Abstract
The study in this issue of the JCI by Tortajada et al. demonstrates that a duplication within the gene complement factor H–related 1 (CFHR1; encoding FHR1) leads to the production of an aberrant larger form of the protein. Elegant in vitro studies of the mutant and normal variants demonstrate an unexpected mechanism of action of FHR1, wherein homodimeration and hetero-oligomerization with FHR2 and FHR5 generates more avid molecules that very effectively compete with FH binding to surfaces and impair its ability to regulate local complement activation. As variants of FHRs are linked to many human inflammatory and autoimmune diseases, these and other recently published structure/function studies of these proteins provide key insights into their complement regulatory activities and likely roles in disease.
Introduction
Major advances have recently been made in our understanding of the biological and pathophysiological roles of the complement system. Genetic association studies, deep sequencing efforts, clinical association findings, results from animal models, and markedly positive results in therapeutic trials in an increasing number of human diseases have refocused attention on the important pathogenic role of inappropriate complement activation in the broad scope of human diseases (1, 2). Complement inhibitors have been successfully developed for therapeutic use in two human genetic deficiency states, hereditary angioedema (HAE; ref. 3) and paroxysmal nocturnal hemoglobinuria (PNH; ref. 4), as well as the rare condition designated atypical hemolytic uremic syndrome (aHUS; ref. 5). Positive initial clinical trial results have been reported in many additional conditions. Notably, major efforts are underway to understand why common polymorphisms and rare variants of complement pathway genes whose products primarily promote activation of the alternative pathway are associated with human age-related macular degeneration (AMD; ref. 6).
The complement system is an evolutionarily ancient member of the innate immune system that is involved in many inflammatory and autoimmune diseases, with functions ranging from modulation of adaptive immunity to generation of potent injurious effector functions when endogenous control mechanisms fail to restrain its activities during tissue injury (7). Complement system components are in general well understood with regard to their structure/function characteristics. The system consists of more than 40 proteins that either function during activation through the three initiation pathways and the amplification loop or act as recognition molecules, receptors, negative regulatory proteins, or stabilizing/activating factors (7). There are, however, a number of fundamental unanswered questions with regard to certain components of the system, and high on the list are the functional roles of complement factor H–related proteins (FHRs).
What are FHRs? As Tortajada et al. note in this issue (8), these proteins have been known for many years to be part of a structurally related family including the larger factor H protein (FH; encoded by CFH). FHRs are encoded by a series of genes adjacent to CFH on human chromosome 1 and also contain short consensus repeat (SCR) domains with homology to subregions of FH (9, 10). As genetic variants characterized by the absence of CFHR1 and CFHR3 (ΔCFHR3-CFHR1), or of CFHR1 and CFHR4 (ΔCFHR1-CFHR4), are relatively common in the human population, the proteins encoded by these genes do not appear to be required for human development or immune competence under normal conditions. Nevertheless, an increasing number of protective or risk associations of deletions or variants of these genes have been reported with human diseases (9, 10). Among these are AMD, for which ΔCFHR3-CFHR1 is a highly penetrant protective factor (11), and aHUS, for which ΔCFHR3-CFHR1 is a disease risk factor and is associated with autoantibodies that interfere with FH regulatory function (12).
Duplication of sequences within CFHR1 is linked to C3 glomerulopathy
Many of the recent advances in the complement field have been driven by study of rare variants and mutations. Tortajada and colleagues follow that approach in the unusual but informative clinical condition designated C3 glomerulopathy (C3G; ref. 8). C3G is a newly classified pathological entity with glomerular immunohistochemistry that is characterized by C3 accumulation in the absence of the substantial concurrent presence of immunoglobulins (13). Notably, C3G is associated with mutations in C3 and complement components that regulate C3 activation (13).
