Abstract
A disintegrin and metalloproteinase 10 (ADAM10), a disintegrin and metalloproteinase that resides in the postsynaptic densities (PSDs) of excitatory synapses, has previously been shown to limit β-amyloid peptide (Aβ) formation in Alzheimer’s disease (AD). ADAM10 also plays a critical role in regulating functional membrane proteins at the synapse. Using human hippocampal homogenates, we found that ADAM10 removal from the plasma membrane was mediated by clathrin-dependent endocytosis. Additionally, we identified the clathrin adaptor AP2 as an interacting partner of a previously uncharacterized atypical binding motif in the ADAM10 C-terminal domain. This domain was required for ADAM10 endocytosis and modulation of its plasma membrane levels. We found that the ADAM10/AP2 association was increased in the hippocampi of AD patients compared with healthy controls. Long-term potentiation (LTP) in hippocampal neuronal cultures induced ADAM10 endocytosis through AP2 association and decreased surface ADAM10 levels and activity. Conversely, long-term depression (LTD) promoted ADAM10 synaptic membrane insertion and stimulated its activity. ADAM10 interaction with the synapse-associated protein-97 (SAP97) was necessary for LTD-induced ADAM10 trafficking and required for LTD maintenance and LTD-induced changes in spine morphogenesis. These data identify and characterize a mechanism controlling ADAM10 localization and activity at excitatory synapses that is relevant to AD pathogenesis.
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia, and at present, therapeutic options to cure the disease are scarce. The underlying neuropathology of AD includes extracellular deposition of β-amyloid peptide (Aβ) and intraneuronal accumulation of aberrant forms of hyperphosphorylated tau as well as synapse dysfunction and neurodegeneration (1–3).
Substantial data indicate accumulation of Aβ in the brain, particularly in its nonfibrillar soluble oligomeric forms, as the primary influence driving synaptic dysfunction and a complex cascade of molecular events leading to dementia (4, 5).
Aβ derives from the concerted action of β-site APP-cleaving enzyme (BACE) (6), which cleaves the amyloid precursor protein (APP) at the Aβ N-terminus, and γ-secretase, which is responsible for the cleavage of the APP C-terminal stub (7). On the other hand, APP can be alternatively processed by a disintegrin and metalloproteinase 10 (ADAM10) pathway that prevents the formation of toxic Aβ (8, 9). These 2 processes are differentially segregated within the cells: ADAM10 activity is localized in the trans-Golgi network or at the plasma membrane (9, 10), whereas BACE activity is mainly confined to the ER and to the endosomal/lysosomal system (11). Hence, understanding the mechanisms that regulate the intracellular localization and trafficking to the neuronal membrane of APP and the related secretases is central for the comprehension of AD pathogenesis and for devising rationale therapeutic approaches.
ADAM10 is an integral component of postsynaptic densities (PSDs) of excitatory synapses (12–14) and binds to synapse-associated protein-97 (SAP97), a member of the MAGUK family of protein scaffolds that governs the trafficking and synaptic anchoring of AMPA- and NMDA-type glutamate receptors. The interaction involves the SH3 domain of SAP97 and an atypical proline-rich domain within the C-terminal region of ADAM10, and favours the forward trafficking of ADAM10 and its localization in synaptic membranes and, thereby, its APP-cleaving activity (12). The ADAM10/SAP97 interaction has been shown to be reduced in brains from AD patients (15), and the disruption of the association between SAP97 and ADAM10 in vivo using peptides leads to the generation of a nontransgenic model of the disease (16). Thus, forward trafficking and membrane insertion are crucial not only for maintaining a pool of ADAM10 at synaptic sites and for ADAM10-shedding activity, but also for healthy neuronal function.
Endocytosis represents another major pathway for controlling the abundance of synaptic proteins (17), and perturbations of clathrin-mediated endocytosis proteins have been reported in numerous human disorders (18). Dendritic spines appear to contain core components of the endocytic machinery (clathrin, AP2, dynamin-2) at endocytic zones located laterally to PSDs (19, 20), and clathrin-mediated endocytosis has been shown to modulate synaptic plasticity; for example, it is required for downregulation of AMPA receptors and expression of long-term depression (LTD) (21, 22).
The relevance of clathrin-mediated endocytosis in AD has been recently highlighted by genome-wide association studies (GWAS), which identified novel loci with significant association to AD (23–25). One of the genes identified is PICALM, an adaptor protein that binds clathrin, phoshoinositol, and the clathrin adaptor protein AP2 to aid the formation of clathrin-coated pits (23). Here, we demonstrate that ADAM10 binds directly to the clathrin adaptor protein AP2 and that this association is instrumental for enzyme internalization, is altered in hippocampi of patients affected by AD, and is regulated by activity-dependent plasticity processes.
Results
ADAM10 binds to the clathrin adaptor AP2, and this interaction is altered in AD patients.
Clathrin-coated pit initiation is triggered by the recruitment of the clathrin adaptor AP2 to cargo proteins in the plasma membrane that can be mediated via direct binding of AP2 to endocytic motifs within cytoplasmic domains of the target proteins (18). To address the role of endocytosis in regulating the membrane levels of ADAM10 and its potential relevance in the etiology of AD (26, 27), we first verified whether the AP2 complex subunits bind ADAM10 in homogenate from human hippocampus. AP2 exists as a heterotetrameric assembly of 2 large subunits (α and β2), a medium subunit termed μ2, and a small subunit, σ (28).
Co-IP experiments using an anti-ADAM10 antibody showed that α-adaptin, β2-adaptin, and μ2 subunits coprecipitate with ADAM10 (Figure 1A). The interaction was specific, as demonstrated by the observation that γ-adaptin, a subunit of AP1 complex, did not bind to ADAM10 (Figure 1A).
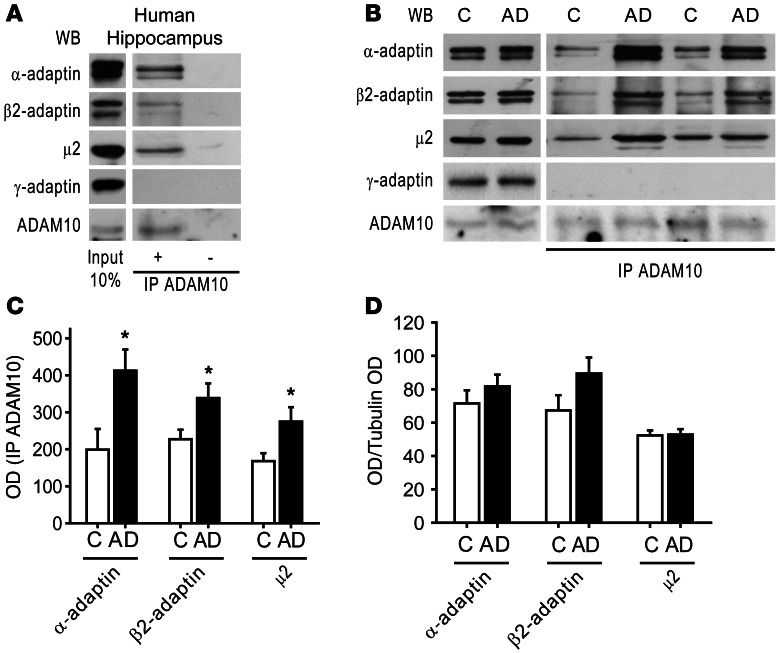
Figure 1. ADAM10 interacts with the clathrin adaptor AP2, and this association is increased in AD patients’ hippocampus.
(A) Human hippocampus homogenate IP with a rabbit anti-ADAM10 antibody. WB analysis revealed the presence of AP2 subunits, α-adaptin, β2-adaptin, and μ2 subunit, but not of the AP1 subunit γ-adaptin, in the immunocomplex. IP ADAM10 was detected by WB with a rat anti-ADAM10 antibody. As shown in the right lanes, no signal is detectable when the sample is precipitated without ADAM10 antibody. Lanes were run on the same gel but were not contiguous. (B) Homogenates from hippocampi of 6 HC and 6 AD patients were IP with anti-ADAM10 antibody and AP2 subunits coprecipitation evaluated. ADAM10/AP2 association is increased in AD compared with HC. γ-adaptin does not precipitate with ADAM10 in both groups. The levels of IP ADAM10 were detected by WB with a rat anti-ADAM10 antibody. Left lanes show WB performed on total homogenate from HC and AD hippocampi. No alterations of α-adaptin, β2-adaptin, μ2 subunit, γ-adaptin, and ADAM10 levels were detected. (C) OD was measured. Quantification of IP experiments in B (*P < 0.05, AD versus HC [C], n = 12). (D) Quantification of total levels of α-adaptin, β2-adaptin, μ2 subunit, shown in B (P < 0.05, AD versus HC, n = 12). All data were normalized on tubulin OD. In this and all subsequent figures, data represent mean ± SEM.
We then searched for a possible alteration of the ADAM10/AP2 complex interaction in hippocampi from AD patients at Braak level 4 (29) and from age-matched healthy controls (HC). Total homogenates from hippocampi of HC and AD patients were IP with anti-ADAM10 antibody, and co-IP with AP2 subunits was evaluated. A large increase in the amount of AP2 subunits bound to ADAM10 was observed in AD patients compared with HC (Figure 1, B and C; α-adaptin: HC = 198.8 ± 55.8, AD = 412.5 ± 57.1, P = 0.023; β2-adaptin: HC = 227 ± 25.7, AD = 338.3 ± 39.5, P = 0.039; μ2: HC = 167.8 ± 21.5, AD = 274.8 ± 38.9, P = 0.037; AD versus HC). As previously demonstrated, the ADAM10 total levels were not affected by the group (15), and no differences were detected between HC and AD patients in α-adaptin, β2-adaptin, and μ2 subunit levels in total homogenates, suggesting that the increased association is not related to the augmented abundance of the partners (Figure 1, B and D; α-adaptin: HC = 71.5 ± 7.8, AD = 81.7 ± 7; β2-adaptin: HC = 67.3 ± 9.1, AD = 89.5 ± 9.5; μ2: HC = 52.3 ± 3, AD = 52.8 ± 3.3; P > 0.05; AD versus HC). Moreover, we confirmed that γ-adaptin does not precipitate with ADAM10 in both HC and AD patients’ hippocampus homogenate and its expression is not affected by the group (γ-adaptin: HC = 21.7 ± 3.1, AD = 20.8 ± 4.9; P > 0.05; AD versus HC).
These data indicate AP2 as a new partner of ADAM10 and reveal a further alteration of ADAM10-binding capability in AD patients. In addition to the reported reduction of ADAM10/SAP97 association (15), we show that the interaction of ADAM10 with AP2 is enhanced in hippocampus at early phases of sporadic AD.
