Background: The methylation status of histone tails is a balance between methylation and demethylation.
Results: Nitric oxide inhibits lysine demethylase 3A and alters cellular histone methylation patterns.
Conclusion: Nitric oxide can significantly modify the epigenetic landscape.
Significance: These results establish nitric oxide as a physiological epigenetic regulator acting through a nonclassical cell signaling mechanism.
Keywords: Cell Biology, Epigenetics, Histones, Iron, Nitric Oxide, Demethylases, Methyltransferases
Abstract
Methylation of lysine residues on histone tails is an important epigenetic modification that is dynamically regulated through the combined effects of methyltransferases and demethylases. The Jumonji C domain Fe(II) α-ketoglutarate family of proteins performs the majority of histone demethylation. We demonstrate that nitric oxide (•NO) directly inhibits the activity of the demethylase KDM3A by forming a nitrosyliron complex in the catalytic pocket. Exposing cells to either chemical or cellular sources of •NO resulted in a significant increase in dimethyl Lys-9 on histone 3 (H3K9me2), the preferred substrate for KDM3A. G9a, the primary methyltransferase acting on H3K9me2, was down-regulated in response to •NO, and changes in methylation state could not be accounted for by methylation in general. Furthermore, cellular iron sequestration via dinitrosyliron complex formation correlated with increased methylation. The mRNA of several histone demethylases and methyltransferases was also differentially regulated in response to •NO. Taken together, these data reveal three novel and distinct mechanisms whereby •NO can affect histone methylation as follows: direct inhibition of Jumonji C demethylase activity, reduction in iron cofactor availability, and regulation of expression of methyl-modifying enzymes. This model of •NO as an epigenetic modulator provides a novel explanation for nonclassical gene regulation by •NO.
Introduction
The accumulation of whole genomic sequence from thousands of individuals has made it clear that epigenetic regulation is as important as nucleotide sequence for determining final phenotypic outcomes (1, 2). For the past 50 years, major advances in our understanding of gene expression have focused on the central dogma of molecular biology (DNA to RNA to protein). More recently, however, the emphasis has shifted toward investigating the importance of epigenetics on phenotypic outcomes. One example is post-translational modifications of histones that have major influences on chromatin structure and gene transcription (3). The most studied of these modifications is acetylation, but it is now recognized that methylation, phosphorylation, ubiquitination, and SUMOylation all have regulatory functions (4, 5). Methylation of lysine residues, once thought to be an irreversible process, has gained prominence over the past decade as one of the most important histone modifications involved in genetic regulation (6). This is due, in part, to the discovery of two families of histone demethylases that catalyze the removal of methyl groups from arginine and lysine residues. The first family is composed of the lysine-specific demethylases LSD1 and LSD2, which are FAD-dependent amine oxidases (7, 8). The other type belongs to a much larger family of the Jumonji C (JMJC)2 domain-containing demethylases. This family is composed of 30 proteins, two-thirds of which target specific Lys residues on the N-terminal tail of histone 3 (H3) (9–18).
The JMJC class of lysine-specific demethylases (KDMs) are mononuclear Fe(II)-dependent dioxygenases that use α-ketoglutarate (α-KG) and oxygen (O2) as cosubstrates to oxidatively demethylate specific Lys residues (11). The common structural motif of this protein family is a 2-histidine-1-carboxylate facial triad that coordinates a non-heme Fe(II) at the catalytic center (19, 20). Although general catalytic mechanisms for these enzymes have been proposed, the details are an ongoing area of research and believed to be enzyme-specific (9). Methylation status of lysine residues on histones can positively or negatively influence gene transcription. These lysine residues can be mono-, di-, or tri-methylated, and the functional consequences depend upon both the location and degree of this methylation. The primary targets on H3 are lysines 4, 9, 27, 36, and 79 with 4 and 9 being the most heavily modified. Methylation at Lys-4 is generally associated with transcriptional activation, although Lys-9 methylation is typically found in silenced chromatin. H4 is also commonly methylated, especially at Lys-20. Methylation is carried out by lysine methyltransferases (KMTs), and methylation status is therefore a dynamic process that represents a balance of KMT and KDM activities. This resulting balance can have both acute and long term impacts on chromatin structure and mRNA expression (6, 9, 21–23).
Nitric oxide (•NO, nitrogen monoxide) is a biological free radical signaling molecule that regulates many physiological processes, including angiogenesis, smooth muscle tone, immune response, apoptosis, and synaptic communication (24). Like iron chelators and divalent metals (Co2+ and Ni2+), •NO has been shown to inhibit mononuclear non-heme iron oxygenases that contain the 2-His-1-carboxylate facial triad structural motif. One of the most studied examples of this interaction occurs with the HIF prolyl hydroxylase (25–27). Its activity is inhibited by Co2+ and Ni2+ by replacing the iron at the catalytic site (28). •NO and iron chelators have also been shown to inhibit prolyl hydroxylase activity by either binding to or removing this catalytic iron (29). Previously, it has been shown that the JMJC domain-containing histone demethylase KDM3A is highly sensitive to inhibition by carcinogenic nickel ions (28, 30, 31). Others have demonstrated that various dioxygenase inhibitors such as α-KG analogs, hypoxia, •NO, reactive oxygen species, CoCl2, and iron chelators like desferrioxamine (DFO) all increase histone methylation, most probably by reducing demethylase activity (32–37).
The common structural similarities between JMJC demethylases and other mononuclear non-heme iron oxygenases spurred us to investigate whether •NO might similarly inhibit this class of demethylases resulting in alterations in histone methylation patterns. We chose to focus on KDM3A as it is the predominant demethylase acting on the di- and monomethylated states of the heavily modified H3K9 residue (12). We demonstrate that •NO directly inhibits KDM3A leading to the accumulation of the H3K9me2. The contribution of methyltransferases to these increases was ruled out. Our studies are the first to provide direct mechanistic evidence for epigenetic regulation by •NO through histone modifications. This is an important finding as it is well known that the multitude of gene expression changes caused by •NO cannot be solely explained by classical •NO-heme interactions or S-nitrosation of protein thiols. Further investigations into •NO-driven changes in histone methylation status at specific promoter sites will provide a model for examining many of the unexplained changes in •NO-associated gene expression.
EXPERIMENTAL PROCEDURES
Chemicals
(Z)-1-[N-(2-Aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate (DETA/NO), (Z)-1-[N-[3-aminopropyl]-N-[4-(3-aminopropylammonio)butyl]-amino]diazen-1-ium-1,2-diolate (Sper/NO), and 1-(hydroxy-NNO-azoxy)-l-proline (Proli/NO) were the generous gifts from Dr. Joseph Hrabie (NCI, National Institutes of Health). Iron(II) sulfate heptahydrate (FeSO4), α-ketoglutaric acid sodium salt, ferric ammonium citrate, DFO, aminoguanidine hydrochloride, lipopolysaccharides (LPS), and 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one (ODQ) were purchased from Sigma. All cell culture reagents were purchased from Invitrogen with the exception of methionine-free DMEM (AthenaES).
Cell Culture
MDA-MB-231 human breast carcinoma cells and RAW 264.7 murine macrophages were cultured in DMEM, and Jurkat T lymphocyte cells were grown in RPMI 1640 medium. In all cases, media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Additionally, media for Jurkat T cells contained 50 μm 2-mercaptoethanol. Cells were grown to 80% confluence, and serum-starved overnight prior to treatment. Cells were incubated in serum-free, methionine-free DMEM for 4 h before addition of DETA/NO wherever necessary. For coculture experiments, RAW 264.7 cells were activated with LPS (1 μg/ml) for 6 h to induce inducible nitric oxide (iNOS) expression. Subsequently, media were aspirated, and Jurkat T cells were counted and added in serum-free media in the presence or absence of 2 mm aminoguanidine. For iron supplementation experiments, cells were treated with ferric ammonium citrate (150 μg/ml) for 16 h and washed with PBS to remove excess iron prior to treatment.
Western Blotting
Histones were extracted as described previously (37). Whole cell lysates were collected using CelLyticTM M cell lysis reagent (Sigma) supplemented with 1% protease inhibitor mixture (Calbiochem) and 1 mm PMSF (Sigma). Protein was transferred to PVDF membranes using the iBlot transfer system (Invitrogen). The membranes were blocked and incubated overnight with suitable primary antibodies as follows: KDM3A (Abcam), H3K9me1 (Abcam), H3K9me2/me3 (Cell Signaling), G9a (Cell Signaling), or β-actin (Cell Signaling). The blots were analyzed in a Fluor Chem HD2 imager (Alpha Innotech) using SuperSignal West Femto maximal sensitivity substrate (Thermo Scientific). Thereafter, membranes were stained with Coomassie Blue to assess histone loading.