In the present article, Tortajada and colleagues describe a mother and son who presented with clinical syndromes and renal biopsies consistent with C3G (8). Extensive genetic and protein analyses demonstrated that an internal duplication within CFHR1 resulted in an aberrant protein containing nine SCRs instead of the normal five, with the first four SCRs duplicated in tandem. The authors purified wild-type nonmutated FHR1 from human plasma, and chromatographic analysis revealed that rather than a single expected peak, multiple peaks contained FHR1. Tortajada et al. showed that FHR1 homo- and hetero-oligomerized with FHR2 and FHR5, but not with FHR3 or FHR4A/B. Next, the authors demonstrated that these complexes compete for the functionally essential FH binding activity to surface-fixed C3 fragments that is encompassed within SCR19–SCR20, impairing the ability of FH to block surface complement activation. Importantly, the FHR1 mutant protein in the patients with C3G assembled into unusually large multimeric complexes that even more effectively impaired FH function. The conclusion of the study was that FHR1, assembled into multimers either alone or in association with FHR2 and FHR5, primarily acts on surfaces to block FH binding, resulting in complement deregulation and enhanced local activation. Presumably, altered function of the FHR1 mutant promoted the development of renal disease in the two patients (8). Copy number variants in FHRs consisting of internal duplication and heterozygous hybrid proteins have been previously identified in patients presenting with renal disease and biopsy findings characteristic of C3G (14, 15). Thus, self-association of FHRs is undoubtedly important in their ability to interfere with FH, which is even more relevant because FH itself forms tetramers (16) and other types of oligomers (17) that appear to promote more avid binding to C3 fragments and other surface determinants, including glycosaminoglycans (GAG).
Other recent, highly informative structural studies of FHRs
Tortajada et al. acknowledge the importance of the recent crystallography and structure/function studies by Goicoechea de Jorge and colleagues (18). Strikingly, that group recently reported that FHR1, FHR2, and FHR5 contain dimerization motifs that are highly conserved and allow head-to-tail dimer formation. They also found predominant homo- and heterodimers of the native proteins in serum and demonstrated more avid interactions of the multimers with C3 fragments, both in vitro and in vivo. Additionally, the complexes exhibited complement deregulation by acting as competitive antagonists of FH binding. Analysis of the variant FHR5 protein, found to be associated with C3G in a Cypriot population (15), similarly demonstrated a substantially increased ability to impair FH binding and deregulate surface complement activation.
What are the primary roles of FHR1 and other FHRs in modulating complement activation?
In contrast to the reports by Tortajada et al. (8) and Goicoechea de Jorge et al. (18), many prior studies of FHRs have focused not on deregulating FH, but on the potential role of a subset of these proteins in directly regulating complement activation and impairing or promoting activation at specific steps of the cascade. For example, FHR2 and FHR4 have been shown to enhance FH cofactor activity, and FHR5 exhibits weak cofactor and decay-accelerating activities (reviewed in ref. 10). Likewise, FHR1 was previously shown to inhibit C5 convertase and terminal complex formation (19), and FHR3 was demonstrated to inhibit C3 convertase and display antiinflammatory activity by blocking C5a generation (20). As the structure/function of FH has become increasingly understood (21), it has become more challenging to reconcile the reported regulatory activities of FHRs with the structural determinants, other than through unusual steric hindrance or allosteric interaction mechanisms. Nevertheless, these prior findings should be taken into account in the overall understanding of FHR family activities.
Additionally, like FH, certain FHRs have been shown to be captured by specific molecules on bacteria and other pathogens and block complement activation, in what has been interpreted to be a protective mechanism to bypass this key innate immune recognition process (reviewed in ref. 10). For example, FHR2 and FHR5 are captured by the complement regulator–acquiring proteins (CRASPs) of serum-resistant Borrelia burgdorferi and modulate complement activation (22). Further work to explore the roles of FHR interactions with pathogens will benefit our understanding of the diverse roles these proteins play in the innate immune response.
Unanswered questions and potential approaches to demonstrating in vivo roles of the FHRs
Unquestionably, the work by Tortajada and colleagues (8), as well as the related report by Goicoechea de Jorge and colleagues (18), have provided important insights into the very likely primary functions of FHR1, FHR2, and FHR5 in deregulating FH on specific binding sites. These observations have built on rapidly increasing understanding of FH structure/function in a way that has allowed investigators to use structural similarities and differences compared with FHRs in order to develop innovative hypotheses. Likewise, the impressive effect on the field of the study of mutant forms of complement molecules that exhibit clinically important phenotypes continues to be validated by this report. Nevertheless, important questions remain with regard to the FHR family.