The clathrin-adaptor protein complex AP2 binds an atypical binding motif in ADAM10 tail.
To identify the domain of ADAM10 responsible for the interaction with AP2, we studied the complex in mouse brain. First, we confirmed the presence of ADAM10/AP2 complex by co-IP assays from total mouse brain homogenate. As shown in Figure 2A, the AP2 subunits, i.e., α-adaptin, β2-adaptin, and μ2 subunit, coprecipitate with ADAM10.
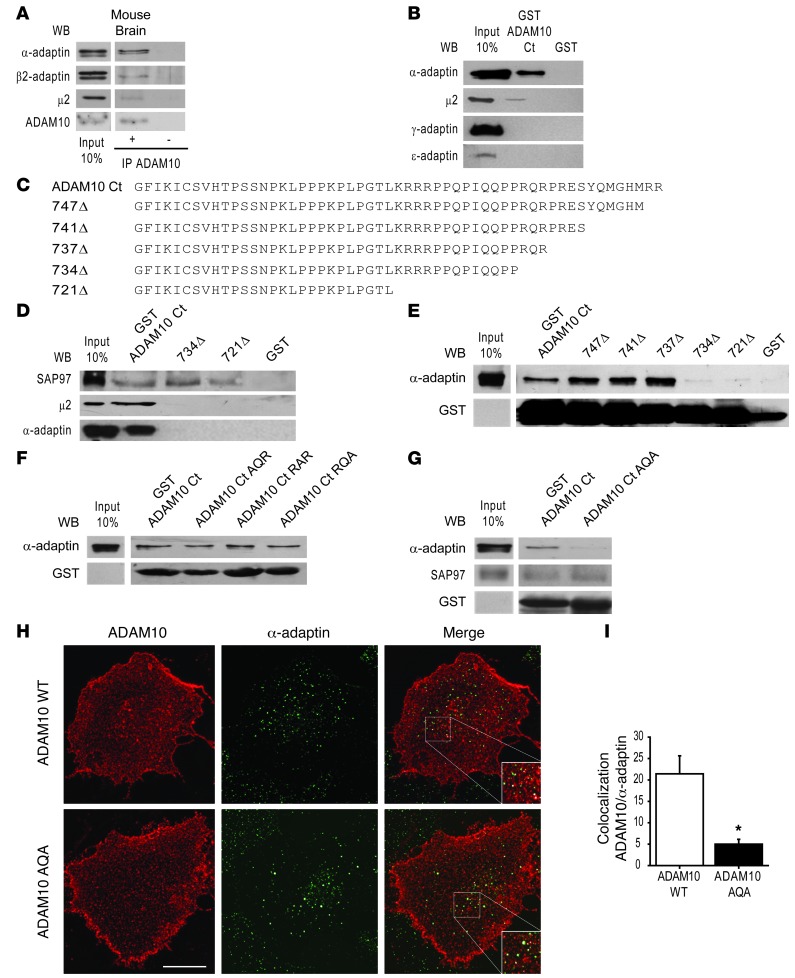
Figure 2. AP2 interacts with 735RQR737 motif of ADAM10 tail.
(A) Mouse brain homogenate was IP with ADAM10 antibody. AP2 subunits coprecipitate with ADAM10. IP ADAM10 was detected by WB. No signal is detectable when the sample is precipitated without ADAM10 antibody. (B) Pulldown assays performed with GST-ADAM10 Ct and GST from mouse brain homogenate. α-adaptin and μ2, but not γ-adaptin and ε-adaptin, are precipitated by ADAM10 Ct. (C) aa sequences of ADAM10 Ct deletion mutants (747Δ, 741Δ, 737Δ, 734Δ, 721Δ). (D) Pulldown assays performed with GST-ADAM10 Ct, 734Δ, and 721Δ mutants. Deletion of the last 15 aa abolishes AP2 subunit binding, whereas SAP97 interacts with the membrane proximal 27 aa of ADAM10 tail. (E) Pulldown assays carried out with all deletion mutants. The 735RQR737 motif is necessary for α-adaptin precipitation. (F) Pulldown assays performed with GST-ADAM10 Ct bearing mutations of 735RQR737 motif. No change in α-adaptin binding was detected. (G) Pulldown assays performed with GST-ADAM10 Ct mutated 735RQR737 to AQA. The mutation of both arginine residues to alanine abolishes α-adaptin binding, but does not affect SAP97 association. (H) Representative images of ADAM10 membrane staining (red) and α-adaptin (green) of COS7 cells expressing ADAM10 WT or ADAM10 AQA. Merged images are shown on the right. Scale bar: 20 μm. (I) Quantification of ADAM10 WT/α-adaptin and ADAM10 AQA/α-adaptin colocalization of experiments in H (*P < 0.05, ADAM10 WT versus ADAM10 AQA, n = 2 experiments, 12 cells per condition). In A and E–G, lanes were run on the same gel but were not contiguous.
To test the role of the cytoplasmic tail (Ct) of ADAM10 in the interaction and identify potential motifs recognizing AP2, we carried out pulldown assays where fusion proteins of glutathione-S-transferase (GST) and GST bound to ADAM10 Ct (GST-ADAM10 Ct) were incubated with mouse brain homogenate. GST-ADAM10 Ct pulled down both the α-adaptin and μ2 subunits, demonstrating that the ADAM10 Ct binds the AP2 complexes (Figure 2B). The interaction was specific because other AP subunits, such as γ-adaptin and ε-adaptin, that compose AP1 and AP4 complexes, did not bind to GST-ADAM10 Ct (Figure 2B).
To define the ADAM10 Ct sequence determinants required for its interaction with the AP2 complexes, we tested a series of deletion mutants of GST-ADAM10 Ct (Figure 2C). Truncation of up to the last 15 aa of ADAM10 tail (GST-734Δ) completely abolished binding to both the α-adaptin and μ2 subunits of the AP2 complex (Figure 2D), indicating that this aa segment is required for ADAM10/AP2 interaction. In contrast, ADAM10 interaction with SAP97 (12) was preserved after deletion of either the last 15 aa (GST-734Δ) or the last 28 aa, including the second proline-rich domain (GST-721Δ) of the ADAM10 tail (Figure 2D). These data demonstrate that our deletion mutants are functional and that the interaction domains responsible for SAP97 and AP2 do not overlap; a distal 15 aa sequence is responsible for the binding to AP2 complex, while SAP97 interacts with the most membrane-proximal proline-rich domain and the second proline stretch is dispensable. AP2 complexes often bind to short, canonical peptide motifs on transmembrane cargo: Yxxϕ motifs (30) or “dileucine” (D/ExxxLL) motifs (31). Other cargo-bearing noncanonical sorting motifs, such as aa stretches enriched in arginine residues, may also bind to AP2 (32–34). The identified 15-aa region contains both a tyrosine residue and several arginines.
To narrow down the region responsible for AP2 binding, we performed pulldown assays with sequential deletion mutants of the last 15 aa of the ADAM10 tail. As shown in Figure 2E, while neither the removal of the last 2 arginines (747Δ) nor of the tyrosine residue (741Δ) affected α-adaptin precipitation, removing the 735RQR737 motif prevented AP2 binding (734Δ). Single mutations of this motif did not compromise the interaction with α-adaptin, as demonstrated by pulldown assays (Figure 2F) but when both the 735R and 737R residues were mutated to alanine in the GST-ADAM10 Ct fusion protein, the binding to α-adaptin was completely abolished (Figure 2G). Furthermore, SAP97 binding to GST-ADAM10 Ct fusion protein was not affected by the mutation of 735R and 737R residues to alanine, suggesting that SAP97 and AP2 do not compete for the binding to ADAM10 Ct (Figure 2G).
To confirm the pulldown assays’ results, we mutated 735R and 737R residues in the context of the ADAM10 full-length protein and compared its colocalization with α-adaptin with that of ADAM10 WT. Because binding to endocytic motifs and AP2 recruitment occur at the plasma membrane, we measured the colocalization degree of α-adaptin and membrane-inserted ADAM10, which was detected in nonpermeabilizing conditions with an antibody against ADAM10 extracellular domain. We found a significant decrease in the colocalization of the ADAM10 AQA mutant when compared with ADAM10 WT (Figure 2, H and I; ADAM10 WT = 21.4% ± 4.2%, ADAM10 AQA = 5% ± 1.1%; P = 0.001, ADAM10 AQA versus ADAM10 WT). Together, these experiments identified a previously uncharacterized atypical AP2-binding motif within the ADAM10 Ct.
The clathrin-adaptor protein complex AP2 is responsible for ADAM10 internalization.
To evaluate the functional significance of the 735RQR737 AP2-binding motif in ADAM10 endocytosis, we performed fluorescence-based “antibody feeding” assays to assess internalization in COS7 cells expressing TacADAM10-RAR, a chimera of the surface reporter protein Tac (human IL-2 receptor α-subunit) (35) with a modified version of the ADAM10 C-terminal tail. The modification consisted in a mutation in the ER retention motif in ADAM10 that allowed its ER exit and permitted delivery to the surface (36). COS7 cells were transfected with either the chimera TacADAM10-RAR-AQA, able to exit the ER and carrying the mutated AP2 binding motif, or the deletion mutant TacADAM10 721Δ, which lacks the last 28 aa.
Surface TacADAM10 chimeras were first “pulse labeled” in live cells, at 4°C to block membrane trafficking by incubation with anti-Tac antibody. Subsequently, cells were either returned to 37°C to resume endocytosis or kept at 4°C as control. The accumulation of antibody-bound TacADAM10 chimeras in intracellular compartments and the amount of antibody-bound TacADAM10 chimeras remaining on the surface were tracked by quantitative immunofluorescence staining under permeabilizing versus nonpermeabilizing conditions. The ratio of intracellular fluorescence/surface fluorescence (internalization index) was used to control for the variable expression levels of TacADAM10 chimeras in different cells.
We found that the internalization indexes of both TacADAM10-RAR-AQA and TacADAM10 721Δ were significantly lower compared with TacADAM10-RAR (Figure 3, A and B; internalization index, TacADAM10-RAR-AQA = –21.7% ± 2.9%, TacADAM10 721Δ = –18.2% ± 3.9%; P < 0.05, TacADAM10-RAR versus TacADAM10-RAR-AQA, TacADAM10 721Δ), demonstrating that the AP2-binding motif 735RQR737 is responsible for the internalization process and no other relevant determinants are contained in the last 28 aa of ADAM10 tail.
