Background: Patients with C1-INH deficiency and streptococcal toxic shock syndrome show similar symptoms.
Results: A streptococcal cysteine protease degrades complement factors and protects bacteria against complement system bactericidal effects.
Conclusion: Streptococcus pyogenes evades complement system eradication by its own cysteine protease.
Significance: Our findings clarify the mechanism shown in previous clinical case reports and are important for future applications.
Keywords: Complement System, Cysteine Protease, Innate Immunity, Protease Inhibitor, Streptococcus pyogenes, C1-Esterase Inhibitor, SpeB, Membrane Attack Complex
Abstract
Streptococcus pyogenes is an important human pathogen that causes invasive diseases such as necrotizing fasciitis, sepsis, and streptococcal toxic shock syndrome. We investigated the function of a major cysteine protease from S. pyogenes that affects the amount of C1-esterase inhibitor (C1-INH) and other complement factors and aimed to elucidate the mechanism involved in occurrence of streptococcal toxic shock syndrome from the aspect of the complement system. First, we revealed that culture supernatant of a given S. pyogenes strain and recombinant SpeB degraded the C1-INH. Then, we determined the N-terminal sequence of the C1-INH fragment degraded by recombinant SpeB. Interestingly, the region containing one of the identified cleavage sites is not present in patients with C1-INH deficiency. Scanning electron microscopy of the speB mutant incubated in human serum showed the abnormal superficial architecture and irregular oval structure. Furthermore, unlike the wild-type strain, that mutant strain showed lower survival capacity than normal as compared with heat-inactivated serum, whereas it had a significantly higher survival rate in serum without the C1-INH than in normal serum. Also, SpeB degraded multiple complement factors and the membrane attack complex. Flow cytometric analyses revealed deposition of C9, one of the components of membrane the attack complex, in greater amounts on the surface of the speB mutant, whereas lower amounts of C9 were bound to the wild-type strain surface. These results suggest that SpeB can interrupt the human complement system via degrading the C1-INH, thus enabling S. pyogenes to evade eradication in a hostile environment.
Introduction
Streptococcus pyogenes is an important Gram-positive bacterium that causes a variety of clinical pathologies, ranging from noninvasive disease, including pharyngitis and impetigo, to more invasive diseases such as necrotizing fasciitis, sepsis, and streptococcal toxic shock syndrome (STSS)2 (1). STSS has been reported to be associated with a high rate of mortality, and its major symptoms include fever, rash, capillary leak syndrome, and disseminated intravascular coagulation (2, 3). Administration of β-lactam agents including penicillin is the standard treatment protocol for this infectious disease, whereas Fronhoffs et al. (4) reported that high dose administration of C1-esterase inhibitor (C1-INH) as adjunctive therapy in STSS patients decreased related STSS symptoms, including circulation disorder, lung edema, and capillary leak syndrome. Furthermore, those authors noted that attenuation of capillary leak syndrome by early inactivation of the complement and contact systems was possibly involved in the favorable outcomes. In another study, disseminated intravascular coagulation was induced by breakdown of the stability of the contact system and coagulation system (5).
C1-INH, a component of the complement immune system, regulates the complement pathway via inactivation of C1r, C1s, and mannose-binding protease-associated serine protease 2 (MASP2). In addition, C1-INH regulates the kallikrein-kinin system by inactivation of activated plasma kallikrein, factor XIIa, and factor XIa, which results in inhibition of bradykinin and plasmin generation (6, 7). In other words, in the contact and coagulation systems, inflammation including leukocyte adherence is modulated by C1-INH. In cases with C1-INH deficiency, the contact system becomes unstable and prone to generate kallikrein, which cleaves high molecular weight kininogen. Thus, the released bradykinin may function as a mediator of increased vascular permeability (8). In the present study, we focused on the similarity of these symptoms and investigated whether a S. pyogenes protease induces collapse of the complement system through C1-INH.
The complement system is a key component of innate immunity and acts as a protective shield in the early phases of infection. Following infection with pathogens, each complement factor is activated in a sequential manner (9) under precise control of complement regulatory factors including C1-INH. In the late complement pathway, the membrane attack complex (MAC) is formed on the bacterial surface. It is thought that the MAC forms pores on the surfaces of Gram-negative bacteria, whereas its function with Gram-positive bacteria is unclear. In previous studies, the complement factors C3b and properdin were cleaved by SpeB, C5a was cleaved by ScpA, and factor H inhibited the function of the protease by binding to streptococcal collagen-like protein 1 of S. pyogenes (10, 11). However, those studies did not examine the correlations of C1-INH and other complement factors with the virulence factors of S. pyogenes. Here, we investigated virulence factors released from S. pyogenes that have effects on C1-INH and other complement factors and aimed to elucidate the mechanism involved in the occurrence of STSS from the aspect of the complement system.
EXPERIMENTAL PROCEDURES
Bacterial Strains
S. pyogenes strain SSI-9 (serotype M1) was isolated from a Japanese patient with STSS (12) Also, its isogenic speB-deficient mutant TR-11 and scpA-deficient mutant TR-9 were constructed with a suicide vector pSF 151, as described previously (13, 14). These strains were grown in Todd Hewitt broth (Difco Laboratories, Detroit, MI) supplemented with 0.2% yeast extract *(THY broth) at 37 °C in CO2 incubator. Furthermore, Escherichia coli strain XL-10 Gold (Stratagene, La Jolla, CA) was grown in Luria-Bertani (LB) broth (Sigma-Aldrich) supplemented with 100 μg/ml ampicillin for stabilization of pGEM-T Easy (Promega, Fitchburg, WI) (15). Also, BL21 (DE3) pLysE (Novagen, Madison, WI) was grown in LB broth supplemented with 30 μg/ml kanamycin to stabilize pGEX-6P-1 (GE Healthcare Bio-Sciences K.K.) or supplemented with 100 μg/ml ampicillin for stabilization of pIVEX2.3-MCS (Roche Applied Science). Also, various S. pyogenes strains were obtained from patients with pharyngitis, impetigo, scarlet fever, and STSS (kindly provided by Dr. T. Murai, the Graduate School of the Japanese Red Cross Akita College of Nursing, Akita, Japan) (15, 16). All streptococcal strains were grown in THY broth.
Construction of Recombinant SpeB and ScpA
recombinant SpeB (rSpeB) and rScpA was prepared as described previously (10). Briefly, the plasmid vector pGEX-6P-1 or pIVEX2.3-MCS was used for producing of rScpA or rSpeB, respectively. The rSpeB protein was produced as a 43-kDa zymogen and converted to the 28-kDa active form by self-processing during purification by nickel-nitrilotriacetic acid-agarose (Qiagen, Düsseldorf, Germany) and diethylaminoethyl column chromatography (Bio-Rad). The rScpA protein was purified by glutathione-Sepharose 4B (GE Healthcare Bio-Sciences) and diethylaminoethyl column chromatography. The purified rSpeB and rScpA (C5a peptidase) proteins were dialyzed against phosphate-buffered saline (PBS).
