Background: TIP150 is a unique microtubule plus-end tracking protein essential for mitotic regulation.
Results: TIP150 cooperates with EB1 to mediate mitotic chromosome segregation and checkpoint control.
Conclusion: The EB1-TIP150-PCAF interaction is essential for kinetochore-microtubule plus-end dynamics and chromosome plasticity in mitosis.
Significance: EB1-TIP150 interaction orchestrated by PCAF governs chromosome dynamics during cell division.
Keywords: Acetyl Coenzyme A, Cell Cycle, Genomic Instability, Mitosis, Mitotic Spindle
Abstract
The microtubule cytoskeleton network orchestrates cellular dynamics and chromosome stability in mitosis. Although tubulin acetylation is essential for cellular plasticity, it has remained elusive how kinetochore microtubule plus-end dynamics are regulated by p300/CBP-associated factor (PCAF) acetylation in mitosis. Here, we demonstrate that the plus-end tracking protein, TIP150, regulates dynamic kinetochore-microtubule attachments by promoting the stability of spindle microtubule plus-ends. Suppression of TIP150 by siRNA results in metaphase alignment delays and perturbations in chromosome biorientation. TIP150 is a tetramer that binds an end-binding protein (EB1) dimer through the C-terminal domains, and overexpression of the C-terminal TIP150 or disruption of the TIP150-EB1 interface by a membrane-permeable peptide perturbs chromosome segregation. Acetylation of EB1-PCAF regulates the TIP150 interaction, and persistent acetylation perturbs EB1-TIP150 interaction and accurate metaphase alignment, resulting in spindle checkpoint activation. Suppression of the mitotic checkpoint serine/threonine protein kinase, BubR1, overrides mitotic arrest induced by impaired EB1-TIP150 interaction, but cells exhibit whole chromosome aneuploidy. Thus, the results identify a mechanism by which the TIP150-EB1 interaction governs kinetochore microtubule plus-end plasticity and establish that the temporal control of the TIP150-EB1 interaction by PCAF acetylation ensures chromosome stability in mitosis.
Introduction
Accurate chromosome segregation requires formation of a bipolar spindle and successful chromosome movement along the spindle (1). Microtubules are fibrous polymers that determine cellular plasticity and dynamics such as mitosis. During cell division, dynamic microtubule attachment to kinetochores is essential for achieving chromosome stability (2, 3). The precise molecular mechanisms of dynamic kinetochore microtubule regulation in mitosis, however, have remained elusive (1, 4, 5).
The end-binding protein, EB1, localizes to kinetochores during mitosis in a microtubule-dependent manner (6–8). EB1 is a regulator of microtubule plus-end interaction networks at growing microtubule ends due to its autonomous tracking of microtubule tips independent of binding partners, most likely by recognizing structural determinants of growing microtubule ends via its N-terminal half (8). As established by reconstitution assays (9), the yeast EB1 homologue, Mal3p, intrinsically tracks the microtubule plus-ends and is sufficient to localize the kinesin, Tea1p, and the CLIP-170 protein, Tip1p, to growing ends. The molecular mechanisms underlying EB1 interaction with its binding proteins, however, have not been established. We have identified a microtubule plus-end tracking protein, TIP150, and have demonstrated that it facilitates the EB1-dependent loading of microtubule-depolymerizing kinesin (MCAK)3 onto microtubule plus-ends (10).
Mounting evidence demonstrates the central importance of acetyl-CoA in governing chromosome stability in yeast during mitosis (11). Our recent study has established the biochemical mechanisms underlying EB1 acetylation by p300/CBP-associated factor (PCAF)/acetyl-CoA in mitosis. However, the functional relevance of this PCAF-regulated TIP150 interaction with the kinetochore microtubule plus-ends has remained elusive. Here, we show that aberrant acetylation of EB1 perturbs EB1-TIP150 association and accurate metaphase alignment, resulting in spindle checkpoint activation. Indeed, suppression of the mitotic checkpoint serine/threonine protein kinase, BubR1, overrides mitotic arrest induced by persistent EB1 acetylation, but cells exhibit whole chromosome aneuploidy. Thus, the results identify a novel mechanism by which the PCAF-acetyl-CoA-EB1-TIP150 axis governs kinetochore microtubule plus-end plasticity and chromosome stability in mitosis.
MATERIALS AND METHODS
cDNA Construction
Human TIP150 complementary DNA (cDNA), previously described (10), was obtained from the Kazuza DNA Research Institute (Chiba, Japan). To generate insect expression of His-GFP-TIP150 for assay with TIRFM, enhanced GFP, and TIP150 were cloned to the pFastBac HTB vector (Invitrogen), followed by generation of the recombinant Bacmid according to the manufacturer's instructions. The purified recombinant His-GFP-TIP150 Bacmid was confirmed by PCR using M13 forward primer, 5′-GTTTTCCCAGTCACGAC-3′, and M13 reverse primer, 5′-CAGGAAACAGCTATGAC-3′. To generate GST-PCAF truncations, cDNA encoding variant truncations of PCAF were amplified by PCR and cloned to the pGEX-6p-1 vector (GE Healthcare). To generate EB1 Lys-220 mutants, EB1 220K was mutated by PCR to Gln-220 and Arg-220. The fragments, containing wild-type and mutated EB1 with NcoI and BamHI restriction sites, were subsequently cloned into the pEGFP-N3 vector (Clontech) to generate pEGFP-N3-EB1, pEGFP-N3-EB1K220R, and pEGFP-N3-EB1K220Q plasmids. The mutants were further subcloned into the pGEX-6P-1 vector by PCR to generate GST-EB1K220R and GST-EB1K220Q. The mCherry-TIP150 constructs were described previously (10).
To generate TAT-GFP proteins to perturb the EB1-TIP150 interaction, we recombined the pET-22b vector with an 11-amino acid TAT sequence followed by a GFP gene and amino acid sequence competing EB1-SXIP interaction of TIP150 (21). TAT-GFP-His fusion proteins were expressed and purified as described previously (21).
Recombinant Protein Production
Purification of recombinant proteins was carried out as described previously (12, 13). Briefly, the GST fusion protein in bacteria in the soluble fraction was purified by using glutathione-agarose chromatography, whereas histidine-tagged protein was purified using nickel-nitrilotriacetic acid-agarose beads.
For introducing TAT-GFP fusion proteins to probe for the functional relevance of EB1-TIP150 interaction, HeLa cells were cultured to 50–60% confluency before experimentation. Just before introduction, the cells were washed with serum-free media and incubated with TAT-GFP fusion peptides at various concentrations. After incubation, the cells were washed with PBS and then examined directly under fluorescence microscopy as detailed previously (21).
Molecular Mass Determination
Size exclusion chromatography was carried out using fast protein liquid chromatography with a Hiload 16/60 Superdex 200 PG column (GE Healthcare) previously equilibrated with PBS. Elution was performed at a flow rate of 1 ml/min. The column was calibrated with ferritin (440 kDa; RS = 6.10 nm), conalbumin (75 kDa; RS = 4.04 nm), ovalbumin (43 kDa; RS = 3.05 nm), carbonic anhydrase (29 kDa; RS = 2.55 nm), and ribonuclease (13.7 kDa; RS = 1.64 nm), which were used as standard proteins according to our recent study (32).
Linear sucrose gradient (5–30%) was prepared in PBS. TIP150-His recombinant protein was sedimented by ultracentrifugation for 12 h at 141,000 × g at 4 °C in an SW28 rotor (Beckman Instruments). The gradients were then fractionated from top to bottom into 28 fractions by a density gradient fraction collector, and equal amounts of fractions were used for further SDS-PAGE analysis. The recombinant TIP150-His was peaked in fraction 12.
Cell Cultures and Transfection of Plasmids and siRNAs
HeLa and 293T cells (American Type Culture Collection) were cultivated as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (HyClone) and 100 units/ml penicillin plus 100 μg/ml streptomycin at 37 °C with 10% CO2. Lipofectamine transfection reagents (Invitrogen) were used for plasmid transfection. Cells were transfected with Lipofectamine 2000 pre-mixed with various plasmids (2 μg/ml) as described above. Stable clones were selected in the presence of 0.5–1 mg/ml G418 (Calbiochem). Synthetic siRNAs were transfected, using Oligofectamine (Invitrogen). siRNA oligonucleotides for TIP150, EB1, and BubR1 were reported recently (10, 21). In pilot experiments, HeLa cells were transfected with different concentrations of siRNA oligonucleotides or control scramble oligonucleotides for different time intervals. The rescue experiment was done using an siRNA targeted to the 3′-UTR of TIP150 (Qiagen) followed by introduction of exogenously expressed TIP150 as described previously (10). Transfected cells were then collected and solubilized in SDS-PAGE sample buffer. The efficiency of this siRNA-mediated protein suppression was judged by Western blotting analyses of target proteins.
Antibodies
Mouse antibodies to TIP150 were generated using full-length recombinant proteins from bacteria using a standard protocol as described previously (10). In addition, a peptide antibody targeted to the C terminus was also developed and described (10). The following antibodies were used: anti-Ac-K rabbit antibody (Cell Signaling; 9814S); anti-FLAG (M2; Sigma; F1804; use 1:1,000); anti-α-tubulin (DM1A; Sigma); anti-BubR1 (BD Biosciences; 612502); anti-PCAF (Santa Cruz Biotechnology; SC-6301); anti-EB1 monoclonal antibody (BD Biosciences; 610534); anti-EB1 Ac-K220 antibody as described previously (14), and human anti-centromere antibody (ACA) (a gift from D. Cleveland, University of California at San Diego, La Jolla, CA; use 1:1,000).
Immunofluorescence Microscopy
For immunofluorescence, cells synchronized by mitotic shake off were seeded onto sterile, acid-treated 18-mm coverslips in 6-well plates (Corning Glass Works, Corning, NY). Two hours after replating, synchronized HeLa cells were transfected with 2 μg/ml Lipofectamine premixed with various oligonucleotides as described previously (15). HeLa cells were seeded onto sterile, acid-treated, 18-mm glass coverslips in 24-well plates. The cells were washed with PHEM (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, pH 6.9). Permeabilization was accomplished by placing cells for 1 min in PHEM buffer containing 0.1% Triton X-100 at 37 °C. Prior to extraction, cells were fixed in 2% paraformaldehyde in PHEM buffer for 10 min. After washing three times with PBS, cells were blocked with 1% bovine serum albumin (Sigma) in PBS containing 0.05% Triton X-100 for 30 min, then incubated with primary antibodies for 1 h at room temperature, followed by three washes to remove unbound antibody. Cells were then incubated with secondary antibody for 1 h. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma).