Quantitative RT-PCR
RNA was extracted using RNAqueous 4PCR kit (Ambion) and reverse-transcribed using the Transcriptor First Strand cDNA synthesis kit (Roche Applied Science). PCR was performed using the Fast SYBR® Green master mix (Applied Biosystems). Standard manufacturer's protocol was followed during each stage. All PCRs were performed on the Step One Plus real time PCR system (ABI). The oligonucleotide primer sequences are as follows: KDM1 forward, 5′-GCCATGGTGGTAACAGGTCT-3′ and reverse, 5′-TGGCCAGTTCCATATTTACTTG-3′; KDM3A forward, 5′-CCAGCCTCAAAGGAAGACCT-3′ and reverse, 5′-ACTGCACCAAGAGTCGGTTT-3′; KDM3B forward, 5′-CATCTGCACAAGTGTCGTGA-3′ and reverse, 5′-CCCTTTTCGAGTGAAGATCAA-3′; KDM4A forward, 5′-GCCGCTAGAAGTTTCAGTGAG-3′ and reverse, 5′-GCGTCCCTTGGACTTCTTATT-3′; KDM4B forward, 5′-AAGTCCTGGTACGCCATCC-3′ and reverse, 5′-TCATCTTATGCCGCAGGAA-3′; KDM4C forward, 5′-AGGCGCCAAGTGATGAAG-3′ and reverse, 5′-GAGAGGTTTCGCCCAAGACT-3′; KDM4D forward, 5′-GGACAAGCCTGTACCACTGAG-3′ and reverse, 5′-CTGCACCCAGAAGCCTTG-3′; PHF8 forward, 5′-GCGTGAGGAGGTTCCTGTC-3′ and reverse, 5′-GCGTCCTCTCTGGACGATAG-3′; KDM7A forward, 5′-GAATTACGCTCTCGAGTCTTCC-3′ and reverse, 5′-CATGTTTCTCCAGATATCTTTGTGTC-3′; G9a forward, 5′-AAGGTCCACCGAGCCCGCAA-3′ and reverse 5′-CCGCTTCTCAGGGACCGGGG-3′; GLP forward, 5′-GGCTGGACACGTGGACATCTG-3′ and reverse, 5′-GCTTCCATCAACGGGGTCCTCT-3′; SETDB1 forward, 5′-GCAAGGACAGCTCAGGACACG-3′ and reverse, 5′-CAGGGCCTTTGCTCCTCACAGC-3′; SETDB2 forward, 5′-CTGTGACTGCTCTGAGGGCTGC-3′ and reverse, 5′-GGGGGAAGTTTTGGCATTCCTTGCT-3′; SUV39h1 forward, 5′-GGCCCGAATGTCGTTAGCCGT-3′ and reverse, 5′-GGCACAGGTCCTGCAGCTGATT-3′; SUV39h2 forward, 5′-CCGCGCCAGTTTGAATGAAAGC-3′ and reverse, 5′-AACTAGGCAAGGCACACACCAAG-3′; PRDM2 forward, 5′-GCTGACTTGAGTGAGAACAAGAGA-3′ and reverse, 5′-AGCGGAGGCTGTAGCTGA-3′; β-actin forward, 5′-CCAACCGCGAGAAGATGA-3′ and reverse 5′-CCAGAGGCGTACAGGGATAG-3′.
KDM3A Activity Assay
Recombinant KDM3A was purchased from BPS Biosciences. 10-μl reactions were conducted at 25 °C in Tris, pH 8.0, containing 80 μm FeSO4, 1 mm α-ketoglutarate, 220 μm O2, 2 mm ascorbate, 40 ng of recombinant KDM3A protein, and 125 nm histone 3 peptide fragment containing a dimethyl lysine 9 (Anaspec). Positive ion MALDI-TOF mass spectra of peptide reactions were acquired using a Voyager DE Pro mass spectrometer (Applied Biosystems, Foster, CA). Preceding the mass spectrometric analysis, each sample was processed with a C18-ZipTip (Millipore, Cambridge, MA) cleanup. A 1-μl aliquot of the desalted peptide solution was mixed with 1 μl of matrix solution composed of a saturated α-cyano-4-hydroxycinnamic acid solution made up in acetonitrile/water (1:1, v/v) acidified with 0.1% (v/v) trifluoroacetic acid. A 1-μl aliquot of the mixture was then spotted on a MALDI-TOF sample stage and air-dried prior to analysis. For each sample, mass spectra from 500 laser shots were acquired in linear mode and signal averaged over the range m/z 500 to 5000.
Electron Paramagnetic Resonance (EPR)
Measurements were performed on a Bruker X-band EMX Plus EPR spectrometer. Samples were frozen and read in liquid N2. Dinitrosyliron complexes (DNIC) were detected at g = 2.03. Settings were as follows: centerfield 3335.25 G, microwave frequency 9.460544 GHz, modulation amplitude 10G, 200 G scan range, 90-s scan time, one scan. For quantification, the double integral of the first derivative spectra was compared with a standard curve generated with synthetic diglutathione DNIC as described previously (38). The CIP was estimated by treating cells with 1 mm DFO for 4 h. The resulting Fe3+-DFO g = 4.3 signal was read with the following settings: center field 1575.65 G, microwave frequency 9.449528 GHz, modulation amplitude of 10 G, 200 G scan range, 30-s scan time, four scans. For quantification, the double integral of the first derivative spectra was compared with that of a standard curve generated with Fe3+-DFO as described previously (38). Settings for measurement of the recombinant KDM3A·•NO complex were as follows: centerfield 3328.15 G, microwave frequency 9.440474 GHz, modulation amplitude 7.5 G, 1000 G scan range, 60-s scan time, 16 scans.
Quantification of NO3−/NO2−, S-Nitrosothiols, and •NO
Quantification was performed by chemiluminescence using the Sievers Nitric Oxide Analyzer 280i. For coculture experiments, 25-μl media aliquots were drawn and injected into a reaction chamber containing vanadium chloride in HCl to determine total NO3−/NO2− concentrations as described previously (39). S-Nitrosothiols were measured as described previously (40, 41). For determining steady-state •NO concentrations, [•NO]ss, 25-μl aliquots were injected with a Hamilton air-tight syringe into a reaction vessel containing 5 m NaOH.
Statistics
Significance was determined using either t test or one-way analysis of variance with Bonferoni post hoc analysis. Error bars indicate S.E.
RESULTS
Nitric Oxide Inhibits KDM3A Activity in Vitro
KDM3A is the predominant demethylase acting upon lysine 9 of histone 3, and the mono- and dimethyl modifications of this residue (H3K9me1 and H3K9me2, respectively) are its preferred substrates. Hypoxia and inducers of “chemical hypoxia,” including nickel (Ni2+), cobalt (Co2+), as well as dioxygenase inhibitors such as N-(methoxyoxoacetyl)glycine methyl ester, are recognized as strong inhibitors of JMJC demethylase activity (28, 30–32, 34, 36). Because•NO is known to inhibit structurally similar mononuclear non-heme iron oxygenases containing the 2-His-1-carboxylate facial triad structural motif, we tested •NO's ability to inhibit KDM3A in vitro. We incubated a histone 3 peptide fragment containing a dimethylated lysine 9 (Mr 2284.1, Fig. 1A) with KDM3A and observed its near complete conversion to the unmethylated state within 60 min (Mr 2256.0, Fig. 1B). When •NO was present (100 μm Sper/NO; [•NO]ss = 1.4 ± 0.8 μm) during identical reaction conditions, the peptide product yields were dramatically different (Fig. 1C). Lesser amounts of the unmethylated peptide were detected, and significant amounts of the monomethyl and dimethyl forms were still present.
FIGURE 1.
Nitric oxide inhibits KDM3A activity. In vitro demethylase reactions were conducted at 25 °C in 10 μl of Tris, pH 8.0, containing 80 μm FeSO4, 1 mm α-ketoglutarate, 220 μm O2, 2 mm ascorbate, 40 ng of recombinant KDM3A protein, and 125 nm histone 3 peptide fragment containing a dimethyl lysine 9 (H3K9me2peptide). Representative MS-MALDI spectra are shown of H3K9me2peptide fragment alone (A), H3K9me2peptide + KDM3A at 60 min (B), H3K9me2peptide + KDM3A + •NO (100 μm Sper/NO; [•NO]ss = 1.4 ± 0.8 μm) at 60 min (C), and H3K9me2peptide + KDM3A + •NO (10 μm Proli/NO; [•NO] = 20 μm bolus) at 60 min (D). E, time and •NO concentration effects on KDM3A activity. Sper/NO was added at 25, 50, 100, and 200 μm to the reaction. H3K9me0 and H3K9me1 product formation was then measured and quantified at the indicated time points. These concentrations of Sper/NO gave a range of initial steady-state •NO concentrations of ≈0.2–2.0 μm. n = 3.
Although •NO inhibited KDM3A enzymatic activity, one possibility is that •NO is simply inducing nonspecific oxidative or nitrosative damage to the protein. To investigate this, we repeated the reaction in the presence of a relatively high amount of •NO, ≈20 μm, given as a bolus (10 μm Proli/NO). This amount of •NO was approximately the same as the total amount used in Fig. 1C, but the exposure time was dramatically shorter (5 s versus 60 min). No inhibition of KDM3A was observed (Fig. 1D). Because the total amount of •NO was similar but the exposure time was different, we took this to indicate that the inhibitory effects of •NO on KDM3A activity are reversible, they require sustained •NO exposure, and that critical protein modifications or destruction of the enzyme most likely do not explain our results. Moreover, in parallel experiments of KDM3A treated with •NO, we could not detect any protein S-nitrosothiols by chemiluminescence, further supporting Fe(II)·•NO complex formation as the mechanism of inhibition (data not shown).
