Background: Wounding renders mosquitoes resistant to human malaria parasites.
Results: Genome-wide transcriptional profiling identified 53 wound response genes, including two transglutaminases. Functional studies revealed the role of AP-1/TGase2 axis in the mosquito resistance to Plasmodium falciparum.
Conclusion: AP-1/TGase2 is a new axis of the mosquito immune responses to malaria parasites.
Significance: Multiple signaling pathways contribute to mosquito resistance to malaria and represent new potential targets for vector control interventions.
Keywords: Fos, Infectious Diseases, Insect Immunity, Malaria, Plasmodium, Transcription Factors, Transcription Regulation, Transcriptomics, Transglutaminases, Mosquito
Abstract
Anopheline mosquitoes are the only vectors of human malaria worldwide. It is now widely accepted that mosquito immune responses play a crucial role in restricting Plasmodium development within the vector; therefore, further dissection of the molecular mechanisms underlying these processes should inform new vector control strategies urgently needed to roll back the disease. Here, using genome-wide transcriptional profiling, bioinformatics, and functional gene analysis, we identify a new axis of mosquito resistance to monoclonal Plasmodium falciparum infections that includes the AP-1 transcription factor Fos and the transglutaminase 2 (TGase2), a cross-linking enzyme with known roles in wound responses. We demonstrate that Fos regulates induction of TGase2 expression after wounding but does not affect expression of the components of the well characterized complement-like system. Silencing of Fos or of TGase2 aborts the wounding-induced mosquito killing of P. falciparum. These results reveal multiple signaling pathways that are required for efficient Plasmodium killing in Anopheles gambiae.
Introduction
Malaria is the most widespread human infectious disease transmitted by an insect vector. It causes high levels of morbidity and mortality in Africa, especially in young children and pregnant women (1). In the current absence of efficient vaccination programs, strategies for malaria elimination should be multifaceted and include drug therapy as well as mosquito vector population control (2, 3). The major malaria vectors in sub-Saharan Africa are mosquitoes of the Anopheles gambiae complex. The mosquito mounts a powerful immune response against Plasmodium infection, which kills most parasites during the first days of invasion (4). Boosting the mosquito immune system is a promising approach for blocking Plasmodium development within the vector to control malaria transmission (5, 6).
Invertebrates rely exclusively on the innate type of immune responses for protection against microbial infections (7, 8). These responses are controlled by signaling pathways that are remarkably well conserved across the animal phyla. Genetic analysis in Drosophila melanogaster revealed four major immune pathways, Toll, Imd, JAK/STAT, and JNK, that contribute to induction of effector molecules, such as antimicrobial peptides and stress response genes (8). In A. gambiae, homologues of the Toll and Imd pathways selectively induce target gene expression through the NF-κB transcription factors Rel1 and Rel2, respectively (9–11). Activation of both the Rel1 and Rel2 pathways results in an efficient immune response against malaria parasites. This response is largely mediated by the mosquito complement-like system that, among others, comprises thioester-containing protein 1 (TEP1) and two leucine-rich repeat proteins, LRIM1 and APL1 (6, 11). Activation of the Rel2 pathway appears to be more efficient at blocking the development of human Plasmodium falciparum parasites, whereas activation of the Rel1 impacts the development of the murine parasites Plasmodium berghei and Plasmodium yoelii, often used as models to study Plasmodium-Anopheles interactions (5, 11).
Previously, we demonstrated that wounding of the mosquito by injection of water or dsRNA promotes TEP1-mediated killing of P. falciparum (12). The efficiency of this response was affected by the parasite genetic clonality because it was predominantly operating in infections with monoclonal, and to a lesser extent with biclonal, parasite isolates but not in infections containing three or more parasite clones. Wound response is a complex process initiated by an injury of epidermis. The hallmarks of this process are coagulation and clot formation that seal the wound site, thereby preventing hemolymph loss and limiting dissemination of infectious agents. In Drosophila, wound healing is similar to well characterized developmental processes that involve epithelial rearrangements, such as dorsal closure during embryonic development (13). These processes involve massive cellular movements that help to establish or to renew cell-cell contacts and are regulated by the JNK pathway (13). Regardless of the trigger, be it developmental reorganization or injury, wound healing elicits an immune response and primes the organism for what is likely to be an increased risk of infection (14).
In invertebrates, coagulation plays essential roles in morphogenesis, wound healing, and immunity and is carried out by transglutaminases (TGases),4 enzymes with protein cross-linking activity. TGases catalyze formation of isopeptide bonds between a free ϵ-amino group and glutamine residue in a Ca2+-dependent manner (15–17) and are essential for a variety of biological processes. For instance, in the nematode Onchocerca volvulus, TGase-catalyzed cross-linking is crucial for the molting of third instar larvae (18), whereas in Drosophila, TGase1 plays an essential role in pupal morphogenesis and cuticle schlerotization brought about by cross-linking of two cuticular chitin-binding proteins, larval serum protein 2 and a putative C-type lectin (19). During immune responses in crayfish, hemolymph coagulation is achieved by TGase-mediated cross-linking of specific clotting proteins (20–22). In the horseshoe crab Tachypleus tridentatus, a proteolytic coagulation cascade activated by septic injury converts coagulogen into insoluble coagulin. Further cross-linking of the coagulin polymers by TGases traps and immobilizes invading pathogens at sites of injury (23, 24). Recently, TGase function in immunity was revealed in Drosophila, where TGase1 mutant larvae displayed high susceptibility to infections with entomopathogenic nematodes harboring the symbiotic bacteria Photorhabdus bacteriophora (25).
In contrast to other insect species that rely on a single TGase, the genome of A. gambiae contains three genes coding for TGases. Phylogenetic analysis of TGases in A. gambiae, D. melanogaster, Culex pipiens, and Aedes aegypti revealed that AGAP009100 is an orthologue of TGase1 conserved in all insects, whereas AGAP009098 is an orthologue of TGase2 identified in mosquitoes of the genera Anopheles, Aedes, and Culex but not in Drosophila (26). The third TGase is encoded by AGAP009099 and is specific to A. gambiae, having no homologues in other species. It is expressed in the male accessory glands and cross-links proteins of the male seminal fluid into a coagulated proteinaceous plug transferred to females during mating (26).
Here, we set out to identify mosquito factors responsible for the wounding-induced P. falciparum killing. A genome-wide transcriptional analysis identified 53 genes whose expression was significantly regulated upon wounding, including TGase1 and TGase2. We demonstrated that depletion of TGase2, but not of TGase1, restored the numbers of developing parasites to those observed in non-injected controls. Using bioinformatics and functional gene analysis by RNAi silencing, we further demonstrated that expression of TGase2 after wounding is regulated by the AP-1 transcriptional factor Fos, the transactivator of the JNK pathway in other animals. Our study identifies the novel role of TGase2 in the elimination of P. falciparum and unravels the importance of the AP-1/Fos-TGase2 axis in immune responses of mosquitoes to human malaria parasites.