Deconvolution Microscopy
Deconvolution images were collected using a Deltavision wide-field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a 100 × 1.35 NA lens was used, and optical sections were taken at intervals of 0.2 μm. Images for display were generated by projecting single optical sections as described previously (16).
Measurement of Inter-kinetochore Distance
The distance between sister kinetochores was measured using LSM-5 imaging (Carl Zeiss), ACA-marked centromeres, and a Zeiss LSM510 confocal microscope as described previously (15). Only sister kinetochores that were in the same focal plane were measured.
GFP-Tubulin Photoactivation in Live Mitosis
The experiment was performed as described previously (26–28). Briefly, HeLa cells were grown on a glass-bottom culture dish (MatTek) at 37 °C with 10% CO2, and after various transfections with siRNA or scrambled (control) oligonucleotides, HeLa cells were transiently transfected with PA-GFP-tubulin and cherry-H2B followed by synchronization, and then mitotic cells were identified by differential interference contrast microscopy. Several pulses from a 405-nm diffraction-limited laser on LSM710 NLO (Carl Zeiss, Germany) were used to photoactivate an area of <2 μm2 within the spindle as described previously. Images were acquired with 63 × 1.4 NA objectives on an LSM710 laser scanning microscope, and images were collected every 30 s.
Analyses of Kinetochore Dynamics in Live Mitosis
For analyzing kinetochore tracking dynamics, aliquots of HeLa cells stably expressing EGFP-CENP-A were transiently transfected to suppress endogenous TIP150 and a scramble siRNA followed by recording with a 100 × 1.35 NA objective on an Olympus DeltaVision microscope (GE Healthcare) at a temporal resolution of 5 s followed by kinetochore tracking assay analysis as we described recently (17). For each time-lapse movie, the position of the metaphase plate was estimated by fitting a plane to the calculated kinetochore positions. To characterize the dynamics of individual sister-kinetochore pairs located on the metaphase plate over time, we monitored the center position of the sister kinetochore along the metaphase plate (spindle axis). The auto-correlation function of sister-kinetochore movements along the spindle axis yielded the periodicity of sister-kinetochore oscillations as analyzed. The displacement intervals correspond to the duration between consecutive directional switches of sister-kinetochore pairs. The average velocity of sister kinetochores along the spindle axis was calculated as the standard deviation of the distribution of all sister-kinetochore frame-to-frame movements.
Total Internal Reflection Fluorescence Microscopic Analyses
Microtubule plus-end tracking experiment was performed as described recently (17), with some modifications. The GMPCPP MT seeds were prepared by polymerizing 30 μm tubulin (at a bovine tubulin/rhodamine tubulin/biotin tubulin ratio of 30:2:1) in the presence of 1 mm GMPCPP (Jena Bioscience, Germany) at 37 °C for 40 min. The seeds were then centrifuged and resuspended in BRB80 buffer (80 mm K-PIPES, pH 6.8, 2 mm MgCl2, 1 mm EGTA). These seeds were sheared with a 25-gauge needle before using to generate short seeds.
Flow chambers were prepared as described previously (18). Chambers were coated with 10% monoclonal anti-biotin antibody (Sigma) followed by blocking with 5% Pluronic F-127 (Sigma). After a brief wash, sheared MT seeds (125 nm) were added into the chamber. Tubulin polymerization mixture (30 μm tubulin in total containing 1:30 rhodamine-labeled bovine tubulin in BRB80, 50 mm KCl, 5 mm DTT, 1.25 mm Mg-GTP, 0.25 mg/ml κ-casein, 0.15% methylcellulose (Sigma), an oxygen-scavenging system, and +TIPs) was introduced into the chamber to initiate polymerization. Unless stated otherwise, the final concentrations of +TIPs were 250 nm EB1 and 250 nm TIP150 or its mutant recombinant proteins. The temperature was kept at 25 °C. Images were collected with a super-resolution microscope configured on an ELYRA system (Carl Zeiss). The laser intensities were kept at a low level to avoid photobleaching. For +TIPs tracking assays, 1 frame was taken/s. Plus-end tracking of GFP-TIP150 and its deletion constructs were analyzed using kymographs in ZEN software (Carl Zeiss).
Real Time Image Acquisition
Cells were maintained at 35–37 °C using a heated stage. Images of cells expressing GFP-histone H2B and GFP-CENP-A were collected on an inverted microscope (Olympus IX-70) and a 60 × 1.4 NA PlanApo objective. 0.2-μm step sections were acquired using a 100 × 1.3 NA U-planApo objective (Olympus) with 1 × 1 binning. Acquisition parameters, including exposure, focus, and illumination, were controlled by Softworx (Applied Precision). Z stack projection, subsequent analysis, and processing of images were performed using Softworx (Applied Precision). For analysis of microtubule attachments, images were deconvolved using the DeltaVision software (Applied Precision). Measurements of the intensity of kinetochore localization were conducted on nondeconvolved images. All images for a specific experiment used identical exposure settings and scaling. For live cell imaging, medium was replaced with CO2-independent medium supplemented with 10% FBS, penicillin/streptomycin, and l-glutamine (Invitrogen) and was covered with mineral oil with a 60 × 1.40 NA PlanApo objective lens together with a filter wheel. Images were analyzed with DeltaVision deconvolution system (Applied Precision).
Analysis of Kinetochore Position Along the Spindle
Kinetochore positions of the TIP150-suppressed and scramble control were determined from maximal intensity projections of confocal images as described previously (9). Based on ACA staining, objects (representing kinetochores) were defined as regions of contiguous pixels with intensity above a threshold. For each object, an internal threshold of 75% of the maximum for that object was used to separate overlapping objects. An average of 10 kinetochore pairs per cell was selected by this method. The position of each kinetochore was recorded as the object centroid, as well as the positions of the two spindle poles, determined from γ-tubulin staining. For each kinetochore, the nearest point on the pole-pole line was determined, and the distance from that point to the nearest pole was calculated and normalized by the pole-pole distance. Aligned kinetochores were defined as those with normalized distance >0.48, whereas normalized distance greater than 0.4 but less than 0.45 was defined as congressed but were not fully aligned kinetochores. Misaligned kinetochores were defined as those with normalized distance <0.2. More than 10 cells (>100 kinetochores) were analyzed for each group expressing either scramble transfected or TIP150 siRNA-treated, respectively. Transfected cells were than synchronized with monastrol and MG132 at prometaphase as described (14). The synchronized cells were then released into metaphase for 35 min followed by fixation and ACA staining. In general, it took an average of 31 ± 2 min for scramble cells to achieve metaphase alignment. However, chromosomes in TIP150-suppressed cells congressed but failed to align.
Data Analyses
All distance and fluorescence intensity measurements were made using MetaMorph software. Analysis of kinetochore movements was performed by measuring the distance from the center of sister kinetochores to a point on the spindle equator along the trajectory of chromosome movement. The bulk of metaphase-aligned chromosomes was used as a reference point for the spindle equator, which occasionally shifted during our analysis. Interkinetochore distances were measured using the centers of the paired CENP-A dots. Kinetochore fluorescence intensities were determined by measuring the integrated fluorescence intensity within a 7 × 7 pixel square positioned over a single kinetochore and subtracting the background intensity of a 7 × 7 pixel square positioned in a region of cytoplasm lacking kinetochores. Maximal projected images were used for these measurements. To determine significant differences between means, unpaired t tests assuming unequal variance were performed; differences were considered significant when p < 0.05.
RESULTS
TIP150 Is Essential for Accurate Chromosome Movements in Mitosis
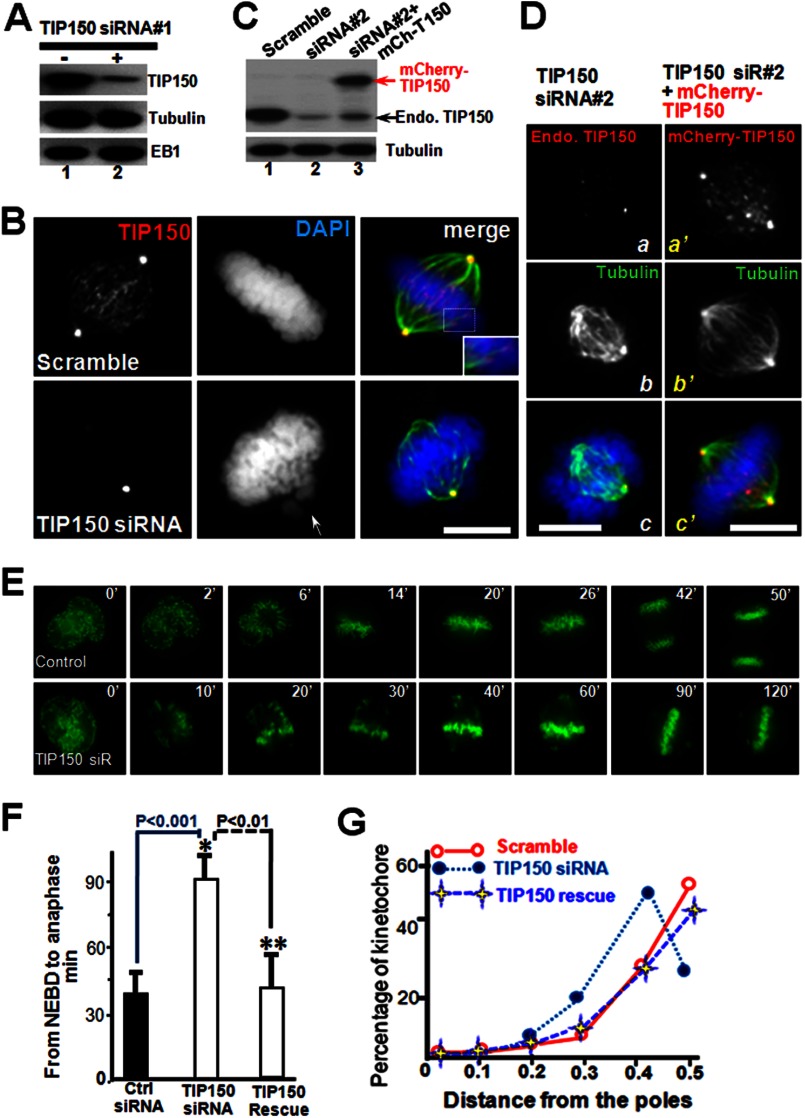
We have identified a microtubule plus-end tracking protein, TIP150, and have established that a hierarchical interaction exists in the EB1-TIP150-MCAK complex (10). However, the physiological function and molecular regulation of TIP150 in mitosis have remained elusive. The level of TIP150 protein expression remains consistent throughout the cell cycle. To elucidate the functional relevance of TIP150 in chromosome alignment during the metaphase, a small interfering RNA (siRNA1) duplex targeted to a region of TIP150 was transfected into HeLa cells. This siRNA targeted specifically to TIP150 suppressed levels of TIP150 protein (Fig. 1A). To determine whether there are phenotypic changes associated with TIP150 knockdown, immunofluorescence microscopic analyses of synchronized HeLa cells transiently transfected with TIP150 siRNA were performed. Cells transfected with TIP150 siRNA displayed misaligned chromosomes; however, for controls transfected with scrambled siRNA, chromosomes were fully aligned (Fig. 1B).