Western Blot Analyses
Western blotting was performed as described previously (10, 17). To examine the cleavage activities of ScpA and SpeB to the C1-INH, S. pyogenes strains SSI-9, TR-9 (ΔscpA), and TR-11 (ΔspeB) were cultured at 37 °C for 18 h in 10 ml of THY broth, and then supernatants were obtained after centrifugation at 4 °C for 6,800 rpm for 5 min. Each supernatant was concentrated 10-fold using 100% ammonium sulfate precipitation and diluted to 1 ml with PBS. Forty μl of concentrated supernatant with 10 μl of PBS or 10 μl of 1 μm recombinant proteins (rScpA and rSpeB) was incubated with 10 μl of human serum (Sigma) at 37 °C for 3 h. The samples were electrophoresed on a 7.5 or 12% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany). The membranes were blocked overnight with 10% membrane blocking agent (Thermo Scientific Pierce) with 10% goat serum (Invitrogen, Life Technologies) at 4 °C. Next, each membrane was incubated with rabbit anti-human C1-INH serum (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoreactive bands were detected with an alkaline phosphatase-labeled goat anti-rabbit IgG and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium alkaline phosphatase substrate solution (Moss, Pasadena, MD). To check the proteolytic activity of SpeB with the C1-INH, rSpeB was serially diluted 2-fold (0.20–50 nm) by PBS and incubated in human serum (without complement inactivation). Following 1 h of incubation at 37 °C, each mixture was examined by Western blot analysis using anti-human C1-INH serum. To investigate SpeB cleavage activities against various complement factors, rSpeB was incubated with recombinant complement factors. Briefly, 1 μm rSpeB was incubated with 25 nm recombinant C2, C3b, C4, C5, C5b, C6, C7, C8, or C9 (Calbiochem) for 3 h at 37 °C. The samples were electrophoresed on 5–20% gradient SDS-PAGE gels and transferred to PVDF membranes and then blocked overnight with 10% membrane blocking agent with 10% goat serum at 4 °C. Following that incubation, each mixture of C2, C3b, or C4 with rSpeB was incubated with goat anti-human C2, C3b, or C4 serum (Calbiochem), and immunoreactive bands were detected with horseradish peroxidase-labeled mouse anti-goat IgG (R&D Systems) and TMB-H peroxidase substrate solution (Moss). Each mixture of C5, C5b, C6, C7, C8, or C9 with rSpeB was then incubated with rabbit anti-human C5a (Calbiochem), C5b (American Research Products, Inc.), C6, C7, C8β, or C9 serum (Atlas Antibodies AB, Stockholm, Sweden) respectively, and immunoreactive bands were detected with an alkaline phosphatase-labeled goat anti-rabbit IgG and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium alkaline phosphatase substrate solution (Moss). The MAC was prepared by mixing 1 μm recombinant C5b6, C7, C8, and C9, and then the resultant mixture was incubated for 1 h at 37 °C. The MAC mixture was then incubated with 1 μm rSpeB, and the MAC or its fragments were detected using an anti-MAC antibody (Calbiochem).
Comparison of C1 Inhibitor-degrading Ability of Different M Type Strains with Amount of SpeB
Thirty different S. pyogenes strains (Table 1) were cultured at 37 °C for 18 h, and the supernatants were obtained. Ten ml of the supernatants was concentrated 10-fold using 100% ammonium sulfate precipitation and then diluted to 1 ml with PBS. SpeB expression in the sample solution was detected by Western blotting using anti-SpeB antibody. Forty μl of each adjusted supernatant or 40 μl of PBS as negative control was incubated with 10 μl of human serum for 3 h at 37 °C. The C1-INH or SpeB was detected by Western blotting using anti-human C1-INH antibody. Optical densities of the detected protein bands were determined using a computer-assisted densitometer (ImageJ version 8.8.7, National Institutes of Health, Bethesda, MD). An intact C1-INH was detected as an ∼105-kDa band in human serum treated with PBS. The highest intensity of SpeB band shown in TW 3532 (obtained from Tokyo Women's Medical College) was defined for the positive control of SpeB. The background of each membrane was subtracted from each obtained band for densitometric analyses.
TABLE 1.
Streptococcal strains used in this study
| Serotype | Strain |
|---|---|
| M1 | SSI-9 |
| M1 | SSI-25 |
| M1 | SSI-127 |
| M2 | SE1013 |
| M2 | SE1181 |
| M2 | SE1207 |
| M3 | SSI-1 |
| M3 | SSI-7 |
| M3 | SSI-8 |
| M4 | TW3392 |
| M4 | TW3398 |
| M4 | TW3400 |
| M6 | SE1387 |
| M6 | SE1303 |
| M6 | TW3420 |
| M12 | NY-5 |
| M12 | TW3337 |
| M12 | TW3344 |
| M18 | T18 |
| M18 | TW3338 |
| M18 | TW3363 |
| M22 | TW3532 |
| M22 | TW3403 |
| M22 | TW3543 |
| M28 | TW3359 |
| M28 | TW3413 |
| M28 | TW3528 |
| M49 | CS101 |
| M49 | TW3425 |
| M49 | TW3424 |
N-terminal Amino Acid Sequencing
The targeted protein was separated using 7.5% SDS-PAGE and transferred to a PVDF membrane. The membrane was stained with 0.1% Coomassie Brilliant Blue R-250 for 1 h and then decolored with 7.5% acetic acid containing 40% methanol and washed with distilled water for 24 h. N-terminal amino acid sequencing was performed using the Edman degradation method with an ABI protein sequencer model 491HT (Applied Biosystems).
Scanning Electron Microscopic Analyses
SSI-9 and TR-11 were cultured until the mid-log phase (optical density at 600 nm = 0.4) and adjusted with PBS to 1.5 × 107 cfu/ml. Next, 90 μl of each sample was incubated with 10 μl of human serum at 37 °C for 15, 30, 45, or 60 min. Fifty μl of each sample was added on a Matrigel (BD Biosciences)-coated cover glass. Next, each bacterial sample was fixed with 2.5% glutaraldehyde on a cover glass for 1 h at room temperature and washed with distilled water. The samples were dehydrated with 100% t-butyl alcohol and freeze-dried. Finally, the samples were coated with platinum and examined with a scanning electron microscope (JSM-6390LVZ, JEOL Ltd., Tokyo, Japan), as described previously (18).
Bactericidal Test
Whole blood in vitro bactericidal assays were performed using a modified version of the bactericidal test described by Lancefield (19). S. pyogenes strains SSI-9 and TR-11 were cultured until the mid-log phase (optical density at 600 nm = 0.4) and adjusted with PBS or culture supernatant to 2,000–10,000 cfu/ml. The obtained samples were incubated with human serum (Sigma), inactivated serum, C1-INH-depleted serum, or C1-INH-depleted serum supplemented with the C1-INH for 3 h at 37 °C. The strains were then grown overnight and quantified after plating on THY agar plates. Depletion of C1-INH from human serum was performed as follows. Protein G antibody affinity chromatography (GE Healthcare) was used for depletion of C1-INH from human serum. The anti-C1-INH antibody (H-300, Santa Cruz Biotechnology Inc.) was captured by protein G using an Econo-column (Bio-Rad). The column was washed with PBS three times, and then human serum was slowly added. The procedure was repeated three times, and depletion was confirmed by Western blotting.