EXPERIMENTAL PROCEDURES
Mosquito Colony
The A. gambiae sensu stricto Ngousso strain was originally established in 2006 at the Institut de Recherche de Yaoundé, Cameroon (OCEAC) from larvae collected in Yaoundé. The colony belongs to the M molecular and Forest chromosomal forms (standard chromosomal arrangements). Mosquitoes were reared in the insectary at 28.0 ± 2 °C and 80.0 ± 5% humidity with a 12/12-h dark/light cycle. Adults were fed on a 6% sugar solution through cotton pads, and the larvae diet consisted of ground fish food (Tetra).
dsRNA Production and Silencing
Fos and TGase2 were PCR-amplified from genomic DNA with the following primers: Fos, AG356 (5′-GAGCCTCCCGATCTACTTTTGCTAC-3′) and AG357 (5′-TGGTTCGGTTTTAATTTTTGGCAGT-3′); TGase2, AG1197 (5′-GGGGAATTCGGCCGAACTAACGATTGAAA-3′) and AG1198 (5′-GGGGCGGCCGCCCGTTAGGCCATCCTCATTA-3′). Amplicons were cloned into pGEM-T Easy Vector (Promega) and subcloned as a 384-bp-long EcoRI-XhoI and 356-bp EcoRI-NotI into pLL10, resulting in pLL323 and pLL556, respectively. Plasmids pLL17 (dsTEP1), pLL100 (dsLacZ), and dsAGAP009100 (kind gift of Dr. D. Rogers, Imperial College London) were used, and the synthesis of dsRNAs was performed as described previously (27).
Gene silencing was achieved by injecting 0.2 μg of dsRNA into the thorax of 1-day-old females using a glass capillary mounted onto a Nanoject II injector (Drummond). A dsRNA targeting a bacterial gene absent from the mosquito genome (dsLacZ) served as a negative control in dsRNA injections. During injection, mosquitoes were immobilized by CO2, and non-injected mosquitoes were also exposed to CO2 treatment.
Transcriptional Profiling Experiments and Data Analysis
To analyze changes in the transcriptome of mosquitoes after dsLacZ injection, we used a microarray approach. At 3 h after injection, 30 mosquitoes were frozen in liquid nitrogen and kept at −80 °C. Two independent experiments were performed using unrelated mosquito populations. Total RNA was extracted using the RNeasy extraction kit (Qiagen) according to the supplier's instructions. RNA was resuspended in water to a final concentration of 0.2 μg/μl, and 10 μg were used for each microarray. The quality and amount of RNA were verified by measuring the optical density (OD) with Nanodrop and by an Affymetrix bioanalyzer. The complementary RNA and hybridization of the GeneChip array (GeneChip® Plasmodium/Anopheles Genome Array, Affymetrix) were performed at the IGBMC platform (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France) according to Affymetrix protocols. The GeneChip arrays, designed in 2002, included probe sets for ∼14,900 A. gambiae transcripts, which may not fully cover the mosquito genome due to incomplete genome annotation. Raw data were deposited in the Array Express archive and are available under the E-MEXP-3677 code.
Intensity data for each probe set were calculated from images generated with the GeneChip scanner and were analyzed with Bioconductor software, which implements packages from the R language. Intensities were normalized according to the robust multiarray analysis method (28) using the Affy package (29). GeneChip array data were quality-controlled using established bioinformatic methods (30) using the Affycore tools package (31). Probe sets with an absolute fold change greater than 2 were filtered, and significantly regulated probe sets (p < 0.05) were identified by conducting a moderated t test, which enhances statistical power compared with the classical t test by borrowing variance information from independent probe sets,5 followed by p value adjustment according to the method of Westfall and Young (33) using the Multtest package (34). Results were visualized in TreeView software after running a hierarchical clustering analysis with Cluster version 3.0 software (35).
To highlight gene ontology (GO) terms that were significantly enriched among the genes regulated by dsLacZ injection, we conducted a GO enrichment test using the DAVID Bioinformatics Resources 6.7 Web site.
To identify genes potentially regulated by the same pathway, we conducted a k-means clustering analysis that groups genes with respect to similarity of their expression profiles. Affymetrix probe set expression data for the genes significantly regulated after dsLacZ injection and for Fos and Jun were retrieved from the A. gambiae Gene Expression Database at the University of California Irvine for the following conditions: BF3 (3 h post-blood feeding), BF24 (24 h post-blood feeding), and NBF (non-fed). The retrieved microarray data were normalized together with our microarray data as described above. k-means clustering analysis was conducted with k = 4 using the Stats package (36).
Quantification of Gene Expression by Quantitative RT-PCR (qRT-PCR)
To validate the results obtained with the GeneChip arrays, we compared the expression levels of 10 genes between microarray and qRT-PCR. Total RNA from 10 mosquitoes (non-injected control and dsLacZ-injected) was extracted 3 h after injection using TRI Reagent® (Molecular Research Centre, Inc.). Three independent biological replicates from different mosquito generations were performed. Reverse transcription was achieved using the RevertAidTM H Minus First Strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. Gene transcript levels were quantified using Fast SYBR Green® master mix (ABI) in an ABI 7500 Fast Real-time PCR machine. Primers are detailed in supplemental Table S2. The housekeeping gene RPL19 was used to normalize transcript levels, and expression levels in dsLacZ-injected mosquitoes were compared with non-injected controls. To determine a correlation between results obtained by qRT-PCR and by microarray, we conducted a linear regression analysis. The p value reported for the regression equation represents the result of the test that estimates the null hypothesis of the slope as being equal to zero, whereas the p value reported after the coefficient of correlation tests for the significance of the correlation coefficient.
To quantify the transcript levels of AGAP009100 (TGase1), AGAP009098 (TGase2), TEP1, APL1C, LRIM1, and CTL4 (supplemental Table S2) at different time points (1.5, 3, 4.5, and 6 h) after dsLacZ and dsFos injection, we used qRT-PCR. Three independent biological experiments were conducted. Fold change after injection of dsLacZ and dsFos was calculated relative to non-injected controls. A two-way analysis of covariance was applied to determine the statistical significance of the effects of dsRNA treatment and time on transcript abundance. The t test was used to identify significant differences in expression levels between control (dsLacZ) and dsFos-injected mosquitoes for each time point separately.
To verify the efficiency of TEP1, TGase1, TGase2, and Fos knockdown, we used qRT-PCR as described above. RNA was extracted from 10 mosquitoes 4 days after injection of dsRNA, reverse-transcribed, and amplified using primers described in supplemental Table S2. Three independent experiments were performed, and expression levels were normalized to RPL19 expression.