FIGURE 1.
TIP150 is essential for chromosome congression in mitosis. A, TIP150 siRNA suppresses accumulation of TIP150 protein. HeLa cells transfected with TIP150 siRNA1 and scrambled control oligonucleotides were harvested 24 h after transfection and subjected to Western blot analyses. B, knockdown of TIP150 impairs chromosome congression. HeLa cells were transfected with TIP150 siRNA or scrambled control oligonucleotides. Immunofluorescence analyses show that TIP150 exhibits a typical plus-end distribution (red) along the kinetochore microtubule (green). TIP150 siRNA treatment prevents TIP150 localization to the plus-ends and chromosome congression (bottom panel, arrow). Bar, 10 μm. C, efficiency of introduction of exogenous TIP150 in HeLa cells treated with siRNA. HeLa cells transfected with TIP150 siRNA2 targeted to 3′-UTR (lane 2), siRNA2 plus mCherry-TIP150 (lane 3), and scrambled control oligonucleotides were harvested 24 h after transfection and subjected to Western blot analyses. D, introduction of exogenously expressed TIP150 rescued the phenotype seen in TIP150-suppressed cells. HeLa cells were transfected with TIP150 siRNA2 (panels a–c) and siRNA2 plus mCherry-TIP150 (panels a′–′). Immunofluorescence analyses show that mCherry-TIP150 exhibits a typical plus-end distribution (panel a′) along the kinetochore microtubule (panel c′). Introduction of mCherry-TIP150 restored the bipolar spindle plasticity and chromosome alignment (panel c′) perturbed in siRNA2-treated cells (panel c). Bar, 10 μm. E, depletion of TIP150 perturbs chromosome alignment and mitotic progression. Real time imaging of chromosome movements in HeLa cells transfected with TIP150 siRNA and scrambled control siRNA is shown. Repression of TIP150 by siRNA resulted in chromosome misalignment and delayed cell division. F, quantitative analysis of the timing of cell division from NEBD to anaphase onset. Anaphase onset is delayed in cells treated with TIP150 siRNA (*, p < 0.01; n = 30 cells for TIP150 siRNA, Ctrl siRNA versus TIP150 siRNA; **, p < 0.01; TIP150 siRNA versus TIP150 rescue). Note that introduction of exogenously expressed TIP150 rescued the phenotype seen in TIP150 siRNA-treated cells. G, schematic illustration showing how kinetochore positions were measured by the distance from the nearest pole along the pole-pole axis and normalized for pole-pole distance as described previously (19, 28). Profiles of kinetochore position of HeLa cells transfected with scramble siRNA or TIP150 siRNA. These data were acquired from synchronized preparations as described under “Materials and Methods.” Positions were annotated in increments of 0.1. Note that introduction of exogenously expressed TIP150 rescued the phenotype seen in TIP150 siRNA-treated cells.
To rule out the off-target effect of siRNA, we employed an independent siRNA targeted to the 3′-UTR of TIP150 (siRNA2) (10) and reintroduced exogenous mCherry-TIP150. As shown in Fig. 1C, siRNA specifically to TIP150 suppressed levels of endogenous TIP150 protein (lane 2) without interference of expression of exogenous mCherry-TIP150 judged by Western blotting using a TIP150 mouse antibody (lane 3). The phenotypic changes associated with TIP150 knockdown in siRNA2-treated cells were similar to those of siRNA1-treated cells (Fig. 1D), which exhibited misaligned chromosomes. Consistent with our hypothesis, expression of exogenous mCherry-TIP150 restored the phenotype of depletion of endogenous TIP150 protein as mCherry-TIP150 exhibited similar distribution patterns to that of endogenous TIP150 (Fig. 1D, panel a′). In those mCherry-TIP150-expressing cells treated with siRNA2, chromosomes are aligned with a typical bipolar spindle (Fig. 1D, panel c′). Thus, we conclude that TIP150 is essential for accurate chromosome movements in mitosis.
To determine how depletion of TIP150 alters the chromosome dynamics in living HeLa cells, a protocol to visualize real time chromosome movements using GFP-tagged H2B was adopted. Typically, real time imaging of siRNA-treated cells began at 24 h after transfection, and chromosome congression was visualized from prophase to anaphase. For scramble-transfected cells to transit from prophase to the anaphase onset of sister chromatid separation required an average of 42 ± 2 min (n = 20 cells) (Fig. 1E, top panel). In TIP150-depleted cells, however, some chromosomes failed to align at the equator even after 120 min (Fig. 1E, lower panel). Statistical analyses of those cells revealed that depletion of TIP150 delayed the onset of anaphase (*, p < 0.001, Fig. 1F).
As the kinetochore position relative to the pole is a reporter for chromosome alignment and compaction (14, 19, 21), this distance, in >100 kinetochore pairs, in which both kinetochores were in the same focal plane, was measured in both scramble-transfected and TIP150-depleted cells, and the results were expressed as the distance from the nearest pole along the pole-pole axis over the pole-pole distance. In TIP150-suppressed cells, most chromosomes congressed but were not fully aligned (Fig. 1G). Significantly, expression of exogenous TIP150 (mCherry-TIP150) rescued the chromosome alignment abnormality seen in TIP150-suppressed cells treated with siRNA2 (Fig. 1G, TIP150 rescue). Thus, TIP150 is essential for accurate chromosome congression and alignment to the metaphase plate.
TIP150 Is Required for Chromosome Bi-orientation and Kinetochore Microtubule Dynamics
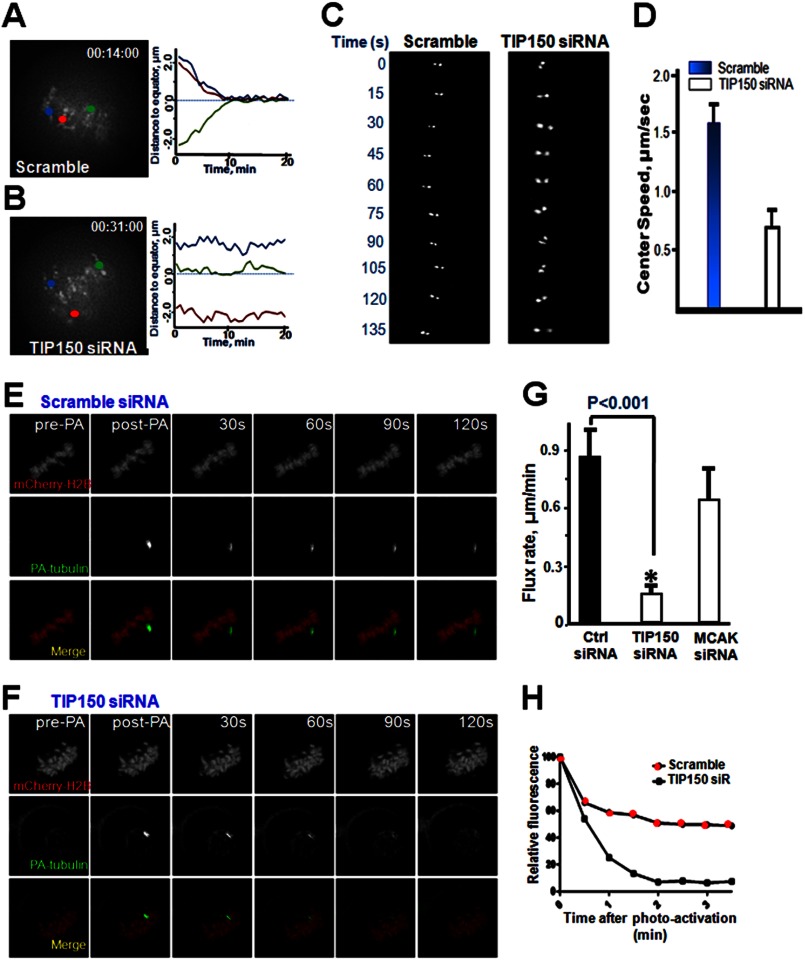
Accurate chromosome alignment is required for equal segregation of sister chromatids in anaphase. To achieve chromosome alignment, bi-oriented paired sister kinetochores generate force to drag chromosomes through regulation of attached microtubule plus-end dynamics. Kinetochore-binding microtubules processively alternate their state between growth and shrinkage, inducing regular kinetochore oscillation along the spindle axis. To characterize the defects in chromosome alignment after TIP150 depletion, kinetochore motility marked by CENP-A at high temporal resolution in the presence or absence of TIP150 was imaged. In control scramble-transfected cells, paired sister kinetochores congressed to the metaphase plate and oscillated until the onset of anaphase, which paired sister kinetochore gave an oscillated curve (Fig. 2, A, right trace, and C). In TIP150-depleted cells, some kinetochore pairs remained at the spindle poles for the duration of the analysis, resulting in a flat curve (Fig. 2, B and C).
FIGURE 2.