Flow Cytometry
S. pyogenes strains SSI-9 and TR-11 in the mid-log growth phase were incubated with human serum at 37 °C for 1 h. Each mixture was incubated with blocking agent (supplemented with 5% BSA and 10% goat serum) for 1 h at room temperature. Next, the bacterial cells were washed twice with PBS and incubated with anti-MAC rabbit serum, anti-C9 rabbit serum, anti-C1-INH rabbit serum, anti-M1 rabbit serum (positive control), or nonimmunized rabbit serum (negative control) for 1 h. After washing with PBS, the samples were incubated with Alexa Fluor 488-labeled anti-rabbit IgG (Life Technologies) for 1 h. Following incubation and washing with PBS, deposition of MAC, C9, or C1-INH on bacterial surfaces was evaluated by flow cytometry, and 10,000 bacterial cells were analyzed. Data acquisition was performed using a CyFlow SL system (Partec GmbH, Münster, Germany), with analyses performed with FlowJo software version 8.8.7 (TreeStar Inc.) as described previously (20).
Statistical Analysis
The significance of differences between the means of groups was evaluated using a nonparametric Mann-Whitney U test. All of the tests were analyzed using StatMate III software (ATMS Co., Ltd., Tokyo, Japan). Differences were considered significant at p < 0.01.
RESULTS
Streptococcal Cysteine Protease SpeB Degrades Human C1 Inhibitor
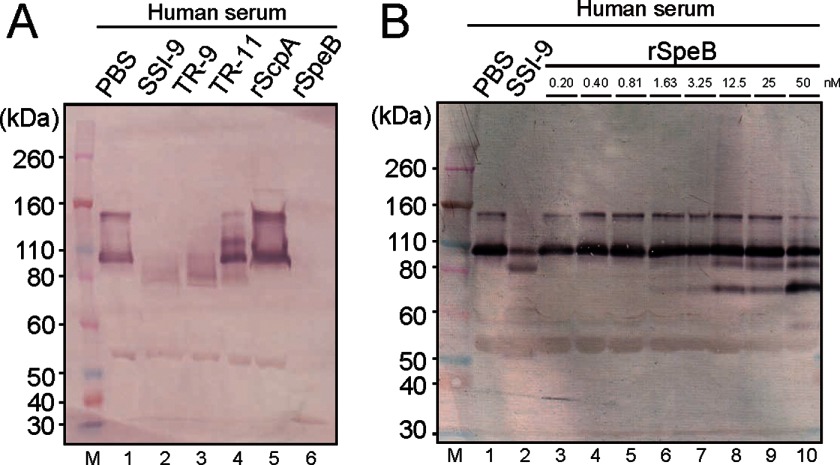
S. pyogenes is known to express major proteases: SpeB, an extracellular cysteine protease (21), and a cell-associated serine protease termed C5a peptidase, also called ScpA (22). We examined the C1-INH-degrading abilities of SpeB and ScpA in human serum. First, each supernatant, as well as bacterial cell samples obtained from three S. pyogenes strains (wild-type SSI-9, ΔscpA isogenic mutant TR-9, and ΔspeB isogenic mutant TR-11), was obtained after 18 h of growth. Western blot analyses demonstrated that the supernatants from both SSI-9 and TR-9 incubated with human serum cleaved the C1-INH (105 kDa, shown in Fig. 1A, lane 1 as positive control) into an ∼80-kDa fragment (Fig. 1A, lanes 2 and 3). However, C1-INH was not degraded in supernatant obtained from TR-11 (Fig. 1A, lane 4). In addition, the bacterial cell samples obtained from the three strains did not cleave the C1-INH (data not shown). Next, we examined the cleavage activity of rScpA and rSpeB for the C1-INH. As shown in Fig. 1A, lane 6, rSpeB degraded the C1-INH, whereas rScpA did not show that activity. Furthermore, analysis of the C1-INH-degrading activity by serially diluted SpeB showed that rSpeB degraded the C1-INH in a concentration-dependent manner (Fig. 1B, lanes 3–10).
FIGURE 1.
Human C1-INH-degrading ability of S. pyogenes SpeB. A, supernatants obtained from strain SSI-9 (lane 2) and isogenic mutant of ΔspeB (lane 3) or ΔscpA (lane 4) and recombinant ScpA (lane 5) and SpeB (lane 6) proteins were separately incubated with human serum (without complement inactivation) for 3 h at 37 °C. The C1-INH was detected by Western blotting. M, molecular mass marker. Lane 1, mixture of human serum and PBS as positive control. B, human serum (without complement inactivation) was incubated with SSI-9 (lane 2) or rSpeB (serially diluted 2-fold with PBS) (lanes 3–10) for 1 h at 37 °C. The C1-INH was detected by Western blotting.
C1 Inhibitor-degrading Activity Correlated with Amount of SpeB
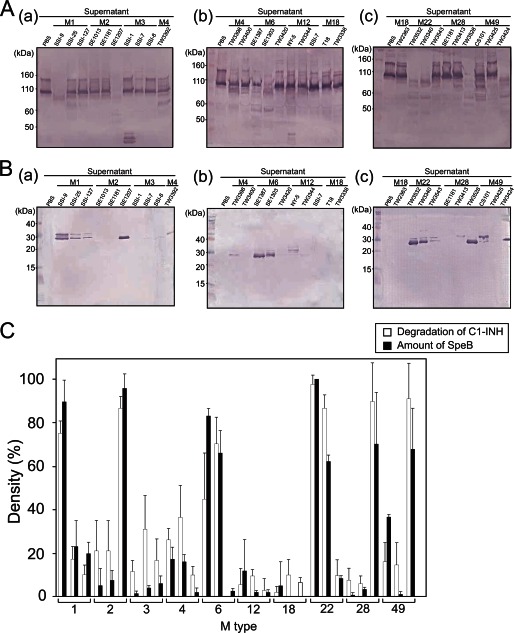
Next, we examined 30 different S. pyogenes strains for their ability to degrade the C1-INH (Fig. 2A) and also detected the amount of SpeB in those streptococcal strains (Fig. 2B). The C1-INH-degrading activities and relative amounts of SpeB were examined using Western blotting. The optical densities of the resultant blot bands were evaluated using computer-assisted densitometry (ImageJ, version 8.8.7), and C1-INH-degrading activity was compared with the amount of SpeB in each strain (Fig. 2C). Strains that showed high levels of C1-INH-degrading activity were likely to produce a large amount of SpeB, whereas there was no relation between C1-INH-degrading ability and M serotypes of S. pyogenes strains.
FIGURE 2.
C1-INH-degrading ability of different M type strains. Culture supernatants obtained from 30 S. pyogenes clinical isolates were incubated with human serum (without complement inactivation) for 3 h at 37 °C. A and B, the C1-INH (A) and SpeB (B) were detected by Western blotting (panels a–c). C, the degradation activity (white bar) and amount of SpeB (black bar) were determined using Western blotting, whereas the optical densities of the detected protein bands were determined using a computer-assisted densitometer (ImageJ version 8.8.7). Data represent the mean values of three technical repeats, and error bars indicate the S.D.
