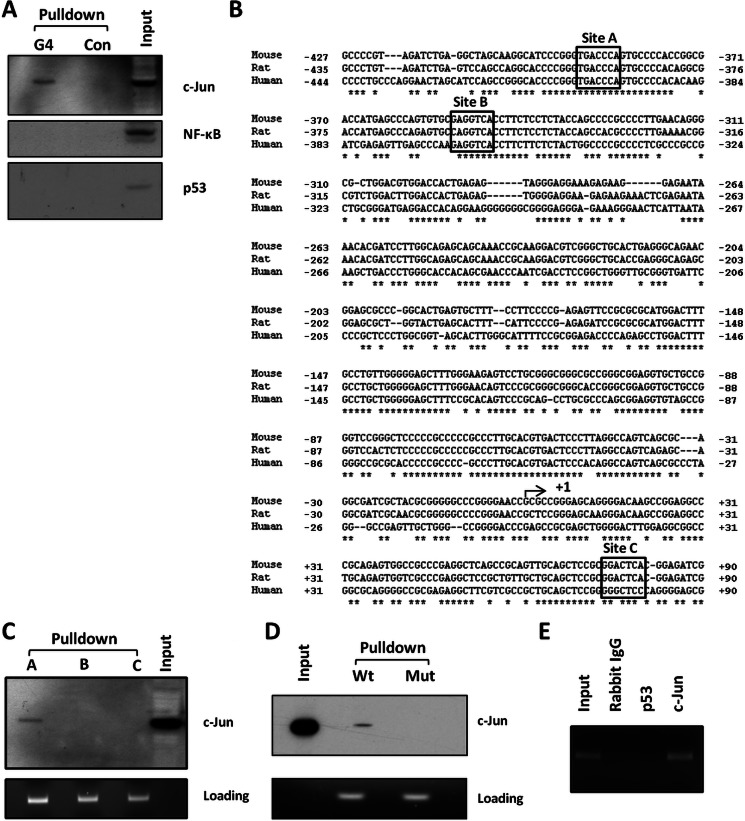
c-Jun Binds to a Canonical AP-1-binding Site within the Redox-regulated GATA-4 Promoter Region
We next sought to identify transcription factor(s) involved in the redox regulation of GATA-4 transcription. Bioinformatic examination of this proximal GATA-4 promoter region (including exon E1a) revealed several potential binding sites for several ubiquitously expressed transcription factors, whose activities are known to be redox-sensitive, including AP-1, NF-κB, and p53 (data not shown).
To test whether these redox-regulated transcription factors bound to the GATA-4 promoter region, we used a biotinylated oligonucleotide “pulldown” assay (32). A 5′-biotinylated double-stranded DNA fragment, comprising the 1.3-kb proximal promoter region together with exon E1a, was generated by PCR and incubated with nuclear extracts from P19 CL6 cells after 4 days of differentiation. The resulting transcription factor-DNA complexes were then affinity-purified on streptavidin-conjugated beads. Bound transcription factors were eluted from the affinity-purified DNA and were screened by means of immunoblot analysis with a panel of antibodies against c-Jun (a subunit of the AP-1 transcription factor complex that is known to be redox-regulated), NF-κB, and p53. Of these transcription factors, only c-Jun was found to bind to the GATA-4 promoter region (Fig. 4A). Input samples (i.e. samples of nuclear extract that had not undergone the pulldown assay) indicated the abundance of these transcription factors in the nuclear extracts from P19 CL6 cells at day 4 of differentiation. Thus, c-Jun and NF-κB were readily detectable, although p53 expression levels were very low (Fig. 4A).
FIGURE 4.
c-Jun binds to a conserved sequence within the GATA-4 promoter region. A, bio-oligonucleotide pulldown assay of a 1.4-kb biotinylated mouse GATA-4 promoter fragment, incubated with nuclear extract from P19 CL6 cells at day 4 of differentiation. Affinity-purified DNA-transcription factor complexes were analyzed for transcription factors c-Jun, p53, and NF-κB by Western blot analyses as indicated (G4). Control (Con) represents equivalent experiment performed in the absence of biotinylated DNA. Input represents equivalent amount of nuclear extract, prior to affinity purification. B, sequence comparison of human, mouse, and rat GATA-4 promoter regions immediately proximal to and including exon E1a. Potential AP-1 (c-Jun)-binding sites are outlined in boxes. Asterisks indicate nucleotide homology between all three species. Nucleotides are numbered relative to the CAP site of each gene (arrowed, +1). C, 100-bp biotinylated DNA fragments comprising individual putative AP-1 sites within the mouse GATA-4 promoter (A–C, respectively) were incubated with nuclear extracts prepared from P19 CL6 cells at day 4 of differentiation. Affinity-purified complexes were analyzed for c-Jun binding by Western blot. Again, nuclear extract was run as a control (input). Loading control denotes (equivalent) levels of oligonucleotides bound to beads, released by incubation with formamide (see under “Experimental Procedures”), and separated by agarose gel electrophoresis. D, oligonucleotides comprising the wild type site A (Wt) or a mutated site A (Mut) AP-1-binding site, subjected to pulldown assay as described in C. E, ChIP analyses of chromatin isolated from P19 CL6 cells after 4 days of differentiation. Equivalent aliquots of chromatin were immunoprecipitated using antibodies to c-Jun, p53, or nonimmune (Rabbit) IgG as indicated. The input fraction corresponds to a 1:100 dilution of initial starting chromatin solution before immunoprecipitation.