TIP150 regulates kinetochore microtubule dynamics in mitosis. A, kinetochore oscillatory dynamics was captured using time-lapse microscopy. HeLa cells were transfected with a scrambled oligonucleotide or with TIP150 siRNA (B) as described under the “Materials and Methods.” Paired sister kinetochores are color-coded to illustrate their dynamics as detailed in the text. Representative kinetochore oscillatory kymograph of scramble siRNA (A, right panel) and TIP150 siRNA-treated cells (B, right panel) are presented. C, magnified image of one kinetochore pair within a cell treated with scrambled siRNA or TIP150 siRNA (right panel). Images were acquired every 15 s as described under “Materials and Methods.” D, analysis of kinetochore velocity of scramble-transfected and TIP150-suppressed cells (n = 100 kinetochore pairs; p < 0.01). E, PA-GFP-tubulin analysis of kinetochore microtubules treated with scrambled oligonucleotide. Images display pre-photoactivated, post-activated (0 s), with 30-s intervals. F, PA-GFP-tubulin analysis of kinetochore microtubules of HeLa cells treated with TIP150 siRNA.G, quantitation of PA-GFP-tubulin pole-ward flux in TIP150-depleted HeLa cells compared with that of control scramble transfected cells. (*, p < 0.001, Ctrl siRNA versus TIP150 siRNA). H, graphical representation of GFP fluorescent decay after photoactivation from kinetochore microtubule of cells treated with TIP150 siRNA. Cells depleted of TIP150 displayed a faster decay rate compared with controls.
To present these data in a vivid way, the motion path was portrayed, and the distance of sister kinetochores was plotted to the metaphase plate over time for these two groups (Fig. 2, A and B, color traces at right). Sister kinetochores were frequently observed moving back and forth between spindle poles and the metaphase plate (Fig. 2C, left panel and tracking). These defects were evident in cells depleted of TIP150. A survey of real time kinetochore oscillation profiles revealed that depletion of TIP150 caused errors in kinetochore orientation, as sister kinetochores exhibited an aberrant rotation (Fig. 2C, right panel, time 0 and 90–135 s). Suppression of TIP150 resulted in reduction of kinetochore velocity during oscillation and congression to the equator (Fig. 2D). Thus, these findings indicate that, upon depletion of TIP150, sister kinetochores are defective in the establishment and maintenance of dynamic attachment with spindle microtubules.
If TIP150 regulates the interaction of spindle microtubules with kinetochores, suppression of TIP150 would reduce the rate of spindle microtubule plus-end dynamics. To test this hypothesis, microtubule dynamics were examined using PA-GFP-tubulin to follow the growth of microtubule plus-ends under various experimental conditions, as the rates of pole-ward microtubule flux and tubulin turnover at plus-ends are usually measured to monitor the dynamics of kinetochore-attached spindle microtubules (20). HeLa cells were co-transfected with mCherry-H2B and PA-GFP-tubulin to allow simultaneous following of the mitotic chromosomes and respective spindle microtubules (Fig. 2, E and F).
To monitor the rate of pole-ward microtubule flux, the spindle microtubules near kinetochores were exposed to UV light to activate GFP and allow tracking of movement of fluorescence-activated GFP-tubulin (17). To monitor tubulin turnover and describe tubulin dynamics at microtubule plus-ends, time-dependent decreases in fluorescence intensity of the activated region were quantified and expressed as the half-life, t½. Pole-ward flux in TIP150-depleted cells was slower relative to that in cells transfected with scrambled siRNA (0.83 ± 0.27 μm/min for control and 0.39 μm/min for TIP150-depleted cells; Fig. 2G), suggesting that TIP150 promotes microtubule plus-end dynamics. Consistent with this observation, tubulin turnover at the plus-ends became slower in TIP150-depleted cells (Fig. 2H). Thus, TIP150 regulates spindle microtubule plus-end dynamics.
Tetrameric TIP150 Exhibits Effective Plus-end Tracking in Vitro
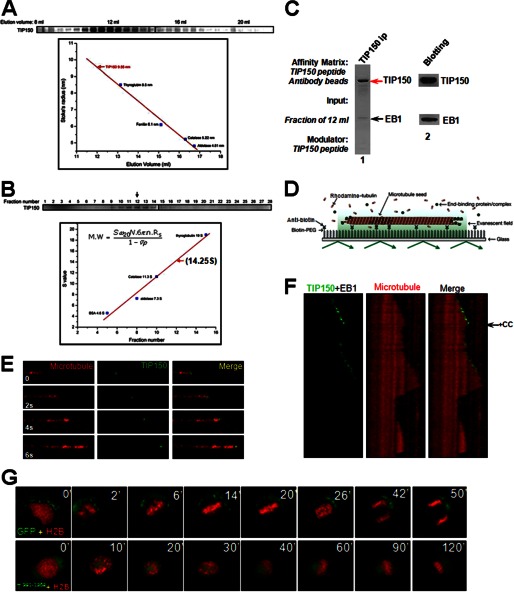
To characterize the properties of the EB1-TIP150 complex and assess its function at microtubule plus-ends, the complex was constructed by use of purified recombinant TIP150 from insect cells and EB1 from bacteria. Western blotting analysis showed that the TIP150-EB1 complex eluted at 12 ml with an estimated Stokes radius of 9.55 nm, which corresponds to the tetrameric form of TIP150 (≈640 kDa; the calculated molecular mass of TIP150 is 153 kDa; Fig. 3A). This is consistent with the elution profile for the endogenous TIP150 complex. An identical value for the sedimentation coefficient (14.25 S) was obtained when the TIP150 complex was analyzed by sucrose gradient centrifugation (Fig. 3B). The hydrodynamic properties of the recombinant EB1-TIP150 complex are identical to those of the endogenous HeLa cell complex and are consistent with a hexameric structure of 2:4 stoichiometry of EB1/TIP150. The hexameric complex was then confirmed using the TIP150 peptide antibody to immunoprecipitate the complex from the fraction of 12 ml shown in Fig. 3A. As shown in Fig. 3C (lane 1), Coomassie Blue staining indicates a major high molecular mass band of 150 kDa with a much less light band around 31 kDa. Western blotting analyses confirmed that the 150-kDa band is TIP150 and the 31-kDa band is EB1. The stoichiometry of TIP150-EB1 molecular association, quantified on the band density shown in Fig. 3C (lane 1), is ∼4:2. Thus, we conclude that endogenous TIP150 forms a hexameric complex with EB1.
FIGURE 3.
TIP150 exhibits an effective microtubule plus-end tracking activity as a tetramer. A, analysis of EB1-TIP150-purified fractions using fast protein chromatography. Constructed EB1-TIP150 complexes were subjected to gel filtration chromatography followed by SDS-PAGE fractionation. A gel stained with Coomassie Blue illustrates that the EB1-TIP150 complex eluted at 12 ml with an estimated Stokes radius of 9.55 nm. B, analysis of TIP150 complex hydrodynamics by sucrose gradient centrifugation, as described previously (29). C, biochemical characterization of TIP150-EB1 hexamer. The fraction of 12 ml from A was incubated with a TIP150 peptide antibody followed by affinity purification using a synthetic peptide (250 μm) for elution. The supernatant was then fractionated on an SDS-polyacrylamide gel followed by Coomassie Blue staining. The stoichiometry of TIP150-EB1 molecular association was calculated based on the band density of respective proteins, which is ∼4:2 (TIP150:EB1), indicating that endogenous TIP150 forms a hexameric complex with EB1. D, schematic representation of a TIRFM assay to visualize plus-end tracking activity of TIP150 using dual color tracking. E, time-lapse fluorescence images of microtubule movement with addition of 50 nm GFP-TIP150 in the presence of EB1 (50 nm). F, dual color kymographs of polymerizing microtubules with EB1 and GFP-TIP150. The histidine-TIP150(981–1368) recombinant protein (CC) was added to perturb the tetramer of TIP150 assembled in vitro (arrow). Note that the GFP-TIP150 tracks only on the growing microtubule plus-end, and addition of TIP150(981–1368) peptide removed GFP-TIP150 from the microtubule plus-end after one growth-shrinkage cycle, which resulted in an increased catastrophic frequency. G, perturbation of TIP150 tetramerization blocks chromosome alignment and mitotic progression in vivo. HeLa cells were transiently transfected to co-express GFP-TIP150(981–1368) protein and mCherry-H2B, and mitotic chromosome movements were followed by real time imaging. Overexpression of GFP-TIP150(981–1368) resulted in chromosome alignment defected and mitotic arrest, suggesting that GFP-TIP150(981–1368) acts as a dominant negative mutant.
To determine how the EB1-TIP150 hexamer interacts with microtubules in vitro, a microscopy flow cell assay was utilized; this assay was previously used to analyze the dynamics of the EB1 protein (9, 17). Short, brightly labeled biotinylated microtubule seeds that had been stabilized with the slowly hydrolyzing GTP analog, GMPCPP, were anchored to the surface of a coverslip via a biotin-anti-biotin antibody interaction (Fig. 3D). After washing to remove unbound seeds, dimly labeled free tubulin (15 μm) was introduced. Microtubules were visualized by TIRFM to eliminate the background fluorescence generated by unincorporated labeled tubulin dimers. Under these conditions, individual microtubules grew from the stable seeds and displayed instability, i.e. the phases of growth were followed by rapid disassembly before another growth phase started.
The behavior of full-length EB1 in the tracking assay was analyzed. EB1-EGFP (70 nm) was added together with free tubulin into the flow chamber. Two-color time-lapse imaging demonstrated that EB1-EGFP preferentially accumulated at microtubule ends during phases of growth but was rapidly lost from the microtubules upon disassembly, as described previously (9). Because the molecular requirements for TIP150 plus-end tracking in vitro have not been established in any defined system, our engineered GFP-TIP150 constructs were tested in the tracking assay to determine whether they associated with the microtubule lattice or with growing ends. Under conditions in which EB1 was present, TIP150 displayed robust plus-end tracking (the final TIP150 concentration is 250 nm; Fig. 3E). This microtubule plus-end tracking activity depended on functional EB1, as no tracking activity was evident in the absence of EB1 (data not shown). Thus, TIP150 interacts with EB1 for microtubule plus-end tracking.
To determine whether the coiled-coil domain of TIP150 perturbs full-length GFP-TIP150 tracking activity on microtubule plus-ends, purified TIP150(981–1368) protein was added to the system after EB1-GFP-TIP150 was loaded onto microtubule plus-ends during a TIRFM assay. Addition of recombinant TIP150(981–1368) protein displaced the GFP-TIP150 assembled onto microtubule plus-ends (Fig. 3F). As a consequence of addition of the TIP150(981–1368) protein, the frequency of catastrophes was increased.