Immunoblotting
Hemolymph of 15 dsRNA-treated females was collected 3–4 days after injection into a Laemmli protein-loading buffer by proboscis clipping. Hemolymph samples were separated on 8% SDS-PAGE. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad). The membrane was blocked by incubation in a 5% milk powder solution for 1 h; rinsed in PBS, 1% Triton X-100; and incubated either with rabbit anti-TEP1 (1:500) or with anti-prophenoloxidase (1:15,000) antibody solutions for 1 h. After PBS washes, the membrane was incubated in a 5% milk powder PBS solution containing a secondary anti-rabbit antibody (1:15,000) tagged with horseradish peroxidase (HRP). The enhanced chemiluminescence kit (Amersham Biosciences) was used to reveal the blot.
Experimental Infections with P. falciparum Gametocytes from Blood Donors
Mosquito infections were carried out in 2007 and in 2010 at OCEAC with the P. falciparum gametocytes collected from the blood of children (5–11 years old) who attended schools in the area of Mfou (3°40 north; 11°35 east), a small city 30 km from Yaoundé. Parasites were detected by Giemsa-stained thick blood smears. Children showing asexual parasitemia were treated with artemisinin-based combination therapy according to national guidelines. Asymptomatic gametocyte-positive children were enrolled as volunteers upon signing of an informed consent form by their legal guardian. The recruitment procedures and experimental protocols were approved by the Cameroonian ethical review committee.
For each volunteer, 4 ml of venous blood were collected by venipuncture in heparinized Vacutainer tubes and centrifuged at 2000 × g for 5 min at 37 °C. The supernatant, containing serum, was replaced by non-immune AB serum to limit transmission-interfering factors (37). The mixture (350 ml) was dispensed into prewarmed glass feeders, on which mosquitoes previously starved for 12 h were allowed to feed for 30 min through a parafilm membrane. Well-fed mosquitoes were maintained in the insectary at 28 °C. After 7 days, midguts were dissected, stained with a 0.4% mercurochrome solution, and examined under light microscopy (×200) for oocyst counts. Significance of differences in oocyst numbers (only for samples with at least one oocyst per midgut) and in prevalence of infection was assessed by computing the standardized mean differences and the odds ratio, respectively, under random effect models using comprehensive meta-analysis software (38).
Genotyping of P. falciparum Gametocytes
Gametocyte filtration and DNA extraction for the samples collected in 2010 were performed as described (12). Purified gametocytes were genotyped at seven microsatellite loci (39): POLYα (chromosome 4, GenBankTM ID G37809), TA87 (chromosome 6, G38838), TA109 (chromosome 6, G38842), ARA2 (chromosome 11, G37848), Pfg377 (chromosome 12, G37851), PfPK2 (chromosome 12, G37852), and TA60 (chromosome 13, G38876). Amplification procedures were performed as described previously (40). The fluorescent PCR products were examined on an AB Prism 3100 Genetic Analyzer (Applied Biosystems) relative to Lys-500 as an internal size standard and examined with GeneMapper software (Applied Biosystems).
RESULTS AND DISCUSSION
Identification of Wound Response Genes by Genome-wide Expression Analysis
We posited that wounding-induced mosquito resistance to human malaria parasites resulted from a transcriptional up-regulation of wound response genes prior to P. falciparum infection. To identify these genes, the transcriptome of A. gambiae females 3 h after injection of dsRNA (dsLacZ) was compared by DNA-based Affymetrix GeneChip arrays with the transcriptome of control age-matched untreated females from sibling cohorts. The choice of this time point was based on our previous observations on TEP1, whose expression was transiently induced by wounding shortly after injection (6). Injections of dsLacZ served as a proxy for wounding and allowed a direct comparison of parasite infection phenotypes with our earlier study (12). Among 141 genes whose expression was modulated (induced or repressed) by injection more than 2-fold, 53 genes displayed statistically significant (p < 0.05) regulation; seven genes were down-regulated (13.2%), whereas 46 were up-regulated (86.8%) (supplemental Fig. S1A and Table S1). Expression profiles of 10 of 53 genes were gauged by qRT-PCR (supplemental Table S2) and compared with the microarray data. Linear regression between the two data sets yielded a coefficient of determination (y = 0.84x + 0.51; P(1,8) = 0.006) and a slope of regression (R2 = 0.64; p = 0.027) close to 1, demonstrating a good correlation between the results obtained by microarrays and by qRT-PCR, thereby validating the values obtained by the microarray approach (supplemental Fig. S2 and Table S3). A previous study identified 81 genes, whose expression was regulated at 1, 6, 12, or 24 h after wounding (41). Only one wound response gene, FBN9 (AGAP011197), was identified by both studies. Methodological differences may explain these results. The previous study was performed with the first generation of cDNA microarrays, representing around 3840 expressed sequence tag clones, and measured transcript abundance at the time points not overlapping with our study. We showed that wounding induces transient changes in gene expression and therefore is sensitive to the timing of the analysis. Additional minor differences may have contributed to the observed discrepancy; the previous study used a different mosquito line (L3-5), wounding consisted of pricking mosquitoes with a needle, and the microarray design was based on expressed sequence tags cloned from a hemocyte-like cell line. Last, distinct statistical methods were used for data analysis.
GO analysis revealed a high proportion of wound response genes that encoded proteins with a potential role in proteolysis-related processes (24.5%). For comparison, the second largest GO class comprised the genes encoding proteins potentially involved in signal transduction, which represented 3.7% of all regulated genes (supplemental Fig. S1B). In line with these data, proteolysis represented the only biological process significantly (p < 0.001) regulated in our data set, and proteolysis-related genes were enriched 5-fold compared with their proportion in the annotated A. gambiae genome. Proteolysis is known to be involved in responses to injury in a number of organisms, where it swiftly activates such processes as blood coagulation, fibrinogenesis, clotting, and angiogenesis (22). Among proteolysis-related genes, we identified 11 clip domain serine proteases (CLIPs), enzymes that trigger extracellular proteolytic cascades in invertebrates (Fig. 1A and supplemental Table S1). Three CLIPs have known functions in mosquito immune responses and/or in interactions with P. berghei. CLIPB15 has been shown to promote parasite killing by lysis (42), whereas CLIPA2 and CLIPA8 are required for ookinete melanization (43, 44). Our analysis identified several non-CLIP serine proteases. One of them, immune-induced serine protease 1 (ISRP1, AGAP005194), was previously reported to be induced by LPS injection (45). In these experiments, LPS-injected mosquitoes were compared with non-injected controls. Therefore, it is possible that expression of this protease is induced by injection only. Moreover, ISRP1 was identified in a transcriptional profiling of genes regulated by mating in the female reproductive organs (46). Expression of ISRP1 was shut down in the female atrium at 24 h postmating. The exact roles of this protease in immunity and reproduction remain to be elucidated. The activity of serine proteases is tightly controlled by serine protease inhibitors (serpins). We detected one wound response gene (AGAP00137) coding for serpin 11 (SRPN11), previously reported to be expressed throughout the mosquito life cycle (47).
