Background: Pif1 helicase plays a variety of roles in both the nucleus and mitochondria.
Results: The kinetic step size for Pif1 is one base pair, and translocation on ssDNA is coupled tightly with ATP hydrolysis.
Conclusion: Hydrolysis of one ATP results in movement of Pif1 by a single nucleotide.
Significance: Pif1 is an active helicase with a uniform stepping mechanism.
Keywords: DNA-binding Protein, DNA Helicase, Kinetics, Protein Translocation, Yeast, DNA Unwinding, Protein Displacement, Step Size
Abstract
Kinetic analysis of the DNA unwinding and translocation activities of helicases is necessary for characterization of the biochemical mechanism(s) for this class of enzymes. Saccharomyces cerevisiae Pif1 helicase was characterized using presteady state kinetics to determine rates of DNA unwinding, displacement of streptavidin from biotinylated DNA, translocation on single-stranded DNA (ssDNA), and ATP hydrolysis activities. Unwinding of substrates containing varying duplex lengths was fit globally to a model for stepwise unwinding and resulted in an unwinding rate of ∼75 bp/s and a kinetic step size of 1 base pair. Pif1 is capable of displacing streptavidin from biotinylated oligonucleotides with a linear increase in the rates as the length of the oligonucleotides increased. The rate of translocation on ssDNA was determined by measuring dissociation from varying lengths of ssDNA and is essentially the same as the rate of unwinding of dsDNA, making Pif1 an active helicase. The ATPase activity of Pif1 on ssDNA was determined using fluorescently labeled phosphate-binding protein to measure the rate of phosphate release. The quantity of phosphate released corresponds to a chemical efficiency of 0.84 ATP/nucleotides translocated. Hence, when all of the kinetic data are considered, Pif1 appears to move along DNA in single nucleotide or base pair steps, powered by hydrolysis of 1 molecule of ATP.
Introduction
Saccharomyces cerevisiae Pif1 is the prototypical member of the Pif1 family of DNA helicases that is conserved from yeast to humans (1). The helicase was discovered by using forward genetic mutation experiments affecting mitochondrial DNA (mtDNA) recombination (2, 3). In the absence of Pif1, yeast cells were sensitive to UV light on a nonfermentable carbon source (4). At higher temperatures, Pif1 null cells lose mtDNA at a high rate and generate respiratory-deficient cells (petites) (4). Later findings led to the discovery of several nuclear functions of Pif1 in addition to the previously known mitochondrial functions (5–7). Pif1 is transcribed from a single open reading frame but has two in-frame translational start codons that determine the localization of the protein to either the nucleus or the mitochondria (8). A full-length protein translated from first start codon (first residue) is targeted to mitochondria, albeit after the cleavage of mitochondrial targeting signal (N-terminal 46 residues). Truncated Pif1 which translates from the second start codon (40th residue) is targeted to the nucleus. In the nucleus, Pif1 is involved in telomere regulation (8, 9), repair of double-stranded DNA (dsDNA) breaks (10, 11), Okazaki fragment maturation (5, 6), and ribosomal DNA replication (12). In the mitochondria, Pif1 functions are required in mtDNA recombination, maintenance of mtDNA by prevention/repair of damage, and mtDNA replication (3, 4, 13–15). All of these functions are driven by the ATPase-dependent motor activity of Pif1 including moving unidirectionally on single-stranded DNA (ssDNA) and unwinding double-stranded nucleic acids into single strands.
Pif1 belongs to the superfamily 1 (SF1)2 class of helicases which includes a diverse group of monomeric and dimeric helicases. SF1 helicases are divided into two groups depending on the directionality of translocation, SF1A (3′ → 5′) and SF1B (5′ → 3′). A few of the SF1 helicases that are relatively well studied include Dda, RecBCD, RecD2, Pif1, PcrA, RepA, UvrD, Dna2, Upf1, and Rrm3. The SF1 helicases are further classified into three families: UvrD/Rep family, Pif1-like family, and Upf1-like family (16). The UvrD/Rep family is SF1A, and the Pif1-like and the Upf1-like families are SF1B. The Pif1-like family includes yeast Pif1 and Rrm3, human Pif1 and HelB, bacteriophage T4 Dda, and Escherichia coli RecD and TraI. Crystal structures of RecD2 and Dda have revealed that the core motor comprises paired RecA-like domains and an SH3 domain (17, 18). Domain 1B, which forms the pin structure, is proposed to split the incoming duplex.
Biochemical studies on Pif1 have revealed that it unwinds DNA with 5′ → 3′ polarity in an ATP and Mg2+-dependent manner (19). It is reported to exist as a monomer in solution and to dimerize upon binding to ssDNA (19, 20). Pif1 prefers forked DNA structures over non-forked structures for unwinding (19). Pif1 does not bind to RNA; however, it preferentially unwinds DNA·RNA duplexes (where the displaced strand is RNA) over DNA·DNA duplexes (21). It also preferentially unwinds G4-DNA structures (22). In vitro telomerase assays showed that the presence of Pif1 reduces the processivity of the telomerase complex (9). A recent report showed that Pif1 stably interacts with a mitochondrial SSB, Rim1, both in vivo and in vitro (23). The presence of Rim1 in helicase assays enhanced Pif1 unwinding activity. Quantitative analysis of the kinetic mechanism of the motor activity of Pif1, including translocation on ssDNA and DNA unwinding, provides the basic foundation to unravel the multiple functions of Pif1 in mitochondrial and nuclear DNA metabolic processes. In this paper we use a series of transient state kinetic experiments to obtain in-depth understanding of the relationship between Pif1 helicase ssDNA translocation and dsDNA unwinding mechanism, including the measurement of its step size.
EXPERIMENTAL PROCEDURES
Materials
HEPES, NaCl, MgCl2, EDTA, 2 mm β-mercaptoethanol (β-ME), BSA, acrylamide, bisacrylamide, KOH, SDS, formamide, xylene cyanol, bromphenol blue, urea, heparin, and glycerol were purchased from Fisher. Heparin was dialyzed extensively against water to remove sodium and any contaminating phosphate. ATP (disodium salt), phosphoenolpyruvate, pyruvate kinase/lactate dehydrogenase, poly(dT), biotin, streptavidin, 7 methyl guanosine, purine nucleoside phosphorylase, and Sephadex G-25 were obtained from Sigma. [γ-32P]ATP was purchased from PerkinElmer Life Sciences. T4 polynucleotide kinase was from New England Biolabs. 7-Diethylamino-3-((((2-maleimidyl)ethyl) amino)carbonyl)coumarin (MDCC) was purchased from Invitrogen. Oligonucleotides were purchased from Integrated DNA Technologies, purified using denaturing polyacrylamide gel electrophoresis (24) and quantified by UV absorbance using calculated extinction coefficients. Chemicals used in phosphate release experiments were ultrapure grade from Sigma. All solutions for phosphate release experiments were prepared in disposable plasticware, and the pH was adjusted by removing small aliquots to check the pH to minimize phosphate contamination from the pH meter.
Recombinant Proteins
Pif1 was purified as described (23). Cells expressing the E. coli A197C phosphate-binding protein (PBP) were grown as described (25), and the A197C-PBP was purified and labeled with MDCC as described (26).