The next goal was to delineate the structural determinant underlying TIP150 tetramerization. If the C-terminal coiled-coil mediates TIP150 tetramerization, overexpression of its C-terminal domain would exhibit a dominant negative effect by forming a heterodimer with endogenous TIP150, thereby depleting the pool of available TIP150. To characterize how overexpression of TIP150(981–1368) alters chromosome segregation in mitosis, we performed real time imaging of HeLa cells transiently transfected for 24 h before imaging to express GFP-TIP150 wild-type and TIP150(981–1368). GFP-expressing HeLa cells required an average of 28 ± 3 min (n = 20 cells) to transit from prophase to the anaphase onset of sister chromatid separation (Fig. 3G, upper panel). In GFP-TIP150(981–1368)-expressing cells, however, some chromosomes failed to align at the equator even after 120 min (Fig. 3G, lower panel). Statistical analyses revealed overexpression of TIP150(981–1368), which delayed anaphase onset, causing 79 ± 8% of the cells to bear misaligned chromosomes. Thus, tetrameric TIP150 forms a complex with EB1 for promoting microtubule plus-end dynamics.
TIP150-EB1 Interaction Is Required for Mitotic Progression
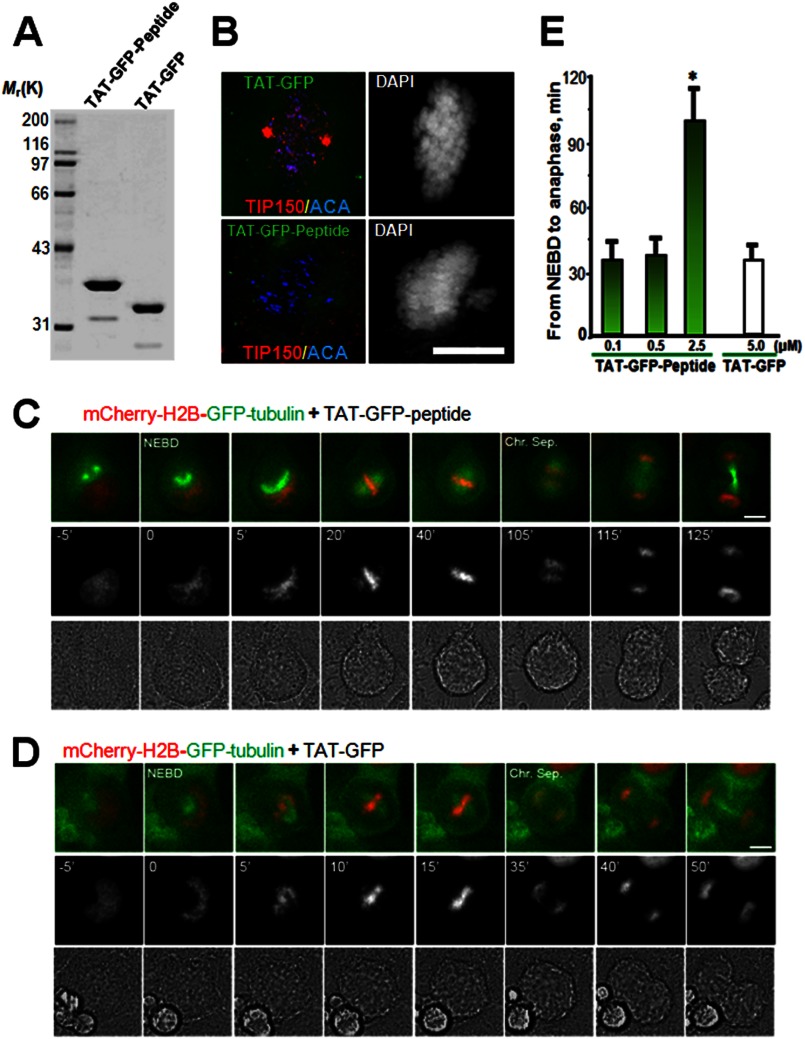
To explore the functional effect of the TIP150-EB1 interaction in mitosis, a membrane-permeable peptide containing TIP150 (862–988 amino acids) was constructed to modulate the endogenous EB1-TIP150 interaction. This was achieved by introducing an 11-amino acid peptide derived from the TAT protein transduction domain into a fusion protein containing amino acids involving binding interface between EB1 and TIP150, as described previously (21, 22). The recombinant protein was histidine-tagged and purified to homogeneity by use of nickel-affinity beads (Fig. 4A, TAT-GFP-peptide). As predicted, the recombinant GFP-TAT-peptide (2.5 μm) disrupted the EB1-TIP150 association in vivo. Neither GFP nor TAT-GFP interfered with the interaction of EB1 with cytoplasmic linker-associated protein 1 (CLASP1), which is involved in regulation of microtubule dynamics. The results demonstrate the effect of TAT-GFP-peptide in competing with full-length TIP150 for association with EB1.
FIGURE 4.
Perturbation of the EB1-TIP150 interaction prevents chromosome alignment and delays anaphase onset. A, Coomassie Blue stain SDS-polyacrylamide gel was used to assess the quality and quantities of the purified TAT-GFP-His6-tagged proteins. Bacteria expressed TAT-GFP-peptide and TAT-GFP-H6 (control) are purified with nickel-nitrilotriacetic acid affinity chromatography and desalted into DMEM. Protein concentration was determined in Bradford assay. B, TAT-GFP-peptide recombinant protein disrupts EB1-TIP150 association. Aliquots of TAT-GFP or TAT-GFP-peptide (2.5 μm) was added into culture HeLa cells for 30 min followed by fixation and examination. Note that incubation of TAT-GFP-peptide librated TIP150 from kinetochore localization and resulted in a phenotype of mitotic arrest with chromosome aberrantly aligned. C and D, HeLa cells expressing mCherry-H2B and enhanced GFP-α-tubulin were synchronized with thymidine and released for 8 h to reach prometaphase. Cells were cultured in DMEM with 500 nm, 1 or 2.5 μm TAT-GFP-TIP150-d-H6 (C) or TAT-GFP-His6 (D) at 37 °C for 1–2 h before images collection. Live cell observation and imaging were performed every 5 min. Representative images are marked especially at the time of NEBD and the time of sister chromosome separation (Chr. Sep.). Bar, 10 μm. E, quantitative analysis of the timing of cell division from NEBD to anaphase onset, which is delayed in cells treated with TAT-GFP-TIP150-d (2.5 μm). The TAT-GFP-TIP150 peptide causes mitotic delay in a dose-dependent manner. (*, p < 0.001, prometa. versus metaphase or anaphase).
To determine whether the EB1-TIP150 interaction is required for mitotic progression, cells expressing mCherry-H2B were synchronized with thymidine and were exposed to TAT-GFP-peptide or TAT-GFP 30 min before nuclear envelope breakdown (NEBD); real time imaging of the cells began 5 min before NEBD (Fig. 4, C and D). The results show that treatment of S- and G2-phase HeLa cells with TAT-GFP-peptide did not alter cell cycle progression into mitosis relative to cells treated with the TAT-GFP control. However, in the presence of 2.5 μm TAT-GFP-peptide, HeLa cells took an average of 99.7 ± 19.3 min (n = 8 cells) to transit from NEBD to the anaphase onset of sister chromatid separation (Fig. 4, C and D). This inhibitory action is relatively specific, as a low concentration of TAT-GFP-peptide did not interfere with mitotic progression (Fig. 4E). However, addition of TAT-GFP peptide (5 μm) did not alter mitotic progression (average 33 ± 3.7 min; n = 8 cells). Thus, the TIP150-EB1 interaction is required for mitotic progression.
EB1-TIP150 Interaction Is Regulated by EB1 Acetylation in Mitosis
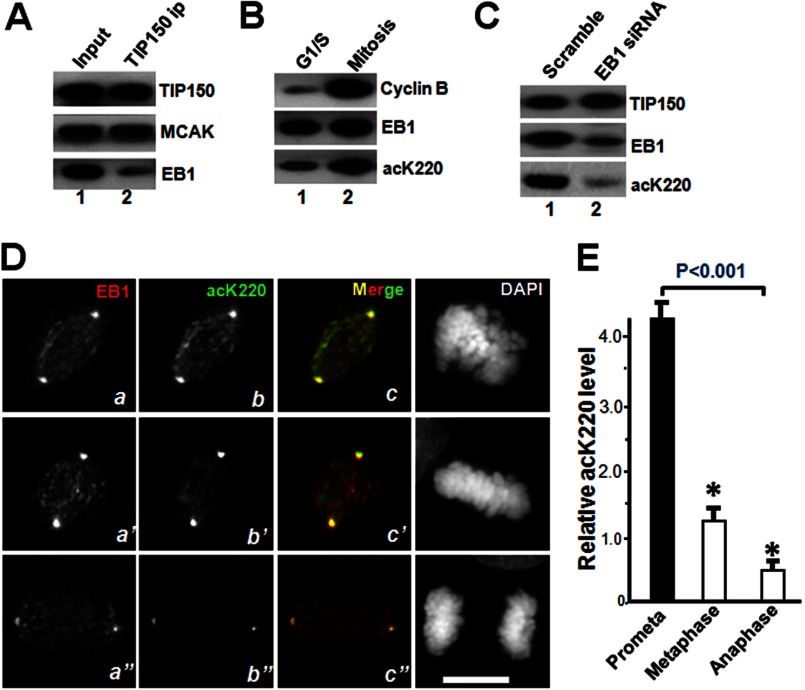
To probe for regulation of TIP150 in mitosis, TIP150 was immunoprecipitated from nocodazole-treated cells, and the immunoprecipitates were screened with mass spectrometric identification of proteins and quantitative Western blotting. The level of EB1 bound to TIP150 was lower relative to that of MCAK from mitotic cells (Fig. 5A). Because acetyl-proteomic studies suggest that lysine acetylation occurs in the highly conserved C terminus of EB1 (14), the acetylated Lys-220 (acK220) level in mitosis was determined by collecting synchronized HeLa cells in G1/S and in mitosis for quantitative Western blotting. The level of cyclin B exhibited a typical 10-fold increase during mitosis (Fig. 5B). In the same preparation, the level of acK220 was elevated to 4.3-fold.
FIGURE 5.