To determine temporal and concentration effects of •NO on enzyme activity, we measured the demethylation products of KDM3A at four different •NO concentrations over time (0–120 min) (Fig. 1E). In solution, •NO reacts with O2 (autooxidation), and O2 is a substrate for the enzyme. Therefore, to eliminate the confounding effect of substrate loss due to O2 scavenging by •NO, we chose concentrations of •NO that would decrease the O2 concentration by no more 10% through the course of the experiment. Under these conditions, even the lowest steady-state concentration of •NO (≈ 200 nm; 25 μm Sper/NO) was able to inhibit product formation by as much as 40%, and ≈2 μm [•NO]ss (200 μm Sper/NO) inhibited this by ≈70%. These experiments were conducted at ambient O2 concentrations (≈220 μm). Although direct comparisons between steady-state •NO concentrations and dissolved O2 concentrations are inexact, we approximate that the •NO to O2 ratios may be as high as 1:1000 under these conditions. Although further experiments will be needed to confirm this, our data suggest a higher enzyme affinity for •NO than for O2. These results indicate both concentration- and duration-dependent effects of •NO exposure on enzyme activity.
Nitric Oxide Forms a Histidyl·Iron·Nitrosyl Complex in the Catalytic Pocket of KDM3A
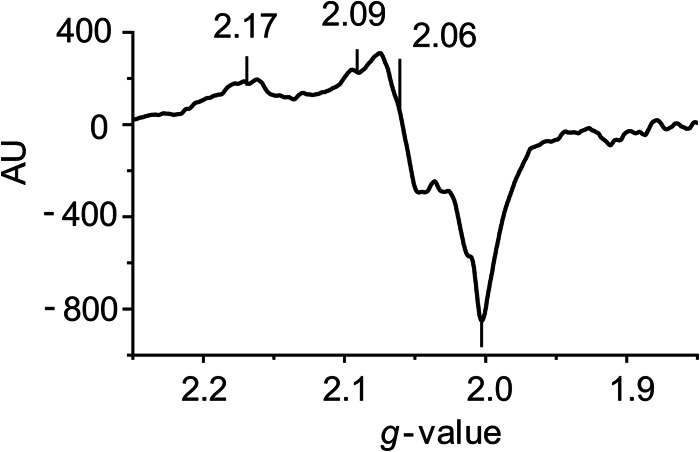
To gain further insight into the mechanism of KDM3A inhibition by •NO, we conducted an EPR analysis of the enzyme. Fig. 2 is an EPR spectrum of KDM3A in the presence of substrates, cofactors, and •NO. Spectra were not observed in the absence of •NO nor was there a signal from •NO combined with all the cofactors in the absence of KDM3A. There appeared to be a superposition of spectra of more than three lines. The major spectrum has g values of 2.17, 2.06, and 2.00 or possibly 2.09, 2.06, and 2.00, with the line at g = 2.17 not assigned. There are weak signals around g = 2.04 assigned to Fe(NO)2(–SR)2 and a line at 1.92 not assigned. Examples of EPR signals assigned to a histidyl·iron·nitrosyl complex have g values of 2.055, 2.033, and 2.015 for the signal from the reaction of ferritin and •NO (42). Furthermore, d7 and d9 forms of histidyl·iron·nitrosyl·aconitase complexes with g values of 2.05 and 2.01 for d7 and 2.032 and 2.004 for d9 were reported (43). g values of 2.07, 2.02, and 1.98 were assigned to a substrate-bound iron-nitrosyl-cysteine dioxygenase site (44). Although the anisotropy in g values is greater than in the examples, an •NO·Fe·KDM3A complex with the 2-His-1-carboxylate facial triad is the most probable source for these spectra under our conditions. The enzymatic inhibition seen in Fig. 1, likely results from the direct binding of •NO to the Fe(II) coordinated by the facial triad.
FIGURE 2.
EPR spectroscopy of •NO in the presence of KDM3A. Recombinant KDM3A protein (25 μg) was incubated at 25 °C in Tris, pH 8.0, in the presence of 80 μm FeSO4, 1 mm α-ketoglutarate, 220 μm O2, 2 mm ascorbate, and •NO (100 μm Sper/NO). The reaction was terminated after 15 min, flash-frozen, and processed for EPR analysis. Representative trace is shown (n = 3).
Physiological •NO Concentrations Alter Histone 3 Lysine 9 Methylation in Cultured Cells
Having established that •NO inhibits KDM3A activity in vitro, we extended these findings into a cellular system to determine whether differential methylation patterns at H3K9 would similarly be observed subsequent to •NO exposure. Initially, we set out to examine changes in H3K9me2 as it is the most abundant of the three methylation states at H3K9 (9), and it is also the primary substrate for KDM3A. We compared differences in methylation patterns between cells exposed to •NO and those exposed to other known inhibitors of protein activity (Ni2+ and DFO). Similar increases in methylation at H3K9me2 were noted for all treatments (Fig. 3A). A 24-h time course of •NO exposure demonstrated that within 8 h increases in H3K9me2 could be observed relative to untreated controls, and at 24 h the degree of methylation was still increasing (1.4-fold, Fig. 3B). Since we observed robust and reproducible responses to •NO at 24 h, this time point was chosen for all subsequent investigations. Next, we investigated concentration-dependent effects of •NO and determined that as little as 100 μm DETA/NO resulted in detectable increases in H3K9me2, but maximal induction was achieved with 500 μm DETA/NO (Fig. 3C). Importantly, these amounts of DETA/NO generated concentrations of •NO well within the physiological range (Fig. 3) (38, 39). Although dimethyl is the principal methyl modification at H3K9, it was also important to measure the relative changes in the mono- and trimethyl states as change in one may be linked to changes in the others. In response to •NO, H3K9me3 was unchanged, whereas H3K9me1 was decreased with respect to untreated controls (Fig. 3D).
FIGURE 3.
Nitric oxide modifies H3K9 methylation patterns in MDA-MB-231 cells. Immunoblots and densitometric quantifications of H3K9 methyl modifications in total histones isolated from cells are shown. A, cells in culture were treated for 24 h with NiSO4 (500 μm), DFO (500 μm), or •NO (500 μm DETA/NO), and changes in H3K9me2 were measured. UT, untreated. B, temporal changes in H3K9me2 were measured after •NO (500 μm DETA/NO) exposure (0–24 h). C, analysis of H3K9me2 after 24 h of exposure to increasing concentrations of •NO (100–1000 μm DETA/NO). D, determination of H3K9 mono-, di-, and trimethylation states after 24 h of treatment with •NO (500 μm DETA/NO). DETA/NO concentrations of 100, 250, 500, and 1000 μm result in [•NO]ss of 16 ± 8, 80 ± 22, 265 ± 21, and 424 ± 42 nm, respectively. All are representative immunoblots of n ≥4. * indicates 0.01<p < 0.05; ** indicates p < 0.01 with respect to untreated controls, which are set arbitrarily to 1.0. AU, arbitrary unit.
Nitric Oxide Increases Demethylase Gene Expression
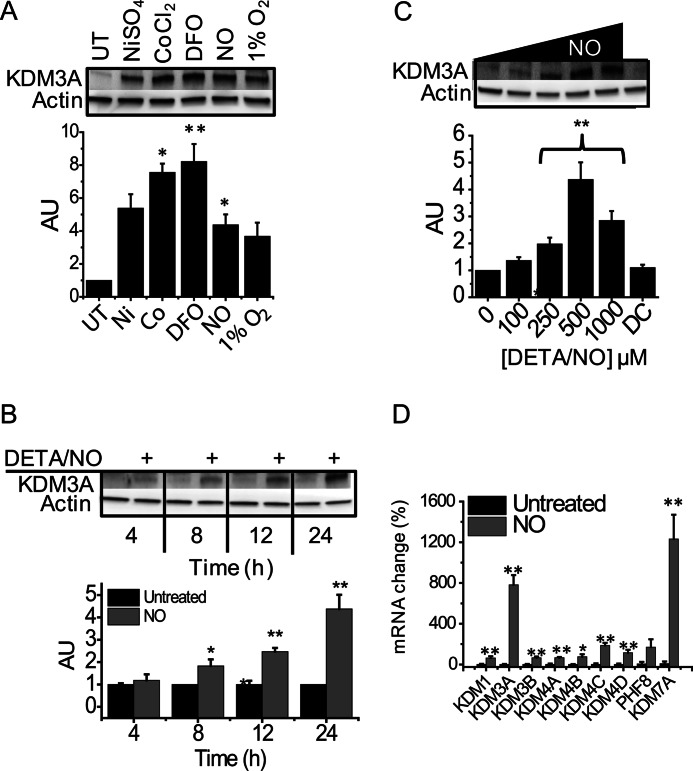
In addition to altering histone methylation patterns, hypoxia and inducers of chemical hypoxia have been shown to strongly induce the expression JMJC domain-containing demethylases. For this reason, we compared the ability of •NO to up-regulate KDM3A to that of Co2+, Ni2+, hypoxia (1% O2, and the metal chelator DFO. Fig. 4A demonstrates that in cells treated with •NO, KDM3A was up-regulated to the same extent as hypoxia, DFO, Co2+, and Ni2+. In response to •NO, KDM3A was up-regulated in both a time- and concentration-dependent manner (Fig. 4, B and C). Interestingly, these responses did not translate into decreases in H3K9me2 levels. In addition to KDM3A, we also examined the steady-state mRNA levels of eight other demethylases that are known to be present in this cell type. Of the nine demethylases, KDM3A and KDM7A were strongly up-regulated in response to 24 h of •NO exposure, and there was a modest up-regulation of the others (Fig. 4D). All of these enzymes can demethylate one of the three methylation states of H3K9 to some extent. The majority of demethylation of H3K9me2, however, is believed to be carried out by KDM3A.