The first question one could ask is whether all FHRs act in the same manner. For example, FHR4 is not a part of the complexes described and has been recently demonstrated to promote complement alternative pathway activation by serving as a platform for activation (23). Indeed, the activities of FHRs may be quite context dependent; one way to consider the various demonstrated and postulated roles of FHRs is outlined in Figure 1. A second question concerns what surface determinants are important in binding of the deregulatory and regulatory FHRs. Why are the kidney, and the retina in AMD, the primary targets of damage in these potentially systemically injurious processes? Why are other organs, such as the liver (where the majority of FH and FHRs are produced) or the joints (where FH plays a key role in controlling activation on the cartilage surface), spared (24)? Does this situation have to do with some as-yet unrecognized unique characteristics in the matrix, cell membrane constituents, or organ physiology that make FHRs particularly important in controlling activation in the kidney and eye? A third question is why a deficiency state, such as ΔCFHR3-CFHR1, is protective in one complement-related disease, like AMD, but a risk factor in another. A fourth is why the FHR family evolved, and, if pathogen interactions were involved, which were the key drivers over eons. Are there additional lessons to be learned by a study of FH and FHRs, in populations with genetic variants, similar to the insights obtained for other complement components?
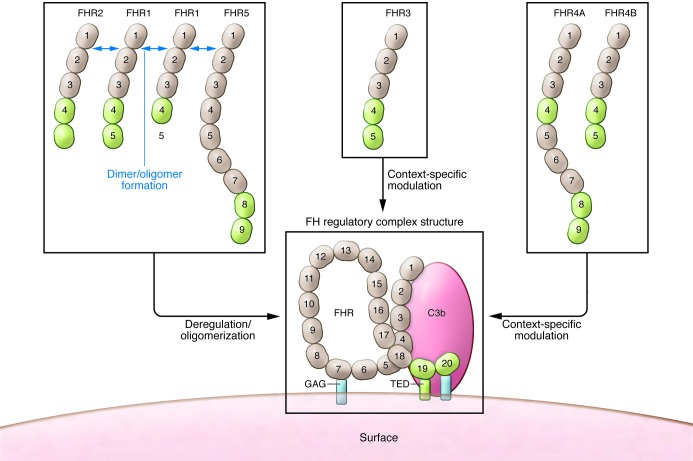
Figure 1. Postulated roles for FHRs based on the current reports discussed herein (8, 18) as well as previous studies (reviewed in refs.
9, 10, 12). Deregulation is manifest by FHR1, FHR2, and FHR5 when these oligomerize and compete with FH for C3b binding. Activities of FHR3 and FHR4A/B are less well defined, but may vary by the context in which they are used. Green color of the C-terminal 2 SCR domains of the FHRs indicates a very high degree of sequence similarity (>90%) with FH SCR19–SCR20. Other domains exhibit lower degrees of homology with FH, but three of the family members (FHR1, FHR2, and FHR5) exhibit conserved dimerization domains (18). The FH regulatory complex model is based on data reported and summarized elsewhere (21): the primary interaction with C3b requires SCR19, and the interaction with GAG occurs within SCR20.
Finally, other than the potential for therapeutic manipulation in humans, how do we learn more about the roles of individual FHRs, such that the various hypotheses illustrated in Figure 1 could be tested in a whole animal system? Mice apparently express a set of FHRs with structural features similar to those of their human counterparts (25, 26). If their activities can be demonstrated to be orthologous, the roles shown in Figure 1 should be able to be effectively tested in models of ophthalmologic, renal, arthritic, and other disease conditions in which alternative pathway activation is essential to develop tissue injury and modulation of FH activities has been shown to affect tissue injury (reviewed in ref. 1). In addition, enforcing tissue-specific inhibition or overexpression will allow us to better understand unique local tissue-specific effects of these proteins on complement activation.
Acknowledgments
The author acknowledges the important contribution to this commentary of many prior and ongoing discussions with Jonathan Hannan, Joshua Thurman, Kevin Marchbank, Nirmal Banda, Alexandra Antonioli, Michael Pangburn, and Viviana Ferreira.
Footnotes
Conflict of interest: V. Michael Holers has received consulting income and research support in the last year from Alexion Pharmaceutics.
Citation for this article: J Clin Invest. 2013;123(6):2357–2360. doi:10.1172/JCI69684.
See the related article beginning on page 2434.