SpeB Cleaves C1 Inhibitor Upstream of Functional Domain
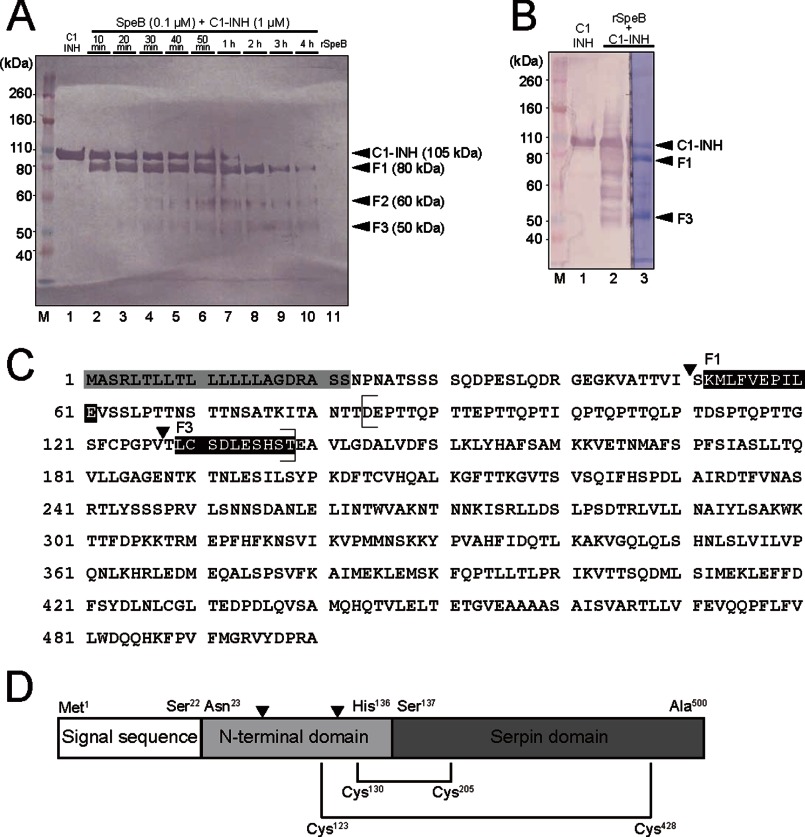
We determined the N-terminal amino acid sequence of the C1-INH fragment after degradation by rSpeB. First, we analyzed SpeB-mediated temporal cleavage of C1-INH. Western blotting combined with electric densitometer analyses revealed that the C1-INH was degraded in a time-dependent manner. The concentrations of F1, F2, and F3 were highest at 50, 60, and 120 min, respectively. As shown in Fig. 3A, the band representing the full length of C1-INH was rarely detected after 2 h. Three fragments were observed in Western blotting of C1-INH cleavage by SpeB, with lengths of 80 (F1), 60 (F2), and 50 (F3) kDa. After 60 min, F1 and F3 were clearly stained with Coomassie Brilliant Blue R-250 (Fig. 3B). Next, the protein bands for the N-terminal and internal residues were subjected to sequencing by Edman degradation to determine the site of C1-INH cleavage by SpeB (Fig. 3, C and D). The sequences of F1 and F3 corresponded with that of the native C1-INH. Also, a part of the C1-INH amino acid sequence of Asp84 to Thr138 corresponded to the sequence that is lost in patients with C1-INH deficiency (Fig. 3C) (23). The second cleavage site between Val127 and Thr128 was also included in the sequence. Therefore, this indicates that the effect of cleavage of the C1-INH by SpeB results in similar symptoms in patients with C1-INH deficiency as those seen in STSS patients with C1-INH deficiency.
FIGURE 3.
Molecular analysis of SpeB cleavage site in C1-INH. A, lane 1, C1-INH (positive control). Lanes 2–10, human serum samples incubated with rSpeB at 37 °C from 10 min to 4 h. Lane 11, rSpeB (negative control). Four black arrowheads show positions of bands representing SpeB-cleaved fragments from of the C1-INH. The right side membrane was stained with Coomassie Brilliant Blue R-250, and then the protein bands were used for N-terminal and internal residue sequencing with the Edman degradation method. Fragments of approximately 80, 60, and 50 kDa were designated as F1, F2, and F3, respectively. M, molecular mass marker. B, human serum was incubated with rSpeB at 37 °C for 1 h. The left side membrane (lanes 1 and 2) was used for Western blot analyses, and the right side membrane (lane 3) was stained with Coomassie Brilliant Blue R-250 and subjected to amino acid sequencing by Edman degradation. C, rSpeB cleavage site of C1-INH. The amino acid sequence of the C1-INH was obtained from the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov/protein/CAA30314.1). The gray box shows the putative signal sequence. N-terminal sequences of the cleaved fragments were in accordance with the sequences shown in the black boxes. Black arrowheads indicate the two cleavage sites of the C1-INH by SpeB. Parentheses indicate the amino acid sequence that is not present in C1-INH deficiency patients. D, schematic diagram of C1-INH protein. Black triangles indicate the two cleavage sites of the C1-INH by rSpeB. The two black lines show disulfide bonds between the N-terminal and serpin domains (Cys123 to Cys428, Cys130 to Cys205).
SpeB Secretion Protects S. pyogenes from Disruption of Architecture on Bacterial Surface when Incubated in Human Serum
In Gram-negative bacteria, the complement pathway is known to form pores on bacterial surfaces and induce a bacteriolytic function. Although S. pyogenes is Gram-positive, it is suspected that SpeB may influence the complement system, although it does not influence the cleavage of the C1-INH. To study the effects of SpeB on the complement system, we observed the surface properties of S. pyogenes SSI-9 and TR-11 after incubation with human serum using a scanning electronic microscope (SEM) (Fig. 4). After 15 min, TR-11 (ΔspeB) showed scant changes of its bacterial surface structure and an oval shape, whereas after 30 min, the structure of the bacterial surface started to change (Fig. 4, C and D). After 45 min of incubation in human serum, TR-11 showed pore structure on its bacterial surface (Fig. 4, E and F). Furthermore, after 60 min, the apparent destroyed superficial architecture and an irregular oval structure of TR-11 were observed (Fig. 4, G and H). On the other hand, SSI-9 showed only subtle changes of its bacterial surface after each time point of incubation with human serum (Fig. 4, A and B) until 60 min. These results suggest that SpeB inhibits the function of bacteriolytic component, such as other complement factors besides C1-INH in human serum.
FIGURE 4.