DNA Unwinding
Partial duplex substrates were prepared and radiolabeled as described (24). All concentrations listed are final, after initiation of the reaction. Pif1 (200 nm, unless otherwise indicated in the figure legends) was preincubated with 2 nm radiolabeled DNA at 20 °C in reaction buffer (25 mm HEPES, pH 7.5, 50 mm NaCl, 0.1 mm EDTA, 2 mm β-ME, and 0.1 mg/ml BSA). Reactions were initiated using a KinTek rapid chemical quench-flow (RQF) by the addition of 5 mm ATP, 10 mm MgCl2, and a DNA trap (60 nm) complementary to the displaced strand to prevent reannealing of the duplex after unwinding. A protein trap (100 μm T50 in nucleotides, unless otherwise noted in the figure legend) was also added at the initiation of the reaction to prevent rebinding of the enzyme to the substrate after dissociation. Aliquots were quenched at various times with 400 mm EDTA and mixed with 0.1% bromphenol blue, 0.1% xylene cyanol, and 6% glycerol. The substrate and ssDNA product were resolved by 20% (w/v) native PAGE and detected using a Typhoon Trio PhosphorImager (GE Healthcare) and ImageQuant software.
Determination of Kinetic Parameters for Unwinding
Unwinding data for substrates of varying duplex length were fit by nonlinear least squares analysis using Scientist (MicroMath Scientific Software) (27–29). The kinetic step size (m) is defined as the number of base pairs that are unwound before each rate-limiting slow step and can be calculated from Equation 1, where LT is the total length of the duplex, L0 is the length of DNA that spontaneously melts, and n is the total number of kinetic steps required to unwind this substrate.
Processivity (P) was calculated from the unwinding rate constant (ku) and the dissociation rate constant (kd) using Equation 2.
Streptavidin Displacement
5′-Radiolabeled 3′-biotinylated homopolymeric oligonucleotide substrates were utilized in the experiment. Substrate sequences were: 3′-biotinylated 9-mer, 5′-T9(BiodT)T-3′; 3′-biotinylated 30-mer, 5′-T30(BiodT)T-3′; 3′-biotinylated 60-mer, 5′-T60(BiodT)T-3′; 3′-biotinylated 90-mer, 5′-T90(BiodT)T-3′. All concentrations given are after mixing. Reactions were performed at 25 °C. Streptavidin (300 nm) was incubated with 5′-radiolabeled 3′-biotinylated substrate (10 nm) for 3 min on ice in reaction buffer containing 10 mm MgCl2. Pif1 was added at the concentrations indicated in the figure legends, and the samples were incubated at 25 °C for 5 min. Reactions were initiated by mixing with 5 mm ATP, 4 mm phosphoenolpyruvate, and 10 units/ml pyruvate kinase/lactate dehydrogenase. Free biotin (6 μm) was added at the initiation of the reaction to prevent displaced streptavidin from rebinding the substrate. The aliquots were quenched at various times by addition of 200 mm EDTA, 0.5% SDS, 100 μm T50 (in nucleotides), 0.1% bromphenol blue, 0.1% xylene cyanol, and 6% glycerol. Samples were separated by 15% (w/v) PAGE and detected using a Typhoon Trio PhosphorImager (GE Healthcare). The quantities of streptavidin-bound and free oligonucleotide were determined using ImageQuant software. The amount of product formed over time was plotted using KaleidaGraph, and the data were fit to a single exponential equation.
Translocation on ssDNA
Translocation experiments were performed as described previously (17). All concentrations are after mixing. Briefly, 100 nm Pif1 was preincubated with 6 μm ssDNA (nucleotides) in reaction buffer without BSA. The reactions were initiated with the addition of saturating concentration of ATP (2.5 mm) and 2 mg/ml heparin sulfate in an SX.18MV stopped flow reaction analyzer (Applied Photophysics). The change in tryptophan fluorescence was monitored using a 320-nm cut-on filter (Newport Optical Filter #FSQ-WG320) with excitation at 280 nm through 1-mm slits. The data were globally fit to a translocation mechanism described in Fig. 6A and supplemental material using KinTek Explorer (30) to obtain the translocation rate and rate constant for dissociation.
FIGURE 6.
Pif1 translocation activity on ssDNA. A, shown is a schematic representation of translocation of Pif1 on ssDNA. The enzyme (E) can unidirectionally translocate (5′ → 3′) on ssDNA (D) in a series of steps defined by translocation rate (vtrans), or it can dissociate from the ssDNA (kd-trans). A change in tryptophan fluorescence is associated with dissociation. B, shown is the change in tryptophan fluorescence of 100 nm Pif1 as it dissociates from 6 μm (nt) T20, T30, T40, T55, T70, and T85 oligonucleotides at 20 °C. The data were globally fit to the scheme in A using KinTek Explorer (30). The observed vtrans and kd-trans for translocation were 89 ± 3 nt/s and 3.8 ± 0.1 s−1, respectively.
Phosphate Release
The rate of ATP hydrolysis during translocation on ssDNA was measured using a coupled assay in which the fluorescence of PBP labeled with MDCC is enhanced upon binding to phosphate (Pi) produced upon hydrolysis of ATP by Pif1 (31). All concentrations listed are final, after mixing. Before each set of reactions, syringes and lines of the stopped flow instrument were allowed to incubate with a Pi mop consisting of 1 unit/ml purine nucleoside phosphorylase and 0.5 mm 7-methyl guanosine in reaction buffer without BSA or β-ME for 30 min to minimize phosphate contamination followed by washing with reaction buffer without BSA or β-ME. ssDNA (400 nm) was preincubated with 36 nm Pif1 and 5 μm MDCC-PBP in buffer without BSA or β-ME in one syringe of an SX.18MV stopped flow reaction analyzer (Applied Photophysics). Reactions were initiated at 20 °C by mixing with 1 mm ATP, 2 mm MgCl2, 2 mg/ml heparin, and 5 μm MDCC-PBP. Also included in the reaction was a Pi mop consisting of 0.01 units/ml purine nucleoside phosphorylase and 0.2 mm 7-methyl guanosine. Both solutions were allowed to incubate for 10 min at room temperature before experiments began to minimize Pi contamination in the solutions. The change in MDCC-PBP fluorescence was monitored using a 450-nm cut-on filter (Newport Optical Filter 10LWF-450-B) with excitation at 425 nm through 1-mm slits.
The fluorescence signal was calibrated by mixing (final concentrations) 5 μm MDCC-PBP, 1 mm ATP, 2 mm MgCl2, 2 mg/ml heparin, 0.01 units/ml purine nucleoside phosphorylase, and 0.2 mm 7-methyl guanosine in buffer without BSA or β-ME with varying concentrations of Pi. The amplitude of the fluorescence increase as a function of Pi concentration was fit to a line, and the equation of the line was used to convert MDCC-PBP fluorescence into Pi concentrations. Pi release during Pif1 translocation on oligonucleotides of different lengths was measured. For each oligonucleotide length, five to seven time courses were averaged and converted to [Pi]/[Pif1]. Data were fit to Equation 3,
where A is the burst amplitude, k is the observed rate constant, t is time, and C is the y intercept.