FIGURE 1.
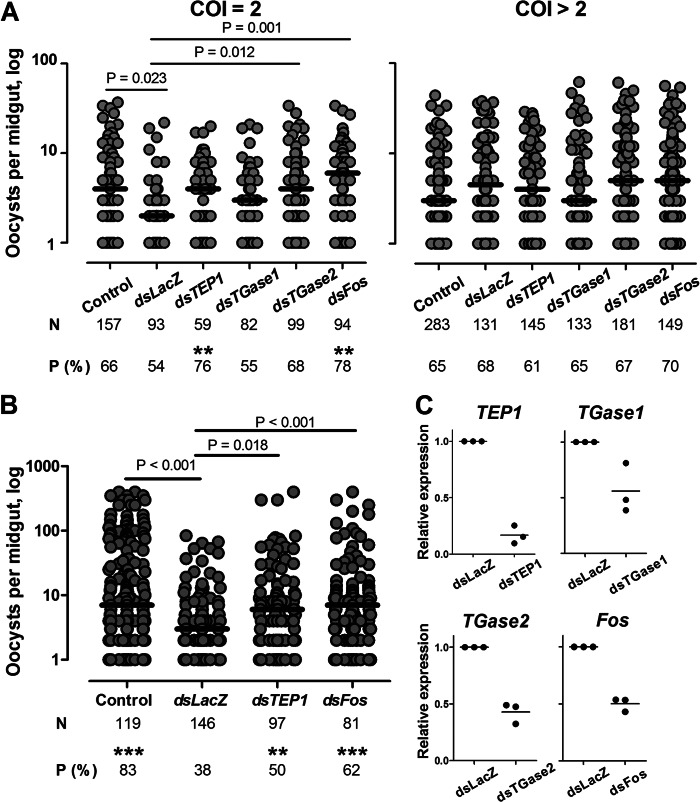
Effect of TGase1, TGase2, and Fos silencing on P. falciparum development in the mosquito. Mosquitoes were injected with dsRNA against TEP1, TGase1, TGase2, and Fos (A) or TEP1 and Fos (B), respectively. Non-injected (Control) and dsLacZ-injected mosquitoes were used as controls. After injection, mosquitoes were infected with four biclonal (COI = 2) and six polyclonal (COI > 2) P. falciparum isolates in A or with four other isolates in B. Mosquito midguts were dissected 7 days after infection. Only samples with at least one oocyst are plotted, and they represent the infection intensity. Horizontal bars indicate medians. N, total numbers of mosquitoes dissected; P (%), percentage of infected mosquitoes (prevalence). Significant differences in prevalence are indicated by two asterisks (p < 0.01) and three asterisks (p < 0.001) above the prevalence values. C, knockdown efficiency was examined by quantitative RT-PCR performed in three independent experiments (black dots) for TEP1, TGase1, TGase2, and Fos. Expression levels of each gene after dsRNA injection were compared with the levels in dsLacZ-injected control mosquitoes. Horizontal bars represent means.
Several other genes with demonstrated functions in anti-Plasmodium responses were identified. Interestingly, most of them are regulated by the members of the NF-κB family, Rel1 and/or Rel2 (11, 48). Fibrinogen domain immunolectins (FBN) are common pattern recognition receptors in invertebrates. Two FBN-encoding genes were up-regulated upon wounding, including FBN9, a known antiplasmodium factor (49). Expression of three TEP genes (TEP1, TEP3, and TEP12) was significantly up-regulated by wounding. TEP1 and TEP3 cooperate to promote phagocytosis of Gram-positive and Gram-negative bacteria (50) and in antiparasitic responses (51, 52). TEP1 circulates in the mosquito hemolymph as a trimeric complex comprising two leucine-rich repeat proteins, APL1C and LRIM1 (53, 54). Levels of APL1C and LRIM1 transcripts were also increased by wounding, confirming our previous observation that the mosquito complement-like system is involved in the wound responses (12). Wounding also induced the transcript abundance of another leucine-rich repeat-encoding gene, APL2 (or LRRD7), which was reported to affect both P. berghei and P. falciparum development, and of two genes encoding C-type lectins, CTLMA2 and CTL4, whose depletion caused melanization of P. berghei ookinetes in the mosquito midguts (55–57).
A subset of the wound response genes encoded diverse proteins with potential functions in tissue repair or cuticle formation. Abundance of AGAP001508 transcripts, a Drosophila homologue of a gene encoding a structural protein of the insect cuticle was 6-fold induced by wounding (supplemental Table S1) (58, 59). Wound response genes also included AGAP003720, a homologue of the Drosophila annexin X (supplemental Table S1), which has been implicated in wounding-induced membrane resealing in Drosophila, and AGAP009145, a homologue of a Drosophila protein involved in DNA repair. In contrast, transcript levels of flightin (AGAP007249), whose homologue in adult Drosophila regulates somatic development of the flight muscles (60–62), were repressed. The gene encoding ICHIT (AGAP006434) contains two putative chitin-binding domains. Although its expression has been reported to be induced by Plasmodium and bacteria infections (63), the ICHIT transcript level was down-regulated by wounding as well as the cuticle protein family-like CPFL6 (AGAP010907). Finally, transcript levels of two genes, AGAP008963 and AGAP012399, that encode α-amylases, were also down-regulated by wounding. These genes are massively expressed in the midgut of males and females and might be involved in glucose degradation during sugar feeding (64).
TGase2 Contributes to the Mosquito Immune Responses against P. falciparum
Among wound response genes, we detected two TGase-encoding genes, AGAP009100 (TGase1) and AGAP009098 (TGase2). To test whether these TGases contributed to mosquito resistance to P. falciparum after wounding, we silenced expression of the corresponding genes by injecting dsRNA into adult females. In these experiments, we used three control groups: (i) non-injected mosquitoes from the same cage as a control for infection levels; (ii) dsLacZ-injected mosquitoes as a control for wounding-induced parasite killing; and (iii) dsTEP1-injected mosquitoes as a positive control (12). Silencing efficiency was validated using qRT-PCR (Fig. 1C). Injected and control mosquitoes were fed 3–4 days after dsRNA injection with the blood of 10 P. falciparum gametocyte carriers. Substitution of donor sera with non-immune sera was performed to eliminate possible effects of donor blood composition and of immune factors on parasite development. Because the genetic complexity of gametocytes impacts the levels of P. falciparum infections and the efficiency of parasite killing by the mosquito immune response, we purified and genotyped gametocytes in the donor blood (12). All 10 gametocyte carriers contained 2–4 P. falciparum clones (supplemental Table S4). Results of experimental infections were analyzed separately for two groups according to the complexity of infection (COI) of the P. falciparum isolates: (i) four biclonal isolates (COI = 2) and (ii) six polyclonal isolates (COI > 2).