TIP150 interaction with EB1 is regulated by acetylation in mitosis. A, Western blot analysis of immunoprecipitates of TIP150, EB1, and MCAK from nocodazole-synchronized HeLa cells. EB1 proteins bind less to TIP150, relative to MCAK. B, EB1 is acetylated in mitosis. Western blot analysis of EB1 acetylation in interphase and mitotic cells. Quantitative Western blotting indicates that EB1 acetylation at Lys-220 is elevated in mitosis. C, characterization of acK220 antibody specificity. Aliquots of HeLa cells were transiently transfected to introduce siRNA and scramble control. Twenty four hours after the transfection, the HeLa cells were blocked with 100 nm nocodazole for 18 h followed by solubilization of cellular proteins. The cell lysates were fractionated by SDS-PAGE followed by Western blotting of TIP150, EB1, and acK220. The acK220 signal was abolished in EB1-suppressed cells (lane 2). D, immunofluorescence analysis showing EB1, acK220, and DAPI for DNA in mitotic HeLa cells of different stages (prometaphase, metaphase, and anaphase). Triple immunofluorescence imaging showed that EB1 acetylation level is highest at prometaphase cells (panel b), and the distribution profile of acK220 is superimposed to that of EB1 (panel c). As cells phase into metaphase and anaphase, the acK220 level decreased (panels b′ and b″). E, fluorescent intensity analysis of acetylated EB1 (acK220) in different mitotic states. EB1 has a highest level of acK220 at prometaphase cells and declines upon metaphase alignment (p < 0.001; n = 30 cells).
Our recent study has established an important enzyme-substrate relationship between PCAF and EB1 (14). Specifically, Lys-220 of EB1 is a substrate of PCAF (Fig. 5B, bottom panel, lane 2) (14). The level of acK220 in endogenous EB1 was minimized when the EB1 level is suppressed by EB1 siRNA (Fig. 5C, lane 2), suggesting that acK220 antibody specifically recognizes the epitope on EB1 protein.
PCAF acetylates the mitotic checkpoint protein, BubR1, increasing its stability (23). To determine the temporal dynamics of Lys-220 acetylation relative to EB1 protein localization, immunofluorescence analyses of mitotic cells at different phases were accomplished. As shown in Fig. 5D, EB1 displayed a typical comet-like distribution along the spindle in prometaphase cells, with some depositions in centrosomes (upper panel). Staining of acK220 was readily apparent in prometaphase cells, which exhibited a typical spindle plus-end distribution (Fig. 5D, top panel, panel b). Merged images showed that the localization of acK220 was superimposed with a subset of EB1 signals at the spindle plus-ends, validating that EB1 Lys-220 was acetylated in prometaphase (Fig. 5D, panel c). The Lys-220 acetylation level was decreased as the cell approached metaphase alignment (Fig. 5D, middle panel, panel b′). The level of superimposition of acK220 onto EB1 staining was reduced. Quantitative imaging analyses of plus-end signals revealed that the level of acK220 typically decreased by 4-fold in metaphase cells compared with that in prometaphase cells (Fig. 5E; p < 0.001, n = 30 cells), demonstrating that acetylation of EB1 at the spindle plus-end is dynamic in mitosis. Consistent with this notion, the acetylation of EB1 at midzone was diminished as sister chromatids separated (Fig. 5, D, bottom panel, b′, and E). Statistical analyses indicated that there was a 9.3-fold reduction of EB1 acetylation in anaphase cells. Those results suggest that reversible acetylation of EB1 is a mechanism regulating the interaction of EB1 and TIP150 to facilitate accurate chromosome movements.
Lys-220 Acetylation Is Necessary for Dynamic Association of EB1 with TIP150
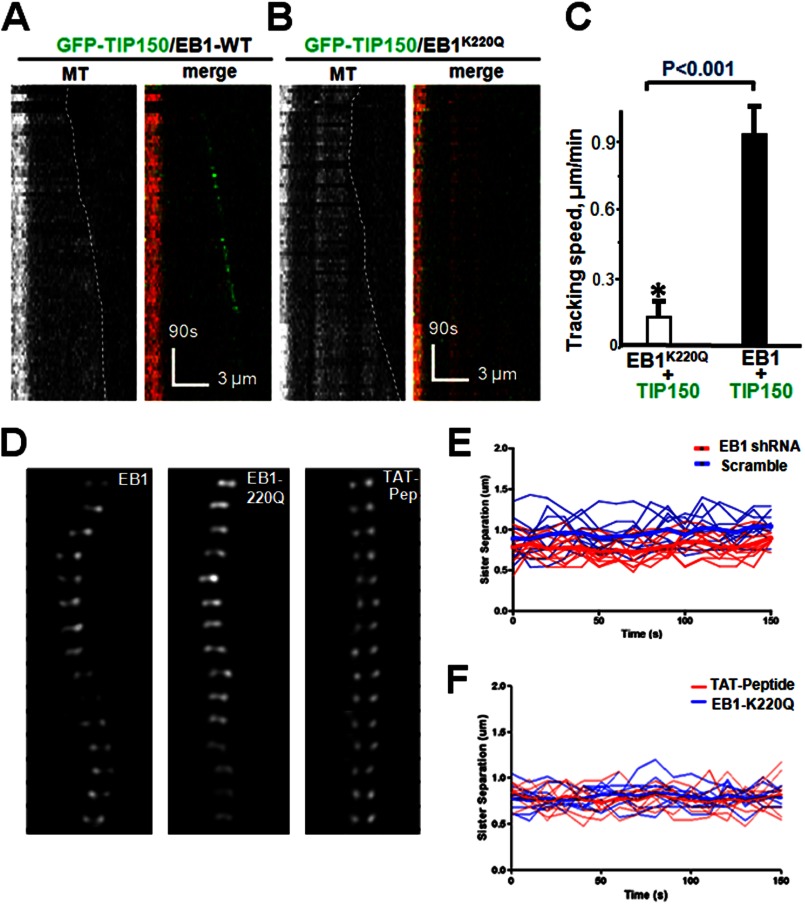
To elucidate the molecular basis for acetyl regulation of the EB1-TIP150 interaction, the crystal structure of EB1 relative to Lys-220 was assessed. An examination of the EB1 crystal structure (PDB 3GJO) suggested that the Lys-220 ϵ-amino group forms three pairs of hydrogen bonds with the backbone carbonyl oxygens of Ile-245, Leu-246, and Ala-248, indicating that acetylation of Lys-220 would perturb those bonds and destabilize the hydrophobic cavity in the EB domain. Molecular modeling of the EB1-SXIP complex showed that Ile-245, Leu-246, and Ala-248 are essential for the plasticity of a hydrophobic cavity responsible for binding to TIP150 (14). To determine whether acetylation of acK220 modulates EB1 interaction with TIP150, an acetylation-mimicking K220Q mutant was generated in bacteria and used in a pulldown assay. The K220Q mutant bound weakly to TIP150 but not CLIP170 in vitro. If acetylation of EB1 perturbs its interaction with TIP150, TIP150 would not track with microtubule plus-ends in the presence of EB1K220Q. To test this hypothesis, the plus-end tracking activity of EB1K220Q was assessed by a TIRFM-based assay. Analyses of kymographs showed that wild-type TIP150 tracked the growing ends of microtubules in the presence of EB1 (Fig. 6A). However, GFP-TIP150 failed to track microtubule plus-ends in the presence of EB1K220Q (Fig. 6B). Statistical analyses indicated that acetylation of Lys-220 negatively modulated the EB1 interaction with TIP150 (Fig. 6C) by reducing TIP150 tracking activity. Thus, acetylation regulates the EB1 molecular dynamics and its interaction with TIP150.
FIGURE 6.
Lys-220 acetylation regulates a dynamic interaction between EB1 and TIP150. A, dual color kymographs of polymerizing microtubules with EB1 and GFP-TIP150 in vitro as illustrated in Fig. 3. B, dual color kymographs of polymerizing microtubules with acetylated EB1 and GFP-TIP150. Note that the GFP-TIP150 failed to track microtubule plus-ends in the presence of acetylated EB1K220Q. C, quantitative analysis of microtubule polymerization with EB1-GFP-TIP150 interaction compared with acetylated EB1-GFP-TIP150. Tracking speed was diminished with acetylated EB1 as determined by single-molecule tracking (*, p < 0.001; n = 30; EB1K220Q+TIP150 versus EB1-TIP150). D, perturbation of EB1 interaction by persistent Lys-220 acetylation or TIP150 peptide addition alters kinetochore oscillatory profiles in mitosis. Kinetochore oscillations of wild-type EB1 (left), K220Q (acetylated-mimicking mutant; center), and TAT-Peptide (right). Oscillations were diminished when EB1 was acetylated. E, representative kinetochore oscillatory kymograph of scramble siRNA (blue line) and EB1siRNA-treated cells (red line). F, representative kinetochore oscillatory kymograph of HeLa cells expressing persistent acetylation mimicking EB1-K220Q (blue line) or treated with TAT-peptide (red line).
To determine whether acetylation of Lys-220 regulates chromosome dynamics during mitosis, kinetochore motility marked by GFP-CENP-A was imaged at high temporal resolution in cells with endogenous EB1 but expressing either the wild-type, the acetylation-mimicking EB1K220Q mutant, or the TAT-peptide. The sister kinetochores exhibited oscillation in wild-type EB1-expressing cells in the absence of endogenous EB1 (Fig. 6D, left panel). Sister kinetochores of K220Q-expressing cells were frequently observed to be less oscillatory but moved back and forth around the metaphase plate (Fig. 6D, middle panel). The sister kinetochores of TAT-peptide-treated cells were more static and never achieved metaphase plate alignment and compaction (Fig. 6D, right panel). The kymograph shows that persistent acetylation of EB1 by overexpression of EB1K220Q mutant or perturbation of EB1-TIP150 interaction by TAT-peptide alters the kinetochore oscillatory behavior during chromosome congression (Fig. 6, E and F).