FIGURE 4.
Changes in KDM3A expression in MDA-MB-231 cells. Immunoblots and densitometric quantifications of KDM3A. A, cells treated for 24 h with 500 μm NiSO4, CoCl2, desferrioxamine, DETA/NO, or hypoxia (1% O2). UT, untreated. B, cells treated for 4–24 h with •NO (DETA/NO 500 μm). C, cells treated with increasing concentrations of •NO (DETA/NO 100–1000 μm) or 500 μm deactivated DETA/NO, “DC,” for 24 h. All are representative immunoblots of n ≥3. D, cells were treated for 24 h with 500 μm DETA/NO, and quantitative RT-PCR analysis was conducted for all known H3K9 demethylase genes (n = 3). * indicates 0.01<p < 0.05; ** indicates p < 0.01 with respect to untreated controls, which are set arbitrarily to 1.0 or 100%. AU, arbitrary unit.
Methyltransferase Activity Does Not Account for Increased H3K9 Methylation by •NO
An increase in histone methylation can result from either a relative decrease in demethylation or a relative increase in methylation. To determine the contribution of increased H3K9 methylation by methyltransferases in response to •NO, we cultured cells in methionine-free media. S-Adenosylmethionine, which is synthesized from methionine and ATP, is the sole methyl donor for methyltransferase reactions. Histone methylation changes detected in cells grown in media depleted of methionine will predominantly reflect alterations in demethylation as opposed to methylation (33, 34, 36, 37). Fig. 5A demonstrates that, even in methionine-free media, H3K9me2 was strongly increased in response to •NO exposure compared with untreated controls.
FIGURE 5.
•NO-mediated changes in H3K9 methylation patterns do not result from methylation. A, immunoblot and densitometric quantification of H3K9me1,2,3. MDA-MB-231 cells were incubated in methionine-free media followed by treatment with 500 μm DETA/NO for 24 h. B, immunoblot and densitometric quantification of the methyltransferase, G9a. MDA-MB-231 cells were grown in standard media and treated with 500 μm DETA/NO for 1–24 h. All are representative immunoblots of n ≥3. UT, untreated. C, quantitative RT-PCR of all known H3K9 methyltransferase genes performed on mRNA isolated from cells treated for 24 h with 500 μm DETA/NO (n = 3). * indicates 0.01 < p < 0.05; ** indicates p < 0.01 with respect to untreated controls, which are set arbitrarily to 1.0 or 100%. AU, arbitrary unit.
Although active methylation did not explain changes in methylation patterns, we went on to examine methyltransferase gene expression in response to •NO. G9a is the major methyltransferase that acts on H3K9 (45, 46). We noted a moderate down-regulation in G9a protein within 2 h of •NO exposure and a marked decrease by 24 h (Fig. 5B). As was seen with KDM3A, changes in G9a expression did not translate into expected changes in H3K9me2. In addition to G9a, at least six more methylating enzymes have been reported to act on this residue. For this reason, we examined changes in steady-state mRNA levels after cellular exposure to •NO for all reported methyltransferases (Fig. 5C). Significant increases were noted for SETDB2 and SUV39h2. These enzymes, however, are primarily responsible for trimethylation of H3K9 (47–49), and increasing their activity would tend to decrease, rather than increase, the dimethyl state of H3K9. PRDM2 was also up-regulated; however, little is known about its substrate specificity (50).
Increasing the Chelatable Iron Pool Can Abrogate the Effect of •NO on Histone Methylation
Iron chelators inhibit 2-His-1-carboxylate non-heme iron oxygenases by depleting the availability of Fe(II) necessary for catalysis. These compounds react with iron in the CIP, which is considered the major source of iron for JMJC domain dioxygenases (51–53). We and others have shown previously that upon cellular exposure to •NO, the CIP is quantitatively converted into paramagnetic DNIC (39, 40, 54). Therefore, in addition to direct enzyme inhibition by •NO, it is likely that KDM3A activity could be further impaired by the loss of iron availability via DNIC formation. To test this, we augmented the CIP, exposed cells to •NO, and measured changes in H3K9 methylation.
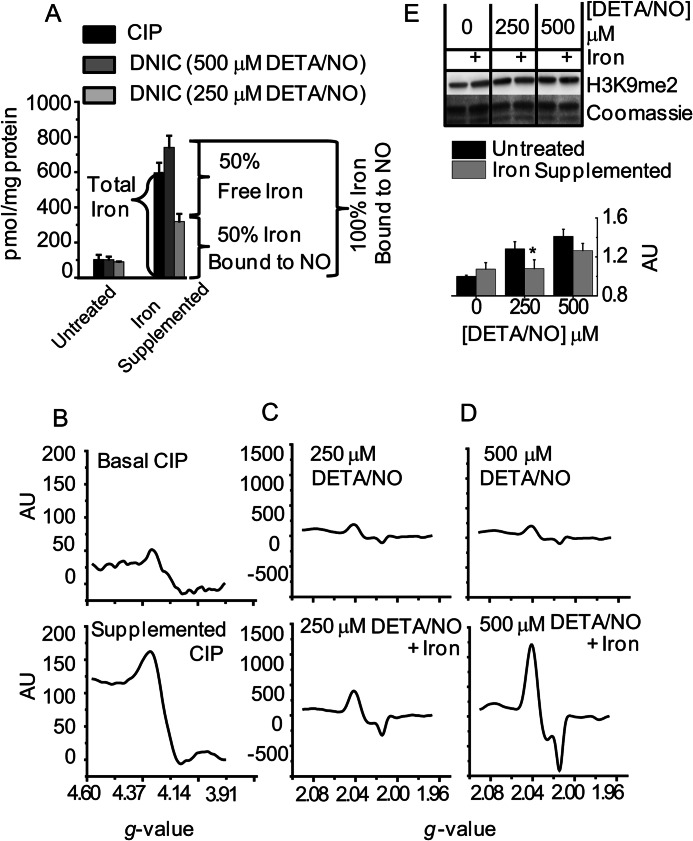
The CIP was increased by exposing cells to media supplemented with ferric ammonium citrate for 16 h. The cells were then treated with two different concentrations of •NO (250 and 500 μm DETA/NO). The resulting increases in CIP and DNIC formation after iron supplementation are shown in Fig. 6A. The CIP was measured by changes in the g = 4.3 signal as detected by EPR (Fig. 6B). DNIC were also quantified with EPR by measuring their characteristic signal centered at g = 2.03 (Fig. 6, C and D). In the absence of iron supplementation, treatment with both 250 and 500 μm DETA/NO formed DNIC at concentrations roughly equal to the CIP. For both concentrations of DETA/NO, supplementation of iron greatly increased the EPR-detectable signal (Fig. 6A). Under iron-supplemented conditions, the higher dose of •NO was able to convert 100% of the CIP into DNIC. At the lower •NO dose, however, only ≈50% the CIP was sequestered in the form of DNIC (Fig. 6A). Changes in H3K9me2 were also measured under identical conditions of iron supplementation and •NO exposure. During iron supplementation, increases in H3K9me2 were only observed with the higher dose of •NO (Fig. 6E). •NO-mediated changes in H3K9me2 were therefore only observed when the CIP was completely converted into DNIC. These results support our hypothesis that sequestration of chelatable iron via DNIC formation contributes to the inhibitory effects of •NO on histone demethylases.
FIGURE 6.
Dinitrosyliron complexes contribute to •NO-mediated histone methyl modifications. MDA-MB-231 cells were either untreated or supplemented with iron (ferric ammonium citrate, 150 μg/ml) for 16 h. Both groups of cells were then exposed to two concentrations of •NO (250 and 500 μm DETA/NO) for 24 h. A, EPR analysis of the CIP and DNIC. n = 3. B–D, representative EPR spectra from A. B are CIP measurements, and C and D are DNIC measurements. E, immunoblot and densitometric quantification of differences in H3K9me2 (n = 3). * indicates 0.01<p < 0.05 with respect to untreated controls, which are set arbitrarily to 1.0. AU, arbitrary unit.