Consistent with our previous results (12), the overall oocyst levels in polyclonal infections were low. In the experiments with biclonal parasites, the infection intensity in non-injected mosquitoes (Control) was significantly higher than in dsLacZ-injected mosquitoes (Fig. 1A and supplemental Table S4). Silencing TGase2 restored the levels of infection intensity to the levels in non-injected mosquitoes, which were significantly higher than after wounding. In contrast, in mosquitoes depleted for TGase1, oocyst numbers per midgut were similar to the ones in dsLacZ-injected mosquitoes. Silencing of TEP1 resulted in a significant increase of the prevalence of infection when compared with dsLacZ mosquitoes and a non-significant increase in infection intensity. Medians of infection intensity were similar between dsTGase2- and dsTEP1-injected mosquitoes. These results suggest that TGase2 mediates wounding-induced killing of malaria parasites. Consistent with our previous study (12), no effect of wounding or of gene silencing was observed in polyclonal infections (COI > 2) (Fig. 1A).
Levels of TGase2 transcripts increased within the first 3 h after dsLacZ-injection. How does this transient TGase2 up-regulation impacts P. falciparum development 3–4 days later? We believe that wounding, similar to Cactus depletion, boosts the efficiency of parasite killing (6). However, better understanding of the TGase2 role, direct or indirect, in parasite killing will be required to answer this question.
Fos Regulates TGase 2 Expression after Wounding
We next sought to identify the signaling pathway(s) that regulate wounding-induced expression of TGase2. Results of our microarray analysis failed to reveal any components of signaling pathways whose expression was induced by wounding (supplemental Table S1). Therefore, we examined the promoter region of TGase2 for transcription factor binding motifs using Tf site scan software (65). A number of potential transcription factor binding sites were identified, including two binding sites for activator protein 1 (AP-1) (TGAGTCA) in the 1-kb region upstream of the TATA box. In Drosophila, AP-1 transcription factors function as heterodimers of Jun and Fos proteins that bind to AP-1 DNA motif and mediate activation of the JNK pathway (66), one of the major regulators of wounding responses in Drosophila (67). Because we did not detect any evidence of Jun or Fos up-regulation 3 h after wounding in the microarray data, we increased the resolution power of our analysis by including additional experimental data retrieved from the University of California Irvine A. gambiae Gene Expression Profile Web site. We then grouped the wounding-induced genes identified here and Fos (AGAP001093) and Jun (AGAP006386) according to their expressional profiles in control, dsLacZ-injected mosquitoes (3 h postinjection) and before and 3 and 24 h after blood feeding in non-injected mosquitoes by applying a k-means clustering analysis (Fig. 2 and supplemental Table S5). Co-regulated genes often belong to the same signaling pathway. Our analysis identified three clusters containing 14–16 genes and one small cluster of three genes. Cluster 1 encompassed the most potent antiparasitic genes, such as TEP1, FBN9, LRIM1, and APL1C, two genes with a reported protective effect on Plasmodium (CTL4 and CTLMA2), and four CLIPA genes (CLIPA1, -4, -7, and -14). In line with these results, TEP1, LRIM1, APL1C, and CTL4 are co-regulated through the Rel1/Cactus and Rel2 pathways (6, 48). Given that co-regulated genes often display similar functions, other genes of the cluster may also have important roles in responses of mosquitoes to malaria parasites. Cluster 2 grouped three genes: ISPR1, Flightin, and ICHIT. Cluster 3 included genes with diverse functions: TEP3 and -12, three genes with potential roles in tissue repair, genes coding for proteolysis-related proteins, such as five CLIP genes (CLIPA2, -A8, -A12, -B15, and -C7), and the two α-amylases repressed by wounding. Cluster 4 contained Tollo, CPFL6, fibrinogen (AGAP005848), and two proteolysis-related CLIP genes (CLIPB17 and -36) but most importantly, TGase1, TGase2, Fos, and Jun, suggesting that these genes may be part of the same signaling pathway.
FIGURE 2.
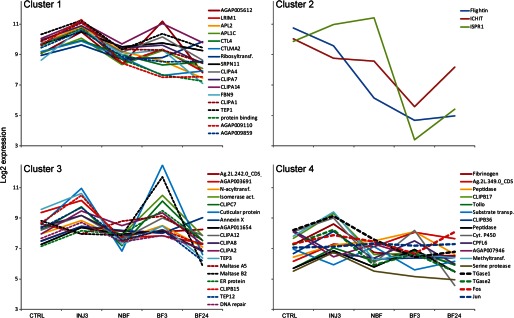
k-means clustering analysis of the expressional profiles of the genes regulated by wounding. Clusters represent genes with similar expressional profiles. Transcript abundance was quantified using microarray data for the following conditions: non-injection, injection (3 h after dsLacZ injection), BF3 (3 h after blood feeding), BF24 (24 h after blood feeding), and NBF (nonfed). Data for the last three conditions were retrieved from the A. gambiae Gene Expression Database at the University of California Irvine.
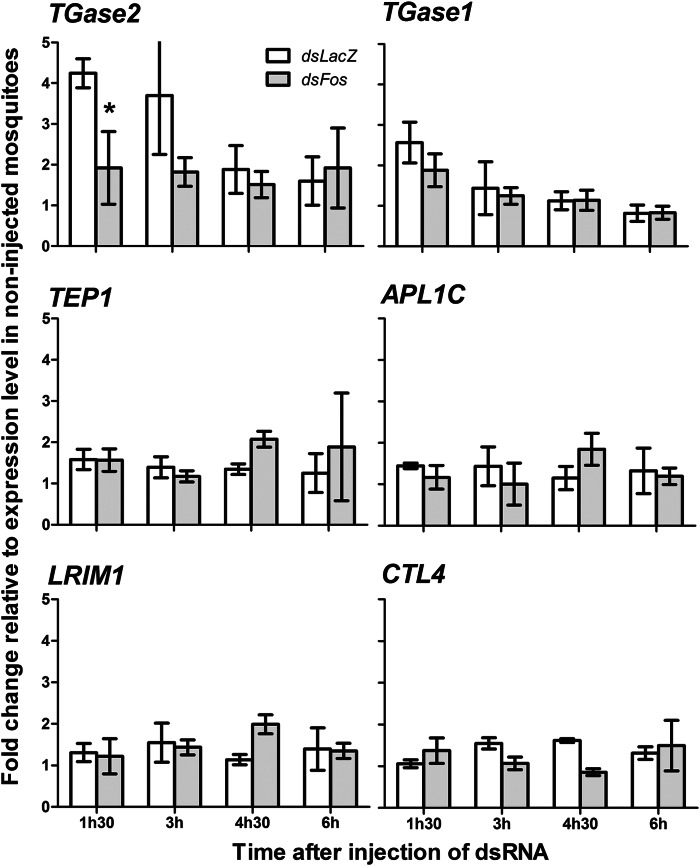
To examine whether expression of the TGases after wounding was indeed regulated by AP-1, we compared expression levels of TGase1, TGase2, TEP1, APL1C, LRIM1, and CTL4 at different time points after injection of dsLacZ and dsFos. Changes in the transcript abundance at the indicated time points after injections were calculated relative to non-injected control mosquitoes. Injection of dsFos significantly decreased levels of TGase2 transcripts after wounding (two-way analysis of covariance, RNAi effect: F(1,20) = 6.83; p = 0.017) but had no effect on transcript abundance of other tested genes (Fig. 3 and supplemental Table S6). Immunoblotting analysis further confirmed at the protein level that silencing of Fos did not affect TEP1 expression at 24 and 48 h after wounding (supplemental Fig. S3). Interestingly, depletion of Fos had no effect on transcript levels of TGase1, although the latter clustered with Fos in the k-means analysis (Fig. 2 and supplemental Table S5) and contained three potential AP-1 binding motifs in its promoter region. Because our analysis also identified one potential NF-κB binding site in its promoter region, AP-1 and Rel1/Cactus modules may cooperate in regulating TGase1 expression after wounding.