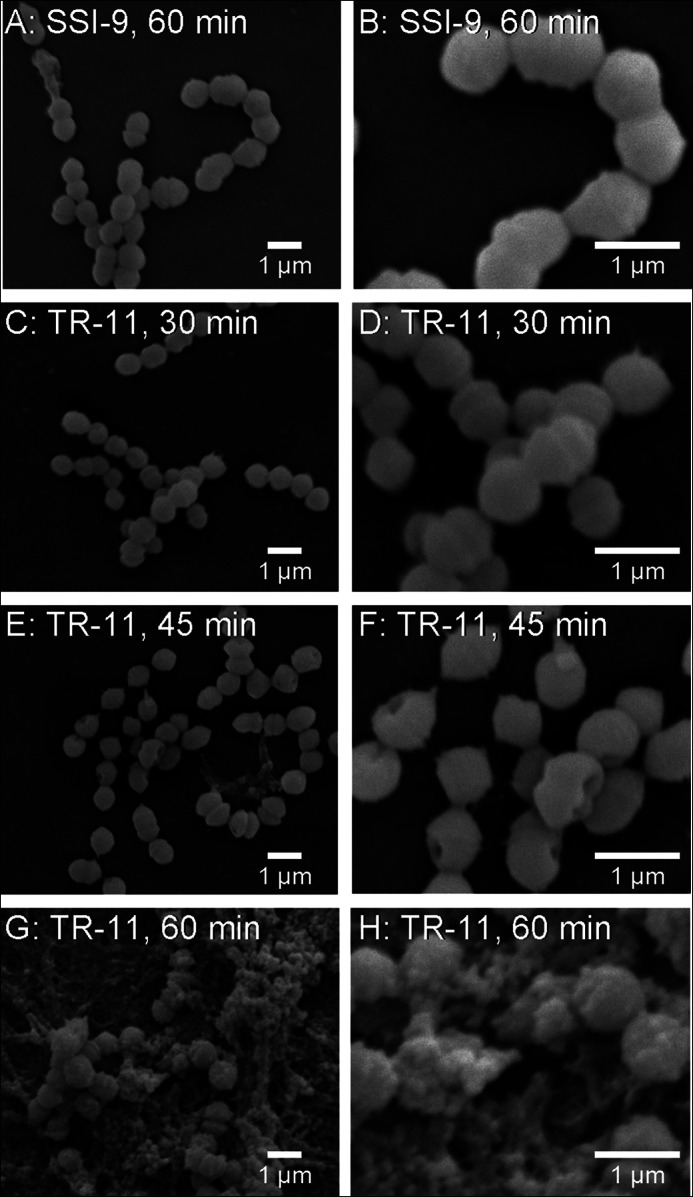
Observation of bacterial surface architecture by SEM. Cultures of GAS strains SSI-9 and TR-11 (ΔspeB) at the mid-log phase were centrifuged and washed with PBS and then incubated with human serum at 37 °C. A and B, SSI-9 after 60 min of incubation. C and D, TR-11 after 30 min of incubation. E and F, TR-11 after 45 min of incubation. G and H, TR-11 after 60 min of incubation. The samples were fixed on glass and observed using an SEM at lower (A, C, E, and G) and higher (B, D, F, and H) magnifications.
SpeB Contributes to S. pyogenes Resistance to Human Complement System
The C1-INH is considered to affect the initial step of the classical complement pathway, whereas it is also possible that a reduction in or deficiency of the inhibitor has a significant effect on S. pyogenes eradication. We examined the survival rate of S. pyogenes organisms in human normal serum or C1-INH-depleted serum. As shown in Fig. 5, both the wild-type and the ΔspeB mutant strains had significantly increased bacterial counts in human serum as compared with the control (p < 0.01), whereas SSI-9 showed a significantly higher growth rate than TR-11 (p < 0.01). In addition, TR-11 showed a significantly greater rate of survival in serum without the C1-INH than in whole serum or in serum supplemented with the C1-INH (p < 0.01 for both cases), whereas there was no significant difference regarding the survival rate of TR-11 between serum and C1-INH-supplemented serum. Furthermore, no significant difference was found for the bacterial survival of strain SSI-9 between serum supplemented with the C1-INH and serum with the C1-INH removed, whereas SSI-9 grown in normal human serum had a higher count as compared with both of those conditions. Next, we tested the effect of complement on the survival of SSI-9 and TR-11 in human serum. Strain SSI-9 showed a higher survival rate in normal serum as compared with inactivated serum (p < 0.01), whereas TR-11 showed a lower survival rate in normal serum than in inactivated serum (p < 0.01). These results suggests that other complement factors besides C1-INH associate with SpeB.
FIGURE 5.
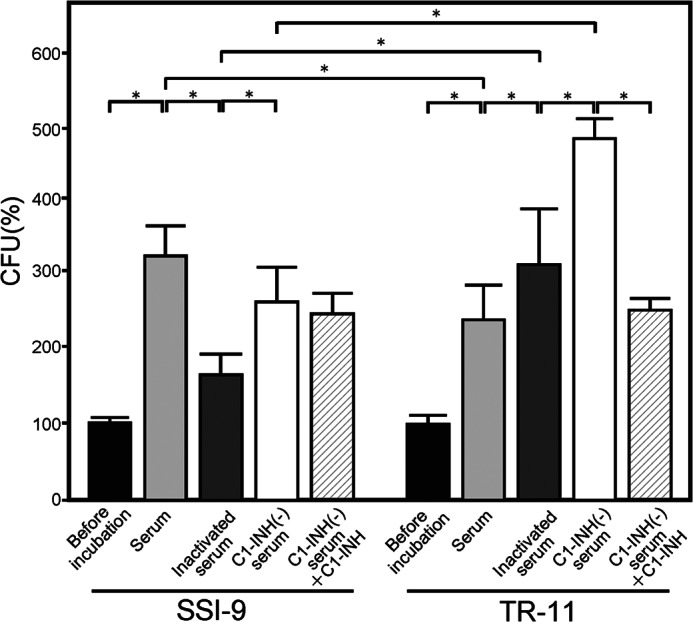
Survival of SSI-9 and TR-11 strains in human serum, inactivated human serum, C1-INH-depleted serum, and C1-INH-depleted serum supplemented with C1-INH. Cultures of GAS strains SSI-9 and TR-11 (ΔspeB) at the mid-log phase were centrifuged and resuspended with PBS to adjust 7.5 × 106-3.8 × 107 cfu/ml to 2,000–10,000 cfu/ml and then incubated with human, inactivated serum C1-INH-depleted serum or C1-INH-depleted serum supplemented with the C1-INH or for 3 h at 37 °C. The strains were grown and quantified after plating on THY agar plates. Representative results from six independent experiments are shown. The CFU of each strain before incubation with serum was set to 100% as compared with the survival rate after incubation with serum. Black, light gray, dark gray, white, and diagonal line bars indicate mean values, and error bars indicate the S.D. of repeated reactions (n = 6). *, p < 0.01, Mann-Whitney U test.