RESULTS
Determination of Optimal Conditions for Pif1 Unwinding of Duplex DNA
Pif1 was previously shown to be a nonprocessive helicase (19); therefore, we sought to determine the optimal conditions to observe unwinding under single turnover conditions of excess enzyme concentrations. We explored three variables that are crucial for determining the unwinding properties of the enzyme: temperature, enzyme concentration, and protein trap using the 28T-16bp substrate (Table 1). At two temperatures, 20 and 25 °C, the amplitude and slope for product formation were identical; however, the lag phase, which is the earliest portion of the progress curve, was longer at 20 °C (Fig. 1A). The lag phase results from the fact that multiple steps of translocation and base pair separation are required before observation of ssDNA products. All subsequent unwinding experiments were performed at 20 °C because it enables experiments to be performed using a shorter duplex substrate (12 bp). We tested three enzyme concentrations, 100, 200, and 500 nm Pif1, which yielded similar results (Fig. 1B). An enzyme concentration of 200 nm was selected for the remaining unwinding experiments. The type of protein trap and its concentration play a very important role in single turnover unwinding experiments. Three types of protein traps were tested: dextran sulfate, poly(dT), and an oligonucleotide, T50. Use of higher concentrations of dextran sulfate (2.5–10 mg/ml) resulted in slightly reduced amplitude for product formation, indicating that this protein trap enhanced dissociation of the enzyme from the substrate (Fig. 1C). On the other hand, a lower concentration of dextran sulfate (1 mg/ml) was not a good protein trap, as a slow increase in product formation was observed after the initial burst phase. Use of poly(dT) as a protein trap resulted in two phases of product formation, the initial burst phase followed by a slow phase of product formation, indicating that it did not trap all the protein after the first turnover (Fig. 1D). Interestingly, a short oligonucleotide (T50) at both 100 and 200 μm nucleotide (nt) served as an adequate protein trap. We selected 100 μm T50 (nt) as the protein trap for all subsequent unwinding experiments.
TABLE 1.
| Substrate | Sequence |
|---|---|
| 28T-16bp | 5′-T28 CGC TGA TGT CGC CTG G-3′ |
| 3′-GCG ACT ACA GCG GAC C 5′ | |
| 7T-16bp non-fork | 5′-T7 CGC TGA TGT CGC CTG G-3′ |
| 3′-GCG ACT ACA GCG GAC C 5′ | |
| 7T/7T-16bp fork | 5′-T7 CGC TGA TGT CGC CTG G-3′ |
| 3′-T7 GCG ACT ACA GCG GAC C-5′ | |
| 14T-16bp non-fork | 5′-T14 CGC TGA TGT CGC CTG G-3′ |
| 3′-GCG ACT ACA GCG GAC C-5′ | |
| 14T/14T-16bp fork | 5′-T14 CGC TGA TGT CGC CTG G-3′ |
| 3′-T14 GCG ACT ACA GCG GAC C-5′ |
FIGURE 1.
Determination of optimal conditions for Pif1 helicase-catalyzed unwinding activity. Single-turnover unwinding of the 28T-16bp DNA substrate (2 nm) under excess helicase conditions ensure that the ssDNA overhang is saturated with Pif1 molecules. Enzyme and substrate were preincubated together, and the reactions were initiated with the addition of ATP (5 mm), MgCl2 (10 mm), DNA trap (60 nm), and protein trap. The reactions were quenched at the indicated times with the addition of 400 mm EDTA. A, shown are varying temperatures (20 °C (squares) and 25 °C (diamonds)). B, shown are varying enzyme concentrations (100 nm (squares), 200 nm (diamonds), and 500 nm (circles)). C–E, shown are different protein traps. C, shown is dextran sulfate: 10 mg/ml (squares), 5 mg/ml (diamonds), 2.5 mg/ml (circles), and 1 mg/ml (triangles). D, shown is poly(dT) 50 μm (squares), and 100 μm (diamonds). E, shown is T50 oligonucleotide: 100 μm (squares), and 200 μm (diamonds).
Pif1 Exhibits a Preference for Unwinding Forked Substrates
DNA substrates containing a non-fork structure or a fork structure were investigated to optimize the substrate. Pif1 unwinding of substrates with 7- and 14-nucleotide overhangs and forks (Table 1) under single turnover conditions were compared to determine which substrate would result in sufficient product formation. Greater product was observed with the fork substrates (Fig. 2), which is consistent with previously reported data under multiple turnover conditions (19). Enhanced product formation was observed with substrates containing 14 nucleotide overhangs as compared with the 7 nucleotide overhangs. Because the 14-nucleotide-forked substrate resulted in the highest levels of product formation, it was chosen for determining the kinetic step size.
FIGURE 2.
Pif1-catalyzed unwinding of forked and non-forked DNA substrates. Single-turnover DNA unwinding was performed as described under “Experimental Procedures.” A, shown is a comparison of Pif1-catalyzed unwinding of a non-forked substrate containing a 7-nt 5′-overhang (circles) with a forked substrate containing 7 nt of both 5′- and 3′-overhangs (squares). B, shown is a comparison of Pif1-catalyzed unwinding of a non-forked substrate containing a 14-nt 5′-overhang (circles) with a forked substrate containing both a 14-nt 5′- and 3′-overhang (squares).
Pif1 Unwinds dsDNA with a Single-base-pair Kinetic Step Size
One of the fundamental parameters associated with helicase-catalyzed unwinding of dsDNA is the number of base pairs unwound per rate-limiting step, the kinetic step size. The scheme depicting the number of steps needed to completely melt a duplex is shown in Fig. 3A. The kinetic step size for Pif1 was determined by measuring DNA unwinding as a function of dsDNA length (32). Four DNA substrates with duplex lengths of 12, 14, 16, and 20 bp were examined under single-turnover conditions (Table 2). The amplitude of product formed decreases with increasing duplex length, which indicates that Pif1 dissociates before complete unwinding of longer duplexes, hence, it is a relatively nonprocessive helicase (Fig. 3B). The lag phase increases with increasing duplex length, which indicates that Pif1 takes more steps to unwind the longer duplexes. All four progress curves were globally fit to the n-step sequential mechanism (Fig. 3A), and the best fit kinetic parameters are listed in Table 3. The results indicate that Pif1 unwinds DNA with a rate constant (ku) of 74.9 ± 12.3 s−1, and the dissociation rate constant (kd) from DNA during unwinding is 8.1 ± 0.6 s−1. The calculated processivity (P) (Equation 2) of Pif1 during DNA unwinding is 0.90. Therefore, the average distance traveled by Pif1 equal to 1/(1-P) is 10 bp of duplex on average before dissociation. This value is associated with relatively nonprocessive helicases, and it is consistent with previous results (19, 21, 23).
FIGURE 3.
Pif1-catalyzed unwinding of DNA substrates with increasing duplex lengths. A, the reaction scheme describes a series of n sequential steps taken by the helicase to catalyze strand separation. ES, enzyme-substrate complex; EI, intermediate; E+P, free enzyme and ssDNA product. Each step is defined by an unwinding rate constant, ku, and a dissociation rate constant, kd. B, unwinding of DNA substrates (2 nm) containing a 14-nt fork structure and varying lengths of duplex, including 14T/14T-12bp (circles), 14T/14T-14bp (squares), 14T/14T-16bp (diamonds), and 14T/14T-20bp (triangles) by 200 nm Pif1. The experiments were performed under single-turnover conditions at 20 °C. Unwinding data were fit by nonlinear least squares analysis using Scientist (MicroMath Scientific Software) to the mechanism in A. Error bars represent the S.D. of three independent experiments. Kinetic steps required for unwinding each of the substrates are plotted in C. The rate constants were 74.9 ± 12.3 and 8.1 ± 0.6 s−1 for unwinding and dissociation, respectively. Error bars represent the S.D. of three independent experiments. C, shown is a plot of the number of steps required for unwinding of each substrate as a function of their dsDNA length. Fitting the data to a linear equation yields an x-intercept of 6, indicating that 6 bp spontaneously melt.