References
- 1.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol. Rev. 2008;223:300–316. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 2.Ricklin D, Lambris JD. Progress and trends in complement therapeutics. Adv Exp Med Biol. 2013;735:1–22. doi: 10.1007/978-1-4614-4118-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varga L, Farkas H. rhC1INH: a new drug for the treatment of attacks in hereditary angioedema caused by C1-inhibitor deficiency. Expert Rev Clin Immunol. 2011;7(2):143–153. doi: 10.1586/eci.11.5. [DOI] [PubMed] [Google Scholar]
- 4.Rother RP, Rollins SA, Mojcik CF, Brodsky RA, Bell L. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat Biotechnol. 2007;25(11):1256–1264. doi: 10.1038/nbt1344. [DOI] [PubMed] [Google Scholar]
- 5.Schmidtko J, Peine S, El-Housseini Y, Pascual M, Meier P. Treatment of atypical hemolytic uremic syndrome and thrombotic microangiopathies: a focus on eculizumab. Am J Kidney Dis. 2013;61(2):289–299. doi: 10.1053/j.ajkd.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 6.Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration — emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38(7):450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tortajada A, et al. C3 glomerulopathy–associated CFHR1 mutation alters FHR oligomerization and complement regulation. . J Clin Invest. 2013;123(6):2434–2446. doi: 10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Cordoba SR, Tortajada A, Harris CL, Morgan BP. Complement dysregulation and disease: from genes and proteins to diagnostics and drugs. Immunobiology. 2012;217(11):1034–1046. doi: 10.1016/j.imbio.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Józsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29(8):380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype,with deletion of CFHR1 and CFHR3,is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38(10):1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 12.Skerka C, Zipfel PF. Complement factor H related proteins in immune diseases. Vaccine. 2008;26(suppl 8):I9–I14. doi: 10.1016/j.vaccine.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Fakhouri F, Frémeaux-Bacchi V, Noël LH, Cook HT, Pickering MC. C3 glomerulopathy: a new classification. Nat Rev Nephrol. 2010;6(8):494–499. doi: 10.1038/nrneph.2010.85. [DOI] [PubMed] [Google Scholar]
- 14.Malik TH, et al. A hybrid CFHR3-1 gene causes familial C3 glomerulopathy. J Am Soc Nephrol. 2012;23(7):1155–1160. doi: 10.1681/ASN.2012020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale DP, et al. Identification of a mutation in complement factor H-related protein 5 in patients of Cypriot origin with glomerulonephritis. Lancet. 2010;376(9743):794–801. doi: 10.1016/S0140-6736(10)60670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pangburn MK, Rawal N, Cortes C, Alam MN, Ferreira VP, Atkinson MA. Polyanion-induced self-association of complement factor H. J Immunol. 2009;182(2):1061–1068. doi: 10.4049/jimmunol.182.2.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins SJ, Nan R, Li K, Khan S, Miller A. Complement Factor H–ligand interactions: Self-association, multivalency and dissociation constants. Immunobiology. 2012;217(2):281–297. doi: 10.1016/j.imbio.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Goicoechea de Jorge E, et al. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci U S A. 2013;110(12):4685–4690. doi: 10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinen S, et al. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood. 2009;114(12):2439–2447. doi: 10.1182/blood-2009-02-205641. [DOI] [PubMed] [Google Scholar]
- 20.Fritsche LG, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration. Hum Mol Genet. 2010;19(23):4694–4704. doi: 10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- 21.Morgan HP, et al. Structural basis for engagement by complement factor H of C3b on a self surface. Nat Struct Mol Biol. 2011;18(4):463–470. doi: 10.1038/nsmb.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel C, et al. Complement factor H-related proteins CFHR2 and CFHR5 represent novel ligands for the infecction-associated CRASP proteins of Borrelia burgdorferi. PLoS One. 2010;5(10):e13519. doi: 10.1371/journal.pone.0013519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebecker M, Józsi M. Factor H-related protein 4 activates complement by serving as a platform for the assembly of alternative pathway C3 convertase via its interaction with C3b protein. J Biol Chem. 2012;287(23):19528–19536. doi: 10.1074/jbc.M112.364471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banda NK, et al. Essential role of surface-bound complement factor H in controlling immune complex-induced arthritis. J Immunol. 2013;190(7):3560–3569. doi: 10.4049/jimmunol.1203271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vik DP, Muñoz-Cánoves P, Kozono H, Martin LG, Tack BF, Chaplin DD. Identification and sequence analysis of four complement factor H-related transcripts in mouse liver. J Biol Chem. 1990;265(6):3193–3201. [PubMed] [Google Scholar]
- 26.Hellwage J, et al. Two factor H-related proteins from the mouse: expression analysis and functional characterization. Immunogenetics. 2006;58(11):883–893. doi: 10.1007/s00251-006-0153-y. [DOI] [PubMed] [Google Scholar]