SpeB Degrades Multiple Complement Factors Including C1 Inhibitor
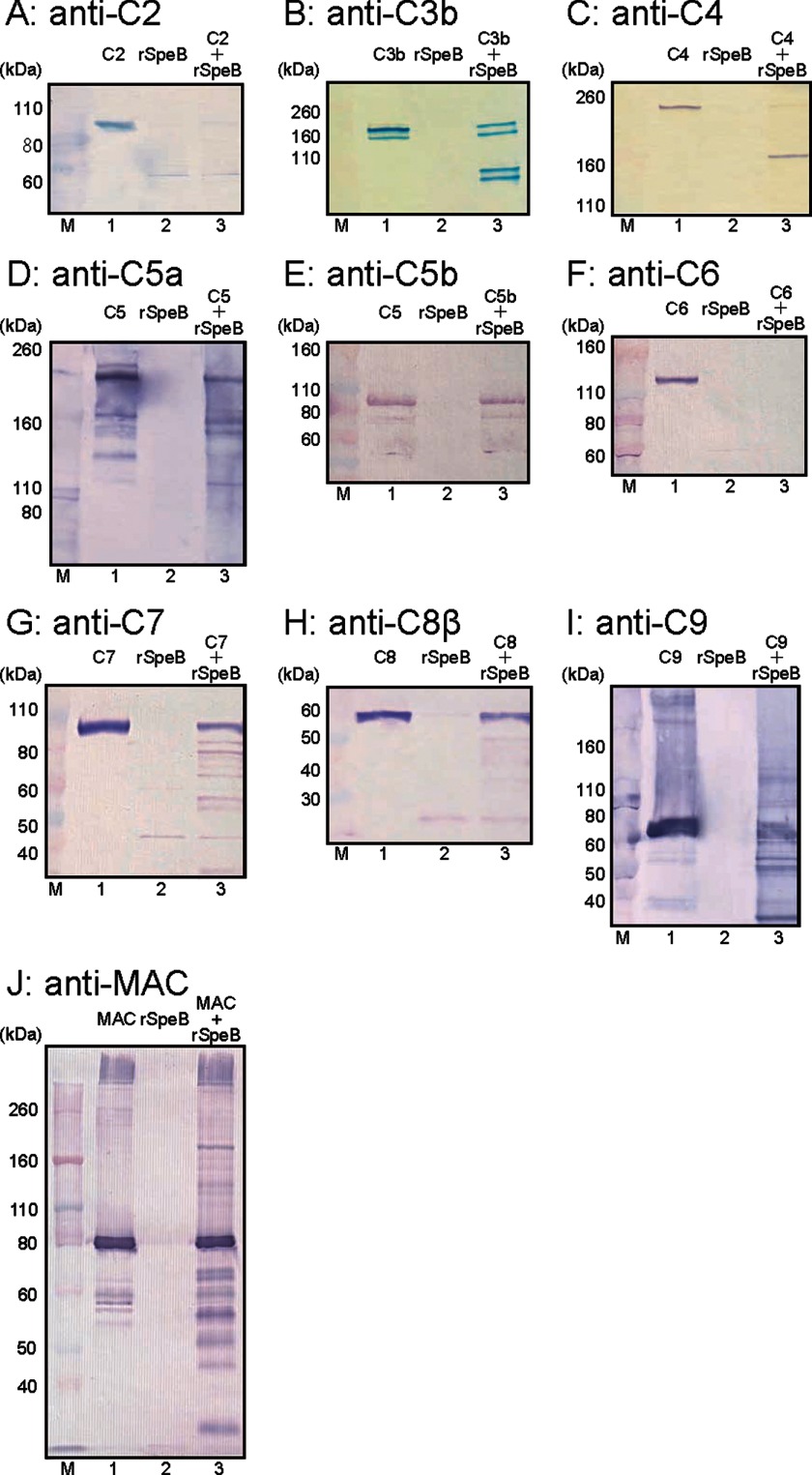
Next, to investigate whether SpeB directly interferes with other complement factors, we examined its cleavage activities with the complement factors C2, C3b, C4, C5a, C5b, C6, C7, C8, and C9. Western blot analyses showed that C2, C3b, C4, C5a, C6, C7, C8, and C9 were cleaved by 1 μm rSpeB at 1 μm (Fig. 6, A–D and F-I), whereas C5b was not degraded (Fig. 6E). In addition, we investigated the cleavage activity of SpeB with the MAC. MAC was detected as an ∼560–1,838-kDa band, whereas C9 was detected as an ∼71-kDa band. A clear blot band considered to be C9 was shown at ∼80 kDa in the control, as well as the MAC and SpeB mixture (Fig. 6J, lane 1 and 3), whereas a large molecular band of at least 260 kDa was considered to correspond to the MAC (Fig. 6J, lane 1 and 3). Small molecule bands of ∼80 kDa were only observed in MAC sample treated with rSpeB (Fig. 6J, lane 3). These findings suggested that SpeB cleaved a partial site in the MAC.
FIGURE 6.
SpeB cleavage activities with various complement factors. rSpeB was incubated with the recombinant complement factors C2 (A), C3b (B), C4 (C), C5 (D), C5b (E), C6 (F), C7 (G), C8 (H), C9 (I), and the formed MAC (J) for 3 h at 37 °C. Proteolysis of each factor was examined by Western blotting. M, molecular mass markers. Lane 1, each recombinant complement was incubated with PBS as a positive control. Lane 2, rSpeB as a negative control. Lane 3, each complement was incubated with rSpeB for 3 h at 37 °C.
SpeB Enables S. pyogenes to Enhance the Binding to MAC in Serum
It has been speculated that the virulence of S. pyogenes such as SpeB induces an inhibitory effect on the MAC in the late complement pathway. However, no clear correlation between S. pyogenes and the MAC or other complement factors has been shown. In addition, it is thought that C9 forms a complex that causes pore development on the bacterial surface in the late complement pathway. Therefore, flow cytometric analyses were performed to evaluate the binding of S. pyogenes strains (SSI-9 and TR-11) and complement factors (MAC, C1-INH, and C9). As shown in Fig. 7, the MAC was recognized in greater amounts on both the SSI-9 and the TR-11 surface by the anti-MAC antibody. On the other hand, a higher amount of C9 was found bound to the surface of TR-11 than the surface of SSI-9. The C1-INH did not attach to the surface of either strain.
FIGURE 7.
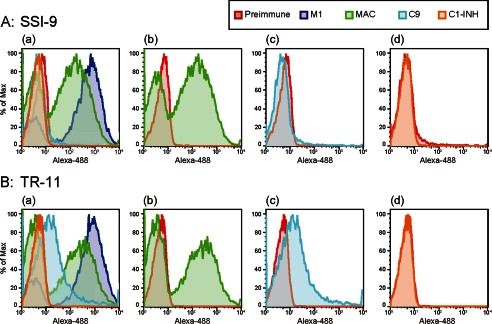
Evaluation of binding ability of complement factors to bacterial surface using flow cytometry. The bacterial strains SSI-9 and TR-11 were separately incubated with blocking agent (supplemented with goat serum) and then reacted with anti-MAC rabbit serum, anti-C9 rabbit serum, anti-C1-INH rabbit serum, anti-M1 rabbit serum (positive control), or nonimmunized rabbit serum (negative control). After washing with PBS, the samples were incubated with Alexa Fluor 488 (Alexa-488)-labeled anti-rabbit IgG, and then deposition of MAC, C9, and C1-INH on bacterial surfaces was evaluated by flow cytometric analyses. A and B, binding of complement factors to SSI-9 (A) and TR-11 (B). Panel a, merge; panel b, MAC; panel c, C9. Red, blue, green, light blue, and orange lines indicate negative control, M1 protein (used as positive control), MAC, C9, and C1-INH, respectively. % of Max, percentage of maximum.
DISCUSSION
C1-INH plays a key role in the complement system that is regulated by inactivation of C1r, C1s, and MASP2. In 2000, Fronhoffs et al. (4) reported that administration of C1-INH at a high dose in STSS patients as adjunctive therapy resulted in positive effects and improved their survival. According to the authors, a possible mechanism related to those results was attenuation of capillary leak syndrome by early inactivation of the complement and contact systems. In addition, several histopathological studies have demonstrated that there are few inflammatory cells at the site of infection in patients with STSS (10, 24, 25). Together, these studies suggest that S. pyogenes inactivates the innate immune system, including complement molecules, although the precise mechanism remains unknown. Thus, we focused on streptococcal inhibition of the complement system in the present study.
It is known that S. pyogenes can evade and manage all aspects of human innate and adaptive immune responses to colonize and infect its human host (10, 11, 26–28). One of the novel major cysteine proteases of S. pyogenes, SpeB, has been reported to contribute to that adaptation in a variety of ways, such as degradation of immunoglobulins (26) and chemokines (27, 28). It is also known that SpeB can cleave the key complement component of the classical and alternative pathway C3b (10), as well as the complement regulator properdin (11). The present study is the first to show that SpeB has cleaving activity toward C1-INH, C2, C4, C5a, C5b, C6, C7, C8, and C9. It is considered that S. pyogenes escapes from the human complement system by the cleaving activity of SpeB.