TABLE 2.
DNA substrates used in Fig. 3
| Fork substrate | Sequence |
|---|---|
| 14T/14T-12bp | 5′-T14 CGC TGA TGT CGC-3′ |
| 3′-T14 GCG ACT ACA GCG-5′ | |
| 14T/14T-14bp | 5′-T14 CGC TGA TGT CGC CT-3′ |
| 3′-T14 GCG ACT ACA GCG GA-5′ | |
| 14T/14T-16bp | 5′-T14 CGC TGA TGT CGC CTG G-3′ |
| 3′-T14 GCG ACT ACA GCG GAC C-5′ | |
| 14T/14T-20bp | 5′-T14 CGC TGA TGT CGC CTG GTA CG-3′ |
| 3′-T14 GCG ACT ACA GCG GAC CAT GC-5′ |
TABLE 3.
Kinetic parameters determined by nonlinear least squares analysis of unwinding data in Fig. 3B

a Final 6 bp separated spontaneously (L0).
The number of steps required to melt the duplex DNA is greater for longer duplexes, and consequently the lag phase increases with increasing duplex lengths. A replot of the number of steps required to melt the duplex versus the length of the substrate was fit to a linear equation (Fig. 3C). The x intercept of 6 bp is indicative of the number of base pairs that melt spontaneously (L0) due to thermal fraying before Pif1 reaches the end of the duplex DNA. Spontaneous melting of 6 bp at 20 °C is in good agreement with previous measurement of 8 bp at 25 °C for a similar sequence (32, 33). The global kinetic step size (m) of 1.0 ± 0.1 bp was calculated using Equation 1. Hence, the calculated rate of unwinding (m × ku) is 75 bp/s.
An Increase in Overhang Length Results in Increased Product Formation by Pif1 without Affecting the Unwinding Rate
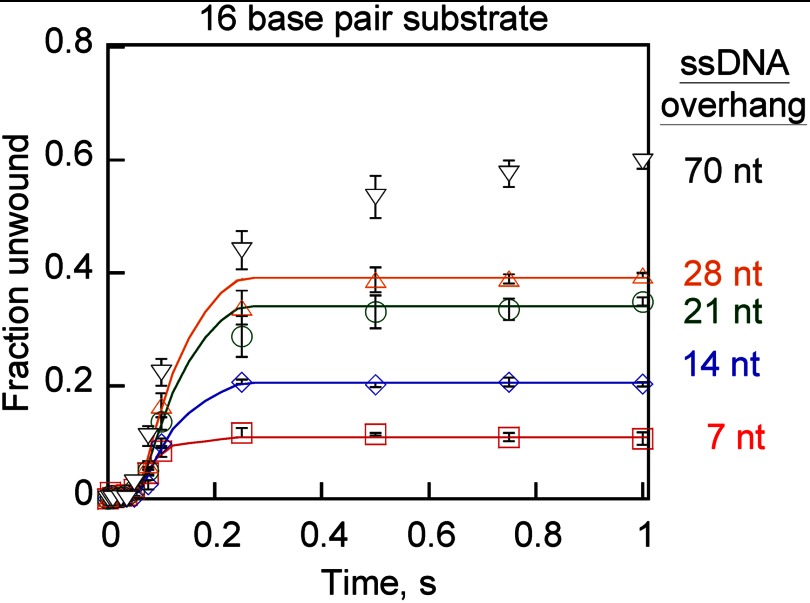
Substrates with varying lengths of 5′-overhang of ssDNA flanking the 16-base pair duplex were utilized to study Pif1 unwinding activity. We expect increasing numbers of Pif1 molecules prealigned on such substrates with increasing overhang length. A recent report on Pif1 demonstrated that the contact site-size for Pif1 is ∼6–8 nt (20). Similarly the crystal structure of a related SF1B helicase, Dda, illustrated a similar contact site-size (17). Therefore, we assume that one molecule of Pif1 sequesters about 7 nucleotides when bound to ssDNA. This implies that substrates containing 7T, 14T, 21T, 28T, and 70T overhangs should favor binding of 1, 2, 3, 4, and 10 molecules of Pif1, respectively. Unwinding of these substrates resulted in an increase in the amplitude of product formation as the length of the single-stranded overhang increased (Fig. 4).
FIGURE 4.
Pif1-catalyzed unwinding of DNA substrates containing varying-length ssDNA overhangs. DNA substrates contained a 16-bp duplex region and varying lengths of 5′-overhangs, including 7T-16bp (squares), 14T-16bp (diamonds), 21T-16bp (circles), 28T-16bp (triangles), and 70T-16bp (inverted triangles). Experiments were performed as described under “Experimental Procedures.” DNA unwinding data were fit to an n-step sequential mechanism containing 10 steps using Scientist (MicroMath Scientific Software), and the resulting kinetic parameters are listed in Table 4. Error bars represent the S.D. of three independent experiments.
To determine the rates for unwinding, each progress curve in Fig. 4 was fit using Scientist (MicroMath Scientific Software) to a 10-step mechanism. The number of steps was based on the fact that 6 bp spontaneously melt, and the remaining 10 bp are unwound in 1-bp steps. The resulting unwinding and dissociation rate constants for four of the substrates are listed in Table 4. These results indicate that the alignment of multiple molecules of Pif1 on the substrate has little effect on the rate of unwinding. The increase in amplitude could be because multiple molecules on the overhang increase the probability of having at least one Pif1 at the fork junction during DNA unwinding.
TABLE 4.
Kinetic patameters determined from fitting the data in Fig. 4A to 10-step kinetic model using Scientist
| Substrate | No. of Pif1 molecules bound | ku | kd |
|---|---|---|---|
| s−1 | s−1 | ||
| 7T-16bp | 1 | 98.3 ± 7.3 | 24.4 ± 1.8 |
| 14T-16bp | 2 | 79.4 ± 4.1 | 13.6 ± 0.7 |
| 21T-16bp | 3 | 80.3 ± 8.4 | 9.1 ± 1.1 |
| 28T-16bp | 4 | 82.8 ± 6.3 | 8.2 ± 0.7 |
For the 70T-overhang substrate, the results deviate significantly from the model used for data fitting; thus, the fit has not been included. The increase in overhang length resulted in multiple phases of product formation including the initial lag and burst phases followed by a slow phase of product formation. The initial lag and burst phases for the 70T-overhang substrate are similar to those for the shorter overhang substrates. The slower phase of product formation results from the fact that Pif1 is a non-processive helicase; therefore, many trailing molecules will dissociate before reaching the duplex. The time required for the trailing molecule to translocate to the junction results in slower product appearance at later stages. Although 21T- and 28T-overhang substrates show this slower phase, it is less pronounced due to the involvement of fewer trailing molecules and the shorter distance they must translocate.