FIGURE 3.
Fos-mediated regulation of TGase2, TGase1, TEP1, APL1, LRIM1, and CTL4 expression after wounding. -Fold change ± S.E. (error bars) relative to non-injected mosquitoes was calculated for TGase1, TGase2, TEP1, LRIM1, APL1C, and CTL4 genes at 1.5, 3, 4.5, and 6 h after injection of dsLacZ and dsFos using qRT-PCR. Non-injected mosquitoes of the corresponding time were used as controls. The RPL19 housekeeping gene was used for normalization. *, significant difference (t test, p < 0.05) in transcript abundance between dsLacZ- and dsFos-injected mosquitoes.
Fos Contributes to the Mosquito Resistance to P. falciparum
To further ascertain the role of Fos in the wounding-induced P. falciparum killing, we compared infection levels in dsLacZ- and dsFos-injected mosquitoes using donor blood infected with field isolates of P. falciparum. Gene silencing was conducted on the 10 polyclonal isolates previously used and on four additional isolates collected earlier in the spring of 2007, for which genotyping was not performed. Several characteristics of these infections (e.g. high infection rates and sensitivity to injection and to TEP1 depletion) were reminiscent of monoclonal infections (12) (Fig. 1B and supplemental Table S7). In both sets of experiments, we observed a significant increase in infection intensity and in oocyst prevalence in dsFos-injected mosquitoes as compared with dsLacZ controls for biclonal isolates and for infections carried out in 2007 (Fig. 1, A and B, respectively). Taken together, our results strongly suggest that AP-1/Fos contributes to the wounding-induced killing of P. falciparum through direct or indirect regulation of TGase2 expression and that Fos/TGase2 constitute a novel axis in the antiparasitic responses of A. gambiae.
CONCLUSIONS
Here we report that AP-1/Fos-regulated transglutaminase 2 restricts development of human malaria parasite in A. gambiae. In Drosophila, TGase1 has been previously implicated in immune defenses against bacteria (25), but this is the first report that demonstrates antiparasitic properties of TGases. How TGase2 contributes to P. falciparum killing is currently unknown. It is also unclear whether TGase2 requires the function of TEP1, which also mediates wounding-induced parasite killing (12). Interestingly, our results suggest that distinct signaling pathways regulate wound responses; for instance, expression of TEP1 is regulated by the Rel1/Rel2 module but not by AP-1/Fos (6). In contrast, induction of TGase2 expression after wounding is controlled by AP-1/Fos. This is the first indication of the role of the component of the AP-1 transcriptional factor in the antiparasitic responses of mosquitoes to the human malaria parasite, P. falciparum. A recent study using a rough eye screen in Drosophila revealed a genetic interaction between the JNK pathway and TGase1 (32). Results reported here extend this observation to mosquito immune responses. In the natural setting, mosquitoes are exposed to a rough environment that causes frequent losses of appendices and exposes mosquitoes to predator insults that could induce wound responses, thereby modulating mosquito vector competence. The identification of pathways responsible for the antiparasitic response in A. gambiae is not only crucial for the design of efficacious vector control strategies based on the release of transgenesis-engineered refractory mosquitoes but also illustrates the diversity of mechanisms that insects employ to cope with diverse pathogens.
Supplementary Material
Acknowledgments
We are grateful to the children of Mfou primary schools and to their parents and guardians for participating in this study. We thank Dr. E. Manga, C. Efemba, and M. Biloa of the Mfou hospital for assistance in the field; S. Kemleu, J.-P. Agbor, and R. Nyambam for blood smear readings; and E. Onana and I. Tchikangwa for mosquito rearing in OCEAC. We thank J. Soichot for help with the mosquito colonies. Drs. F. Catteruccia and S. Mitchell are gratefully acknowledged for critical reading of the manuscript.
This work was supported by funds from CNRS (UPR 9022), INSERM (U963), and l'Institut de Recherche pour le Développement (IRD); by grants from EC FP7 MALVECBLOK (grant agreement number 223601) and EVIMalar (grant agreement number 242095); by the IRD-CNRS collaboration (Convention 6774.00); by a fellowship from the Fondation pour la Recherche Médicale (FRM) (to J. P.); by a DSFIRD scholarship (to S. E. N.); and by a BIOMALPAR Ph.D. fellowship (to A. R.).

This article contains supplemental Tables S1–S3 and Figs. S1–S7.
D. M. Witten and R. Tibshirani, unpublished data.
- TGase
- transglutaminase
- GO
- gene ontology
- qRT-PCR
- quantitative RT-PCR
- CLIP
- clip domain serine protease
- COI
- complexity of infection.