Three potential binding sites for AP-1 (and therefore c-Jun) were identified by our bioinformatic analyses within the GATA-4 promoter region (sites A–C, Fig. 4B), which all show significant similarity to the AP-1 core consensus sequence (TGA(G/C)TCA). Site A and B are upstream of the transcriptional start site, whereas site C is located within exon E1a (Fig. 4B). To determine which site(s) can bind c-Jun in vitro, the pulldown assay was repeated using smaller (100 bp) promoter fragments, each encompassing an individual putative binding site. Of the three potential sites, c-Jun was only detected bound to site A (Fig. 4C). To confirm the identity of this binding site, 21-bp biotinylated double-stranded oligonucleotides were generated, comprising the wild type site A, or an equivalent sequence in which the core binding domain was mutated (tgaccca to agacctg). As shown in Fig. 4D, mutation of the putative AP-1 core binding site ablated the binding of c-Jun. It is of note that site A lies in the center of a region of DNA that displays exceptionally high homology between mammalian species (22 contiguous nucleotides identical between human mouse and rat), strongly suggesting functional significance, whereas sites B and C are less well conserved between species (Fig. 4B).
To demonstrate the binding of c-Jun to the GATA-4 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) analyses on P19 CL6 cells at day 4 of cardiac differentiation. As shown in Fig. 4E, c-Jun was found to bind to GATA-4 promoter sequence spanning site A in vivo. By contrast, no amplification of the putative AP-1 site was found in ChIP analyses using nonimmune IgG or an antibody to p53 (which was previously shown not to bind to this region of the GATA-4 promoter; Fig. 4A).
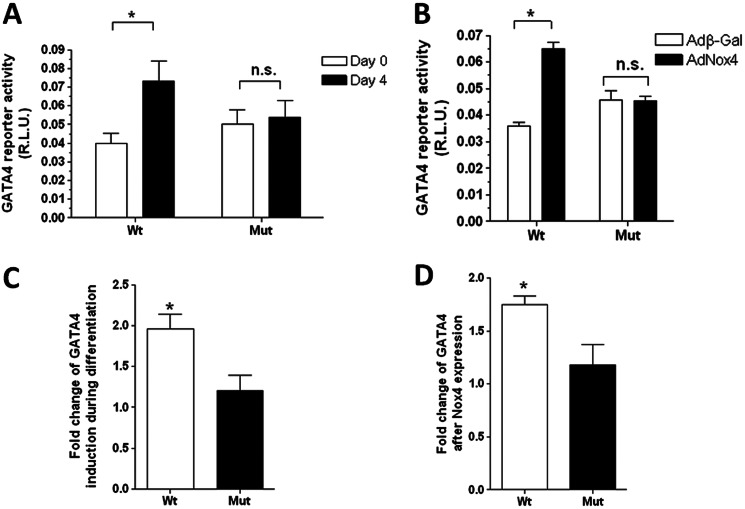
Nox4 Up-regulates GATA-4 Transcription in P19 CL6 Cells via Redox Activation of c-Jun through JNK Signaling
To investigate whether the binding of c-Jun to the AP-1 site in the GATA-4 promoter had functional significance during differentiation, we mutated specifically the core AP-1 binding consensus sequence of site A, within the luciferase reporter gene construct (described under “Experimental Procedures”). Wild type and mutant promoter constructs were introduced into P19 CL6 cells, and the resulting reporter activity was assessed both in undifferentiated P19 CL6 cells and in cells after 4 days of differentiation. In undifferentiated cells, mutation of the AP-1-binding site tended to give a small increase in reporter expression (see under “Discussion”). However, as shown in Fig. 5A, this mutation clearly acted to ablate the increase in reporter activity that is normally apparent between undifferentiated cells and cells after 4 days of differentiation (Fig. 5, A and C). Similarly, mutation of the AP-1-binding site also ablated the increase in reporter activity observed in undifferentiated P19 CL6 cells upon ectopic expression of Nox4 (Fig. 5, B and D).
FIGURE 5.
Functional analyses of the AP-1 site in the GATA-4 promoter. A, luciferase activity resulting from undifferentiated (day 0; open bars) or differentiated (day 4; black bars) P19 CL6 cells, transfected with either a WT GATA-4 promoter-luciferase plasmid (Wt) or the same plasmid containing a mutated AP-1-binding site (Mut) as indicated. B, luciferase activity resulting from undifferentiated P19CL6 cells, again transfected with either the WT or mutated promoter-luciferase constructs and additionally transduced with AdNox4 or AdβGal. In all cases, luciferase activity was measured 24 h after transfection and transduction and was normalized to that of the co-transfected SV40-Renilla luciferase vector and expressed as RLU. C and D, histograms depicting the fold induction, relative to controls, of the WT and mutated GATA-4 promoter activities upon either differentiation or Nox4 transduction, respectively. All data are presented as mean ± S.E. *, p < 0.05. n.s., not significant.