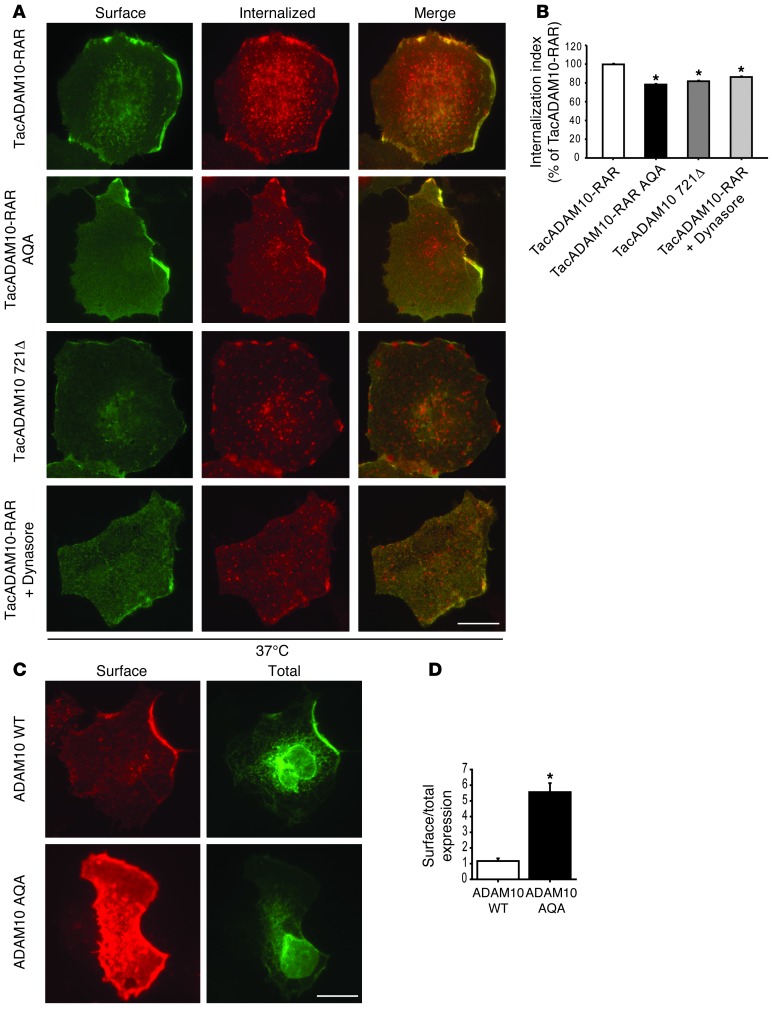
Figure 3. ADAM10/AP2 interaction is relevant for ADAM10 endocytosis and membrane expression.
(A) Antibody uptake assays were performed on COS7 cells transfected with either TacADAM10-RAR or the mutant TacADAM10-RAR AQA (lacking AP2-binding motif) or the deletion mutant Tac721Δ. TacADAM10-RAR internalization was also analyzed after treatment with dynamin inhibitor dynasore (TacADAM10-RAR+dynasore, 80 μM for 30 minutes before the internalization assay). Representative images of cells returned to 37°C to allow endocytosis. A decrease in the number of internalization puncta of TacADAM10-RAR AQA and of the deletion mutant Tac721Δ, of TacADAM10-RAR+dynasore is detected when compared with TacADAM10-RAR. Scale bar: 20 μm. (B) Quantification of the internalization index of experiments in A (*P < 0.05, TacADAM10-RAR AQA versus TacADAM10-RAR, Tac721Δ versus TacADAM10-RAR, TacADAM10-RAR+dynasore versus TacADAM10-RAR, n = 3 experiments, 27 cells per condition). (C) ADAM10 WT and the mutant ADAM10 AQA, lacking the AP2-binding motif, were transfected into COS7 cells and surface/total labeling was analyzed. ADAM10 WT was faintly localized at the surface despite intense intracellular labeling. In contrast, the mutant tested displayed strong surface staining. Scale bar: 20 μm. (D) Quantification of surface expression ratios in C (*P < 0.05, ADAM10 WT versus ADAM10 AQA, n = 18–20 cells per condition).
To confirm the involvement of clathrin-dependent endocytosis in ADAM10 internalization, we performed the experiment in the presence of the dynamin inhibitor dynasore (80 μM for 30 minutes before the internalization assay). As shown in Figure 3, A and B, dynasore treatment led to a subtle but significant reduction of the TacADAM10-RAR internalization index to the same extent as that of the mutation of the AP2-binding domain, proving that clathrin-dependent endocytosis is responsible for ADAM10 removal from the plasma membrane (Figure 3, A and B; TacADAM10-RAR+Dynasore = –13.9% ± 4.5%; P < 0.05, TacADAM10-RAR+Dynasore versus TacADAM10-RAR). As expected, at 4°C there was no internalization (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI65401DS1).
To test whether the lack of the AP2-binding motif and the consequent decrease of internalization rate affected ADAM10 membrane levels, ADAM10 WT or ADAM10 AQA was transfected in COS7 cells and their surface expression was analyzed by immunofluorescence. Both ADAM10 WT and ADAM10 AQA showed a similar intracellular pattern staining. The mutant ADAM10 AQA displayed stronger surface labeling (Figure 3, C and D; ADAM10 WT = 1.16 ± 0.17; ADAM10 AQA = 5.56 ± 0.58; P = 4.7 × 10–9, ADAM10 WT versus ADAM10 AQA).
Since it has been demonstrated that ADAM10 endocytosis affects the ectodomain shedding of ADAM10 itself (37), we checked the ectodomain shedding of the AP2-binding–deficient ADAM10 mutant. In agreement with previous studies (38, 39), we observed a fragment of approximately 16 kDa (ADAM10 CTF) in cell lysates from HEK293 cells transfected with ADAM10 WT and ADAM10 AQA. However, in our experimental system, no differences in the ADAM10 CTF levels were detected (Supplemental Figure 1B; ADAM10 WT, ADAM10 CTF/ADAM10 = 0.62 ± 0.14; ADAM10 AQA, ADAM10 CTF/ADAM10 = 0.38 ± 0.05; P > 0.05, ADAM10 WT versus ADAM10 AQA). These experiments demonstrate that the AP2-binding motif within ADAM10 Ct controls ADAM10 endocytosis and plays a role in limiting its surface expression.
The C-terminal tail is therefore the domain responsible for the regulation of ADAM10 binding to its intracellular partners, i.e., AP2 and SAP97, and these interactions modulate ADAM10 membrane levels and are altered in AD patients’ hippocampus (ref. 15 and see Figure 1, B and C). In light of these considerations and to verify whether genetic modulation of protein/protein interaction might occur, screening of ADAM10 C-terminal tail was carried out. A sample of 200 patients with possible AD (mean age = 68.0 ± 7.2 yr, 67.7% female) was screened for ADAM10 exon 15 and exon 16 genetic polymorphisms. In the examined 400 alleles, no genetic variations were found. Despite the importance of ADAM10 C-terminal tail, no genetic variations associated with AD have been found, suggesting that other cellular mechanisms might be responsible for the altered interactions between ADAM10 and its partners in AD patients.
Structural characterization of ADAM10/AP2 interaction.
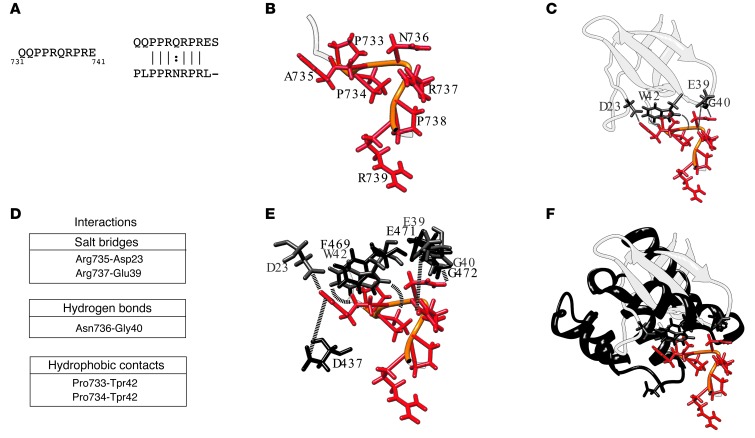
As demonstrated by the results obtained through the analysis of the ADAM10 mutants (Figure 2), the region of ADAM10 important for the interaction with AP2 contains 2 positively charged residues, R735 and R737, and a hydrophilic amino acid, Q736, crucial for binding (Figure 4A). A database search for similar sequences in the Protein Data Bank (PDB) (40) identified a peptide with a similar sequence (PLPPRNRPRL; Figure 4A) cocrystallized with the c-Src SH3 domain and derived from a phage display library experiment (PDB Id 1QWE) (41). This peptide does not have a regular secondary structure, and the side chains of its R735 and R737 residues (numbering as in the PDB entry) are oriented in opposite direction with respect to N736 (Figure 4B). The analysis of the structure of the c-Src SH3-peptide complex shows that the 2 arginines interact through electrostatic interactions with D23 and E39 of its cognate protein, while N736 establishes a hydrogen bond with G40 (Figure 4, C and D). The 2 conserved prolines, P733 and P734, establish hydrophobic contacts with W42 (Figure 4, C and D).
Figure 4. Structural characterization of ADAM10/AP2 binding.
(A) ADAM10 sequence involved in binding the AP2 complex and its alignment with the most similar peptide sequence found in the PDB (Id: 1QWE) (B) Ribbon representation of the 1QWE peptide structure (PDB). Conserved aa are shown as red sticks with their backbone in orange. (C) Representation of the peptide–c-Src SH3 complex. The c-Src SH3 domain is represented in light gray ribbon and the interacting residues in gray stick. The interactions established by the 2 molecular partners are highlighted by dashed lines. (D) List of the interactions between the peptide and the c-Src SH3 domain. (E) Structural superposition between the residues of the c-Src SH3 domain and the β2 subunit of the AP2 complex colored in gray and black, respectively. The same color code in B was used for the peptide. (F) Ribbon representation of the structures of the c-Src SH3 domain and a portion of the β2 subunit of the AP2 complex, reported in light and dark gray, respectively. The interacting residues and the peptide are also shown.
Given the high degree of sequence similarity between the peptide and the C-terminal region of ADAM10, we can hypothesize a similar binding mode in the 2 systems. However, AP2 does not contain a SH3 domain nor a region with a structure resembling it (Figure 4C). Therefore, we searched the AP2 structure for regions similar to that interacting with the peptide in the c-Src SH3 complex. We found that the only exposed region with a backbone similar to that of the region 39–42 of the SH3 protein was the 469–472 stretch of the β2 subunit of AP2 (Supplemental Figure 2 and ref. 42) that can be superimposed with a root mean square deviation (RMSD) of 1.6 Å (Figure 4E).