Strains that show higher ability to degrade C1-INH are likely to produce a large amount of SpeB. On the other hand, some strains, such as SSI-1, SSI-7, and SSI-8, demonstrated C1-INH-degrading activity without adequate secretion of SpeB, as shown in Fig. 2C. Thus, other factors may be involved in the degradation of C1-INH by those strains. In previous studies, S. pyogenes was found to possess the cysteine protease IdeS (29, 30). Further investigations are needed to examine cysteine proteases that possess C1-INH-degrading activities. At the same time, we recognize the accuracy limitations of the semiquantification method used in our study.
The present findings revealed that the C1-INH was cleaved by SpeB. As shown in Fig. 3D, C1-INH is composed of three domains: a signal sequence (Met1 to Ser22), an N-terminal domain (Asp23 to His136), and a serpin (Ser137 to Ala500) domain. According to the results of Edman degradation analyses, C1-INH was cleaved by SpeB at two sites, as shown in Fig. 3C. Bos et al. (23) reported that two disulfide bonds maintain the conformation of C1-INH, whereas a mutation lacking both disulfide bonds conformed to the multimer and lost that activity. In our study, the SpeB cleavage site was shown to be one of those disulfide bonds. In addition, that cleavage site was included in the amino acid sequence missing in a heredity angioedema type II patient (23). Together, these results suggest that cleavage of C1-INH causes structural change and dysfunction to the protein. C1-INH administration to STSS patients may supplement intact C1-INH, which could precede reactivation of the complement system.
Western blot analyses revealed that rSpeB degraded purified C2, C4, C5a, C6, C7, C8, and C9, of which C2 and C4 are known to contribute to suppression of both the classical and the lectin pathways. Sim et al. (31) reported that target recognition by the classical and lectin pathways leads to activation of serine proteases, which cleave the complement components C2 and C4, leading to formation of the protease complex C4b2a, which cleaves C3 into C3a and C3b. The complement pathway maintains the balance of activation because the amount of each complement factor is regulated by complement control proteins. When the balance is disrupted, complement activation will be enhanced, resulting in excessive consumption of complement components and breaking down of the complement system. Our results suggest that S. pyogenes escapes from human complement killing through disrupting the balance of the amount of complement factors by SpeB (Fig. 6). If the complement system was overactivated, the complement pathway would not work homeostatically in the immune system. In addition, if C1-INH, C2, C3b, and C4 were degraded or C2, C3b and C4 were overused by degradation of C1-INH, MAC would not be formed, and the immune system would not work.
There was a significantly higher number of TR-11 organisms that survived in C1-INH-depleted serum as compared with C1-INH-supplemented serum (Fig. 5). Previous studies have shown that C1-INH is important to maintain the balance of complement components (28, 32). Thus, serum without C1-INH is more favorable for survival of the TR-11 strain as compared with whole or C1-INH-supplemented serum. As for SSI-9, there was no significant difference between survival rates in C1-INH-depleted and -supplemented serum. These results indicate that SpeB allows S. pyogenes to survive with an advantage in serum containing C1-INH.
Our results demonstrated that SpeB cleaved some of the late complement factors (C6, C7, C8, and C9), whereas C5b was not cleaved. In addition, Western blotting showed that SpeB degraded assembled MAC. It has been shown that the MAC forms pores only on the surface of Gram-negative bacteria. The bacterial surface of the TR-11 strain was seen as rough and irregular, which might be interpreted as pore formation by MAC activity in SEM observation analyses. Further detailed investigation is required to elucidate this issue, although our findings imply an interaction by the MAC with Gram-positive bacteria.
As shown in Fig. 5, the survival rate of SSI-9 in human serum was significantly higher than that of TR-11 in the same condition, indicating that TR-11 survived better in inactivated serum than in human serum. From those results, we speculated that complement factors in serum with SSI-9 are degraded, thus inhibiting complement activation in the immune system. On the other hand, TR-11 could evade the complement system only in inactivated serum and not in normal serum. S. pyogenes SpeB was previously reported to degrade immunoglobulins (26, 33, 34); thus, the survival rates of these strains may have been influenced by that effect.
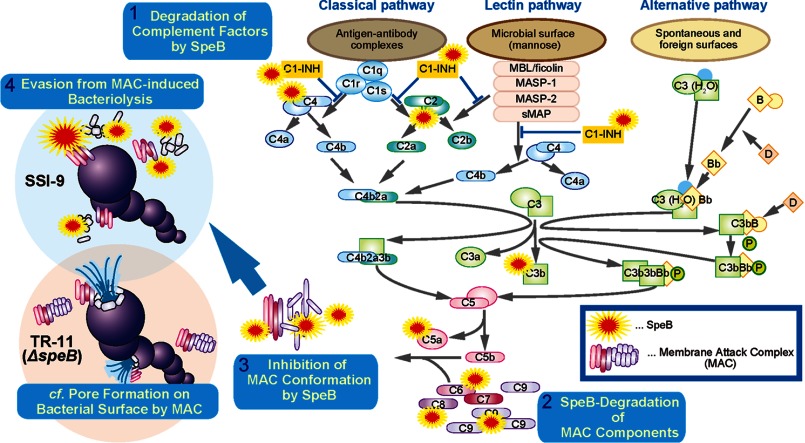
In flow cytometric analyses, the MAC was detected as a clear peak on the surface of both strains. On the other hand, C9 was more likely to bind to the surface of TR-11 than SSI-9. In the present study, the MAC was attached to the surface of SSI-9 with an imperfect form (lack of C9), whereas that attached to the TR-11 surface had pore-forming activity, resulting in a lower survival rate for the same reason. Taken together, we concluded that SpeB has the ability to interrupt the human complement system via degrading C1-INH, thus enabling S. pyogenes to evade eradication in a hostile environment, as shown in Fig. 8.
FIGURE 8.
Schematic diagram of the complement inhibitory system by SpeB. SpeB degrades various complement factors including C1-INH, and thus, it would disrupt the balance of activation in the complement pathway. Furthermore, the MAC complex might be inactivated by SpeB. In particular, degradation of C9 could inhibit MAC formation. Consequently, SpeB enables S. pyogenes to evade MAC-dependent bacteriolysis. MBL, mannose binding lectin. sMAP, small MBL-associated protein.
This study was supported in part by grants-in-aid for scientific research (B), young scientists (B), and challenging exploratory research through the Japan Society for the Promotion of Science (JSPS).
- STSS
- streptococcal toxic shock syndrome
- C1-INH
- C1-esterase inhibitor
- SpeB
- streptococcal pyogenic exotoxin B
- rSpeB
- recombinant SpeB
- rScpA
- recombinant ScpA
- MAC
- membrane attack complex.