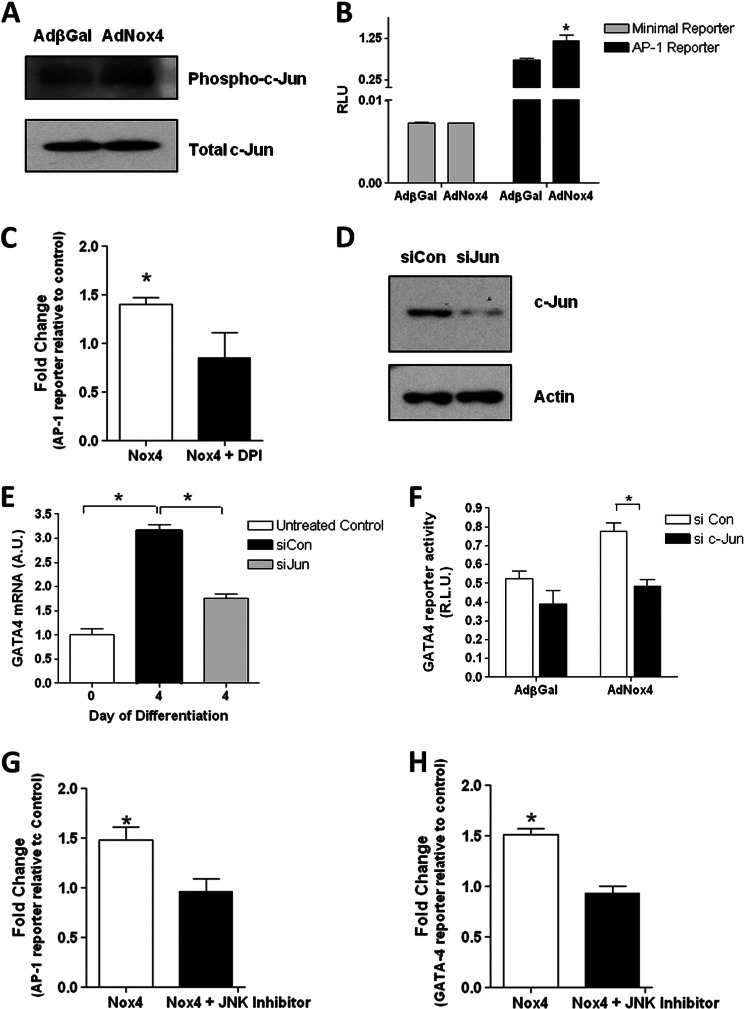
We therefore sought to investigate whether Nox4-generated ROS could regulate the activity of c-Jun in P19 CL6 cells. Undifferentiated P19 CL6 cells were transduced with an adenoviral vector encoding either Nox4 (AdNox4) or control β-galactosidase (AdβGal) and incubated for 48 h. Adenovirus-mediated Nox4 overexpression significantly increased the phosphorylation of c-Jun within the nucleus, whereas the levels of total c-Jun nuclear protein were not affected (Fig. 6A). Consistent with these findings, Nox4 overexpression also specifically increased the expression from an AP-1-dependent luciferase reporter gene construct while not affecting that resulting from a similar construct containing a minimal promoter insert (Fig. 6B). Furthermore, this increase in the activity of the AP-1-dependent luciferase reporter was ablated in the presence of the NADPH oxidase inhibitor, DPI (Fig. 6C).
FIGURE 6.
Nox4 increases GATA-4 expression via activation of c-Jun. A, Western blot analyses of phosphorylated (Phospho-) and total c-Jun levels in nuclear extracts from undifferentiated P19 CL6 cells, transduced with either AdβGal or AdNox4 for 24 h prior to harvesting. B, luciferase activity resulting from undifferentiated P19 CL6 cells transfected with an AP-1-luciferase reporter construct (black bars) or minimal promoter-containing control plasmid (open bars), and additionally transduced with AdβGal or AdNox4 as indicated for 24 h prior to harvesting. C, fold induction of Nox4-induced increase in AP-1 luciferase reporter activity in the presence or absence of DPI. Cells were transduced with AdNox4 for 24 h and treated overnight with 1 μm DPI before assaying. D, Western blot analysis of c-Jun protein levels in undifferentiated P19 CL6 cells, 48 h after transfection with control siRNA (siCon) or siRNA directed against c-Jun (siJun). β-Actin is included as a loading control. E, Q-PCR analysis of GATA-4 mRNA levels in undifferentiated P19 CL6 cells (untreated control; open bar) or P19 CL6 cells transfected with control siRNA (siCon; black bar) or siRNA directed against c-Jun (siJun) for 24 h, and subsequently induced to differentiate for 4 days as indicated. F, luciferase activity resulting from undifferentiated P19 CL6 cells transfected with control siRNA (si Con; open bar) or siRNA directed against c-Jun (si c-Jun; black bar), and incubated for 24 h. All cells were subsequently transfected with a GATA-4 reporter plasmid and transduced with either AdβGal or AdNox4 as indicated and cultured for a further 24 h. G, fold induction of Nox4-induced increase in AP-1 luciferase reporter activity in the presence or absence of a JNK inhibitor. Cells were transduced with AdNox4 for 24 h and treated overnight with 20 μm JNK inhibitor II before assaying. H, fold induction of Nox4-induced increase in GATA-4 luciferase reporter activity in the presence or absence of a JNK inhibitor. Cells were transduced with AdNox4 for 24 h and treated overnight with 20 μm JNK inhibitor II before assaying. For all reporter assays luciferase activity was normalized to that of a co-transfected SV40-Renilla luciferase vector and is expressed as RLU or fold change as indicated. All data are presented as mean ± S.E. *, p < 0.05.
To confirm the involvement of c-Jun in the redox activation of GATA-4 transcription during early cardiomyocyte differentiation, P19 CL6 cells were differentiated in the presence of siRNAs directed against c-Jun or control siRNAs. c-Jun knockdown was confirmed by Western blot (Fig. 6D). Silencing of c-Jun significantly decreased the activation of GATA-4 in differentiating P19 CL6 cells at day 4 (Fig. 6E). Furthermore, silencing of c-Jun decreased slightly the activity of the GATA-4 promoter in undifferentiated P19 CL6 cells, and it completely ablated the increase in the activity of this promoter upon ectopic expression of Nox4 (Fig. 6F).
The coordinated involvement of mitogen-activated protein kinases (MAPKs) in the cardiotypic differentiation of P19 cells has been demonstrated previously (38). In addition, c-Jun is the basic mediator of the transcriptional response to JNK signaling and has been shown to be phosphorylated in response to JNK activation in these cells (38). To determine whether JNK signaling was involved in the ROS-dependent activation of GATA-4 transcription, Nox4 was overexpressed in the P19CL6 cells in the presence or absence of the JNK inhibitor, JNK inhibitor II. In the presence of this inhibitor, the Nox4-dependent increase in the activity of the AP-1 luciferase reporter construct was blocked (Fig. 6G). Furthermore, the inhibition of JNK signaling completely ablated the increase in the activity of the GATA-4 promoter fragment in response to Nox4 overexpression (Fig. 6H).