Cellular Production of •NO Changes Global Methylation Status
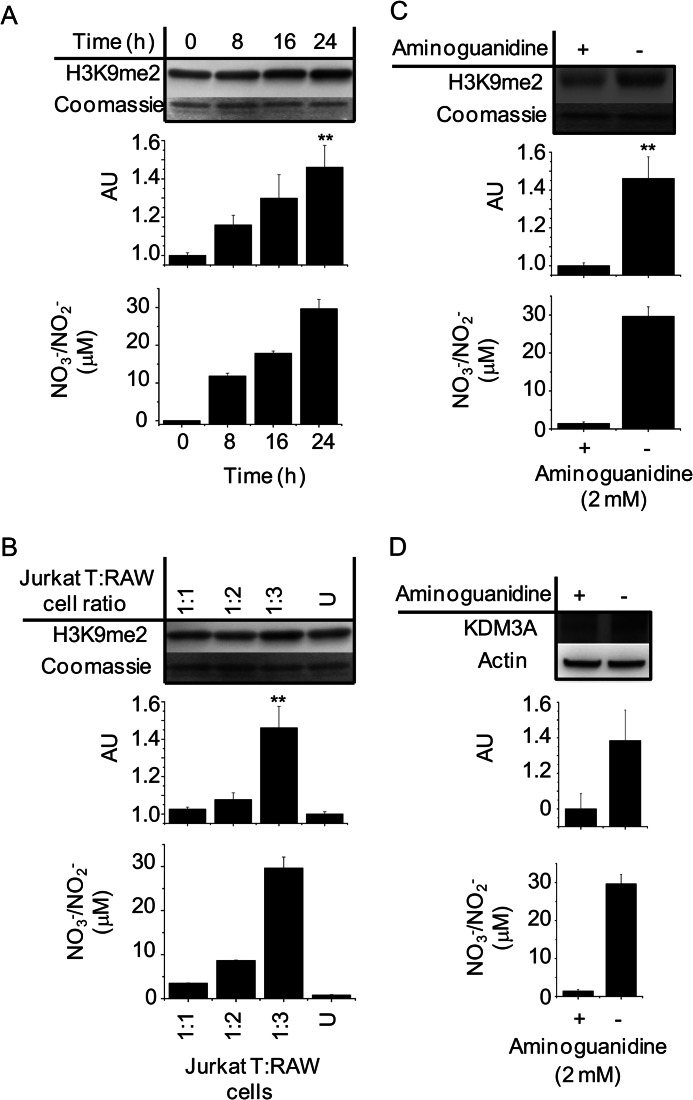
Having established that histone methylation patterns were altered by physiological •NO concentrations, we sought to replicate these findings under conditions of endogenous •NO synthesis. We developed a coculture assay that allowed for measurements of global levels of H3K9me2 in Jurkat T cells grown in suspension over a monolayer of •NO-producing RAW 264.7 macrophages. Using this technique, we observed significant time-dependent increases in macrophage-derived •NO production (NO3−/NO2−) and in Jurkat T cell H3K9me2 levels (Fig. 7A). Fig. 7B demonstrates that increases in H3K9me2 correlated to increases in the total amount of •NO to which the Jurkat T cells were exposed. By changing the ratio of Jurkat T/RAW 264.7 cells, we were able to modulate the amount of •NO exposure. Addition of the iNOS inhibitor aminoguanidine prevented NO3−/NO2− accumulation in the media and confirmed that changes in Jurkat T H3K9me2 only occurred in the presence of RAW 264.7-derived •NO (Fig. 7C). Finally, we measured changes in KDM3A protein in Jurkat T cells following 24 h of coculture (Fig. 7D). The total amount of KDM3A increased in response to •NO and was comparable with the change seen with 100 μm DETA/NO. This effect was also abrogated by aminoguanidine. Overall, H3K9me2 and KDM3A changes in Jurkat T cells correlated strongly with •NO synthesis, and they were indistinguishable from what was observed utilizing •NO-donor compounds.
FIGURE 7.
Paracrine regulation of H3K9me2 methylation and KDM3A expression in Jurkat T cells cocultured with •NO-synthesizing RAW 264.7 cells. RAW 264.7 cells in monolayer were stimulated with LPS for 6 h. Thereafter, media were replaced with LPS-free media followed by the addition of Jurkat T cells in suspension. Nitric oxide synthesis was verified in each experiment by chemiluminescent measurements of NO3−/NO2− accumulation in the media of the cocultured cells. Immunoblot and densitometric quantifications are shown in A. Temporal changes in H3K9me2 from Jurkat T cell total histone extracts after 0–24 h coculture are shown. B, H3K9me2 measurements from Jurkat T cell total histone extracts after 24 h coculture with increasing concentrations of activated RAW 264.7 cells. U indicates incubation with nonactivated RAW 264.7 cells 1:3. C, changes in H3K9me2 from Jurkat T cell total histone extracts after 24 h coculture ± the iNOS inhibitor aminoguanidine. D, changes in KDM3A protein in whole cell lysates of Jurkat T cells after 24 h coculture ± the iNOS inhibitor aminoguanidine. All are representative immunoblots and chemiluminescent measurements of n ≥3. ** indicates p < 0.01 with respect to untreated controls, which are set arbitrarily to 1.0. AU, arbitrary unit.
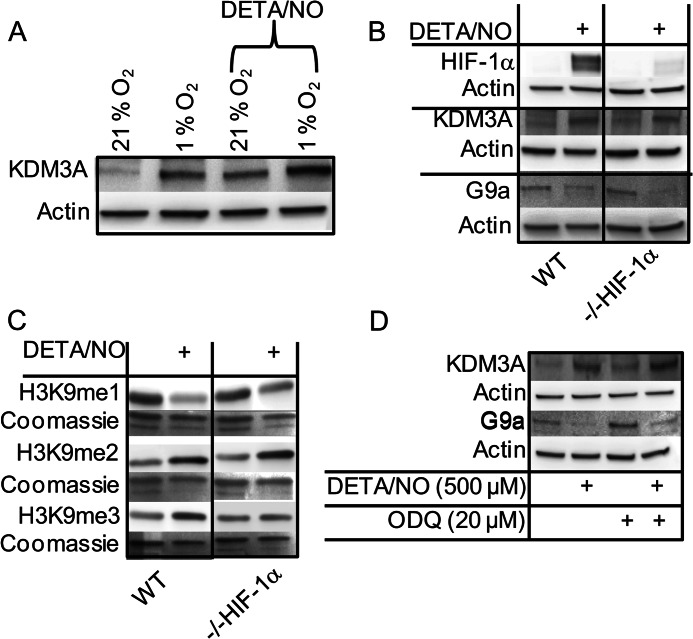
HIF-1α and sGC Are Not Required for •NO-mediated Regulation of Histone Modifications
Several reports indicate that KDM3A is up-regulated by hypoxia-inducible factor 1α (HIF-1α) accumulation (55–57). Other studies have demonstrated that G9a is up-regulated under hypoxia (33). It is also well known that •NO can strongly induce HIF-1α accumulation under normoxic conditions (58). To investigate this pathway, we treated cells with •NO at 1 and 21% O2 and measured KDM3A expression (Fig. 8A). •NO up-regulated KDM3A regardless of oxygen concentration. In fact, expression was higher at 1% O2 when •NO is predicted to destabilize HIF-1α (59). Using a HIF-1α knockdown cell line (38), we further probed the role of HIF-1α in the up-regulation of KDM3A and down-regulation of G9a by •NO. Fig. 8B demonstrates that in the absence of HIF-1α, KDM3A was still up-regulated to the same extent, and G9a was down-regulated in response to •NO. These data strongly suggest that •NO effects these changes in a HIF-1α-independent manner. When •NO-mediated changes in histone methylation patterns were examined in the HIF-1α knockdown cell line, they were indistinguishable from what was observed in the wild type (Fig. 8C).
FIGURE 8.
•NO-mediated changes in histone methylation are independent of HIF-1α and soluble guanylyl cyclase. MDA-MB-231 cells (WT) or MDA-MB-231 HIF-1α knockdown cells (−/− HIF-1α) were treated with ±500 μm DETA/NO for 24 h followed by extraction of total protein or histones. A, WT cells cultured at 21 or 1% O2 in the presence or absence of •NO. Immunoblot for KDM3A protein is shown. B, immunoblot demonstrating that •NO-mediated changes in KDM3A and G9a protein expression are independent of •NO-mediated HIF-1α accumulation is shown. C, immunoblot demonstrating that •NO-mediated changes in H3K9me1, -2, and -3 are independent of cellular HIF-1α status. D, immunoblot demonstrating that KDM3A is up-regulated and G9a is down-regulated by •NO in a guanylyl cyclase-independent manner. WT cells were treated with the guanylyl cyclase inhibitor ODQ (20 μm) prior to treatment with •NO. All are representative immunoblots of n = 3.
The activation of soluble guanylyl cyclase (sGC) by •NO is largely considered its most important biological function (24). To determine whether changes in KDM3A and G9a protein expression were a result of sGC activation by •NO, we pretreated cells with the sGC inhibitor ODQ. Inhibition of sGC did not affect the ability of •NO to up-regulate KDM3A or down-regulate G9a (Fig. 8D)
DISCUSSION
The identification of endogenously produced, epigenetic regulatory molecules would significantly alter our understanding of gene expression in health and disease. This study provides compelling evidence that •NO falls into this category of molecules. Until now, direct inhibition of histone demethylase activity by any endogenously produced small molecule has not been reported. Our data indicate that •NO is capable of directly inhibiting the lysine-specific histone demethylase KDM3A, and likely other JMJC demethylases, resulting in variations of histone methylation patterns. As these changes dramatically influence chromatin structure and gene transcription, this model provides a direct mechanism for the regulation of a multitude of genes by •NO. Classical mechanisms of •NO signaling result from its ability to regulate enzyme function by binding to the heme center of proteins or through covalent or redox modifications of key protein residues. Our results highlight the importance of non-heme, iron·nitrosyl complexes and provide a direct link between •NO and significant epigenetic modifications. Thus, this alternative mode of •NO signaling is unique in that it does not invoke much of the commonly relied upon complex chemistry necessary to explain phenotypic consequences of •NO synthesis.