Pif1 Displaces Streptavidin from Biotinylated Oligonucleotides
To investigate the mechanism by which multiple Pif1 molecules on ssDNA overhang increase the unwinding amplitude, we investigated the kinetics of Pif1 displacing streptavidin from biotinylated ssDNA. Our previous studies have shown that multiple molecules of T4 Dda helicase bound to ssDNA function cooperatively to displace streptavidin from biotinylated ssDNA (34). The kinetics of streptavidin displacement were measured under multiple turnover conditions in which Pif1 molecules can rebind the substrate; however, rebinding of displaced streptavidin was prevented by including excess biotin trap in the reaction (Fig. 5A). This assay measures the appearance of free DNA, which migrates faster on a polyacrylamide gel compared with streptavidin-bound DNA (Fig. 5B). Streptavidin displacement was not observed in the absence of ATP, indicating that translocation of Pif1 on ssDNA is required for protein displacement (Fig. 5B), which is consistent with a recent report (35); however, this report did not address the role of multiple Pif1 molecules. DNA substrates of increasing length included a 9-, 30-, 60-, and 90-mer. Displacement was determined in the presence of increasing concentrations of Pif1 from each of the 3′-biotinylated substrates (10 nm) (Fig. 5, C–F). The rate constants for streptavidin displacement increased modestly with increasing enzyme concentrations (Fig. 5G). If multiple Pif1 molecules function together in a cooperative manner during streptavidin displacement, then the rate constants for displacement should increase in a sigmoidal manner. However, as seen in Fig. 5G, the rate constants for displacement increase in a linear manner as the Pif1 concentration increases, suggesting that multiple Pif1 molecules exhibit a modest cooperativity for streptavidin displacement. Similarly, plotting the rate constants as a function of the length of the oligonucleotide at 500 nm Pif1 shows a linear 4.5-fold increase in the rate constants for displacement from a 9-mer to a 90-mer (Fig. 5H). This rate enhancement is significantly less than that observed for T4 Dda helicase (1000-fold enhancement) with the same length oligonucleotides, where the sizable increase in rate of streptavidin displacement was attributed to cooperative function of multiple Dda molecules aligning on the substrate and pushing one another (34). The modest increase in the Pif1 rate of streptavidin displacement on longer ssDNAs indicates modest cooperativity and is attributed to the presence of a greater number of trailing Pif1 molecules that can rapidly translocate forward and take the place of the leading molecule participating in displacement.
FIGURE 5.
Displacement of streptavidin from 3′-biotinylated oligonucleotides with increasing concentrations of enzyme. A, shown is a schematic representation of streptavidin displacement from 3′-biotinylated oligonucleotides. Biotinylated oligonucleotides were prebound to streptavidin and Pif1, and the streptavidin displacement reaction was initiated with the addition of ATP and an excess of free biotin. Biotin prevents rebinding of displaced streptavidin to the biotinylated oligonucleotide. B, streptavidin (SA)-bound biotinylated substrate was resolved from free oligonucleotide product by native 15% PAGE gels. No streptavidin displacement was observed in the absence of ATP. C–F, shown is streptavidin displacement from 10 nm 3′-biotinylated 9-mer (C), 3′-biotinylated 30-mer (D), 3′-biotinylated 60-mer (E), and 3′-biotinylated 90-mer (F) by 25 nm (inverted triangles), 50 nm (triangles), 100 nm (diamonds), 250 nm (squares), and 500 nm (circles) Pif1. The data were fit to a single exponential equation to obtain rate constants for streptavidin displacement. G, shown is a replot of the displacement rate constants from C–F as a function of concentration of enzyme. H, rate constants for streptavidin displacement at 500 nm Pif1 were 0.20 ± 0.01, 0.40 ± 0.03, 0.53 ± 0.04, and 0.89 ± 0.10 min−1 from 3′-biotinylated 9-, 30-, 60-, and 90-mer, respectively. The data were fit to linear equation. Error bars represent the S.D. of three independent experiments.
The Unwinding Rate of Pif1 Is Similar to Its Rate of Translocation on ssDNA
Comparison of the rate of translocation on ssDNA with the rate of dsDNA unwinding provided insight into the active versus passive mechanism (36, 37). The rate of translocation of Pif1 along ssDNA was determined by measuring the change in tryptophan fluorescence upon dissociation of protein from varying lengths of ssDNA. Pif1 protein dissociation from the end of ssDNA requires more time as the length of the ssDNA increases because it must translocate farther to reach the end of oligonucleotide. The translocation data were fit to a scheme described in Fig. 6A using KinTek Explorer (30). In this scheme, the enzyme is equally distributed along all the DNA binding sites (ED1 to EDL-7), where L is the length of the ssDNA and the enzyme binding site size is 7 nucleotides (supplemental material). The enzyme can translocate on the ssDNA substrate in a series of identical steps defined by the translocation rate (vtrans), and at each step the enzyme can dissociate from the substrate with a dissociation rate constant (kd-trans). Homopolymeric oligonucleotide substrates T20, T30, T40, T55, T70, and T85 were used to measure the translocation rates. Enzyme and ssDNA substrate were preincubated together under conditions where only a single Pif1 molecule would be bound to the ssDNA, and the reaction was initiated by rapid mixing with ATP and heparin sulfate. The change in tryptophan fluorescence due to dissociation of Pif1 from the ssDNA substrates was measured (Fig. 6B). All the translocation data were globally fit to the scheme described in the Fig. 6A and supplemental material. The resulting translocation rate is 89 ± 3 nt/s, and the rate constant for dissociation is 3.8 ± 0.1 s−1 at 20 °C (Fig. 6B). The rate of translocation (89 ± 3 nt/s) is very similar to the rate of unwinding (75 ± 12 bp/s). These results indicate that Pif1 unwinds dsDNA by an active mechanism similar to the T4 Dda helicase, which also belongs to the Pif1 subfamily of helicases and is also a highly active helicase even though Dda unwinds and translocates at much higher speeds (about 250 bp or nt/s) (17, 37). Processivity for Pif1 translocation was 0.96, indicating that it translocates an average length of ∼25 nt before dissociation. This is higher than the processivity for unwinding (10 bp), indicating that Pif1 remains bound to the DNA longer when translocating on an unobstructed ssDNA lattice than when displacement of a complimentary strand is required even though both translocation and unwinding occur at similar rates.
The unwinding experiments in this paper were performed at 20 °C to accurately measure rates with short duplex regions, which was necessary due to the low processivity of Pif1. However, most published experiments with Pif1 have been performed at 25 °C. To allow comparison of the translocation activity of Pif1 to previously published unwinding data, the translocation experiments were also performed at 25 °C (supplemental Fig. 1). The observed vtrans and kd-trans for translocation were 119 ± 4 nt/s and 6.8 ± 0.2 s−1, respectively, at 25 °C.