Nox4 Overexpression Activates c-Jun in Neonatal Rat Cardiomyocytes but Does Not Affect Transcription of GATA-4
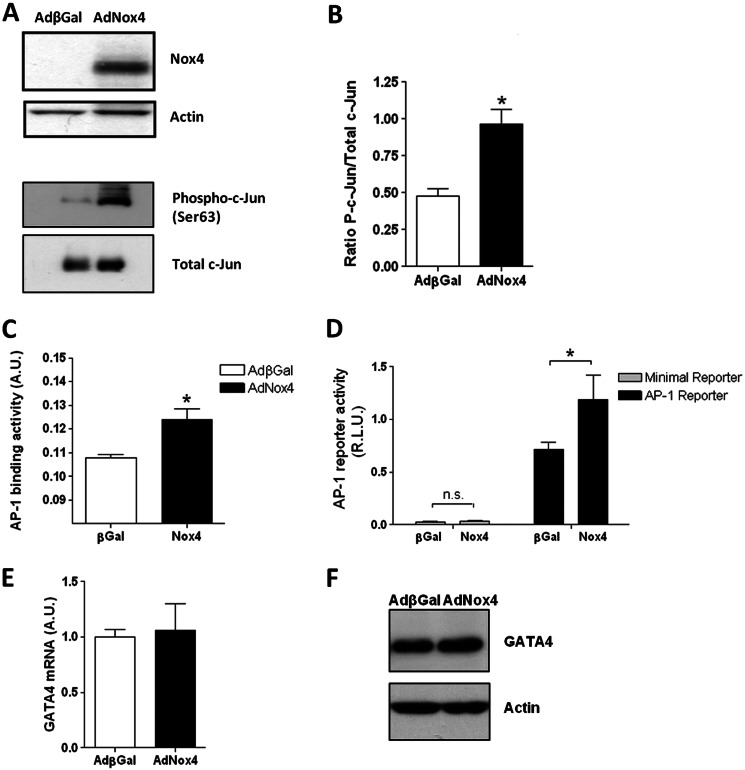
To investigate whether Nox4-generated ROS and c-Jun activation can act to regulate the expression of GATA-4 transcription in fully differentiated cardiomyocytes, Nox4 was ectopically expressed in freshly isolated NRC. NRCs were transduced with AdNox4 or control Adβgal and incubated for 24 h. Consistent with findings in P19 CL6 cells, Nox4 overexpression strongly increased the phosphorylation of c-Jun in nuclear extracts from NRC, although the levels of total c-Jun nuclear protein were not affected (Fig. 7, A and B). Additionally, an ELISA-based c-Jun/AP-1 binding assay also demonstrated significantly higher levels of c-Jun binding to an immobilized consensus AP-1-binding motif in nuclear extracts prepared from Nox4-overexpressing cells compared with βGal-expressing controls (Fig. 7C). As expected from these findings, Nox4 overexpression also specifically increased the expression from an AP-1-dependent luciferase reporter gene without affecting that resulting from a similar construct containing a minimal promoter insert (Fig. 7D). However, by contrast to the observations in P19 CL6 cells, Nox4 expression did not significantly affect the expression of GATA-4 mRNA levels in NRCs (Fig. 7E).
FIGURE 7.
Nox4 activates c-Jun in NRCs but does not affect GATA-4 mRNA expression levels. A, representative Western blot analyses of Nox4 protein levels in whole cell lysates or phospho- and total c-Jun protein levels in nuclear extracts of NRCs transduced with either AdβGal or AdNox4 for 24 h. Actin is included as a loading control. B, densitometric analysis of the proportion of c-Jun phosphorylation, derived from three separate Western blot analyses. C, ELISA-based analyses of binding activity of c-Jun in nuclear extracts of NRCs transduced with AdβGal-transduced (open bar) or AdNox4-transduced (black bar) as in A. D, luciferase activity resulting from NRCs transduced for 24 h with either AdβGal or AdNox4 as indicated and subsequently transfected with an AP-1-luciferase reporter construct (black bars) or minimal promoter-containing control plasmid (open bars) and cultured for a further 24 h. Luciferase activity was normalized to that from the co-transfected SV40 Renilla luciferase control vector and is expressed as RLU. E, Q-PCR analysis of GATA-4 mRNA levels in NRCs transduced with AdβGal or AdNox4 for 24 h. F, representative Western blot analyses of GATA-4 protein levels in whole-cell lysates of NRCs transduced with either AdβGal or AdNox4 for 24 h. Actin is included as a loading control. All data are presented as mean ± S.E. *, p < 0.05; n.s., not significant.
DISCUSSION
The data presented here provide novel insights into the redox-based mechanisms that underlie the differentiation of pluripotent cells into cardiotypic cells. We demonstrate that in P19 CL6 EC cells, endogenous Nox4 expression increases during the early stages of cardiac differentiation, and ablation of Nox4 acts to decrease the level of differentiation to the cardiac phenotype. We further elucidate a molecular mechanism underlying the initial activation of transcription of an early determinant of the cardiotypic phenotype, GATA-4. Thus, we have identified a cis-acting, AP-1 consensus binding element within the proximal GATA-4 promoter that binds to c-Jun and mediates the redox-dependent increase in GATA-4 transcription during the differentiation process. The ROS-dependent activation of c-Jun involves its phosphorylation that is regulated, at least in part, by the JNK-dependent signaling pathway. Thus, we demonstrate the transcriptional activation of the (cardiac-expressed) factor GATA-4, in a pluripotent cellular background, by redox-mediated activation of a widely expressed transcription factor (c-Jun). We suggest also that this may prove to be a more general type of molecular mechanism that potentially underlies the differentiation to other cell types during embryonic development.