As the inter-kinetochore distance on sister chromatids has been proposed as an accurate reporter for judging tension developed across the kinetochore pair, this distance was measured in >100 pairs, in which both kinetochores were in the same focal plane, in cells that were EB1-depleted but expressing various exogenous EB1 proteins (wild type, or K220Q; Table 1). Control kinetochores exhibited a separation of 1.41 ± 0.21 μm, whereas the distance between the sister kinetochores in K220Q-expressing cells was 1.13 ± 0.16 μm, which is significantly reduced relative to those of cells expressing wild-type EB1 (p < 0.05). The inter-kinetochore distance of EB1-depleted cells was 1.07 ± 0.15 μm. As determined from four separate experiments, the distance in nocodazole-treated TIP150-suppressed cells, in which polymerization of microtubules was inhibited and kinetochore pairs were presumably under no tension, was 1.01 ± 0.13 μm. Thus, perturbation of Lys-220 acetylation alters the accurate attachment of spindle microtubules to the kinetochores.
TABLE 1.
EB1-TIP150 interaction controls the inter-kinetochore tension
Data were obtained from >150 kinetochore pairs in which kinetochores were in the same focal plane. Note that depletion of TIP150 and EB1 abolishes the tension between sister kinetochores.
| Treatment | Distancea |
|---|---|
| μm | |
| Scramble/DMSO-treated | 1.41 ± 0.21 |
| TIP150 siRNA (misaligned kinetochore) | 1.12 ± 0.16 |
| EB1 siRNA (misaligned kinetochore) | 1.07 ± 0.15 |
| TAT-peptide (misaligned kinetochore) | 1.09 ± 0.17 |
| EB1K220Q (misaligned kinetochore) | 1.13 ± 0.16 |
| TSA (misaligned kinetochore) | 1.09 ± 0.15 |
| TIP150 siRNA + nocodazole treatment | 1.01 ± 0.13 |
a Distance measured between ACA-marked sister kinetochore from same focal plane.
Perturbation of the EB1-TIP150 Interaction Activates the Spindle Checkpoint
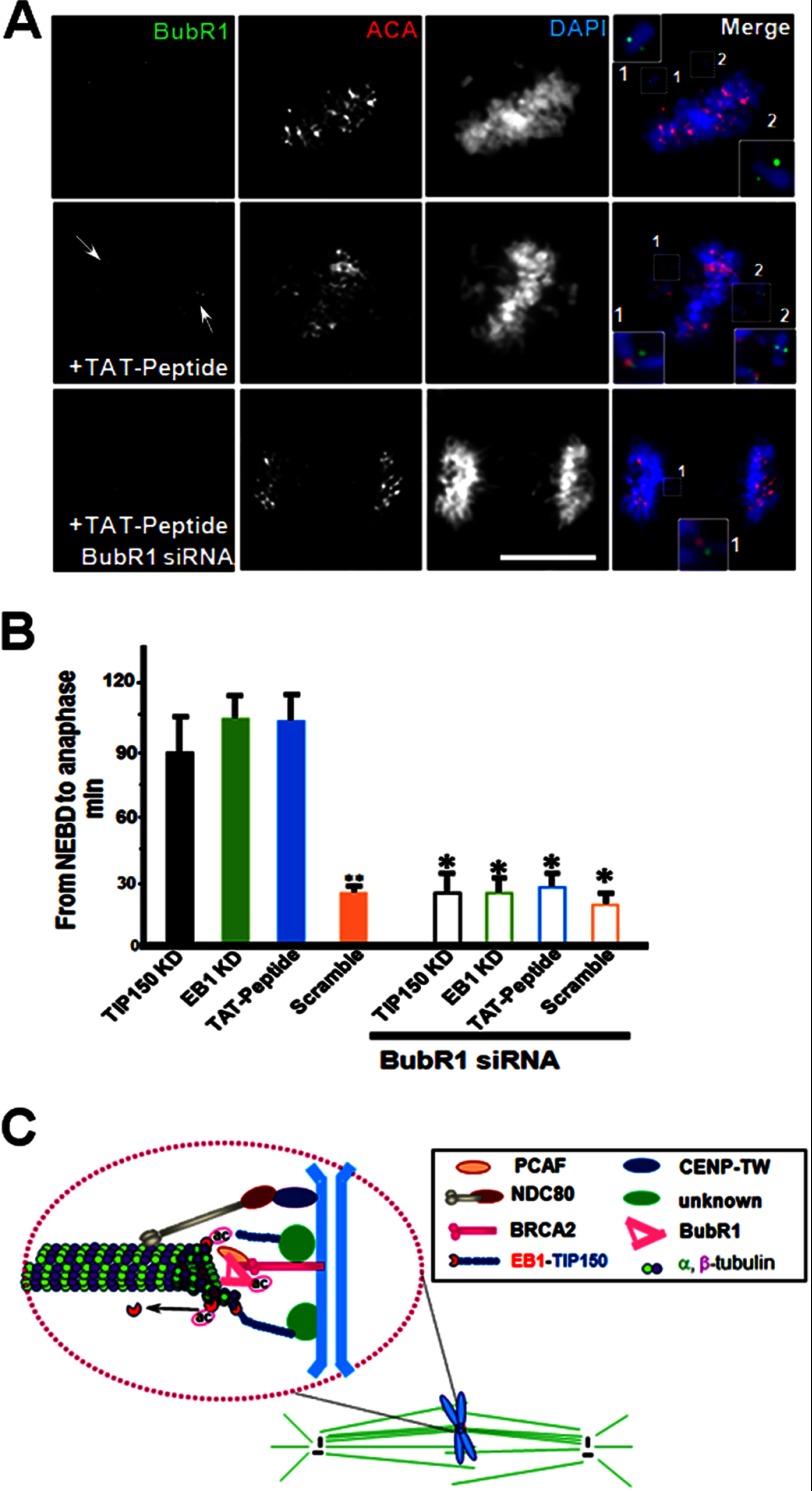
The dynamic acetylation in mitosis prompted us to determine the role of acK220 in chromosome dynamics. To determine whether the spindle assembly checkpoint (SAC) is activated by perturbation of the EB1-TIP150 interaction, the distribution the spindle checkpoint kinase, BubR1, in cells treated with the TAT-GFP-peptide was assessed. BubR1 distribution in cells treated with TAT-GFP binds to unattached kinetochores and to those that are attached but not under tension (Fig. 7A, top panel and enlarged insets). Consistent with the concept of continued activation of the mitotic checkpoint from misaligned chromosomes, high levels of kinetochore-associated BubR1 were apparent in cells treated with TAT-GFP-peptide (Fig. 7A, middle panel, arrows), particularly on kinetochores misaligned but not apparently aligned chromosomes. The attachment of BubR1 to the kinetochores of cells treated with the TAT-GFP-TIP150 peptide suggests that interaction between EB1 and TIP150 orchestrates accurate kinetochore-microtubule interactions and the SAC.
FIGURE 7.
Perturbation of EB1-TIP150 interaction activates the spindle checkpoint. A, immunofluorescence analysis shows BubR1, ACA, and DAPI for DNA. In SAC-activated cell BubR1 siRNA overrides the induced mitotic arrest by acetylated EB1-TIP150. B, quantitative analysis of the timing of cell division from NEBD to anaphase onset. Anaphase onset is delayed in cells with TIP150 knockdown, EB1 knockdown, and TAT-GFP-peptide (2.5 μm). This condition is rescued by treatment with BubR1 siRNA. **, p < 0.001, Scramble versus TP150 KD or EB1 KD or TA-peptide; *, p > 0.05, BubR1 siRNA (TIP150 KD, EB1 KD, TAT-peptide or Scramble) versus without BubR1 siRNA. C, model representation of PCAF-TIP150-EB1 interaction. In this model, PCAF is localized to kinetochore via interaction with BRCA2, and PCAF is critical for chromosome stability in mitosis (23). EB1-TIP150 interaction is required for efficient and accurate kinetochore-microtubule interactions and for achieving faithful alignment at the equator. Development of normal level of tension across bioriented kinetochore pairs, as reflected in the spatial separation of sister kinetochores, requires a temporal and dynamic regulation of EB1 acK220. The effect of temporal control of acK220 is 2-fold. First, TIP150 cooperates with EB1 to maintain a stable kinetochore-microtubule interaction, and acK220 weakens the tension across the sister kinetochores to facilitate attachment error correction. Second, the BubR1 (a checkpoint kinase that is bound to EB1/APC (adenomatous polyposis coli) and is essential for metaphase alignment (14) and disruption of EB1-TIP150 interaction) results in a prolonged activation of spindle checkpoints, as marked by BubR1 enrichment at the kinetochore, preventing activation of the APC/C (anaphase-promoting complex/cyclosome) and onset of anaphase.
To validate SAC activation in cells with perturbed EB1-TIP150 interaction, HeLa cells were transiently transfected with BubR1 siRNA to suppress BubR1, followed by treatment with TAT-GFP-peptide (2.5 μm) for 24 h after the transfection. Treatment of BubR1-suppressed cells with the TAT-GFP-peptide resulted in an increase in premature anaphase, as sister chromatids separated in the presence of misaligned chromosomes, a phenotype characteristic of SAC inactivation (Fig. 7A, lower panel). Quantitative analyses showed that BubR1 knockdown overrode the mitotic arrest induced by perturbation of the EB1-TIP150 interaction, as there was no significant difference in the interval between NEBD and anaphase onset in TAT-GFP-treated cells compared with that of TAT-GFP-peptide-treated and BubR1-suppressed cells (Fig. 7B, p > 0.01). This suggests that interaction of EB1 and TIP150 is coupled to SAC in mitotic cells. This interaction is dependent on acetylation activity. Thus, during mitosis, the PCAF-EB1-TIP150 axis is associated with chromosome stability.
DISCUSSION
The kinetochore is a complex structure that functions as a molecular machine to power chromosome movement along dynamic microtubule polymers; it is also a signaling device governing chromosome segregation and controlling the cell cycle. We recently identified a novel microtubule plus-end tracking protein, TIP150. TIP150 cooperates with EB1 in microtubule plus-end tracking and forms a functional hexamer with EB1. This TIP150-EB1 interaction tracks microtubule plus-ends. Consequently, mitotic cells overexpressing a TIP150 C-terminal mutant are delayed in metaphase alignment due to depletion of functional TIP150 dimers. Our findings identify a molecular mechanism orchestrating the dynamics of kinetochore-microtubule attachment during mitotic chromosome segregation and provide a unifying view of a regulatory mechanism underlying an array of functionally and structurally diversified plus-end tracking proteins dependent on EB1.