REFERENCES
- 1. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brosnahan A. J., Schlievert P. M. (2011) Gram-positive bacterial superantigen outside-in signaling causes toxic shock syndrome. FEBS J. 278, 4649–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lappin E., Ferguson A. J. (2009) Gram-positive toxic shock syndromes. Lancet Infect. Dis. 9, 281–290 [DOI] [PubMed] [Google Scholar]
- 4. Fronhoffs S., Luyken J., Steuer K., Hansis M., Vetter H., Walger P. (2000) The effect of C1-esterase inhibitor in definite and suspected streptococcal toxic shock syndrome. Report of seven patients. Intensive Care Med. 26, 1566–1570 [DOI] [PubMed] [Google Scholar]
- 5. Pixley R. A., De La Cadena R., Page J. D., Kaufman N., Wyshock E. G., Chang A., Taylor F. B., Jr., Colman R. W. (1993) The contact system contributes to hypotension but not disseminated intravascular coagulation in lethal bacteremia. In vivo use of a monoclonal anti-factor XII antibody to block contact activation in baboons. J. Clin. Invest. 91, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schapira M., Despland E., Scott C. F., Boxer L. A., Colman R. W. (1982) Purified human plasma kallikrein aggregates human blood neutrophils. J. Clin. Invest. 69, 1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Graaf F., Koedam J. A., Bouma B. N. (1983) Inactivation of kallikrein in human plasma. J. Clin. Invest. 71, 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castelli R., Zanichelli A., Cugno M. (2013) Therapeutic options for patients with angioedema due to C1-inhibitor deficiencies: from pathophysiology to the clinic. Immunopharmacol. Immunotoxicol. 35, 181–190 [DOI] [PubMed] [Google Scholar]
- 9. Müller-Eberhard H. J., Schreiber R. D. (1980) Molecular biology and chemistry of the alternative pathway of complement. Adv. Immunol. 29, 1–53 [DOI] [PubMed] [Google Scholar]
- 10. Terao Y., Mori Y., Yamaguchi M., Shimizu Y., Ooe K., Hamada S., Kawabata S. (2008) Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J. Biol. Chem. 283, 6253–6260 [DOI] [PubMed] [Google Scholar]
- 11. Tsao N., Tsai W. H., Lin Y. S., Chuang W. J., Wang C. H., Kuo C. F. (2006) Streptococcal pyrogenic exotoxin B cleaves properdin and inhibits complement-mediated opsonophagocytosis. Biochem. Biophys. Res. Commun. 339, 779–784 [DOI] [PubMed] [Google Scholar]
- 12. Terao Y., Kawabata S., Kunitomo E., Murakami J., Nakagawa I., Hamada S. (2001) Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42, 75–86 [DOI] [PubMed] [Google Scholar]
- 13. Okamoto S., Kawabata S., Terao Y., Fujitaka H., Okuno Y., Hamada S. (2004) The Streptococcus pyogenes capsule is required for adhesion of bacteria to virus-infected alveolar epithelial cells and lethal bacterial-viral superinfection. Infect. Immun. 72, 6068–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Terao Y., Yamaguchi M., Hamada S., Kawabata S. (2006) Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 281, 14215–24223 [DOI] [PubMed] [Google Scholar]
- 15. Kunitomo E., Terao Y., Okamoto S., Rikimaru T., Hamada S., Kawabata S. (2008) Molecular and biological characterization of histidine triad protein in group A streptococci. Microbes Infect. 10, 414–423 [DOI] [PubMed] [Google Scholar]
- 16. Murakami J., Kawabata S., Terao Y., Kikuchi K., Totsuka K., Tamaru A., Katsukawa C., Moriya K., Nakagawa I., Morisaki I., Hamada S. (2002) Distribution of emm genotypes and superantigen genes of Streptococcus pyogenes isolated in Japan, 1994–9. Epidemiol. Infect. 128, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Terao Y., Kawabata S., Nakata M., Nakagawa I., Hamada S. (2002) Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 277, 47428–47435 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi M., Terao Y., Mori Y., Hamada S., Kawabata S. (2008) PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. J. Biol. Chem. 283, 36272–36279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lancefield R. C. (1957) Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J. Exp. Med. 106, 525–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogawa T., Terao Y., Okuni H., Ninomiya K., Sakata H., Ikebe K., Maeda Y., Kawabata S. (2011) Biofilm formation or internalization into epithelial cells enable Streptococcus pyogenes to evade antibiotic eradication in patients with pharyngitis. Microb. Pathog. 51, 58–68 [DOI] [PubMed] [Google Scholar]
- 21. Bohach G. A., Hauser A. R., Schlievert P. M. (1988) Cloning of the gene, speB, for streptococcal pyrogenic exotoxin type B in Escherichia coli. Infect. Immun. 56, 1665–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C. C., Cleary P. P. (1990) Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J. Biol. Chem. 265, 3161–3167 [PubMed] [Google Scholar]
- 23. Bos I. G., Lubbers Y. T., Roem D., Abrahams J. P., Hack C. E., Eldering E. (2003) The functional integrity of the serpin domain of C1-inhibitor depends on the unique N-terminal domain, as revealed by a pathological mutant. J. Biol. Chem. 278, 29463–29470 [DOI] [PubMed] [Google Scholar]
- 24. Cockerill F. R., 3rd, Thompson R. L., Musser J. M., Schlievert P. M., Talbot J., Holley K. E., Harmsen W. S., Ilstrup D. M., Kohner P. C., Kim M. H., Frankfort B., Manahan J. M., Steckelberg J. M., Roberson F., Wilson W. R. (1998) Molecular, serological, and clinical features of 16 consecutive cases of invasive streptococcal disease. Clin. Infect. Dis. 26, 1448–1458 [DOI] [PubMed] [Google Scholar]
- 25. Hidalgo-Grass C., Dan-Goor M., Maly A., Eran Y., Kwinn L. A., Nizet V., Ravins M., Jaffe J., Peyser A., Moses A. E., Hanski E. (2004) Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet 363, 696–703 [DOI] [PubMed] [Google Scholar]
- 26. Collin M., Olsén A. (2001) EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20, 3046–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang D., Chen Q., Hoover D. M., Staley P., Tucker K. D., Lubkowski J., Oppenheim J. J. (2003) Many chemokines including CCL20/MIP-3α display antimicrobial activity. J. Leukoc. Biol. 74, 448–455 [DOI] [PubMed] [Google Scholar]
- 28. Egesten A., Olin A. I., Linge H. M., Yadav M., Mörgelin M., Karlsson A., Collin M. (2009) SpeB of Streptococcus pyogenes differentially modulates antibacterial and receptor activating properties of human chemokines. PLoS One 4, e4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fiedler T., Sugareva V., Patenge N., Kreikemeyer B. (2010) Insights into Streptococcus pyogenes pathogenesis from transcriptome studies. Future Microbiol. 5, 1675–1694 [DOI] [PubMed] [Google Scholar]
- 30. von Pawel-Rammingen U., Björck L. (2003) IdeS and SpeB: immunoglobulin-degrading cysteine proteinases of Streptococcus pyogenes. Curr. Opin. Microbiol. 6, 50–55 [DOI] [PubMed] [Google Scholar]
- 31. Sim R. B., Tsiftsoglou S. A. (2004) Proteases of the complement system. Biochem. Soc. Trans. 32, 21–27 [DOI] [PubMed] [Google Scholar]
- 32. Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Complement activation and inhibition: a delicate balance. Trends Immunol. 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 33. Collin M., Olsén A. (2001) Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 69, 7187–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eriksson A., Norgren M. (2003) Cleavage of antigen-bound immunoglobulin G by SpeB contributes to streptococcal persistence in opsonizing blood. Infect. Immun. 71, 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]