Our observations in P19 CL6 EC cells are in agreement with and build on previous in vitro studies that demonstrate an involvement of ROS, and in particular ROS generated by Nox4, in mammalian cardiotypic differentiation from ES cells (12). In the ES cell model, the requirement for Nox4 was demonstrated to be during very early differentiation. Thus, a pulse of low concentrations of H2O2 at a time concomitant with the activation of expression of the earliest cardiac transcription factors (GATA-4 and Nkx2.5) could restore cardiotypic differentiation in Nox4-deficient ES cells (12). Accordingly, in our study Nox4 expression increased before or co-incident with expression of cardiac-restricted molecular markers. Furthermore, we demonstrate that shRNA to Nox4 inhibits the initial increase in expression of GATA-4 mRNA, whereas ectopic expression of Nox4 per se was sufficient to activate GATA-4 transcription in undifferentiated ECs. In vivo, GATA-4 expression is known to be a critical early determinant of cardiogenic differentiation. Thus, GATA-4 is found in the precardiac splanchnic mesoderm as early as embryonic day 7.0 (39, 40), prior to the migration of cardiac progenitors to form the heart tube. Furthermore, ablation of GATA-4 on a GATA-6-null genetic background resulted in embryos that completely lacked heart tissue (41). Our data therefore suggest an involvement of ROS in very early stages of cardiac cellular determination during mammalian embryogenesis. We also investigated the effect of Nox4 ablation upon the expression of other early expressed cardiac markers in this system, and we found that the initial transcriptional activation of neither GATA-6 nor Nkx2.5 was significantly affected. This therefore indicates promoter specificity in the redox-mediated activation of gene expression during cardiogenesis. The expression of MEF2C was, however, shown to be decreased. Further experiments will be required to determine the molecular mechanism(s) and trans-acting factors involved in this regulation and how they relate to the redox-mediated, c-Jun activation of GATA-4.
The AP-1 transcription factor complex can be composed of different combinations of multiple subunits, each with varying affinities for AP-1-binding sites (42). AP-1 binding activity has been shown previously to be activated during, and to be necessary for, the cardiac differentiation of P19 cells, and in one report this binding activity was found to comprise c-Jun, Jun-D, and Fra-2 proteins (38). Consistent with this, we found all three subunits (c-Jun, Fra-2, and Jun-D) to be expressed in P19 CL6 cells (data not shown). In addition, specificity in the relative affinities of the different AP-1 subunits to the binding site within the GATA-4 promoter was demonstrated. Thus, in addition to c-Jun, Jun-D was also found to bind, albeit less strongly, whereas the binding of Fra-2 could not be detected (data not shown). It has been demonstrated previously in cardiomyocytes that although c-Jun acts as a positive regulator of transcription, Jun-D acts as a transcriptional repressor (43). Binding of the repressive factor Jun-D could therefore account for the slight increase in the activity of the GATA-4 promoter that was observed upon mutation of the AP-1-binding site in undifferentiated cells (Fig. 4A). Thus, competitive binding of different factor subunits to this site may be involved in the full regulation of GATA-4 transcription during the differentiation process, and this is under further investigation in our laboratory.
The positive involvement of c-Jun in this cellular differentiation has also been demonstrated as ectopic expression of c-Jun per se was sufficient to drive P19 cell differentiation into a mixed mesodermal and endodermal population (44). Conversely, overexpression of a dominant-negative c-Jun blocked cardiomyocyte differentiation from P19 cells (38). The significance of c-Jun in cardiogenesis and heart development in vivo has also been demonstrated. Thus, c-Jun null-mice die at E13, in part due to heart defects, including significant outflow tract malformations (45). Recently, the intrinsic importance of c-Jun in cardiotypic differentiation and development, specifically within the Isl1-expressing embryonic progenitor cells, has been demonstrated. Isl1 is a molecular marker of the secondary heart field, and mouse embryos lacking c-Jun expression within these cells displayed multiple heart defects, including ventricular septal defects, double outlet right ventricle, and semilunar valve hyperplasia (46).
Pathways that lead to the activation of the MAPKs are known to be the primary mediators of c-Jun activation (47–49). ROS are well known to induce the activation of members of the MAPK family (50), and Nox enzymes, including Nox4, have been shown to modulate multiple MAPK family members within cardiovascular cells (19, 51). In P19 cells, the JNK, ERK, and p38 MAPKs have been shown previously to be activated in a coordinated manner to effect an increase in the DNA binding and transactivation potential of c-Jun during cardiomyocyte differentiation (38). p38 MAPK was also found to be critical in the Nox4-regulated cardiogenic differentiation from ES cells and specifically in the nuclear translocation of MEF2C (12). We demonstrate here the requirement for JNK-mediated signaling in the c-Jun-dependent, redox-mediated up-regulation of GATA-4 by Nox4. It is now believed that protein phosphatases, including PTP1B, PP2A, and PTEN, are the likely primary targets of oxidation-mediated inhibition in these processes (52–55). Further experiments will be needed in the future to identify the precise target(s) of ROS in cardiotypic cells through cellular differentiation and development.