We used the relative orientation of the peptide/SH3 interacting regions as a template to model the interaction between the ADAM10 C-terminal tail and the AP2 structure (42). Interestingly, after superposition, the same pattern of side-chain interactions observed in the PDB Id complex could be reconstructed with the ADAM10 residues P733, P734, Q736, and R737, interacting with residues F469, G472, and E471 of AP2 (Figure 4E). Although there is no acidic residue corresponding to D23 in the examined region of AP2, residue D437 of the β2 subunit is properly positioned to establish an electrostatic interaction with R735 (distance between [R735-NHC(NH2)2+] and (D437-COO–) = 3.6 Å). In summary, although the fold of the binding region identified on the β2 subunit of AP2 is different from that of the c-Src–SH3 domain, the orientation of the residues potentially involved in the binding is conserved, as well as the network of interactions they can establish (Figure 4, E and F).
AP2 complex mediates long-term potentiation–induced ADAM10 internalization.
At excitatory synapse, the relative rates of insertion/removal of glutamate receptors control synaptic abundance of glutamate receptors and, thereby, are tightly regulated by activity dependent synaptic plasticity (43). The major routes to govern synaptic insertion/removal processes are forward trafficking mechanisms, which require interactions with specific proteins, and clathrin-mediated endocytosis (44–46). In light of this consideration and of the relevance of synaptic failure in AD pathogenesis (47), we analyzed the effect of synaptic plasticity on ADAM10 binding to its partners, i.e., AP2 and SAP97. To this, we chose 2 main paradigms of activity-dependent synaptic plasticity, i.e., long-term potentiation (LTP) and LTD.
We induced LTP and LTD in primary hippocampal neuronal cultures using highly validated chemical treatments (48, 49) and analyzed ADAM10 binding to the AP2 complex and to SAP97, using co-IP experiments. Chemical LTD (cLTD) was induced with a bath application of NMDA (50 μM for 10 minutes) (48), and neurons were recovered in normal ACSF for 20 minutes (Figure 5A). To induce LTP, we used a chemical stimulation protocol with forskolin plus rolipram for 16 minutes that results in prolonged NMDA receptor-dependent LTP (cLTP) (ref. 49 and Figure 5B).
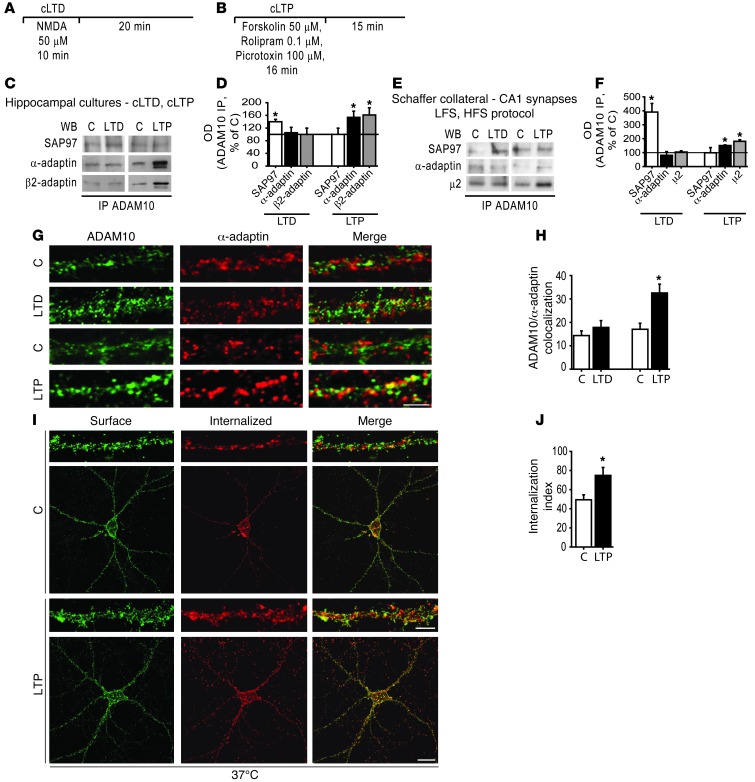
Figure 5. LTP enhances ADAM10/AP2 association and ADAM10 internalization.
Schematic representation of the chemical protocol used to induce cLTD (A) and cLTP (B). (C) ADAM10/SAP97 and ADAM10/AP2 co-IP assays performed on homogenates of control and either cLTD or cLTP-treated neurons. cLTD increases ADAM10/SAP97 coprecipitation, while cLTP enhances ADAM10/AP2 complex. (D) Quantification of OD of experiments in C (*P < 0.05 cLTP versus C, n = 6, cLTD versus C, n = 4). (E) LTP/LTD were induced by electrophysiological stimulation in hippocampal slices. ADAM10/SAP97 and ADAM10/AP2 co-IP assays were performed in CA1 homogenates. LTD fosters ADAM10/SAP97 association without affecting ADAM10/α-adaptin and ADAM10/μ2 precipitation. LTP stimulates ADAM10 association to both α-adaptin and μ2, while it does not change ADAM10/SAP97 complex levels. (F) Quantification of OD of experiments in E (*P < 0.05 LTP versus C, LTD versus C, n = 4). (G) ADAM10 (green) and α-adaptin (red) staining in dendritic compartment after either cLTD or cLTP induction in hippocampal neurons. Merged images are shown on the right. Scale bar: 4 μm. (H) Quantification of ADAM10/α-adaptin colocalization of experiments in G (*P < 0.05 cLTP versus C, n = 2 experiments, 10 neurons per condition). (I) Internalization assays were performed on neurons expressing TacADAM10-RAR either exposed or not exposed to cLTP treatment. Representative images of surface (green) and internalized (red) TacADAM10-RAR from control and cLTP induced neurons are shown. Insets show higher magnification examples of internalized puncta in dendrites. Scale bars: 20 μm; 4 μm (insets). (J) Quantification of the internalization index of experiments in I (*P < 0.05 cLTP versus C, n = 2 experiments, 10 neurons per condition).
cLTP induction significantly increased ADAM10 precipitation with the α-adaptin and β2-adaptin subunits of AP2 complexes without affecting SAP97 binding (Figure 5, C and D; α-adaptin = +53.7% ± 19.6%, P = 0.04; β2-adaptin = +61.3% ± 19.6%, P = 0.04; SAP97 = +0.3% ± 19.3%, P > 0.05; cLTP versus control). On the contrary, cLTD fostered the formation of ADAM10/SAP97 without altering the interaction with the α-adaptin and β2-adaptin subunits of the AP2 complexes (Figure 5, C and D; SAP97 = +40% ± 7.6%, P = 0.013; α-adaptin = +5.3% ± 17.0%, β2-adaptin = +0.5% ± 20.3%, P > 0.05; cLTD versus control).
To confirm these results, LTP and LTD were induced in hippocampal slices by electrophysiological stimulation: LTD was induced with low frequency stimulation (LFS) protocol and LTP by means of high frequency stimulation (HFS), and after 30 minutes the CA1 region was dissected out to perform the IP assays. Quantitative analyses showed a significant increase in ADAM10/AP2 complex levels after LTP induction, while the ADAM10/SAP97 complex was not affected (Figure 5, E and F; α-adaptin = +50.9% ± 4.7%, P = 0.008; β2-adaptin = +81% ± 11.7%, P = 0.01; SAP97 = –1% ± 37.2%, P > 0.05; LTP versus control). In contrast, LTD induction promoted ADAM10 binding to SAP97 without modifying the ADAM10/AP2 interaction (Figure 5, E and F; SAP97 = +291% ± 61.6%, P = 0.04; α-adaptin = –19.2% ± 26.6%; β2-adaptin = +4.6% ± 8.4%, P > 0.05; LTD versus control).
To analyze whether LTP induces AP2-mediated ADAM10 endocytosis, we first quantified the colocalization of ADAM10 and α-adaptin after either cLTP or cLTD induction in hippocampal neurons. A quantitative analysis of dendritic ADAM10 staining showed a significant increase in ADAM10 colocalization with α-adaptin in cLTP-treated neurons when compared with control cultures (Figure 5, G and H; C = 17.0% ± 2.6%, cLTP = 32.5% ± 3.8%; P = 0.003, cLTP versus control). No modifications of colocalization of ADAM10 and α-adaptin were detected after cLTD induction (Figure 5, G and H; C = 14.4% ± 2.0%, cLTD = 17.8% ± 2.9%; P > 0.05, cLTD versus control).
We then used direct fluorescence-based “antibody feeding” assays to measure ADAM10 internalization (50, 51). Neuronal cultures were transfected with TacADAM10-RAR, and the assays were carried out as described in Figure 3, A and B.
A significant increase in TacADAM10-RAR internalization was observed in cLTP-induced neurons when compared with control cultures (Figure 5, I and J; internalization index, C37°C = 49.4 ± 5.1, cLTP37°C = 74.9 ± 8.3; P = 0.02, C37°C versus cLTP37°C), indicating that cLTP promotes TacADAM10-RAR removal from the plasma membrane and increases its intracellular accumulation. No significant differences between control and cLTP were observed when trafficking was arrested at 4°C (Supplemental Figure 3) and the internalization index was significantly increased at 37°C compared with the 4°C condition (P = 0.003, C4°C versus C37°C; P = 0.0004, cLTP4°C versus cLTP37°C), confirming the reliability of our assay. These data demonstrate that the AP2 complex is responsible for LTP-induced ADAM10 endocytosis, whereas it is not involved in LTD mechanisms, which implicate a modulation of the interaction with SAP97.
LTP and LTD have opposite effects on the plasma membrane expression of ADAM10.
To demonstrate that ADAM10 endocytosis mediated by AP2 could influence ADAM10 membrane levels, we assessed ADAM10 surface expression in primary hippocampal cultures by biochemical assays after induction of cLTP and cLTD. To carry out the crosslinking method (52), control and cLTD hippocampal cultures were treated with the BS3, a membrane-impermeable, irreversible, amine-reactive crosslinker reagent. The BS3 caused the crosslinking of surface ADAM10 and the formation of high–molecular weight aggregates that barely entered the gel. Therefore, ADAM10 surface pool was not detectable and the modifications of ADAM10 intracellular pool reflected alterations of ADAM10 surface expression (12). In cLTD-treated cultures, we observed a significant decrease in the ADAM10 intracellular pool (Figure 6, A and B; ADAM10 = –27.7% ± 7.1%; P = 0.008, cLTD versus control), indicating that cLTD promotes ADAM10 insertion in the cell membranes.