REFERENCES
- 1. World Health Organization (2010) World Malaria Report. World Health Organization, Geneva [Google Scholar]
- 2. Breman J. G., Alilio M. S., Mills A. (2004) Conquering the intolerable burden of malaria. What's new, what's needed. A summary. Am. J. Trop. Med. Hyg. 71, Suppl. 2, 1–15 [PubMed] [Google Scholar]
- 3. Collins F. H., Sakai R. K., Vernick K D.., Paskewitz S., Seeley D. C., Miller L. H., Collins W. E., Campbell C. C., Gwadz R. W. (1986) Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science 234, 607–610 [DOI] [PubMed] [Google Scholar]
- 4. Michel K., Kafatos F. C. (2005) Mosquito immunity against Plasmodium. Insect Biochem. Mol. Biol. 35, 677–689 [DOI] [PubMed] [Google Scholar]
- 5. Dong Y., Das S., Cirimotich C., Souza-Neto J. A., McLean K. J., Dimopoulos G. (2011) Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 7, e1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frolet C., Thoma M., Blandin S., Hoffmann J. A., Levashina E. A. (2006) Boosting NF-κB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 25, 677–685 [DOI] [PubMed] [Google Scholar]
- 7. Fearon D. T., Locksley R. M. (1996) The instructive role of innate immunity in the acquired immune response. Science 272, 50–53 [DOI] [PubMed] [Google Scholar]
- 8. Lemaitre B., Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 9. Barillas-Mury C., Charlesworth A., Gross I., Richman A., Hoffmann J. A., Kafatos F. C. (1996) Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. EMBO J. 15, 4691–4701 [PMC free article] [PubMed] [Google Scholar]
- 10. Christophides G. K., Zdobnov E., Barillas-Mury C., Birney E., Blandin S., Blass C., Brey P. T., Collins F. H., Danielli A., Dimopoulos G., Hetru C., Hoa N. T., Hoffmann J. A., Kanzok S. M., Letunic I., Levashina E. A., Loukeris T. G., Lycett G., Meister S., Michel K., Moita L. F., Muller H.-M., Osta M. A., Paskewitz S. M., Reichhart J.-M., Rzhetsky A., Troxler L., Vernick K. D., Vlachou D., Volz J., von Mering C., Xu J., Zheng L., Bork P., Kafatos F. C. (2002) Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165 [DOI] [PubMed] [Google Scholar]
- 11. Garver L. S., Dong Y., Dimopoulos G. (2009) Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 5, e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nsango S. E., Abate L., Thoma M., Pompon J., Fraiture M., Rademacher A., Berry A., Awono-Ambene P. H., Levashina E. A., Morlais I. (2012) Genetic clonality of Plasmodium falciparum affects the outcome of infection in Anopheles gambiae. Int. J. Parasitol. 42, 589–595 [DOI] [PubMed] [Google Scholar]
- 13. Wood W., Jacinto A., Grose R., Woolner S., Gale J., Wilson C., Martin P. (2002) Wound healing recapitulates morphogenesis in Drosophila embryos. Nat. Cell Biol. 4, 907–912 [DOI] [PubMed] [Google Scholar]
- 14. Stramer B., Winfield M., Shaw T., Millard T. H., Woolner S., Martin P. (2008) Gene induction following wounding of wild-type versus macrophage-deficient Drosophila embryos. EMBO Rep. 9, 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furie B., Furie B. C. (1988) The molecular basis of blood coagulation. Cell 53, 505–518 [DOI] [PubMed] [Google Scholar]
- 16. Kalinin A., Marekov L. N., Steinert P. M. (2001) Assembly of the epidermal cornified cell envelope. J. Cell Sci. 114, 3069–3070 [DOI] [PubMed] [Google Scholar]
- 17. Lorand L., Graham R. M. (2003) Transglutaminases. Crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 18. Lustigman S., Brotman B., Huima T., Castelhano A. L., Singh R. N., Mehta K., Prince A. M. (1995) Transglutaminase-catalyzed reaction is important for molting of Onchocerca volvulus third-stage larvae. Antimicrob. Agents Chemother. 39, 1913–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shibata T., Ariki S., Shinzawa N., Miyaji R., Suyama H., Sako M., Inomata N., Koshiba T., Kanuka H., Kawabata S. (2010) Protein crosslinking by transglutaminase controls cuticle morphogenesis in Drosophila. PLoS ONE 5, e13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kopàcek P., Hall M., Söderhäli K. (1993) Characterization of a clotting protein, isolated from plasma of the freshwater crayfish Pacifastacus leniusculus. Eur. J. Biochem. 213, 591–597 [DOI] [PubMed] [Google Scholar]
- 21. Scherfer C., Qazi M. R., Takahashi K., Ueda R., Dushay M. S., Theopold U., Lemaitre B. (2006) The Toll immune-regulated Drosophila protein Fondue is involved in hemolymph clotting and puparium formation. Dev. Biol. 295, 156–163 [DOI] [PubMed] [Google Scholar]
- 22. Theopold U., Schmidt O., Söderhäll K., Dushay M. S. (2004) Coagulation in arthropods. Defence, wound closure and healing. Trends Immunol. 25, 289–294 [DOI] [PubMed] [Google Scholar]
- 23. Matsuda Y., Koshiba T., Osaki T., Suyama H., Arisaka F., Toh Y., Kawabata S. (2007) An arthropod cuticular chitin-binding protein endows injured sites with transglutaminase-dependent mesh. J. Biol. Chem. 282, 37316–37324 [DOI] [PubMed] [Google Scholar]
- 24. Osaki T., Kawabata S. (2004) Structure and function of coagulogen, a clottable protein in horseshoe crabs. Cell Mol. Life Sci. 61, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z., Wilhelmsson C., Hyrsl P., Loof T. G., Dobes P., Klupp M., Loseva O., Mörgelin M., Iklé J., Cripps R. M., Herwald H., Theopold U. (2010) Pathogen entrapment by transglutaminase. A conserved early innate immune mechanism. PLoS Pathog. 6, e1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogers D. W., Baldini F., Battaglia F., Panico M., Dell A., Morris H. R., Catteruccia F. (2009) Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLoS Biol. 7, e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blandin S., Moita L. F., Köcher T., Wilm M., Kafatos F. C., Levashina E. A. (2002) Reverse genetics in the mosquito Anopheles gambiae. Targeted disruption of the Defensin gene. EMBO J. 3, 852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 29. Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004) affy. Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 [DOI] [PubMed] [Google Scholar]
- 30. Burgoon L. D., Eckel-Passow J. E., Gennings C., Boverhof D. R., Burt J. W., Fong C. J., Zacharewski T. R. (2005) Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 33, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. MacDonald J. W. (2008) Affycoretools: Functions Useful for Those Doing Repetitive Analyses with Affymetrix GeneChips. R package, version 1.26.0, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 32. Umehara M., Ichikawa A., Sakamoto H., Yamada A., Yoshioka Y., Yamaguchi M., Ikura K. (2010) Over-expression of transglutaminase in the Drosophila eye imaginal disc induces a rough eye phenotype. Mol. Cell Biochem. 342, 223–232 [DOI] [PubMed] [Google Scholar]
- 33. Westfall P. H., Young S. S. (1993) Resampling-based Multiple Testing: Examples and Methods for p-Value Adjustment, John Wiley & Sons, Inc., New York [Google Scholar]