It is clear that transcriptional responses to changes in redox status, including those mediated by Nox4, are cell type-specific. Indeed, although we show that ectopic expression of Nox4 in undifferentiated P19 CL6 cells is sufficient per se to induce GATA-4 transcription, an increase in GATA-4 transcription was not observed upon transduction of Nox4 into differentiated neonatal cardiomyocytes. This is in agreement with our in vivo data, where (post-natal) cardiomyocyte-specific overexpression of a Nox4 transgene did not result in increased GATA-4 mRNA within the heart (56). However, we demonstrate here that Nox4-generated ROS act to increase the phosphorylation, binding capacity, and transactivation capability of c-Jun in these post-natal cardiomyocytes. We therefore suggest that the nuclear environment, including the epigenetic status of the pluripotent cells, renders them a competency to elicit a ROS-dependent increase in GATA-4 transcription through c-Jun activation that is subsequently lost in more differentiated cells. Thus, once commitment to the cardiac phenotype has been established, the signature of cardiac gene expression is maintained by other mechanisms. Nonetheless, the ectopic expression of Nox4 in differentiated cardiomyocytes in vivo has been shown to have profound effects upon their phenotype, as exemplified by the significant changes in the resultant transcriptome of the heart (56), many of which are likely to be mediated via c-Jun activation.
An increasing body of work suggests the involvement of redox changes in the regulation of mammalian heart development (57–60). The developing heart is known to be differentially sensitive to oxidants in a spatial and temporally defined way in vivo (61). The challenge ahead is to delineate the precise sources of ROS and opposing antioxidant systems within the developing embryo and to determine their roles in coordinating the diversity of cellular transcriptional programs during mammalian development. We demonstrate here that ROS produced by Nox4 acts to drive cardiotypic differentiation through activation of GATA-4 via c-Jun activation. Nox4-null mice have been generated, and their hearts apparently develop normally; however it is not clear whether other sources of ROS, including other Nox isoforms, may act to compensate for the lack of Nox4 in this setting. The temporal and spatial expression patterns of Nox4, together with other potential sources/scavengers of ROS in the early precardiac mesoderm, now need to be determined. Furthermore, the targeted ablation of ROS within the early cardiac mesoderm (for instance by the generation of inducible catalase/superoxide dismutase transgenic mouse lines, driven by the Mesp2 or Isl1 promoters) will inform the importance of different ROS upon cardiac differentiation in vivo (62). This will better our understanding of the roles of ROS in heart development in vivo and inform strategies to drive more efficient cardiac differentiation in vitro. This in turn may result in more effective cell-based therapies to aid cardiac repair in the future.
Acknowledgment
We are grateful to Robin Davisson for providing adenoviral shRNAs.
This work was supported by British Heart Foundation Grants PG/08/110/26228 and RG/08/110/25922 and British Heart Foundation Centre of Excellence Award 2E/08/003.
- ES
- embryonic stem
- m.o.i.
- multiplicity of infection
- ROS
- reactive oxygen species
- EC
- embryonal carcinoma
- MHC
- myosin heavy chain
- Q-PCR
- quantitative PCR
- EB
- embryoid body
- DPI
- diphenyliodonium
- RLU
- relative luminescence unit
- NRC
- neonatal rat cardiomyocyte
- F
- forward
- R
- reverse
- Nox
- NADPH oxidase.
REFERENCES
- 1. Zaffran S., Frasch M. (2002) Early signals in cardiac development. Circ. Res. 91, 457–469 [DOI] [PubMed] [Google Scholar]
- 2. Boheler K. R., Czyz J., Tweedie D., Yang H. T., Anisimov S. V., Wobus A. M. (2002) Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 91, 189–201 [DOI] [PubMed] [Google Scholar]
- 3. van der Heyden M. A., Defize L. H. (2003) Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc. Res. 58, 292–302 [DOI] [PubMed] [Google Scholar]
- 4. Brewer A. C., Alexandrovich A., Mjaatvedt C. H., Shah A. M., Patient R. K., Pizzey J. A. (2005) GATA factors lie upstream of Nkx 2.5 in the transcriptional regulatory cascade that effects cardiogenesis. Stem Cells Dev. 14, 425–439 [DOI] [PubMed] [Google Scholar]
- 5. Brown C. O., 3rd, Chi X., Garcia-Gras E., Shirai M., Feng X. H., Schwartz R. J. (2004) The cardiac determination factor, Nkx2–5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J. Biol. Chem. 279, 10659–10669 [DOI] [PubMed] [Google Scholar]
- 6. Lien C. L., Wu C., Mercer B., Webb R., Richardson J. A., Olson E. N. (1999) Control of early cardiac-specific transcription of Nkx2–5 by a GATA-dependent enhancer. Development 126, 75–84 [DOI] [PubMed] [Google Scholar]
- 7. Molkentin J. D., Antos C., Mercer B., Taigen T., Miano J. M., Olson E. N. (2000) Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev. Biol. 217, 301–309 [DOI] [PubMed] [Google Scholar]
- 8. Dodou E., Verzi M. P., Anderson J. P., Xu S. M., Black B. L. (2004) Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development 131, 3931–3942 [DOI] [PubMed] [Google Scholar]
- 9. Ishibashi T., Yokura Y., Ohashi K., Yamamoto H., Maeda M. (2011) Conserved GC-boxes, E-box, and GATA motif are essential for GATA-4 gene expression in P19CL6 cells. Biochem. Biophys. Res. Commun. 413, 171–175 [DOI] [PubMed] [Google Scholar]
- 10. Schmelter M., Ateghang B., Helmig S., Wartenberg M., Sauer H. (2006) Embryonic stem cells utilize reactive oxygen species as transducers of mechanical strain-induced cardiovascular differentiation. FASEB J. 20, 1182–1184 [DOI] [PubMed] [Google Scholar]