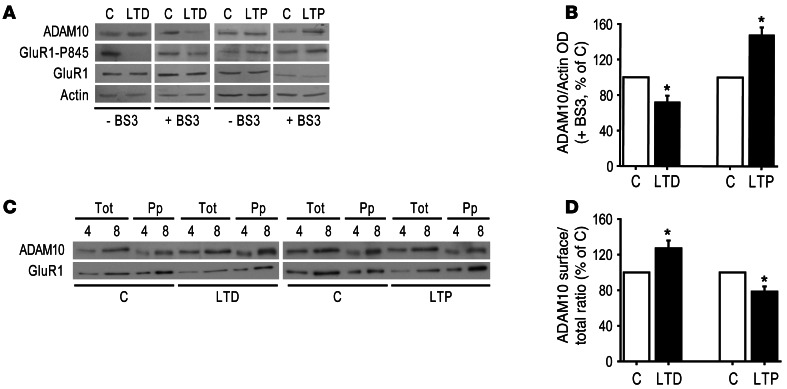
Figure 6. LTD promotes ADAM10 insertion into the membranes, whereas LTP reduces ADAM10 membrane levels.
(A) Immunoblot of ADAM10, GluR1-P845, GluR1, and actin from control and cLTD/cLTP-treated hippocampal cultures either exposed or not exposed to the crosslinker BS3. cLTD leads to a significant decrease in ADAM10 intracellular pool, which reflects an augment in ADAM10 surface expression. cLTP significantly increases ADAM10 intracellular pool because cLTP results in a decrease in ADAM10 membrane localization. As expected, cLTD induction reduces and cLTP increases the Ser-845 phosphorylation of GluR1. (B) Quantification of the ADAM10/Actin OD ratio of BS3-treated neurons of experiments in A (*P < 0.05 cLTD versus C, n = 7, cLTP versus C, n = 3). (C) After either cLTD or cLTP induction, hippocampal cultures were biotinylated and the extracts were precipitated with neutravidin. To avoid saturation of band signal in order to carry out precise quantitative analyses, samples of extracts (Tot) and neutravidin-precipitated samples (Pp) were loaded such that each lane represents a percentage of the total material per plate. Representative immunoblot of ADAM10 and GluR1 from control and treated cultures. (D) ADAM10 OD in Pp samples (surface) was measured and normalized to ADAM10 OD in tot samples (total) to calculate the surface/total ratio. Quantification of the surface/total ratio of experiments in C (*P < 0.05 cLTD versus C, n = 5, cLTP versus C, n = 6).
As internal control, we measured the surface levels of the AMPA receptor subunit GluR1 and its phosphorylation at Ser-845. Phosphorylation of Ser-845 by cAMP-dependent PKA (53) regulates the surface expression of AMPARs by increasing the pool of GluR1 recycled back to the surface after their endocytosis (17, 46). Surface levels of phosphorylated GluR1 were significantly reduced (Figure 6A; GluR1 Ser-845 = –29.7% ± 6.6%; P = 0.045, cLTD versus control), while GluR1 surface levels were unchanged probably due to recycling to the plasma membrane (Figure 6A; GluR1 = –9.9% ± 10.6%; P > 0.05, cLTD versus control). In fact, when BS3 assays were performed immediately after LTD induction, without recovering neurons, a slight but significant increase of GluR1 intracellular pool was detected (GluR1 = +6% ± 1.5%; P = 0.02, cLTD versus control), demonstrating GluR1 internalization.
On the other hand, after cLTP induction, a significant increase in ADAM10 intracellular pool was detected (Figure 6, A and B; ADAM10 = +48% ± 8.4%; P = 0.03, cLTP versus control), indicating that cLTP triggers ADAM10 removal from the membranes. As expected, GluR1 intracellular levels were significantly decreased because of membrane insertion (Figure 6A; GluR1 = –19% ± 3.3%; P = 0.005, cLTP versus control) and phosphorylation of Ser-845 was significantly enhanced (Figure 6A; GluR1 Ser-845 = +66% ± 17.4%; P = 0.03, cLTP versus control).
These results were confirmed by surface biotinylation assays. Quantitative analyses showed a significant increase in ADAM10 surface levels in hippocampal cultures after cLTD induction (Figure 6, C and D; ADAM10 = +27.3% ± 8.7%; P = 0.035, cLTD versus control), whereas cLTP significantly decreased ADAM10 surface levels (Figure 6, C and D; ADAM10 = –21.5% ± 5.7%; P = 0.013, cLTP versus control). As expected, GluR1 membrane expression was significantly increased by cLTP (Figure 6C; GluR1 = +31% ± 8.2%; P = 0.019, cLTP versus control), while no changes were noted after cLTD stimulation (Figure 6C; GluR1 = +25.9% ± 19.6%; P > 0.05, cLTP versus control). The efficiency and specificity of our assay was verified (Supplemental Figure 4). The precipitation was efficient because no ADAM10 was detected after a second precipitation. Moreover, virtually no actin was revealed in the neutravidin-precipitated material, indicating that only cell surface proteins were recovered. Taken together, these results suggest that activity-dependent synaptic plasticity paradigms have an opposite effect on ADAM10 membrane expression: LTP induces ADAM10 removal whereas LTD promotes ADAM10 membrane insertion.
LTD but not LTP stimulates ADAM10 forward trafficking.
Since ADAM10 is located at the glutamatergic PSDs (12), we asked whether activity-dependent synaptic plasticity modulates ADAM10 synaptic availability. To test this, synaptic Triton insoluble fraction (TIF, a fraction highly enriched in all categories of PSD proteins and absent of presynaptic markers; ref. 54) was purified from control and either cLTD- or cLTP-treated neurons, and protein levels were measured by Western blot (WB). cLTD stimulation caused a significant increase in both ADAM10 and SAP97 levels in TIF (Figure 7, A and B; ADAM10 = +28.5% ± 6.0%, P = 0.018; SAP97 = +24.8%± 7.7%; P = 0.04, cLTD versus control), a reduction in the phosphorylation of Ser-845 of GluR1 (Figure 7A; GluR1 Ser-845 = –14% ± 2.4%; P = 0.001, cLTD versus control), and no modifications of GluR1 synaptic levels (Figure 7A; GluR1 = +14% ± 9%; P > 0.05, cLTD versus control). In contrast, cLTP induction led to no significant alterations in ADAM10 and SAP97 synaptic levels (Figure 7C; ADAM10 = –21% ± 13.3%, SAP97 = –1.75% ± 13.5%; P > 0.05, cLTP versus control), but induced phosphorylation of Ser-845 of GluR1 (Figure 7C; GluR1 Ser-845 = +381.9% ± 37.1%; P = 0.005, cLTP versus control) and increased GluR1 synaptic levels (Figure 7C; GluR1 = +43.7% ± 12.0%, P = 0.035, cLTP versus control). No significant changes in ADAM10 and SAP97 levels in total homogenate after either cLTD or cLTP induction were observed (Figure 7, A and C; cLTD, ADAM10 = +37.8% ± 44.3%, SAP97 = +6.5% ± 24.1%; P > 0.05, cLTD versus control; cLTP, ADAM10 = –26.8% ± 25%, SAP97 = +47.8% ± 26.8%, P > 0.05, cLTP versus control), pointing toward a trafficking rather than a transcriptional effect.
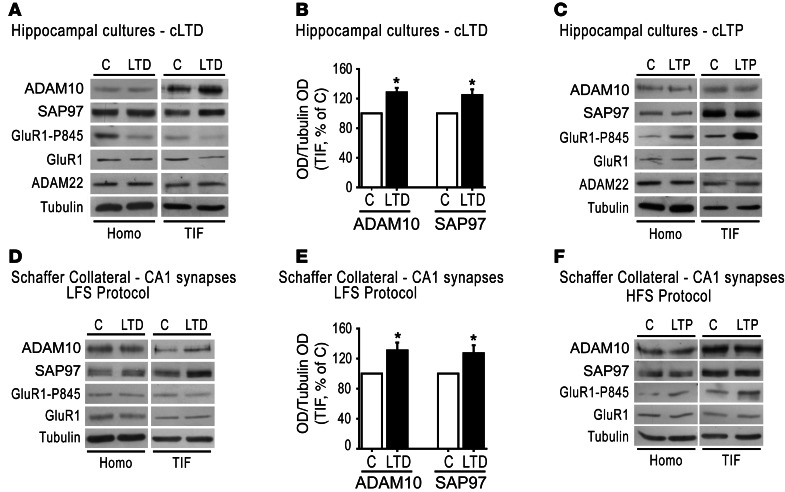
Figure 7. LTD induces ADAM10 trafficking toward the postsynaptic membranes.
(A) WB analysis of homogenate and TIF from control or cLTD-treated hippocampal cultures. cLTD leads to a redistribution of SAP97 and ADAM10 in TIF but not in homogenate and to a decrease in Ser-845 phosphorylation of GluR1. ADAM22 localization is not affected. (B) Quantification of the ratio of ADAM10/tubulin and SAP97/tubulin OD of experiments in A (*P < 0.05, cLTD versus C, n = 4). (C) WB of homogenate and TIF from control and cLTP-treated cultures. Ser-845 phosphorylation of GluR1 is enhanced, whereas ADAM10, SAP97, and ADAM22 protein levels and localization are not affected. (D) LTD was induced by LFS and TIF was purified from CA1 region. WB shows an increased localization of ADAM10 and SAP97 in TIF, while total protein levels are not changed. (E) Quantification of the ratio of ADAM10/tubulin and SAP97/tubulin OD of experiments in D (*P < 0.05, LTD versus C, n = 4). (F) WB analysis of homogenate and TIF from CA1 region of control and LTP-induced slices shows no changes in ADAM10 and SAP97 synaptic localization and total protein levels.
To verify the specificity of the effect, we evaluated the postsynaptic levels of ADAM22, another member of the ADAM family localized at synapses (55). WB experiments showed no significant modifications of ADAM22 levels in total homogenate and TIF after either cLTD or cLTP induction (Figure 7, A and C; cLTD, ADAM22 = +13.5% ± 18.4%; P > 0.05, cLTD versus control; cLTP, ADAM22 = +0.8% ± 11.1%; P > 0.05, cLTP versus control).
Finally, similar results were obtained when LTP and LTD were induced in hippocampal slices by electrophysiological stimulation, providing further validation of our results. Thirty minutes after LTD induction, the TIF was purified from the CA1 region of hippocampal slices and quantitative WB analyses showed a significant increase in both ADAM10 and SAP97 levels after 30 minutes (Figure 7, D and E; ADAM10 = +31.0% ± 10.4%, P = 0.03; SAP97 = +27.1% ± 10.6%; P = 0.04, LTD versus control). In contrast, LTP induction did not change ADAM10 and its binding partner SAP97 synaptic levels (Figure 7F, ADAM10 = –7.5% ± 19.3%, SAP97 = –13.8% ± 13.4%; P > 0.05, LTP versus control). No significant alterations of total ADAM10 and SAP97 levels after either LTD or LTP induction were detected (Figure 7, D and F; LTD, ADAM10 = +32% ± 28.1%, SAP97 = +11% ± 7.6%, P > 0.05, LTD versus control; LTP, ADAM10 = –22% ± 13.3%, SAP97 = +3% ± 15%, P > 0.05, LTP versus control). These results indicated that LTD, but not LTP, increases ADAM10 membrane expression by stimulating SAP97-mediated ADAM10 trafficking, revealing that activity-dependent plasticity exerts a bidirectional control of ADAM10 synaptic levels in the postsynaptic compartment.