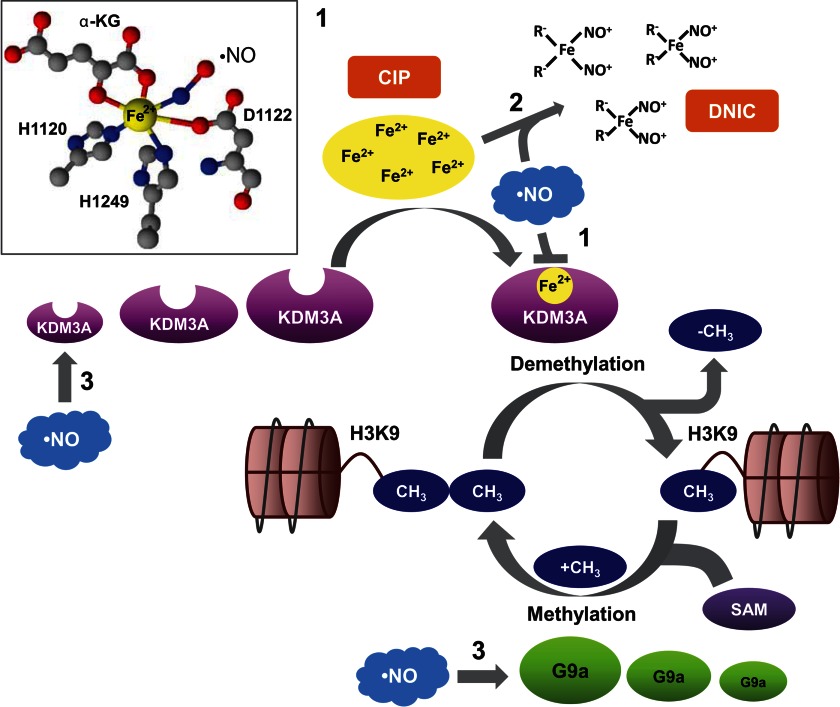
KDM3A, like all JMJC domain mononuclear Fe(II)-dependent dioxygenases, contains the 2-His-1-carboxylate facial triad. Unlike heme proteins, this enzyme coordinates iron at only three sites, leaving three additional sites available for substrate and cofactor binding (19, 20). Although there is no crystal structure for KDM3A, it is thought that His-1120, Asp-1122, and His-1249 are most likely to participate in coordinating the iron. During catalysis, α-KG is bound in a bidentate fashion, which results in the formation of a five-coordinate Fe(II) center. At this stage, the enzyme is poised to bind O2 (19, 20) or, as we propose, •NO at the sixth coordination site (Fig. 9, inset). Although KDM3A contains 39 cysteine residues, S-nitrosothiol formation was not detected following •NO exposure. Moreover, removal of •NO restored KDM3A activity indicating that it binds to the enzyme reversibly. The inhibitory effect of •NO is therefore caused by neither covalent modification nor oxidative destruction of the enzyme. This reversibility may be important under physiological conditions by tying the regulation of •NO synthesis to the simultaneous fine-tuning of demethylase activity. Interestingly, all experiments in this study were conducted at ambient O2 concentrations (≈220 μm O2), yet nanomolar steady-state •NO concentrations significantly inhibited enzyme function. Kinetic studies on demethylases such as KDM4E demonstrated that the enzyme·Fe(II)·α-KG·substrate complex reacts slowly with O2, and linear responses to increasing O2 concentrations have been observed up to 21% O2 (60, 61). This is agreement with our results that suggest that the affinity of this enzyme for •NO is greater than for O2. Although this requires further confirmatory studies, we hypothesize that, over a range of physiological O2 tensions, the inhibitory effect of •NO would be even more profound.
FIGURE 9.
Mechanisms of •NO-mediated epigenetic regulation. 1, nitric oxide directly binds to the iron atom that is coordinated by the 2-His-1-carboxylate facial triad in the active center of KDM3A (inset). This inhibits demethylase activity by preventing the binding of O2. 2, nitric oxide reacts with iron in the CIP and two anions (typically thiols, “R-,” like glutathione or cysteine) to form DNIC. Because the CIP is the major source of iron required as a cofactor by JMJC demethylases like KDM3A, the accumulation of DNIC results in iron sequestration and indirectly contributes to enzyme inhibition. 3, major H3K9 methyl-modifying enzymes are the methyltransferase, G9a, and the demethylase, KD3MA. G9a adds a methyl group from the donor compound, S-adenosylmethionine, to yield mono- and dimethylated products. Nitric oxide up-regulates KDM3A and concomitantly down-regulates G9a. Expression changes in methyl-modifying enzymes will influence the overall steady-state levels of H3K9 methylation status.
Cells treated with physiological •NO concentrations demonstrated changes in H3K9 methylation patterns in a time- and concentration-dependent manner. Collectively, we have identified three unique mechanisms that explain how •NO can mediate these changes (Fig. 9). The JMJC class of demethylases contains a non-heme iron as a cofactor, and the chelatable iron pool is generally considered the source of this iron (51–53). Similarly, the iron utilized for DNIC assembly is also derived from this pool (39, 40, 54). Just as chemical iron chelators dramatically inhibit enzyme function by starving the cell of iron (62), we observed analogous effects of DNIC formation that likely result from shifts in the enzyme equilibrium toward its iron-free apoenzyme form (Fig. 9). Therefore, in addition to direct inhibition of demethylase activity by •NO, our data suggest that formation of DNIC indirectly inhibits enzyme activity by reducing the overall pool of available iron. This is significant as functional roles for DNIC assembly have remained obscure. With continued •NO exposure, KDM3A protein levels increased while the methyltransferase G9a protein levels decreased. Logic would predict that increasing demethylating enzymes and decreasing methylating enzymes would favor an overall decrease in histone methylation. What we observed, however, was significant increases in methylation at H3K9. We suspect that increased methylation subsequent to KDM3A inhibition by •NO results in compensatory changes in gene expression that attempt to override this inhibitory effect by modulating the relative concentrations of methyl-modifying enzymes.
Whether JMJC demethylase inhibition by •NO is an important physiological regulatory mechanism or associated more with •NO pathologies is also not clear. Both changes in histone methylation patterns and changes in methyl-modifying enzymes occurred in response to low physiological concentrations of •NO. Although both responses exhibited significant dose dependence on •NO concentration, as little as 16 ± 8 nm steady-state •NO was sufficient. Under both normal and disease conditions, there are numerous locations where •NO is synthesized. Although •NO may be derived from enzymatic, dietary (NO3−/NO2−), and pharmacological origins, the biological activities of •NO are independent of its source. As JMJC demethylases are ubiquitous in nature, the manner by which •NO inhibits these enzymes in a tumor cell will be identical to how it inhibits these enzymes in a neuron, myocyte, hepatocyte, macrophage, or bacteria. Thus, like most •NO-mediated biological responses, epigenetic effects will largely be a function of the cellular microenvironment that ultimately dictates the concentration and duration of •NO exposure (63). Therefore, although our studies examined global methylation patterns, it is likely that subtle changes in methylation will be localized around areas of basal •NO synthesis.
It is becoming increasing clear that the regulation of methyl-modifying enzymes and post-translational histone modifications play important roles in tumor biology. For example, KDM3A expression was found to be elevated in certain types of cancer (64, 65). In one particular study, immunohistochemical analysis revealed higher levels of KDM3A near the vessels of renal cell carcinomas, implicating a possible role in regulation of VEGF (65). Furthermore, inhibition of KDM3A improved outcomes of the anti-angiogenic VEGF antibody, bevacizumab (66). Thus, paradoxically, a combination of VEGF inhibitors and •NO might prove beneficial despite the fact that •NO drives up-regulation of angiogenesis through VEGF (67). Expression of KDM2B promotes differentiation of more aggressive forms of pancreatic cancer (68). KDM4A promotes cellular transformation through transcriptional repression of the tumor suppressor CHD5 (69), and KDM6B has been found to promote the epithelial-mesenchymal transition (70). Conversely, others have demonstrated that KDM6B acts as a tumor suppressor by regulating p53 nuclear stabilization (71). In colorectal cancer, depletion of KDM5B induced cellular senescence (72). In breast cancer, the methyltransferase G9a plays a critical role in the epigenetic regulation of epithelial-mesenchymal transition (73). G9a has also been shown to specifically methylate p53 at Lys-373, which correlates with inactivation of the protein (74). Thus, down-regulation of G9a by •NO may serve a tumor-suppressive function. As a whole, these studies underscore the complexity of epigenetic regulation in cancer biology and emphasize the importance of tumor type and specific epigenetic makeup. Furthermore, these novel epigenetic regulatory effects of •NO may have an important yet unrealized influence on cancer etiology.
Methyl-modifying enzymes are also involved in numerous other physiological and pathological conditions. During neurogenesis, methylation of H3K27 by KDM6B plays a critical role in the maintenance of the embryonic respiratory neuronal network (75). H3K4 demethylation by KDM5A drives differential transcriptional silencing during development (76). In human mesenchymal stem cells derived from bone marrow, histone demethylases KDM4B and KDM6B promote osteogenic differentiation (77). In hematopoietic stem cells, the H3K27 demethylase KDM6A regulates stem cell migration and hematopoiesis (78). These are just a few of the numerous examples that demonstrate the critical importance and the diversity of function of the methyl-modifying enzymes and suggest a potential role for regulation by •NO as well. In fact, a recent study has shown dendritic outgrowth in neuronal cells arising from the decreased trimethylation at H3K9 was due to nitric oxide-dependent degradation of the methyltransferase SUV39h1 (79).