- 11. Serena E., Figallo E., Tandon N., Cannizzaro C., Gerecht S., Elvassore N., Vunjak-Novakovic G. (2009) Electrical stimulation of human embryonic stem cells: cardiac differentiation and the generation of reactive oxygen species. Exp. Cell Res. 315, 3611–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J., Stouffs M., Serrander L., Banfi B., Bettiol E., Charnay Y., Steger K., Krause K. H., Jaconi M. E. (2006) The NADPH oxidase NOX4 drives cardiac differentiation: Role in regulating cardiac transcription factors and MAP kinase activation. Mol. Biol. Cell 17, 3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sauer H., Rahimi G., Hescheler J., Wartenberg M. (2000) Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 476, 218–223 [DOI] [PubMed] [Google Scholar]
- 14. Buggisch M., Ateghang B., Ruhe C., Strobel C., Lange S., Wartenberg M., Sauer H. (2007) Stimulation of ES cell-derived cardiomyogenesis and neonatal cardiac cell proliferation by reactive oxygen species and NADPH oxidase. J. Cell Sci. 120, 885–894 [DOI] [PubMed] [Google Scholar]
- 15. Pucéat M., Travo P., Quinn M. T., Fort P. (2003) A dual role of the GTPase Rac in cardiac differentiation of stem cells. Mol. Biol. Cell 14, 2781–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santos C. X., Anilkumar N., Zhang M., Brewer A. C., Shah A. M. (2011) Redox signaling in cardiac myocytes. Free Radic. Biol. Med. 50, 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cave A., Grieve D., Johar S., Zhang M., Shah A. M. (2005) NADPH oxidase-derived reactive oxygen species in cardiac pathophysiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bedard K., Krause K. H. (2007) The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
- 19. Anilkumar N., Weber R., Zhang M., Brewer A., Shah A. M. (2008) Nox4 and Nox2 NADPH oxidases mediate distinct cellular redox signaling responses to agonist stimulation. Arterioscler. Thromb. Vasc. Biol. 28, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 20. Zhang M., Brewer A. C., Schröder K., Santos C. X., Grieve D. J., Wang M., Anilkumar N., Yu B., Dong X., Walker S. J., Brandes R. P., Shah A. M. (2010) NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18121–18126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krijnen P. A., Meischl C., Hack C. E., Meijer C. J., Visser C. A., Roos D., Niessen H. W. (2003) Increased Nox2 expression in human cardiomyocytes after acute myocardial infarction. J. Clin. Pathol. 56, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K. H. (2007) NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 406, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. von Löhneysen K., Noack D., Jesaitis A. J., Dinauer M. C., Knaus U. G. (2008) Mutational analysis reveals distinct features of the Nox4-p22 phox complex. J. Biol. Chem. 283, 35273–35282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takac I., Schröder K., Zhang L., Lardy B., Anilkumar N., Lambeth J. D., Shah A. M., Morel F., Brandes R. P. (2011) The E-loop is involved in hydrogen peroxide formation by the NADPH oxidase Nox4. J. Biol. Chem. 286, 13304–13313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martyn K. D., Frederick L. M., von Loehneysen K., Dinauer M. C., Knaus U. G. (2006) Functional analysis of Nox4 reveals unique characteristics compared with other NADPH oxidases. Cell. Signal. 18, 69–82 [DOI] [PubMed] [Google Scholar]
- 26. Ding L., Liang X. G., Hu Y., Zhu D. Y., Lou Y. J. (2008) Involvement of p38MAPK and reactive oxygen species in icariin-induced cardiomyocyte differentiation of murine embryonic stem cells in vitro. Stem Cells Dev. 17, 751–760 [DOI] [PubMed] [Google Scholar]
- 27. Cross J. V., Templeton D. J. (2006) Regulation of signal transduction through protein cysteine oxidation. Antioxid. Redox Signal. 8, 1819–1827 [DOI] [PubMed] [Google Scholar]
- 28. Haddad J. J. (2002) Antioxidant and prooxidant mechanisms in the regulation of redox(y)-sensitive transcription factors. Cell. Signal. 14, 879–897 [DOI] [PubMed] [Google Scholar]
- 29. Peterson J. R., Burmeister M. A., Tian X., Zhou Y., Guruju M. R., Stupinski J. A., Sharma R. V., Davisson R. L. (2009) Genetic silencing of Nox2 and Nox4 reveals differential roles of these NADPH oxidase homologues in the vasopressor and dipsogenic effects of brain angiotensin II. Hypertension 54, 1106–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones D. H., Howard B. H. (1991) A rapid method for recombination and site-specific mutagenesis by placing homologous ends on DNA using polymerase chain reaction. BioTechniques 10, 62–66 [PubMed] [Google Scholar]
- 31. Andrews N. C., Faller D. V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buzek J., Latonen L., Kurki S., Peltonen K., Laiho M. (2002) Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 30, 2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexandrovich A., Arno M., Patient R. K., Shah A. M., Pizzey J. A., Brewer A. C. (2006) Wnt2 is a direct downstream target of GATA6 during early cardiogenesis. Mech. Dev. 123, 297–311 [DOI] [PubMed] [Google Scholar]
- 34. Mazaud Guittot S., Bouchard M. F., Robert-Grenon J. P., Robert C., Goodyer C. G., Silversides D. W., Viger R. S. (2009) Conserved usage of alternative 5′-untranslated exons of the GATA4 gene. PloS One 4, e8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazaud Guittot S., Tétu A., Legault E., Pilon N., Silversides D. W., Viger R. S. (2007) The proximal Gata4 promoter directs reporter gene expression to Sertoli cells during mouse gonadal development. Biol. Reprod. 76, 85–95 [DOI] [PubMed] [Google Scholar]
- 36. Ohara Y., Atarashi T., Ishibashi T., Ohashi-Kobayashi A., Maeda M. (2006) GATA-4 gene organization and analysis of its promoter. Biol. Pharm. Bull. 29, 410–419 [DOI] [PubMed] [Google Scholar]