SAP97-mediated LTD-induced ADAM10 trafficking is required for LTD.
To clarify the role of SAP97 in LTD-induced ADAM10 trafficking, we took advantage of the cell-penetrating Pro peptide to disrupt the ADAM10/SAP97 interaction (Figure 8A and ref. 12). Treatments with Ala peptide were always performed as control. Hippocampal cultures exposed or not exposed to either Pro or Ala peptide (1 μM, 30 minutes) were then treated with NMDA (50 μm, 10 minutes) to induce cLTD.
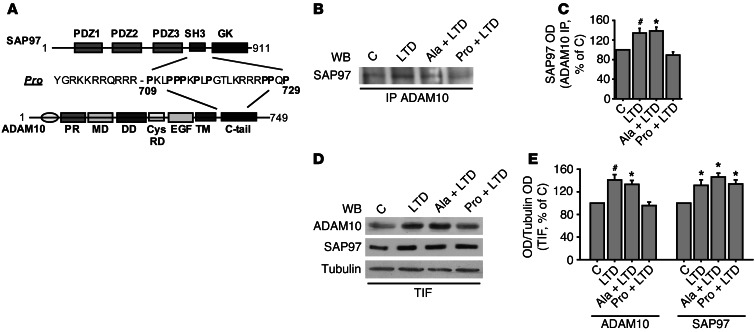
Figure 8. ADAM10/SAP97 interaction is required for LTD-induced ADAM10 trafficking and LTD maintenance.
(A) Schematic representation of SAP97, ADAM10, and Pro peptide. (B) Primary hippocampal cultures were either exposed or not exposed to either Pro or Ala peptide, and, after 30 minutes, they were either left untreated or cLTD treated. ADAM10/SAP97 co-IP assays were carried out from total homogenate. cLTD fosters ADAM10/SAP97 association, while Pro peptide pretreatment prevents this effect. Ala peptide incubation does not affect cLTD-induced ADAM10 trafficking. (C) Quantification of OD of experiments in B (*P < 0.05 Pro+LTD versus Ala+LTD, #P < 0.05 Pro+LTD versus LTD, n = 4). (D) Immunoblot of TIF obtained from above-described treated cultures. cLTD results in a redistribution of ADAM10 into TIF, whereas Pro, but not Ala peptide, preincubation prevents this process. SAP97 localization is not affected by peptide incubation. (E) Quantification of OD of experiments in D (ADAM10, *P < 0.05 Pro+LTD versus Ala+LTD, #P < 0.05 Pro+LTD versus LTD; SAP97, *P < 0.05 Pro+LTD, Ala+LTD, LTD versus C, n = 4).
To test the efficacy of the treatment, cells were lysed after 20 minutes of recovering, and co-IP assays were performed. As shown in Figure 5C, cLTD induction stimulates ADAM10/SAP97 complex formation, significantly raising ADAM10/SAP97 co-IP (Figure 8, B and C; LTD = +34.3% ± 9.6%; P < 0.05, LTD versus control). Preincubation with the Pro peptide prevented this effect (Figure 8, B and C; Pro+LTD = –10.8% ± 6.5%; P < 0.05 Pro+LTD versus Ala+LTD, Pro+LTD versus LTD), thus proving this strategy useful for verifying the role of SAP97 in LTD-induced ADAM10 trafficking to the postsynaptic membrane. Treatment with the control Ala peptide did not affect LTD-induced ADAM10/SAP97 association (Figure 8, B and C; Ala+LTD = +38.5% ± 7.7%; P > 0.05, Ala+LTD versus LTD).
To analyze ADAM10 and SAP97 localization, we purified the PSD protein-enriched TIF from neuronal cultures exposed or not exposed to either Pro or Ala peptide and then treated with NMDA (50 μm, 10 minutes). As previously shown (Figure 7, A and B), quantification of ADAM10 staining revealed a significant increase in both ADAM10 and SAP97 levels in synaptic membranes after cLTD induction (Figure 8, D and E; LTD, ADAM10 = +41% ± 9.5%, SAP97 = +31% ± 9.8%; P < 0.05, LTD versus control). Conversely, when neuronal cultures were preincubated with Pro, cLTD failed to induce the increase in ADAM10 localization at the synapse, whereas SAP97 levels were not affected (Figure 8, D and E; Pro+LTD, ADAM10 = –4.3% ± 6.1%, SAP97 = +34% ± 7.1%; P < 0.05, Pro+LTD versus Ala+LTD, Pro+LTD versus LTD). No effect was induced by treatment with the Ala control peptide on ADAM10 and SAP97 localization (Figure 8, D and E; Ala+LTD, ADAM10 = +33.3 5 ± 6.8%, SAP97 = +46.3% ± 6.8%; P > 0.05, Ala+LTD versus LTD). Hence, the cell-permeable peptide, which interferes with ADAM10/SAP97 association, is able to prevent cLTD-induced ADAM10 trafficking to the postsynaptic compartment, indicating the pivotal role of SAP97 in LTD-induced ADAM10 trafficking.
Since LTD is bearing ADAM10 to the postsynaptic compartment, we attempted to determining whether the hampering of ADAM10 trafficking occludes LTD. Hippocampal slices were incubated for 30 minutes with either Pro or Ala peptide before recording field excitatory postsynaptic potentials (fEPSPs) in CA1 region and inducing LTP or LTD. As shown in Figure 9A, treatment with the Pro peptide was sufficient to abolish the maintenance of LTD induced by LFS (P < 0.001, Pro versus Ala). On the contrary, we found that exposure of hippocampal slices to the Pro peptide had no detectable effect on the ability to elicit robust LTP by HFS when compared with slices treated with Ala control peptide (Figure 9B, P > 0.05 Pro versus Ala). No effect of the Pro peptide on baseline transmission was observed. The specificity and efficacy of the peptide has been proven as previously described (12, 13, 16) and as shown in Supplemental Figure 5.
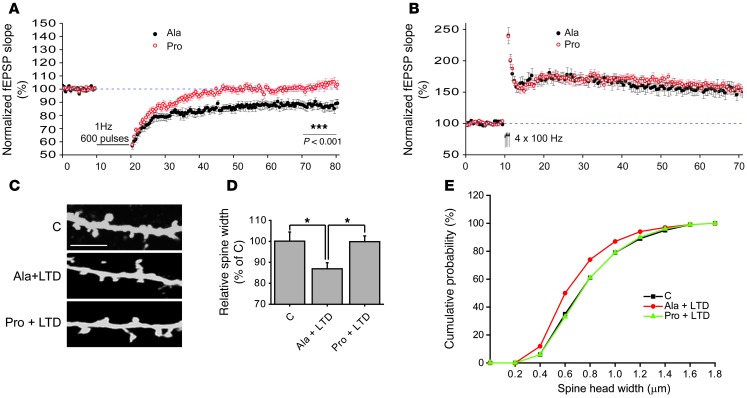
Figure 9. SAP97-mediated ADAM10 trafficking is necessary for LTD maintenance and LTD-induced spine shrinkage.
(A) Time course of the normalized fEPSP slope recorded in slices treated with Pro and Ala peptides. Notably, in Pro-treated slices, the LFS protocol fails to induce the LTD, whereas in Ala-treated slices, this form of synaptic plasticity is expressed (P < 0.001, Ala n = 13, Pro n = 12). Sample traces of fEPSPs acquired before the application of the LFS protocol and 60 minutes after LFS, in Ala-treated slices and in Pro-treated slices. (B) Time course of the normalized fEPSP slope recorded in slices treated with Pro and Ala peptides. In both groups, an HFS protocol (arrows) produces a robust LTP. Traces of fEPSP acquired before HFS and 60 minutes after HFS indicating a similar fEPSP increase in Ala-treated slices and in Pro-treated slices. (C) Primary hippocampal neurons were transfected with GFP construct at DIV7 and, at DIV14, cLTD was induced after incubation with either Ala or Pro peptides (1 μm, 30 minutes). Cells were fixed and immunolabeled for GFP. Representative images show dendrites from C, Ala+LTD, and Pro+LTD neurons. Scale bar: 5 μm. (D) Diagram showing relative average spine head width (*P < 0.05 Pro+LTD vs. Ala+LTD, Ala+LTD vs. C; n > 700 spines from 14 different neurons for each group). (E) Cumulative frequency plots from C, Ala+LTD, Pro+LTD neurons.
These results indicate that SAP97-mediated ADAM10 trafficking is required for hippocampal LTD expression. LTD is associated with a marked shrinkage of the spine head (56, 57), but the cellular mechanisms underlying this event are still not completely clear. Since we have previously demonstrated ADAM10 cleavage activity role in spine remodeling (13), we tested whether ADAM10 could be involved in LTD-induced spine morphology changes.
GFP-transfected hippocampal neurons were incubated with either Pro or Ala peptide (1 μM, 30 minutes), and then cLTD was induced by NMDA bath (50 μM, 10 minutes). After 20 minutes of recovering, cells were fixed and immunolabeled with an anti-GFP antibody; average spine head width and number of dendritic spines were evaluated. Statistical analysis revealed a significant decrease in spine head width in the Ala+LTD-treated neurons when compared with the controls (Figure 9, C and D; Ala+LTD = –13.2% ± 2.3%; P < 0.05, Ala+LTD versus control). On the other hand, in neurons incubated with the Pro peptide, LTD induction did not lead to a reduction of spine head width (Figure 9, C and D; Pro+LTD = –0.1% ± 2.2%; P > 0.05, Pro+LTD versus control, Pro+LTD versus Ala+LTD), demonstrating the relevance of ADAM10 for LTD-induced spine shrinkage. Cumulative frequency plots of spine head width confirmed a significant shift toward smaller spine size in Ala+LTD-treated neurons that was rescued by preincubation with the Pro peptide (Figure 9E). No significant changes in the mean spine density were found (C = 3.85 ± 0.33 spines/10 μm, Ala+LTD = 3.58 ± 0.20 spines/10 μm, Pro+LTD = 3.64 ± 0.27 spines/10 μm, P > 0.05).
Activity-dependent synaptic plasticity modulates APP shedding.
Synaptic activity has been shown to regulate APP-processing, leading to alterations in Aβ production (58). Therefore, we determined whether activity-dependent synaptic plasticity could affect APP trafficking and processing.