Although we have focused on lysine 9 methylation of histone 3, this is just one of a vast number of possible modifications. Understanding how histone methylation status is controlled at specific DNA sequences by •NO and how these modifications affect local protein interactions will be required to elucidate the ultimate role of •NO in epigenetic regulation. This study revealed the following three distinct mechanisms whereby •NO could affect histone methylation patterns in general: direct inhibition of JMJC demethylases, reduction in iron cofactor availability, and regulation of KDM and KMT gene expression (Fig. 9). As we have discussed, both the methylation status as well as changes in steady-state levels of methyl-modifying enzymes result in a multitude of phenotypic consequences. Our results revealed the dramatic effects •NO could have on both of these aspects suggesting that the magnitude of •NO-attributable responses may be much greater than previously thought. Moreover, in addition to methylation, other histone lysine modifications such as acetylation can occur on the same residue. This is a reciprocal relationship, however, and each histone modification is mutually exclusive for a specific residue (i.e. a methylated lysine cannot be acetylated and vice versa). This means that •NO-mediated increases in methylation may have additional unrealized and far-reaching consequences such as precluding the formation of other critical regulatory histone marks.
Based on our current data, we classify •NO as an endogenously produced, epigenetic regulatory molecule. Although these epigenetic mechanisms do not eliminate or contradict currently accepted, well defined paths of •NO signaling, they may turn out to be equally as important under a diverse set of cellular conditions. Furthermore, these mechanisms address some fundamental problems in •NO chemical biology by providing a simple and plausible alternative hypothesis explaining the pleiotropic nature of a complex signaling molecule. The ability of •NO to change the topography of the epigenetic landscape reveals a new dimension to its list of regulatory functions and allows us to rethink classical •NO-signaling mechanisms.
Acknowledgment
MS-MALDI was performed by the University of Illinois at Chicago-Research Resources Center mass spectrometry, metabolomics, and proteomics facility.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM085232 and 1S10RR027848-01A1 from NIGMS. This work was also supported by the University of Illinois at Chicago Cancer Center.
This article was selected as a Paper of the Week.
- JMJC
- Jumonji C
- α-KG
- α-ketoglutarate
- CIP
- chelatable iron pool
- DETA/NO
- (Z)-1-[N-(2-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate
- DFO
- desferrioxamine
- DNIC
- dinitrosyliron complex
- FeSO4
- ferrous sulfate
- H3K9me1/2/3
- mono/di/trimethylated lysine 9 on histone 3
- HIF-1α
- hypoxia-inducible factor 1α
- iNOS
- inducible nitric-oxide synthase
- KDM
- lysine demethylase
- KMT
- lysine methyltransferase
- NO3−
- nitrate
- NO2−
- nitrite
- •NO
- nitric oxide
- ODQ
- 1H-[1,2,4]oxadiazolo[4,3-a]quinoxalin-1-one
- PRDM2
- PR domain-containing 2, with ZNF domain
- Proli/NO
- 1-(hydroxy-NNO-azoxy)-l-proline
- SETDB1
- SET domain bifurcated 1
- SETDB2
- SET domain bifurcated 2
- Sper/NO
- (Z)-1-[N-[3-aminopropyl]-N-[4-(3-aminopropylammonio)butyl]-amino]diazen-1-ium-1,2-diolate
- sGC
- soluble guanylyl cyclase
- SUV39h1
- suppressor of variegation 3–9 homolog 1 (Drosophila)
- SUV39h2
- suppressor of variegation 3–9 homolog 2 (Drosophila).
REFERENCES
- 1. Frazer K. A., Murray S. S., Schork N. J., Topol E. J. (2009) Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251 [DOI] [PubMed] [Google Scholar]
- 2. Roberts N. J., Vogelstein J. T., Parmigiani G., Kinzler K. W., Vogelstein B., Velculescu V. E. (2012) The predictive capacity of personal genome sequencing. Sci. Transl. Med. 4, 133ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 4. Kuo M. H., Allis C. D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays 20, 615–626 [DOI] [PubMed] [Google Scholar]
- 5. Khan S. N., Khan A. U. (2010) Role of histone acetylation in cell physiology and diseases: An update. Clin. Chim. Acta 411, 1401–1411 [DOI] [PubMed] [Google Scholar]
- 6. Martin C., Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 7. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 8. Karytinos A., Forneris F., Profumo A., Ciossani G., Battaglioli E., Binda C., Mattevi A. (2009) A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 284, 17775–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kooistra S. M., Helin K. (2012) Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 [DOI] [PubMed] [Google Scholar]
- 10. Klose R. J., Kallin E. M., Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 11. Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 12. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 13. Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 14. Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 15. Fodor B. D., Kubicek S., Yonezawa M., O'Sullivan R. J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cloos P. A., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K. H., Helin K. (2006) The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311 [DOI] [PubMed] [Google Scholar]
- 17. Zhu Z., Wang Y., Li X., Wang Y., Xu L., Wang X., Sun T., Dong X., Chen L., Mao H., Yu Y., Li J., Li J., Chen P. A., Chen C. D. (2010) PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 20, 794–801 [DOI] [PubMed] [Google Scholar]
- 18. Huang C., Xiang Y., Wang Y., Li X., Xu L., Zhu Z., Zhang T., Zhu Q., Zhang K., Jing N., Chen C. D. (2010) Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165 [DOI] [PubMed] [Google Scholar]
- 19. Loenarz C., Schofield C. J. (2011) Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 36, 7–18 [DOI] [PubMed] [Google Scholar]
- 20. McDonough M. A., Loenarz C., Chowdhury R., Clifton I. J., Schofield C. J. (2010) Structural studies on human 2-oxoglutarate-dependent oxygenases. Curr. Opin. Struct. Biol. 20, 659–672 [DOI] [PubMed] [Google Scholar]
- 21. Yoshimi A., Kurokawa M. (2011) Key roles of histone methyltransferase and demethylase in leukemogenesis. J. Cell. Biochem. 112, 415–424 [DOI] [PubMed] [Google Scholar]
- 22. Lohse B., Kristensen J. L., Kristensen L. H., Agger K., Helin K., Gajhede M., Clausen R. P. (2011) Inhibitors of histone demethylases. Bioorg. Med. Chem. 19, 3625–3636 [DOI] [PubMed] [Google Scholar]
- 23. Lim S., Metzger E., Schüle R., Kirfel J., Buettner R. (2010) Epigenetic regulation of cancer growth by histone demethylases. Int. J. Cancer 127, 1991–1998 [DOI] [PubMed] [Google Scholar]
- 24. Ignarro L. J. (2002) Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J. Physiol. Pharmacol. 53, 503–514 [PubMed] [Google Scholar]
- 25. Metzen E., Zhou J., Jelkmann W., Fandrey J., Brüne B. (2003) Nitric oxide impairs normoxic degradation of HIF-1α by inhibition of prolyl hydroxylases. Mol. Biol. Cell 14, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fandrey J., Gorr T. A., Gassmann M. (2006) Regulating cellular oxygen sensing by hydroxylation. Cardiovasc. Res. 71, 642–651 [DOI] [PubMed] [Google Scholar]
- 27. Berchner-Pfannschmidt U., Tug S., Kirsch M., Fandrey J. (2010) Oxygen-sensing under the influence of nitric oxide. Cell. Signal. 22, 349–356 [DOI] [PubMed] [Google Scholar]
- 28. Chen H., Costa M. (2009) Iron- and 2-oxoglutarate-dependent dioxygenases: an emerging group of molecular targets for nickel toxicity and carcinogenicity. Biometals 22, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chowdhury R., Flashman E., Mecinović J., Kramer H. B., Kessler B. M., Frapart Y. M., Boucher J. L., Clifton I. J., McDonough M. A., Schofield C. J. (2011) Studies on the reaction of nitric oxide with the hypoxia-inducible factor prolyl hydroxylase domain 2 (EGLN1). J. Mol. Biol. 410, 268–279 [DOI] [PubMed] [Google Scholar]
- 30. Chen H., Giri N. C., Zhang R., Yamane K., Zhang Y., Maroney M., Costa M. (2010) Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 285, 7374–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen H., Kluz T., Zhang R., Costa M. (2010) Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis 31, 2136–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tausendschön M., Dehne N., Brüne B. (2011) Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine 53, 256–262 [DOI] [PubMed] [Google Scholar]
- 33. Chen H., Yan Y., Davidson T. L., Shinkai Y., Costa M. (2006) Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 66, 9009–9016 [DOI] [PubMed] [Google Scholar]
- 34. Zhou X., Sun H., Chen H., Zavadil J., Kluz T., Arita A., Costa M. (2010) Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 70, 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chervona Y., Costa M. (2012) The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic. Biol. Med. 53, 1041–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q., Ke Q., Costa M. (2009) Alterations of histone modifications by cobalt compounds. Carcinogenesis 30, 1243–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen H., Ke Q., Kluz T., Yan Y., Costa M. (2006) Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell. Biol. 26, 3728–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hickok J. R., Sahni S., Mikhed Y., Bonini M. G., Thomas D. D. (2011) Nitric oxide suppresses tumor cell migration through N-Myc downstream-regulated gene-1 (NDRG1) expression: role of chelatable iron. J. Biol. Chem. 286, 41413–41424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hickok J. R., Sahni S., Shen H., Arvind A., Antoniou C., Fung L. W., Thomas D. D. (2011) Dinitrosyliron complexes are the most abundant nitric oxide-derived cellular adduct: biological parameters of assembly and disappearance. Free Radic. Biol. Med. 51, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hickok J. R., Vasudevan D., Thatcher G. R., Thomas D. D. (2012) Is S-nitrosocysteine a true surrogate for nitric oxide? Antioxid. Redox Signal. 17, 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. MacArthur P. H., Shiva S., Gladwin M. T. (2007) Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescence. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 851, 93–105 [DOI] [PubMed] [Google Scholar]