- 37. Brewer A., Gove C., Davies A., McNulty C., Barrow D., Koutsourakis M., Farzaneh F., Pizzey J., Bomford A., Patient R. (1999) The human and mouse GATA-6 genes utilize two promoters and two initiation codons. J. Biol. Chem. 274, 38004–38016 [DOI] [PubMed] [Google Scholar]
- 38. Eriksson M., Leppä S. (2002) Mitogen-activated protein kinases and activator protein 1 are required for proliferation and cardiomyocyte differentiation of P19 embryonal carcinoma cells. J. Biol. Chem. 277, 15992–16001 [DOI] [PubMed] [Google Scholar]
- 39. Heikinheimo M., Scandrett J. M., Wilson D. B. (1994) Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev. Biol. 164, 361–373 [DOI] [PubMed] [Google Scholar]
- 40. Arceci R. J., King A. A., Simon M. C., Orkin S. H., Wilson D. B. (1993) Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 13, 2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao R., Watt A. J., Battle M. A., Li J., Bondow B. J., Duncan S. A. (2008) Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev. Biol. 317, 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hess J., Angel P., Schorpp-Kistner M. (2004) AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117, 5965–5973 [DOI] [PubMed] [Google Scholar]
- 43. Hilfiker-Kleiner D., Hilfiker A., Castellazzi M., Wollert K. C., Trautwein C., Schunkert H., Drexler H. (2006) JunD attenuates phenylephrine-mediated cardiomyocyte hypertrophy by negatively regulating AP-1 transcriptional activity. Cardiovasc. Res. 71, 108–117 [DOI] [PubMed] [Google Scholar]
- 44. de Groot R. P., Kruyt F. A., van der Saag P. T., Kruijer W. (1990) Ectopic expression of c-jun leads to differentiation of P19 embryonal carcinoma cells. EMBO J. 9, 1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eferl R., Sibilia M., Hilberg F., Fuchsbichler A., Kufferath I., Guertl B., Zenz R., Wagner E. F., Zatloukal K. (1999) Functions of c-Jun in liver and heart development. J. Cell Biol. 145, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang T., Liu J., Zhang J., Thekkethottiyil E. B., Macatee T. L., Ismat F. A., Wang F., Stoller J. Z. (2013) Jun is required in isl1-expressing progenitor cells for cardiovascular development. PloS One 8, e57032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Humar M., Loop T., Schmidt R., Hoetzel A., Roesslein M., Andriopoulos N., Pahl H. L., Geiger K. K., Pannen B. H. (2007) The mitogen-activated protein kinase p38 regulates activator protein 1 by direct phosphorylation of c-Jun. Int. J. Biochem. Cell Biol. 39, 2278–2288 [DOI] [PubMed] [Google Scholar]
- 48. Son Y., Cheong Y. K., Kim N. H., Chung H. T., Kang D. G., Pae H. O. Mitogen-activated protein kinases and reactive oxygen species: How can ROS activate MAPK pathways? J. Signal Transduct. 2011, 792639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shaulian E., Karin M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 50. McCubrey J. A., Lahair M. M., Franklin R. A. (2006) Reactive oxygen species-induced activation of the MAP kinase signaling pathways. Antioxid. Redox Signal. 8, 1775–1789 [DOI] [PubMed] [Google Scholar]
- 51. Xiao L., Pimentel D. R., Wang J., Singh K., Colucci W. S., Sawyer D. B. (2002) Role of reactive oxygen species and NAD(P)H oxidase in α(1)-adrenoceptor signaling in adult rat cardiac myocytes. Am. J. Physiol. Cell Physiol. 282, C926–C934 [DOI] [PubMed] [Google Scholar]
- 52. Tanner J. J., Parsons Z. D., Cummings A. H., Zhou H., Gates K. S. (2011) Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid. Redox Signal. 15, 77–97 [DOI] [PubMed] [Google Scholar]
- 53. Luo L., Kaur Kumar J., Clément M. V. (2012) Redox control of cytosolic Akt phosphorylation in PTEN null cells. Free Radic. Biol. Med. 53, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 54. Lee S. R., Yang K. S., Kwon J., Lee C., Jeong W., Rhee S. G. (2002) Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336–20342 [DOI] [PubMed] [Google Scholar]
- 55. Leslie N. R., Bennett D., Lindsay Y. E., Stewart H., Gray A., Downes C. P. (2003) Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 22, 5501–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brewer A. C., Murray T. V., Arno M., Zhang M., Anilkumar N. P., Mann G. E., Shah A. M. (2011) Nox4 regulates Nrf2 and glutathione redox in cardiomyocytes in vivo. Free Radic. Biol. Med. 51, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ufer C., Wang C. C., Borchert A., Heydeck D., Kuhn H. (2010) Redox control in mammalian embryo development. Antioxid. Redox Signal. 13, 833–875 [DOI] [PubMed] [Google Scholar]
- 58. Drenckhahn J. D. (2011) Heart development: mitochondria in command of cardiomyocyte differentiation. Dev. Cell 21, 392–393 [DOI] [PubMed] [Google Scholar]
- 59. Patterson A. J., Zhang L. (2010) Hypoxia and fetal heart development. Curr. Mol. Med. 10, 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee Y. M., Jeong C. H., Koo S. Y., Son M. J., Song H. S., Bae S. K., Raleigh J. A., Chung H. Y., Yoo M. A., Kim K. W. (2001) Determination of hypoxic region by hypoxia marker in developing mouse embryos in vivo: a possible signal for vessel development. Dev. Dyn. 220, 175–186 [DOI] [PubMed] [Google Scholar]
- 61. Raddatz E., Thomas A. C., Sarre A., Benathan M. (2011) Differential contribution of mitochondria, NADPH oxidases, and glycolysis to region-specific oxidant stress in the anoxic-reoxygenated embryonic heart. Am. J. Physiol. Heart Circ. Physiol. 300, H820–H835 [DOI] [PubMed] [Google Scholar]
- 62. Yoshida T., Vivatbutsiri P., Morriss-Kay G., Saga Y., Iseki S. (2008) Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 125, 797–808 [DOI] [PubMed] [Google Scholar]