First of all, we analyzed APP total and synaptic levels after either cLTD or cLTP induction in hippocampal cultures. As shown in Figure 10A, cLTP significantly reduced APP synaptic localization, as previously reported (59), while cLTD did not modify APP levels in the TIF (Figure 10, A and B; LTP = –44.2% ± 13%, P = 0.027, LTP versus control; LTD = –0.5% ± 13%, P > 0.05, LTD versus control). Since ADAM10 synaptic levels are modulated by activity-dependent synaptic plasticity (Figures 5 and 6) and the enzyme is active toward its substrates when localized at the plasma membrane (9), we determined whether the shedding of APP was affected by activity-dependent synaptic plasticity.
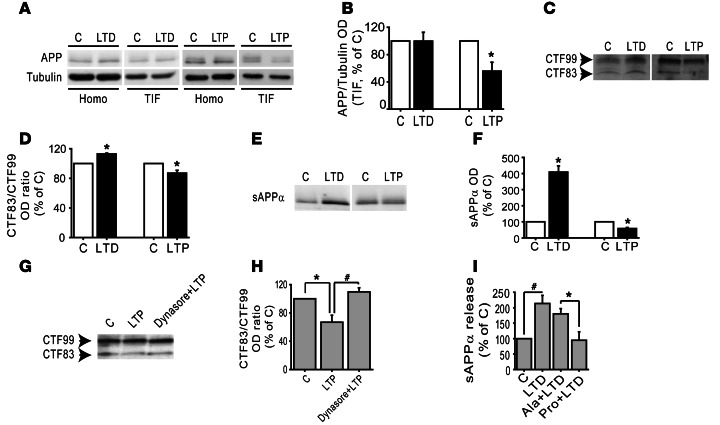
Figure 10. LTD and LTP modulate APP trafficking and processing.
(A) WB analysis of APP in homogenate and TIF from control and either cLTD- or cLTP-treated hippocampal cultures. cLTP leads to a reduction in APP levels in TIF but not in homogenate. cLTD does not affect APP in both homogenate and TIF. (B) Quantification of the ratio of APP/tubulin of experiments in A (*P < 0.05, cLTP versus C, n = 5). (C) LTD enhances whereas LTP reduces CTF83/CTF99 ratio in hippocampal neurons. APP CTFs were revealed by immunoblotting with 4G8 antibody. (D) Quantification of CTF83/CTF99 ratio OD of experiments in C (*P < 0.05 cLTD versus C, cLTP versus C, n = 3). (E) WB analysis of sAPPα released in 1 hour from control and cLTD/cLTP-treated hippocampal neuronal cultures. cLTD increases while cLTP reduces sAPPα secretion in primary hippocampal cultures. (F) Quantification of sAPPα OD of experiments in E (*P < 0.05 cLTD versus C, cLTP versus C, n = 3). (G) Incubation with the dynamin inhibitor dynasore (80 μM, 30 minutes) prevents the cLTP-induced decrease in CTF83/CTF99 ratio. (H) Quantification of OD of experiments in G (*P < 0.05 LTP versus C, #P < 0.05 Dynasore+LTP versus LTP, n = 4). (I) Quantification of sAPPα release in the medium of hippocampal neurons either exposed or not exposed to either Pro or Ala peptide; after 30 minutes, cLTD was induced. After 20 minutes of recovering, the medium was collected and sAPPα levels measured by ELISA assay (*P < 0.05 Pro+LTD versus Ala+LTD, #P < 0.05 LTD versus C, n = 5).
APP-processing products were further investigated in hippocampal neuronal cultures after either cLTD or cLTP induction. In these experimental conditions, both sAPPα release in the medium and APP CTFs — CTF99 (for BACE-cleavage) and CTF83 (for ADAM10 cleavage) — were evaluated in the TIF. In neuronal cultures, the CTF83/CTF99 ratio was significantly increased by cLTD and decreased after cLTP induction (Figure 10, C and D; cLTD, CTF83/CTF99 = +13.1% ± 1.0%, P = 0.006, cLTD versus control; cLTP, CTF83/CTF99 = –13.0% ± 3.8%, P = 0.04, cLTP versus control), suggesting that LTD enhances while LTP reduces ADAM10 activity modulating APP metabolism.
As further proof of these results, we detected a significant increase in sAPPα release in the neuronal medium collected after 1 hour recovery from cLTD induction (Figure 10, E and F; LTD = +309% ± 38.4%, P = 0.02, LTD versus control). In contrast, LTP causes a significant reduction in sAPPα release (Figure 10, E and F; LTP = –42% ± 6.2%, P = 0.02, LTP versus control).
To prove the involvement of ADAM10 endocytosis in the shifting of APP processing toward amyloidogenesis, we took advantage of the dynamin inhibitor dynasore, which interferes with ADAM10 endocytosis as shown in Figure 3, A and B. In neuronal cultures preincubated with dynasore, cLTP did not lead to the expected decrease in the CTF83/CTF99 ratio (Figure 10, G and H; LTP = –33.2% ± 9.9%, dynasore+LTP = +9.7% ± 5.8%, P < 0.05 LTP versus control, LTP versus dynasore+LTP).
Since SAP97 is responsible for LTD-induced ADAM10 delivery to the postsynaptic compartment, we determined whether the treatment with Pro peptide was able to affect LTD-induced sAPPα release. Hippocampal cultures exposed or not exposed to either Pro or Ala peptide (1 μM, 30 minutes) were then treated with NMDA (50 μm, 10 minutes) to induce cLTD. The medium was collected after 20 minutes of recovering, and the amount of sAPPα released was measured by ELISA assays. As shown in Figure 10I, the treatment with Pro peptide prevented the LTD-induced marked increase in sAPPα release, whereas the control peptide Ala had no effect (Figure 10I, LTD = +113.7% ± 26.1%, Ala+LTD = +80.2% ± 17%, Pro+LTD = –5.2% ±27.5%, P < 0.05, LTD versus control, Pro+LTD versus Ala+LTD, Pro+LTD versus LTD). Overall, these data suggest that activity-dependent synaptic plasticity exerts a bidirectional effect on APP shedding through the modulation of ADAM10 endocytosis/trafficking.
Discussion
Several studies highlighted the key role of the disintegrin and metalloproteinase family member ADAM10 in health and disease, due to its shedding activity toward a number of functional membrane proteins such as APP and N-cadherin (13, 60). In fact, through its shedding activity, ADAM10 has been shown to regulate key cellular functions including cell growth, adhesion, and migration and spine stabilization in excitatory neurons (13, 61, 62).
Hence, a finely balanced membrane level of ADAM10 is an essential prerequisite to control enzyme activity and its functions (63). A possible approach to modulating enzyme membrane availability can be to interfere with mechanisms regulating its intracellular trafficking. In this context, ADAM10 protein interactors regulating its forward trafficking have been previously characterized (12, 64). Less is known about mechanisms regulating ADAM10 membrane retrieval, although a balance between these 2 processes may represent another way to influence ADAM10 strength and shedding activity.
Here, we report that ADAM10 removal from the plasma membrane is mediated by clathrin-dependent endocytosis and represents what we believe is a novel mechanism to finely tune its synaptic availability. The clathrin adaptor AP2, a heterotetrameric assembly that initiates the endocytosis process, directly interacts with ADAM10 C-terminal domain. Here, we describe a pathological role of the ADAM10/AP2 complex in AD patients and a physiological role of the same complex in activity-dependent synaptic plasticity. In particular, in addition to a defect in ADAM10 binding to SAP97 (15), we report the concomitant increase in ADAM10 association to the clathrin adaptor AP2 in the hippocampus of AD patients.
Our results suggest that in early stages of the disease, the reduction of α-secretase activity (15) could be ascribed to a defect in ADAM10 exocytosis/endocytosis processes rather than to an alteration of its expression. Furthermore, this alteration is not related to the presence of genetic variations in the C-teminal domain of ADAM10. Indeed, modifications of endo/exocytosis mechanisms in AD have been put forward by several observations. First, GWAS studies have identified a number of genes coding for endocytic factors, such as PICALM, as susceptibility loci for sporadic AD (23–25). Second, neurons obtained by reprogramming AD patients’ fibroblasts have early endosome phenotypes (65). Third, Aβ oligomers have been reported to affect clathrin-dependent endocytosis (26).
These studies pointed to a role of endocytic mechanisms in AD pathogenesis primarily focusing on the presynaptic site, i.e., synaptic vesicles release, APP recycling mechanisms (26). Our findings highlight the importance of exo/endocytosis mechanisms also in the postsynaptic compartment.
The structural determinants of ADAM10/AP2 interaction have been defined in minute details using several approaches. In particular, we have identified an atypical AP2-binding motif in ADAM10 Ct: the 735RQR737 sequence is necessary for AP2 binding and ADAM10 internalization. The lack of AP2 interaction significantly affects ADAM10 plasma membrane levels, underlining the relevance of clathrin-mediated endocytosis in the modulation of ADAM10 surface expression.
Moreover, we carried out a careful mapping of the ADAM10 tail, demonstrating that the domains responsible for the interaction with AP2 complex and SAP97 do not overlap. The last 15-aa sequence, containing the 735RQR737 motif, is responsible for the binding to AP2 complex, while SAP97, previously reported to bind the 2 proline-rich domains of ADAM10 tail (12), mainly interacts with the most membrane-proximal proline-rich domain and the second proline stretch is not essential.
We also propose a physical model for the interaction between the tail region of ADAM10 and AP2 based on structural similarity with a known protein-peptide complex. Clearly the hypothesized mode of interaction is only speculative; we present it here as it can be used as a guide for site directed mutagenesis experiment.
Furthermore, we demonstrate that SAP97 and clathrin adaptor AP2 are critical links between activity-dependent synaptic plasticity and ADAM10. In particular, here we show that the 2 main paradigms of activity-dependent synaptic plasticity, i.e., LTP and LTD, have opposite effects on ADAM10 association to its binding partners, i.e., SAP97 and AP2, thus affecting ADAM10 localization and activity in hippocampal neurons.
It turned out that ADAM10 undergoes a dynamic regulation of its membrane levels by activity-dependent synaptic plasticity: LTD boosts ADAM10 membrane insertion by fostering its SAP97-mediated forward trafficking to synaptic membrane, whereas LTP reduces the enzyme membrane levels by inducing AP2-mediated endocytosis.