- 42. Lee M., Arosio P., Cozzi A., Chasteen N. D. (1994) Identification of the EPR-active iron-nitrosyl complexes in mammalian ferritins. Biochemistry 33, 3679–3687 [DOI] [PubMed] [Google Scholar]
- 43. Kennedy M. C., Antholine W. E., Beinert H. (1997) An EPR investigation of the products of the reaction of cytosolic and mitochondrial aconitases with nitric oxide. J. Biol. Chem. 272, 20340–20347 [DOI] [PubMed] [Google Scholar]
- 44. Pierce B. S., Gardner J. D., Bailey L. J., Brunold T. C., Fox B. G. (2007) Characterization of the nitrosyl adduct of substrate-bound mouse cysteine dioxygenase by electron paramagnetic resonance: electronic structure of the active site and mechanistic implications. Biochemistry 46, 8569–8578 [DOI] [PubMed] [Google Scholar]
- 45. Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang H., An W., Cao R., Xia L., Erdjument-Bromage H., Chatton B., Tempst P., Roeder R. G., Zhang Y. (2003) mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12, 475–487 [DOI] [PubMed] [Google Scholar]
- 48. Falandry C., Fourel G., Galy V., Ristriani T., Horard B., Bensimon E., Salles G., Gilson E., Magdinier F. (2010) CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 285, 20234–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 50. Wu H., Min J., Lunin V. V., Antoshenko T., Dombrovski L., Zeng H., Allali-Hassani A., Campagna-Slater V., Vedadi M., Arrowsmith C. H., Plotnikov A. N., Schapira M. (2010) Structural biology of human H3K9 methyltransferases. PLoS One 5, e8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cyr A. R., Domann F. E. (2011) The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid. Redox Signal. 15, 551–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 53. Knowles H. J., Mole D. R., Ratcliffe P. J., Harris A. L. (2006) Normoxic stabilization of hypoxia-inducible factor-1α by modulation of the labile iron pool in differentiating U937 macrophages: effect of natural resistance-associated macrophage protein 1. Cancer Res. 66, 2600–2607 [DOI] [PubMed] [Google Scholar]
- 54. Toledo J. C., Jr., Bosworth C. A., Hennon S. W., Mahtani H. A., Bergonia H. A., Lancaster J. R., Jr. (2008) Nitric oxide-induced conversion of cellular chelatable iron into macromolecule-bound paramagnetic dinitrosyliron complexes. J. Biol. Chem. 283, 28926–28933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wellmann S., Bettkober M., Zelmer A., Seeger K., Faigle M., Eltzschig H. K., Bührer C. (2008) Hypoxia up-regulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 372, 892–897 [DOI] [PubMed] [Google Scholar]
- 56. Pollard P. J., Loenarz C., Mole D. R., McDonough M. A., Gleadle J. M., Schofield C. J., Ratcliffe P. J. (2008) Regulation of Jumonji domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem. J. 416, 387–394 [DOI] [PubMed] [Google Scholar]
- 57. Beyer S., Kristensen M. M., Jensen K. S., Johansen J. V., Staller P. (2008) The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283, 36542–36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomas D. D., Espey M. G., Ridnour L. A., Hofseth L. J., Mancardi D., Harris C. C., Wink D. A. (2004) Hypoxic inducible factor 1α, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 101, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mateo J., García-Lecea M., Cadenas S., Hernández C., Moncada S. (2003) Regulation of hypoxia-inducible factor-1α by nitric oxide through mitochondria-dependent and -independent pathways. Biochem. J. 376, 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cascella B., Mirica L. M. (2012) Kinetic analysis of iron-dependent histone demethylases: α-ketoglutarate substrate inhibition and potential relevance to the regulation of histone demethylation in cancer cells. Biochemistry 51, 8699–8701 [DOI] [PubMed] [Google Scholar]
- 61. Sánchez-Fernández E. M., Tarhonskaya H., Al-Qahtani K., Hopkinson R. J., McCullagh J. S., Schofield C. J., Flashman E. (2013) Investigations on the oxygen dependence of a 2-oxoglutarate histone demethylase. Biochem. J. 449, 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Richardson D. R. (2005) Molecular mechanisms of iron uptake by cells and the use of iron chelators for the treatment of cancer. Curr. Med. Chem. 12, 2711–2729 [DOI] [PubMed] [Google Scholar]
- 63. Thomas D. D., Ridnour L. A., Isenberg J. S., Flores-Santana W., Switzer C. H., Donzelli S., Hussain P., Vecoli C., Paolocci N., Ambs S., Colton C. A., Harris C. C., Roberts D. D., Wink D. A. (2008) The chemical biology of nitric oxide: implications in cellular signaling. Free Radic. Biol. Med. 45, 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cho H. S., Toyokawa G., Daigo Y., Hayami S., Masuda K., Ikawa N., Yamane Y., Maejima K., Tsunoda T., Field H. I., Kelly J. D., Neal D. E., Ponder B. A., Maehara Y., Nakamura Y., Hamamoto R. (2012) The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int. J. Cancer 131, E179–E189 [DOI] [PubMed] [Google Scholar]
- 65. Guo X., Shi M., Sun L., Wang Y., Gui Y., Cai Z., Duan X. (2011) The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 58, 153–157 [DOI] [PubMed] [Google Scholar]
- 66. Osawa T., Tsuchida R., Muramatsu M., Shimamura T., Wang F., Suehiro J. I., Kanki Y., Wada Y., Yuasa Y., Aburatani H., Miyano S., Minami T., Kodama T., Shibuya M. (2013) Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor associated macrophages. Cancer Res., in press [DOI] [PubMed] [Google Scholar]
- 67. Hickok J. R., Thomas D. D. (2010) Nitric oxide and cancer therapy: the emperor has NO clothes. Curr. Pharm. Des. 16, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tzatsos A., Paskaleva P., Ferrari F., Deshpande V., Stoykova S., Contino G., Wong K. K., Lan F., Trojer P., Park P. J., Bardeesy N. (2013) KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J. Clin. Invest. 123, 727–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mallette F. A., Richard S. (2012) JMJD2A promotes cellular transformation by blocking cellular senescence through transcriptional repression of the tumor suppressor CHD5. Cell Rep. 2, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 70. Ramadoss S., Chen X., Wang C. Y. (2012) Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 287, 44508–44517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ene C. I., Edwards L., Riddick G., Baysan M., Woolard K., Kotliarova S., Lai C., Belova G., Cam M., Walling J., Zhou M., Stevenson H., Kim H. S., Killian K., Veenstra T., Bailey R., Song H., Zhang W., Fine H. A. (2012) Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One 7, e51407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ohta K., Haraguchi N., Kano Y., Kagawa Y., Konno M., Nishikawa S., Hamabe A., Hasegawa S., Ogawa H., Fukusumi T., Uemura M., Nishimura J., Hata T., Takemasa I., Mizushima T., Noguchi Y., Ozaki M., Kudo T., Sakai D., Satoh T., Fukami M., Ishii M., Yamamoto H., Doki Y., Mori M., Ishii H. (2013) Depletion of JARID1B induces cellular senescence in human colorectal cancer. Int. J. Oncol. 42, 1212–1218 [DOI] [PubMed] [Google Scholar]
- 73. Dong C., Wu Y., Yao J., Wang Y., Yu Y., Rychahou P. G., Evers B. M., Zhou B. P. (2012) G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 122, 1469–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang J., Dorsey J., Chuikov S., Pérez-Burgos L., Zhang X., Jenuwein T., Reinberg D., Berger S. L. (2010) G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 285, 9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Burgold T., Voituron N., Caganova M., Tripathi P. P., Menuet C., Tusi B. K., Spreafico F., Bévengut M., Gestreau C., Buontempo S., Simeone A., Kruidenier L., Natoli G., Casola S., Hilaire G., Testa G. (2012) The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2, 1244–1258 [DOI] [PubMed] [Google Scholar]
- 76. Beshiri M. L., Holmes K. B., Richter W. F., Hess S., Islam A. B., Yan Q., Plante L., Litovchick L., Gévry N., Lopez-Bigas N., Kaelin W. G., Jr., Benevolenskaya E. V. (2012) Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 18499–18504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ye L., Fan Z., Yu B., Chang J., Al Hezaimi K., Zhou X., Park N. H., Wang C. Y. (2012) Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thieme S., Gyarfas T., Richter C., Ozhan G., Fu J., Alexopulou D., Muders M. H., Michalk I., Jakob C., Dahl A., Klink B., Bandola J., Bachmann M., Schrock E., Buchholz F., Stewart A. F., Weidinger G., Anastassiadis K., Brenner S. (2013) The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 121, 2462–2473 [DOI] [PubMed] [Google Scholar]
- 79. Sen N., Snyder S. H. (2011) Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 108, 20178–20183 [DOI] [PMC free article] [PubMed] [Google Scholar]