Phosphate Release Kinetics of Pif1
The kinetics of ATP hydrolysis while Pif1 was translocating along ssDNA were measured using a real time coupled fluorescent assay that monitors the release of inorganic phosphate, Pi (Fig. 7A) (31, 38). The reaction conditions were similar to those used during the ssDNA translocation assays so that the rates of ATP hydrolysis and ssDNA translocation would be directly comparable. Pif1 was preincubated with oligonucleotides of various lengths, and the change in fluorescence of MDCC-PBP upon binding of the Pi released during ATP hydrolysis was monitored (Fig. 7B). The quantity or the amplitude of Pi released in a single turnover of Pif1 translocation on ssDNA increases with increasing oligonucleotide length. This is consistent with a model where Pi release is coupled to translocation along ssDNA. The amplitude of Pi produced per Pif1 versus oligonucleotide length shows a linear relationship with a slope of 0.42 ± 0.05 (Fig. 7C). Assuming random binding of Pif1 to the ssDNA, Pif1 translocates on average half the length of the oligonucleotide, resulting in 0.84 ± 0.10 ATP hydrolyzed per nucleotide translocated by Pif1 or 1.2 nucleotides translocated per ATP hydrolyzed.
FIGURE 7.
Phosphate release after ATP hydrolysis during translocation by Pif1. A, shown is illustration of the Pi release reaction. Pif1 hydrolyzes ATP during translocation on ssDNA, producing ADP and Pi. The released Pi is bound by MDCC-PBP, resulting in an enhancement in fluorescence. B, shown are progress curves for Pi release during translocation along oligonucleotides of increasing length. C, shown are burst amplitudes of time courses in B plotted as a function of oligonucleotide length. Error bars represent the S.D. of three experiments. Fitting the data to a line provides a slope of 0.42 ± 0.05.
DISCUSSION
Although Pif1 has been demonstrated to unwind short duplex substrates with 5′ → 3′ polarity (19, 21), the kinetic mechanism of the helicase activity was not well characterized. Consistent with previous reports on Pif1 helicase activity, we show here that DNA substrates containing a fork structure yield higher product formation even under single-turnover conditions (Fig. 2) (19). Therefore, fork substrates with varying duplex lengths were used to measure the unwinding kinetics for Pif1 to determine the kinetic step size, chemical step-size, unwinding processivity, base pair unwinding rate, and protein dissociation rate constants. The movement of helicases along nucleic acids can be described in terms of the kinetic, physical, and chemical step size. The kinetic step size refers to the average number of bases traversed between successive rate-limiting kinetic steps. The physical step size is a measure of the number of base pairs that are unwound simultaneously, and the chemical step size refers to the number of bp unwound or nt translocated per ATP hydrolyzed. Depending on the mechanism of unwinding, the kinetic step size can differ from the physical and/or chemical step size. Also, the step size for translocation on ssDNA may not match the step size for DNA unwinding.
Reported kinetic step sizes for unwinding by SF1 helicases include 4.4 bp for UvrD (27), 3–4 bp for RecBCD (28, 39), 4 bp for PcrA (40), 3.4 bp for Dda (32, 33), and 7–8 bp for TraI (42). For many of these helicases the kinetic step size is larger than the chemical step size, which has been interpreted to result from a non-uniform stepping mechanism in which ATP cycles are not rate-limiting. We demonstrate here that Pif1 unwinds DNA with a kinetic step size of 1 bp (Fig. 3). This is the first non-hexameric helicase for which a single base pair kinetic step size has been reported. The hexameric helicase, DnaB, was reported to unwind with a kinetic step size of 1.4 bp (16).
Experiments measuring the rate of phosphate release upon ATP hydrolysis by Pif1 indicate an ATP hydrolysis rate of 0.84 ATP hydrolyzed per nucleotide translocated by Pif1 (Fig. 7). This is consistent with the measured kinetic step size of 1 base pair (Fig. 3) and suggests tight coupling between ATP hydrolysis and directional movement. This value is also similar to that obtained for other SF1 and SF2 helicases including 0.98 ATP hydrolyzed per nucleotide translocated for UvrD (43), 1 ATP per nucleotide for PcrA (38), 1.1 ATP per nucleotide for NS3-4A (44), and 0.625–1.1 ATP per nucleotide for RecQ (45, 46). In contrast, one of the least efficient helicases reported to date is NS3h, with a coupling efficiency of 5 ATP/nt (44).
Here we have shown that Pif1 exhibits both a 1-base-pair kinetic step size for unwinding and chemical step size during translocation on ssDNA. These values are consistent with the single-base-pair physical step size suggested by structural studies of several helicases including PcrA (47), RecD2 (18), and UvrD (48). The data presented here suggest that Pif1 unwinds dsDNA with a uniform mechanism where hydrolysis of a single ATP molecule results in translocation of the helicase by one nucleotide in the 5′ → 3′ direction with concomitant unwinding of a single base pair of dsDNA. However, for many other SF1 helicases, including Dda (33) and UvrD (43), a non-uniform mechanism with several rapid steps that are followed by a rate-limiting slow step that repeats every few steps has been suggested. Thus, Pif1 moves along DNA with a physical step size of 1 bp that is rate-limited by the ATPase cycle.
SF1 helicases exhibit varying processivity of DNA unwinding ranging from as low as 10 bp for UvrD (27) to as high as 30,000 bp for RecBCD (49). Most SF1 helicases have low processivity except for RecBCD and TraI. RecBCD contains multiple motor domains that bind to both strands of DNA, which results in an increased processivity. TraI is a unique helicase known to function as a monomer but has a processivity of ∼850 bp in vitro (42). Dda is one of the most closely related helicases to Pif1 based on its sequence similarity in the helicase domains (16). Dda exhibits relatively low processivity of 25 bp (32). Similarly, the calculated processivity for Pif1 is 10 bp per binding event, which is based on the measured intrinsic dissociation rate constant (8.1 s−1) and rate constant for unwinding (75 s−1). Thus, Pif1 ranks among the SF1B helicases with the lowest processivity. For some helicases, such as PcrA, interaction with accessory proteins dramatically increases the processivity for unwinding (50, 51). Hence, regulation of helicase processivity can readily occur through protein-protein interactions. Some functions of Pif1 such as mitochondrial DNA recombination and replication may require the enzyme to be more processive. Recently we reported that Pif1 stably interacts with the mitochondrial SSB, Rim1 (23). The presence of Rim1 in Pif1-catalyzed unwinding experiments significantly increased product formation, indicating a possible increase in processivity of the enzyme, which remains to be investigated.
Some helicases exhibit higher activity when multiple molecules can align on the same DNA substrate (32). We found that multiple Pif1 molecules bound to the DNA loading site results in increased unwound DNA product formation (Fig. 4). The increase in product could be explained by an increase in the unwinding rate where multiple molecules aligned on the substrate push one another forward in a cooperative manner. Alternatively, the increase in product could be explained by multiple molecules increasing the probability that at least one molecule completes the unwinding process (32, 52). Fitting the data indicates no increase in the rate for unwinding with increasing ssDNA overhang length; hence, the increased amplitude is likely due to increasing the opportunity for at least one molecule to move through the DNA. Previously we have reported similar observations with T4 Dda helicase (32) and HCV NS3h (52).
The longest overhang tested in this report was 70 nt. In vivo, longer ssDNA regions loaded with Pif1 could yield enhanced DNA unwinding. Pif1 exhibits low processivity, so even if long stretches of ssDNA are loaded with Pif1, only a small increase in DNA unwinding would be expected in the absence of a processivity factor. For example, the 70T overhang is more than double the length of the 28T overhang; however, the observed product formation increased by less than 20%. If Pif1 is able to cycle on and off of the ssDNA and work with a single-stranded binding protein, then much longer duplexes could be melted (45).