- 34. Pollard K. S., Gilbert H. N., Ge Y., Taylor S., Dudoit S. (2004) Multtest: Resampling-based Multiple Hypothesis Testing. R package, version 2.10.0, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 35. de Hoon M. J., Imoto S., Nolan J., Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 36. R Development Core Team (2011) R. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 37. Boudin C., Diop A., Gaye A., Gadiaga L., Gouagna C., Safeukui I., Bonnet S. (2005) Plasmodium falciparum transmission blocking immunity in three areas with perennial or seasonal endemicity and different levels of transmission. Am. J. Trop. Med. Hyg. 73, 1090–1095 [PubMed] [Google Scholar]
- 38. Borenstein M., Hedges L., Higgins J., Rothstein H. (2005) Comprehensive Meta-analysis, version 2 Biostat, Englewood, NJ [Google Scholar]
- 39. Anderson T. J., Haubold B., Williams J. T., Estrada-Franco J. G., Richardson L., Mollinedo R., Bockarie M., Mokili J., Mharakurwa S., French N., Whitworth J., Velez I. D., Brockman A. H., Nosten F., Ferreira M. U., Day K. P. (2000) Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17, 1467–1482 [DOI] [PubMed] [Google Scholar]
- 40. Annan Z., Durand P., Ayala F. J., Arnathau C., Awono-Ambene P., Simard F., Razakandrainibe F. G., Koella J. C., Fontenille D., Renaud F. (2007) Population genetic structure of Plasmodium falciparum in the two main African vectors, Anopheles gambiae and Anopheles funestus. Proc. Natl. Acad. Sci. U.S.A. 104, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dimopoulos G., Christophides G. K., Meister S., Schultz J., White K. P., Barillas-Mury C., Kafatos F. C. (2002) Genome expression analysis of Anopheles gambiae. Responses to injury, bacterial challenge, and malaria infection. Proc. Natl. Acad. Sci. U.S.A. 99, 8814–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volz J., Osta M. A., Kafatos F. C., Muller H. M. (2005) The roles of two clip domain serine proteases in innate immune responses of the malaria vector Anopheles gambiae. J. Biol. Chem. 280, 40161–40168 [DOI] [PubMed] [Google Scholar]
- 43. Schnitger A. K., Kafatos F. C., Osta M. A. (2007) The melanization reaction is not required for survival of Anopheles gambiae mosquitoes after bacterial infections. J. Biol. Chem. 282, 21884–21888 [DOI] [PubMed] [Google Scholar]
- 44. Volz J., Müller H.-M., Zdanowicz A., Kafatos F. C., Osta M. A. (2006) A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 8, 1392–1405 [DOI] [PubMed] [Google Scholar]
- 45. Oduol F., Xu J., Niare O., Natarajan R., Vernick K. D. (2000) Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. U.S.A. 97, 11397–11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rogers D. W., Whitten M. M., Thailayil J., Soichot J., Levashina E. A., Catteruccia F. (2008) Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc. Natl. Acad. Sci. U.S.A. 105, 19390–19395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suwanchaichinda C., Kanost M. R. (2009) The serpin gene family in Anopheles gambiae. Gene 442, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meister S., Kanzok S. M., Zheng X.-L., Luna C., Li T.-R., Hoa N. T., Clayton J. R., White K. P., Kafatos F. C., Christophides G. K., Zheng L. (2005) Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 102, 11420–11425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dong Y., Dimopoulos G. (2009) Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 284, 9835–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moita L. F., Wang-Sattler R., Michel K., Zimmermann T., Blandin S., Levashina E. A., Kafatos F. C. (2005) In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity 23, 65–73 [DOI] [PubMed] [Google Scholar]
- 51. Blandin S., Shiao S.-H., Moita L. F., Janse C. J., Waters A. P., Kafatos F. C., Levashina E. A. (2004) Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670 [DOI] [PubMed] [Google Scholar]
- 52. Povelones M., Upton L. M., Sala K. A., Christophides G. K. (2011) Structure-function analysis of the Anopheles gambiae LRIM1/APL1c complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 7, e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riehle M. M., Markianos K., Niaré O., Xu J., Li J., Touré A. M., Podiougou B., Oduol F., Diawara S., Diallo M., Coulibaly B., Ouatara A., Kruglyak L., Traoré S. F., Vernick K. D. (2006) Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 312, 577–579 [DOI] [PubMed] [Google Scholar]
- 54. Riehle M. M., Xu J., Lazzaro B. P., Rottschaefer S. M., Coulibaly B., Sacko M., Niare O., Morlais I., Traore S. F., Vernick K. D. (2008) Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE 3, e3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dong Y., Aguilar R., Xi Z., Warr E., Mongin E., Dimopoulos G. (2006) Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Osta M. A., Christophides G. K., Kafatos F. C. (2004) Effects of mosquito genes on Plasmodium development. Science 303, 2030–2032 [DOI] [PubMed] [Google Scholar]
- 57. Cohuet A., Osta M. A., Morlais I., Awono-Ambene P. H., Michel K., Simard F., Christophides G. K., Fontenille D., Kafatos F. C. (2006) Anopheles and Plasmodium. From laboratory models to natural systems in the field. EMBO Rep. 7, 1285–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Togawa T., Augustine Dunn W., Emmons A. C., Willis J. H. (2007) CPF and CPFL, two related gene families encoding cuticular proteins of Anopheles gambiae and other insects. Insect Biochem. Mol. Biol. 37, 675–688 [DOI] [PubMed] [Google Scholar]
- 59. Ren N., Zhu C., Lee H., Adler P. N. (2005) Gene expression during Drosophila wing morphogenesis and differentiation. Genetics 171, 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abreu-Blanco M. T., Verboon J. M., Parkhurst S. M. (2011) Single cell wound repair. Dealing with life's little traumas. Bioarchitecture 1, 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adams M. D., McVey M., Sekelsky J. J. (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265–267 [DOI] [PubMed] [Google Scholar]
- 62. Schnorrer F., Schönbauer C., Langer C. C., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A., Keleman K., Dickson B. J. (2010) Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464, 287–291 [DOI] [PubMed] [Google Scholar]
- 63. Dimopoulos G., Seeley D., Wolf A., Kafatos F. C. (1998) Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. EMBO J. 17, 6115–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marinotti O., James A. A. (1990) An α-glucosidase in the salivary glands of the vector mosquito, Aedes aegypti. Insect Biochem. 20, 619–623 [Google Scholar]
- 65. Ghosh D. (2000) Object-oriented transcription factors database (ooTFD). Nucleic Acids Res. 28, 308–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zeitlinger J., Kockel L., Peverali F. A., Jackson D. B., Mlodzik M., Bohmann D. (1997) Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila Fos mutants. EMBO J. 16, 7393–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rämet M., Lanot R., Zachary D., Manfruelli P. (2002) JNK signaling pathway is required for efficient wound healing in Drosophila. Dev. Biol. 241, 145–156 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.