A characteristic of the kinetochore-spindle interface is its capacity to orchestrate stable and dynamic associations while bound microtubules are polymerizing or depolymerizing. Such properties would be best coordinated by several distinct but cooperative kinetochore-microtubule-binding sites regulated by signaling cascades (24, 25). In mitotic cells, EB1, together with its cargo proteins, such as MCAK and TIP150, localize to various subcellular structures, including outer kinetochores and are involved in mitotic chromosome segregation. The diversified array of EB1 cargo proteins implies a complex but perhaps coordinated regulation in mitosis to ensure accurate chromosome segregation. We and others have shown that mitotic kinases, such as PLK1 and Aurora B, form a feedback loop to orchestrate MCAK activity as a microtubule regulator (26, 27). In budding yeast, EB1 protein is regulated by Aurora B-mediated phosphorylation on six serine residues located in the flexible linker connecting the CH and EB1 domains. It would be of interest to determine whether the EB1-TIP150 interaction is regulated by mitotic phosphorylation.
The central activities of microtubule plus-end tracking are orchestrated by their common properties, such as EB1 binding to TIP150. We have shown that, in the absence of functional TIP150, like EB1 depletion, cells remain in mitosis for extended periods, failing to satisfy the SAC. The failure of stable bipolar attachment indicates that the EB1-based protein, Hub, functions in the kinetochore-microtubule interaction, including initial capture and microtubule stabilization or error correction. These findings indicate that EB1-TIP150 is an essential linking factor that mediates stability of spindle microtubules at kinetochores and/or linkage of kinetochore bundles to the bulk microtubules of the spindle without affecting the number of bound microtubules. It is uncertain, however, if the bi-oriented chromosomes seen in TIP150-depleted and/or TIP150(981–1368)-overexpressing cells capture polar microtubules. Furthermore, it is not known whether microtubule bundles attached either to mono-oriented or bi-oriented chromosomes have lifetimes that are comparable with those of typical kinetochore fibers, or whether the absence of TIP150 generates unstable kinetochore fibers, producing chromosomes that cycle between mono- and bi-orientation.
It seems apparent that, after initial activation of the mitotic checkpoint as the nuclear envelope is disassembled, one or more kinetochore-associated, microtubule-binding components must be involved in linking microtubule attachment to SAC silencing. Indeed, suppression of TIP150 leads to chronic activation of the SAC and continued association of high levels of BubR1 with misaligned kinetochores.4 The fact that knockdown of BubR1 overrides the persistent SAC activation seen in TIP150-depleted cells or TIP150(981–1368)-overexpressing cells supports the function of TIP150 in SAC satisfaction. It is worth noting that the mitotic dynamics of acK220 support the concept that a high level of acetylation promotes correction of aberrant attachments through inhibition of kinetochore plus-end growth. Upon error correction, reduced acetylation allows for stabilizing the interactions of EB1 with its cargo proteins at the kinetochore and switches the centromere to a mode for proper chromosome alignments at the equator so that anaphase begins. In human breast and liver cancers, hyperacetylation promotes genomic instability via centrosome amplification (31). This finding indicates the importance of temporal regulation of acetylation dynamics in ensuring the fidelity of chromosome segregation and maintaining chromosome stability in mitosis.
The kinetochore machinery is highly dynamic, both in the remodeling of a subset structure and turnover of its components during cell division cycle. The kinetochore plasticity and macromolecular assembly dynamics are orchestrated by cell cycle machinery through protein covalent modifications, such as phosphorylation, acetylation, and methylation (21, 30, 32, 33). It will be very challenging and exciting to visualize and illustrate the precise mechanism of action of the aforementioned protein modifications in orchestrating dynamic kinetochore assembly and signaling circuitry at the individual molecular level of different chromosomes relative to the global consequence of chromosome plasticity in cell division.
In summary, the present results reveal the importance of TIP150 in HeLa cells during mitosis and demonstrate the function of TIP150 dimerization in mitotic chromosome congression toward the metaphase plate. The observation that EB1 forms a functional hexamer with TIP150 provides a better understanding of the dynamics and plasticity of plus-end tracking proteins. These results present a unified view of previously uncharacterized molecular mechanisms underlying dynamic regulation of the TIP150-EB1 complex by PCAF-mediated acetylation in kinetochore-microtubule attachment during mitosis.
Acknowledgments
We thank the members of our groups for insightful discussions during the course of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants DK56292, CA164133, G12RR03034, UL1 RR025008, and CA132389. This work was also supported by Chinese 973 Project Grants 2010CB912103, 2012CB917204, 2012CB945002, and 2002CB713700; Chinese Academy of Science Grant KSCX2-YW-H-10; Anhui Province Key Project Grant 08040102005; International Collaboration Grant 2009DFA31010; Chinese Natural Science Foundation Grants 90508002, 91129714, 31071184, 81270466, 90913016, and MOE20113402130010, Chinese Postdoctoral Fellowship 2012M510210; and Fundamental Research Funds for Central Universities Grants WK2060190018 and WK2340000021.
T. Ward, X. Liu, Y. Huang, and X. Yao, unpublished observations.
- MCAK
- mitotic centromere-associated kinesin, a microtubule-depolymerizing kinesin
- PCAF
- p300/CBP-associated factor
- NEBD
- nuclear envelope breakdown
- TIRFM
- total internal reflection fluorescence microscopy
- ACA
- anti-centromere antibody
- PA
- photo-activated
- EGFP
- enhanced GFP
- SAC
- spindle assembly checkpoint
- GMP-CPP
- guanosine 5′-(α,β-methylene)triphosphate, sodium salt.
REFERENCES
- 1. Cleveland D. W., Mao Y., Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 2. Siegel J. J., Amon A. (2012) New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 28, 189–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon D. J., Resio B., Pellman D. (2012) Causes and consequences of aneuploidy in cancers. Nat. Rev. Genet. 13, 189–203 [DOI] [PubMed] [Google Scholar]
- 4. Cheeseman I. M., Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 5. Santaguida S., Musacchio A. (2009) The life and miracles of kinetochores. EMBO J. 28, 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su L. K., Burrell M., Hill D. E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B., Kinzler K. W. (1995) APC binds to the novel protein EB1. Cancer Res. 55, 2972–2977 [PubMed] [Google Scholar]
- 7. Fodde R., Kuipers J., Rosenberg C., Smits R., Kielman M., Gaspar C., van Es J. H., Breukel C., Wiegant J., Giles R. H., Clevers H. (2001) Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 3, 433–438 [DOI] [PubMed] [Google Scholar]
- 8. Akhmanova A., Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 9. Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T. (2007) Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100–1105 [DOI] [PubMed] [Google Scholar]
- 10. Jiang K., Wang J., Liu J., Ward T., Wordeman L., Davidson A., Wang F., Yao X. (2009) TIP150 interacts with and targets MCAK at the microtubule plus-ends. EMBO Rep. 10, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakamura T., Pluskal T., Nakaseko Y., Yanagida M. (2012) Impaired coenzyme A synthesis in fission yeast causes defective mitosis, quiescence-exit failure, histone hypoacetylation, and fragile DNA. Open Biol. 2, 120117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A. W., Teng M., Cleveland D. W., Goldberg M. L., Niu L., Yao X. (2004) Human zwint-1 specifies localization of zeste white 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279, 54590–54598 [DOI] [PubMed] [Google Scholar]
- 13. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., Yao X. (2004) CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J. Biol. Chem. 279, 20049–20057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., Yao X. (2012) EB1 acetylation by p300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 16. Kops G. J. (2009) Dividing the goods: co-ordination of chromosome biorientation and mitotic checkpoint signalling by mitotic kinases. Biochem. Soc. Trans. 37, 971–975 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., Ding X., Huang Y., Wang D., Yao X. (2012) Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287, 39380–39390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dixit R., Ross J. L. (2010) Studying plus-end tracking at single molecule resolution using TIRF microscopy. Methods Cell Biol. 95, 543–554 [DOI] [PubMed] [Google Scholar]
- 19. Du J., Cai X., Yao J., Ding X., Wu Q., Pei S., Jiang K., Zhang Y., Wang W., Shi Y., Lai Y., Shen J., Teng M., Huang H., Fei Q., Reddy E. S., Zhu J., Jin C., Yao X. (2008) The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene 27, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitchison T. J. (1989) Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109, 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu J., Wang Z., Jiang K., Zhang L., Zhao L., Hua S., Yan F., Yang Y., Wang D., Fu C., Ding X., Guo Z., Yao X. (2009) PRC1 cooperates with CLASP1 to organize central spindle plasticity in mitosis. J. Biol. Chem. 284, 23059–23071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou R., Guo Z., Watson C., Chen E., Kong R., Wang W., Yao X. (2003) Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell 14, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi E., Choe H., Min J., Choi J. Y., Kim J., Lee H. (2009) BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 28, 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 [DOI] [PubMed] [Google Scholar]
- 25. Huang Y., Wang W., Yao P., Wang X., Liu X., Zhuang X., Yan F., Zhou J., Du J., Ward. T., Zou H., Zhang J., Fang G., Ding X., Dou Z., Yao X. (2012) CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu Y., Yao P. Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., Yao X. (2011) Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell. Biol. 3, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G., Ding X., Yao X. (2011) PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286, 3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lampson M. A., Kapoor T. M. (2005) The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7, 93–98 [DOI] [PubMed] [Google Scholar]
- 29. Zhao L., Jin C., Chu Y., Varghese C., Hua S., Yan F., Miao Y., Liu J., Mann D., Ding X., Zhang J., Wang Z., Dou Z., Yao X. (2010) Dimerization of CPAP orchestrates centrosome cohesion plasticity. J. Biol. Chem. 285, 2488–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu C., Yan F., Wu F., Wu Q., Whittaker J., Hu H., Hu R., Yao X. (2007) Mitotic phosphorylation of PRC1 at Thr-470 is required for PRC1 oligomerization and proper central spindle organization. Cell Res. 17, 449–457 [DOI] [PubMed] [Google Scholar]
- 31. Kim H. S., Vassilopoulos A., Wang R. H., Lahusen T., Xiao Z., Xu X., Li C., Veenstra T. D., Li B., Yu H., Ji J., Wang X. W., Park S. H., Cha Y. I., Gius D., Deng C. X. (2011) SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., Yao X. (2012) SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J. Mol. Cell. Biol. 4, 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yao X., Fang G. (2009) Visualization and orchestration of the dynamic molecular society in cells. Cell Res. 19, 152–155 [DOI] [PubMed] [Google Scholar]