We previously found that multiple molecules of Dda helicase work together to efficiently displace streptavidin from biotinylated DNA (34). The fact that the streptavidin block prevented forward movement of the leading enzyme molecule allowed the trailing molecules to push forward, greatly increasing the rate of streptavidin displacement. For Pif1 helicase, the rate of streptavidin displacement from 3′-biotinylated oligonucleotides increases in a linear manner as the length of ssDNA substrate increases (Fig. 5). Hence, Pif1 can produce force on proteins blocking its path. Pif1 exhibits a 4.5-fold increase in rate of displacement as the length of the substrate increases from a 9-mer to a 90-mer biotinylated oligonucleotide. A similar phenomenon was previously observed with T4 Dda helicase (34). However, in case of Dda, a 1000-fold enhancement in the rate of displacement was observed over a similar substrate range. Rrm3, a Pif1 homolog in S. cerevisiae is known to disrupt non-nucleosomal protein-DNA complexes in vivo (53, 54). The absence of Rrm3 results in pausing of replication forks and often breakage at various sites in the genome. Although Rrm3 has not been studied in vitro to show its protein displacement function, we speculate that it would displace streptavidin from the biotinylated oligonucleotides in a manner similar to Pif1.
Although multiple molecules of Dda exhibit much higher displacement rates than Pif1 when compared directly with a corresponding length of ssDNA, this is not the case when results from a short substrate of 9-mer are compared. Under similar conditions, comparison between Dda and Pif1 streptavidin displacement rates from a 90-mer substrate indicates a 60-fold higher rate of displacement by Dda (34). However, from a 9-mer oligonucleotide, ∼4-fold higher displacement rates were observed with Pif1. A 9-mer oligonucleotide can only occupy one molecule of Dda. A recent report showed that Pif1 exists as monomer in solution and dimerizes upon binding to ssDNA (20). More efficient streptavidin displacement from the 9-mer by Pif1 could be due to the combined effects of the dimer molecule. As the substrate length increases, allowing multiple enzyme molecules to bind and function together, Dda becomes significantly more efficient and is able to displace streptavidin from the longest oligonucleotides much faster than Pif1.
Pif1 negatively regulates the telomere lengths by removal of telomerase holoenzyme from telomeres (1). Two models are proposed for this function. The first model postulates that Pif1 releases TLC1 (telomerase RNA) from the telomere ends, which would abolish the association of the telomerase complex with the telomeres. This model is based on the observation that Pif1 preferentially unwinds DNA·RNA duplexes (21). The second model postulates that Pif1 displaces Est2 (catalytic subunit of the telomerase complex) from the telomere ends or disrupts the interaction of telomeric DNA with telomerase. This model is based on the observation that in vivo overexpression of Pif1 in yeast reduced association of Est1 and Est2 at telomere ends (9). It is uncertain whether the duplex region containing the telomeric DNA and TLC1 would be exposed for Pif1 unwinding without steric hindrance by the telomerase complex. Therefore, our studies showing Pif1 ability to displace streptavidin from DNA, indicating that Pif1 can push/displace proteins from DNA, would be consistent with the protein-pushing model, although it does not argue against the favored unwinding of DNA·RNA duplexes.
Our measurements of the ssDNA translocation rate and unwinding rate allows us to access the degree of coupling of translocation to unwinding for differentiating an active helicase from a passive helicase (36). The observed rate of translocation (vtrans) of 89 ± 3 nt/s (Fig. 6) was very similar to the rate of unwinding (vun) of 74.9 ± 12 bp/s (Fig. 3). As we were completing this manuscript, another manuscript reported studies of Pif1 translocation on ssDNA (35). Using a fluorescence-based method to measure helicase translocation, they described two populations of Pif1, one of which translocated at a rate of 81 nt/s, which is similar to the rates reported here. We have measured the average rate of Pif1 translocation on ssDNA by measuring dissociation of the enzyme from varying lengths of ssDNA. An active helicase destabilizes the duplex through direct interaction at the junction of single- and double-stranded nucleic acids, whereas a passive helicase relies on transient fraying of the duplex at the junction. The ratio of vun/vtrans can be used to distinguish between active and passive helicases, where a highly active helicase has a value of ∼1. The ratio of vun/vtrans for Pif1 is 0.85, which makes it an active helicase. Several SF1 helicases are reported to function as active helicases. Recently we reported an optimally active helicase, Dda, with vun/vtrans of 0.96 (37). Similar observations were reported for E. coli RecQ helicase with a vun/vtrans of 0.9 (36).
The coupling ratio of one ATP hydrolyzed per bp unwound may appear to be energetically inefficient because the free energy available from hydrolysis of one ATP could separate multiple base pairs depending on reaction conditions (55). However, the biological function of Pif1 may include processes that are more energetically demanding than melting of duplex DNA. For example, Pif1 has been shown to regulate telomere length by displacing telomerase from DNA. The energetic requirement for this process is unknown but is likely to be greater than melting of DNA alone because protein-nucleic interactions must be disrupted in addition to base pairing. Another function of Pif1 is to unwind G4 quadruplex DNA. The mechanism for this process is not known, but quadruplex structures can be highly stable (41, 56), so melting such structures may be facilitated by a one-nt step size driven by hydrolysis of 1 ATP so that the quadruplex structure is melted gradually in an energetically favorable manner.
In addition to its role in telomerase displacement and quadruplex unwinding, Pif1 reportedly plays a role in Okazaki fragment maturation. All these in vivo functions of Pif1 are not expected to require a highly processive enzyme. However, its processivity can be enhanced through interaction with accessory proteins such as the mitochondrial SSB, Rim1 (23). To perform its diverse functions in the cell, it may be advantageous for Pif1 to be non-processive but be capable of increasing processivity when necessary through interaction with other proteins.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM098922 (to K. D. R.), Translational Research Institute (TRI) grant UL1TR000039 through the NIH National Center for Research Resources (NCRR) and National Center for Advancing Translational Sciences (NCATS), and GM51966 (to S. S. P.).

This article contains supplemental material and Fig. 1.
- SF1
- superfamily 1
- β-ME
- β-mercaptoethanol
- MDCC
- 7-diethylamino-3-((((2-maleimidyl)ethyl) amino)carbonyl)coumarin
- PBP
- phosphate-binding protein
- nt
- nucleotide.
REFERENCES
- 1. Bochman M. L., Sabouri N., Zakian V. A. (2010) Unwinding the functions of the Pif1 family helicases. DNA Repair 9, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Foury F., Kolodynski J. (1983) pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 80, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Foury F., Dyck E. V. (1985) A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 4, 3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Dyck E., Foury F., Stillman B., Brill S. J. (1992) A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 11, 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase δ. Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paeschke K., Capra J. A., Zakian V. A. (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou J., Monson E. K., Teng S. C., Schulz V. P., Zakian V. A. (2000) Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289, 771–774 [DOI] [PubMed] [Google Scholar]
- 9. Boulé J. B., Vega L. R., Zakian V. A. (2005) The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 10. Schulz V. P., Zakian V. A. (1994) The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76, 145–155 [DOI] [PubMed] [Google Scholar]
- 11. Myung K., Chen C., Kolodner R. D. (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411, 1073–1076 [DOI] [PubMed] [Google Scholar]
- 12. Ivessa A. S., Zhou J. Q., Zakian V. A. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100, 479–489 [DOI] [PubMed] [Google Scholar]
- 13. O'Rourke T. W., Doudican N. A., Mackereth M. D., Doetsch P. W., Shadel G. S. (2002) Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 22, 4086–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng X., Dunaway S., Ivessa A. S. (2007) The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion 7, 211–222 [DOI] [PubMed] [Google Scholar]
- 15. Cheng X., Ivessa A. S. (2010) Association of the yeast DNA helicase Pif1p with mitochondrial membranes and mitochondrial DNA. Eur. J. Cell Biol. 89, 742–747 [DOI] [PubMed] [Google Scholar]
- 16. Fairman-Williams M. E., Guenther U. P., Jankowsky E. (2010) SF1 and SF2 helicases. Family matters. Curr. Opin. Struct. Biol. 20, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He X., Byrd A. K., Yun M. K., Pemble C. W., 4th, Harrison D., Yeruva L., Dahl C., Kreuzer K. N., Raney K. D., White S. W. (2012) The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation. Structure 20, 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saikrishnan K., Powell B., Cook N. J., Webb M. R., Wigley D. B. (2009) Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell 137, 849–859 [DOI] [PubMed] [Google Scholar]
- 19. Lahaye A., Leterme S., Foury F. (1993) PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 268, 26155–26161 [PubMed] [Google Scholar]
- 20. Barranco-Medina S., Galletto R. (2010) DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry 49, 8445–8454 [DOI] [PubMed] [Google Scholar]
- 21. Boulé J. B., Zakian V. A. (2007) The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 35, 5809–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ribeyre C., Lopes J., Boulé J. B., Piazza A., Guédin A., Zakian V. A., Mergny J. L., Nicolas A. (2009) The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramanagoudr-Bhojappa R., Blair L. P., Tackett A. J., Raney K. D. (2013) Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 41, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tackett A. J., Morris P. D., Dennis R., Goodwin T. E., Raney K. D. (2001) Unwinding of unnatural substrates by a DNA helicase. Biochemistry 40, 543–548 [DOI] [PubMed] [Google Scholar]
- 25. Wang Q., Arnold J. J., Uchida A., Raney K. D., Cameron C. E. (2010) Phosphate release contributes to the rate-limiting step for unwinding by an RNA helicase. Nucleic Acids Res. 38, 1312–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salins L. L., Deo S. K., Daunert S. (2004) Phosphate binding protein as the biorecognition element in a biosensor for phosphate. Sens. Actuators B Chem. 97, 81–89 [DOI] [PubMed] [Google Scholar]
- 27. Ali J. A., Lohman T. M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 [DOI] [PubMed] [Google Scholar]
- 28. Lucius A. L., Vindigni A., Gregorian R., Ali J. A., Taylor A. F., Smith G. R., Lohman T. M. (2002) DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 324, 409–428 [DOI] [PubMed] [Google Scholar]
- 29. Lucius A. L., Maluf N. K., Fischer C. J., Lohman T. M. (2003) General methods for analysis of sequential “n-step” kinetic mechanisms. Application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson K. A. (2009) Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 467, 601–626 [DOI] [PubMed] [Google Scholar]
- 31. Brune M., Hunter J. L., Corrie J. E., Webb M. R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 [DOI] [PubMed] [Google Scholar]
- 32. Eoff R. L., Raney K. D. (2010) Kinetic mechanism for DNA unwinding by multiple molecules of Dda helicase aligned on DNA. Biochemistry 49, 4543–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eoff R. L., Raney K. D. (2006) Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat. Struct. Mol. Biol. 13, 242–249 [DOI] [PubMed] [Google Scholar]
- 34. Byrd A. K., Raney K. D. (2004) Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 11, 531–538 [DOI] [PubMed] [Google Scholar]
- 35. Galletto R., Tomko E. J. (2013) Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 41, 4613–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manosas M., Xi X. G., Bensimon D., Croquette V. (2010) Active and passive mechanisms of helicases. Nucleic Acids Res. 38, 5518–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Byrd A. K., Matlock D. L., Bagchi D., Aarattuthodiyil S., Harrison D., Croquette V., Raney K. D. (2012) Dda helicase tightly couples translocation on single-stranded DNA to unwinding of duplex DNA. Dda is an optimally active helicase. J. Mol. Biol. 420, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dillingham M. S., Wigley D. B., Webb M. R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase. Measurement of step size and translocation speed. Biochemistry 39, 205–212 [DOI] [PubMed] [Google Scholar]
- 39. Lucius A. L., Wong C. J., Lohman T. M. (2004) Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339, 731–750 [DOI] [PubMed] [Google Scholar]
- 40. Yang Y., Dou S. X., Ren H., Wang P. Y., Zhang X. D., Qian M., Pan B. Y., Xi X. G. (2008) Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res. 36, 1976–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhakal S., Cui Y., Koirala D., Ghimire C., Kushwaha S., Yu Z., Yangyuoru P. M., Mao H. (2013) Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 41, 3915–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sikora B., Eoff R. L., Matson S. W., Raney K. D. (2006) DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J. Biol. Chem. 281, 36110–36116 [DOI] [PubMed] [Google Scholar]
- 43. Tomko E. J., Fischer C. J., Niedziela-Majka A., Lohman T. M. (2007) A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell 26, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rajagopal V., Gurjar M., Levin M. K., Patel S. S. (2010) The protease domain increases the translocation stepping efficiency of the hepatitis C virus NS3-4A helicase. J. Biol. Chem. 285, 17821–17832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rad B., Kowalczykowski S. C. (2012) Efficient coupling of ATP hydrolysis to translocation by RecQ helicase. Proc. Natl. Acad. Sci. U.S.A. 109, 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sarlós K., Gyimesi M., Kovács M. (2012) RecQ helicase translocates along single-stranded DNA with a moderate processivity and tight mechanochemical coupling. Proc. Natl. Acad. Sci. U.S.A. 109, 9804–9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., Wigley D. B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 [DOI] [PubMed] [Google Scholar]
- 48. Lee J. Y., Yang W. (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roman L. J., Eggleston A. K., Kowalczykowski S. C. (1992) Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J. Biol. Chem. 267, 4207–4214 [PubMed] [Google Scholar]
- 50. Soultanas P., Dillingham M. S., Papadopoulos F., Phillips S. E., Thomas C. D., Wigley D. B. (1999) Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res. 27, 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noirot-Gros M. F., Soultanas P., Wigley D. B., Ehrlich S. D., Noirot P., Petit M. A. (2002) The β-propeller protein YxaL increases the processivity of the PcrA helicase. Mol. Genet. Genomics 267, 391–400 [DOI] [PubMed] [Google Scholar]
- 52. Levin M. K., Wang Y. H., Patel S. S. (2004) The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279, 26005–26012 [DOI] [PubMed] [Google Scholar]
- 53. Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A. (2002) Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16, 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., Zakian V. A. (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12, 1525–1536 [DOI] [PubMed] [Google Scholar]
- 55. Lohman T. M., Bjornson K. P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214 [DOI] [PubMed] [Google Scholar]
- 56. Lane A. N., Chaires J. B., Gray R. D., Trent J. O. (2008) Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.