We then asked why ADAM10 surface expression is finely tuned by activity-dependent synaptic plasticity. We demonstrate that SAP97-mediated ADAM10 forward trafficking represents a process necessary for LTD expression. LTD induction enhances ADAM10/SAP97 complex formation. This interaction is necessary for LTD-induced ADAM10 trafficking, since the treatment of hippocampal neurons with the cell-permeable Pro peptide mimicking the proline-rich region of ADAM10 and disrupting SAP97/ADAM10 interaction counteracts the capability of LTD to foster SAP97/ADAM10 complex to the membrane. Besides interfering with LTD-induced ADAM10 trafficking, the uncoupling of ADAM10 and SAP97 also hampers LTD maintenance and LTD-induced spine shrinkage.
In light of the above, ADAM10 delivery to the postsynaptic compartment seems to be critical for synaptic activity–induced spine remodeling. In fact, ADAM10 cleavage of synaptic adhesion molecules may allow the activated synapse to rapidly modulate the spine size during induction of activity-dependent synaptic plasticity (13, 14).
Activity-dependent synaptic plasticity also represents an important mechanism of regulation of ADAM10 activity toward APP at synaptic membranes. According to previous studies (59), APP synaptic levels are reduced by LTP, whereas LTD does not affect its localization at the postsynaptic compartment. The measurement of both sAPPα release and of the levels of APP CTFs — CTF99 for BACE cleavage and CTF83 for ADAM10 cleavage — in the postsynaptic compartment demonstrated that LTP decreases whereas LTD stimulates ADAM10 cleavage of APP. It has been reported that neuronal activity dynamically influences Aβ levels in vivo (66) and modulates APP processing at the β-secretase site (58). Our data demonstrate that neuronal activity, and in particular, activity-dependent synaptic plasticity, also affects ADAM10 activity and, thereby, modulates APP processing. Moreover, interfering with the mechanisms underlying ADAM10 trafficking/endocytosis prevents the modulation of APP processing triggered by activity-dependent synaptic plasticity. Regulated interaction of ADAM10 with SAP97 and AP2 discloses a physiological mechanism of ADAM10 activity regulation at the synapses. This phenomenon produces a situation whereby synaptically regulated ADAM10 activity is positioned to modulate synaptic functioning. The balance between these 2 partners’ association is impaired in AD patients’ hippocampus at early stages of disease, leading to a reduction of ADAM10 levels at the postsynaptic compartment (15). In light of the role of ADAM10 in Aβ production and synapse function, this loss of balance could affect both APP processing and activity-dependent synaptic plasticity in AD.
Together, these results put forward an innovative and comprehensive vision of AD pathogenesis recapitulating the amyloid cascade in the synaptic context and taking into account the strict interdependence of amyloid cascade and synaptic activity.
Methods
Human studies.
Hippocampi from 6 AD patients and 6 HC were obtained from the Netherlands Brain Bank (NBB). Established Braak and Braak criteria were used to categorize AD tissues (29). AD patients fulfilled Braak 4 stage. Accordingly, in AD cases, there were tangles and neuritic plaques in hippocampus. HC had no history of psychiatric or neurological disease and no evidence of age-related neurodegeneration.
Genetic sequencing of ADAM10 Ct.
Patients were consecutively enrolled from a tertiary referral centre for memory disturbances, University of Brescia, and fulfilled current clinical criteria for AD. Total genomic DNA was prepared from peripheral blood according to standard procedures. Exon 15 and exon 16 of ADAM10 and at least 30 bp of their flanking introns were evaluated by PCR. Primers were designed for exon 15 (F:5′-CAGTGGAAAAATCTGTTTATTT-3′, R:5′-TATAATTCAATTCTACCTGCTAAA-3′) and exon 16 (F:5′-TTTCTTATTGAAACAGCAGCTTTGC-3′, R:5′-CTCTTTGGAGTGAAGTTTTCCCAT -3′). PCR annealing temperatures were 51°C and 58°C, respectively. The PCR products were separated on a 2% agarose gel, and the fragments were evaluated by denaturing HPLC (dHPLC) analysis (Transgenomic).
Neuronal culture treatment, TIF preparation, crosslinking assay.
To induce chemical LTP, hippocampal neuronal cultures at 14 DIV were first incubated in artificial cerebrospinal fluid (ACSF) for 30 minutes: 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 33 mM D-glucose, and 25 mM HEPES (pH 7.3; 320 mosM final), followed by stimulation with 50 μM forskolin, 0.1 μM rolipram, and 100 μM picrotoxin (Tocris) in ACSF (no MgCl2). After 16 minutes of stimulation, neurons were replaced in regular ACSF for 15 minutes and then subjected to TIF extraction or surface-expression assays (48, 49). To induce cLTD, neuronal cultures were incubated in ACSF for 30 minutes, followed by stimulation with 50 μM NMDA (Sigma-Aldrich) in ACSF. After 10 minutes of stimulation, neurons were replaced in regular ACSF for 20 minutes and then subjected to TIF extraction or surface-expression assays (48, 49). TIF was isolated from either neurons or acute hippocampal slices as previously described (54). Crosslinking experiments by means of BS3 (Pierce) were performed as previously described (12, 52, 67).
Surface biotinylation.
Hippocampal cultures were washed once with PBS/Ca2+/Mg2+ (10 mM phosphate buffer, 2.7 mM KCl, 137 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, pH 7.4) at 37°C and then cooled gradually to 4°C before they were washed twice with cold PBS/Ca2+/Mg2+. Cultures were incubated with biotinylation reagent (1.5 mg/ml NHS-SS-biotin [Pierce]in PBS/ Ca2+/Mg2+) at 4°C for 20 minutes, and then washed 3 times with cold PBS/Ca2+/Mg2+ including 50 mM glycine. Cells were scraped into cold lysis buffer, and total membrane fraction (Tot) was extracted and mixed with UltraLink immobilized neutravidin beads (Pierce) and rotated for 2 hours at 4°C. The beads were washed 5 times, and after the addition of SDS-PAGE sample buffer to the beads, the resulting slurry (Pp) was boiled for 3 minutes. To evaluate the efficiency of the biotinylation reaction, the first supernatant was reprecipitated with neutravidin beads, and the additional neutravidin-precipitated material was loaded as well (Pp2) (68). Proteins were resolved by SDS-PAGE and probed with the anti-ADAM10, anti-GluR1 or anti-actin antibodies. To avoid saturation of band signal in order to carry out precise quantitative analyses, samples of Tot and Pp were loaded such that each lane represented a percentage of the total material per plate. Proteins were visualized with ECL (GE Healthcare).
Internalization assays.
To label surface TacADAM10-RAR, TacADAM10-RAR AQA, and Tac ADAM10 721Δ, live COS7 cells were incubated with anti-Tac antibody for 45 minutes in medium at 4°C, while live neurons were labeled by incubation in ACSF with anti-Tac antibody for 30 minutes at 10°C. After brief washing in DMEM, COS7 cells were returned to 37°C for 10 minutes. Neurons were washed with cold ACSF, and internalization was allowed at 37°C in response to cLTP induction (16 minutes plus 15 minutes to allow internalization). A parallel set of controls was kept at 4–8°C to stop trafficking. Cells were then fixed with 4% PFA and 4% sucrose in PBS, pH 7.4 and blocked with normal serum; remaining surface Tac chimeras were labeled with 488 secondary antibody (nonpermeabilized), and internalized receptors were labeled with 555-conjugated secondary antibody after Triton permeabilization. In some experiments, dynasore (80 μM; Tocris Bioscience) was added for 30 minutes to block receptor internalization. For COS7 internalization experiments, wide-field fluorescence images were acquired with a Zeiss ×40 objective and a CoolSnap CCD camera. For neurons, fluorescence images were acquired by using the confocal LSM510 Meta system (Zeiss) with a ×63 objective and a sequential acquisition setting at 1024 × 1024 pixel resolution; for each image, 2 to 3 sections of 0.5 μm were acquired, and z-projection was obtained.
Electrophysiological experiments.
Acute hippocampal slices (400 μm) were placed into an interface recording chamber where they were continuously perfused with oxygenated ACSF maintained at 29°C and at a flowing rate of 1.6–1.8 ml/min. Extracellular fEPSPs were recorded in stratum radiatum of CA1 with glass electrodes filled with 3 M NaCl coupled to the input stage of a Warner IE-210 amplifier as previously described (16). Briefly, stimuli were delivered to the Schaffer collateral/commissural afferents with bipolar concentric stainless steel electrodes. Basal stimulation frequency was 0.1 Hz. LTP was induced by a HFS protocol: 4 trains of 100 Hz for 1 second, separated by 30 seconds LTD was induced by delivering an LFS protocol: 600 pulses at 1 Hz. The slope of the rising phase of the fEPSP was calculated as a measure of the synaptic response.
Data analysis.
Quantification of WB analysis was performed by means of computer-assisted imaging (ImageJ) after normalization on tubulin or actin levels, and values were expressed as mean ± SEM. For internalization or surface/total ratio assays, colocalization, and morphological analysis, cells were chosen randomly for quantification from 4 different coverslips (2 independent experiments), images were acquired using the same settings/exposure times, and at least 10 cells for each condition were analyzed. Colocalization analysis was performed using AIM 4.2 software (Zeiss). For quantification of internalization, all images were analyzed using ImageJ, the average fluorescence intensities in the green (total) and red (internalized) channels were measured, background subtracted, and the internalized/fluorescence ratio was calculated for each cell. The average intensity of surface fluorescence staining (red) was determined after cell tracing and normalized to the total intensity (green) to correct for differences in expression. Surface ratios were obtained by dividing the background subtracted fluorescence intensities. Morphological analysis was performed with ImageJ software to measure spine head width. For electrophysiological experiments, data are presented as means ± SEM. LTP or LTD value was calculated for each single experiment as the percentage of increase in the slope of the fEPSP during minutes 55 to 60 after the end of HFS or LFS, respectively, compared with control baseline.
Statistics.
Statistical evaluations were performed by using 2-tailed Student’s t test (a P value less than 0.05 was considered significant.) or, when appropriate, by using 1-way ANOVA followed by Bonferroni’s post hoc test.
Study approval.
All experimental procedures were carried on with care to minimize discomfort and pain to treated animals in accordance with the guidelines of the European Communities Council (Directive of November 24, 1986, 86/609/EEC) and were approved by the Italian Ministry of Health (as indicated in Digs n. 295/2012-A). For the experiments carried on human brain samples, all procedures were in accordance with the NIH Guide for Care and Use of Laboratory Human Tissues and were approved by the Ethics Committee of the University of Milan. For the genetic analysis, written informed consent (from the subject or from the responsible guardian if the subject was incapable) was obtained, before study initiation, for blood collection and genotyping. The work conformed with the Declaration of Helsinki and was approved by the local Ethics Committee.
See Supplemental Methods for details on antibodies and cell-permeable peptide, DNA constructs, acute hippocampal slices, cell cultures and transfection methods, SDS-PAGE, determination of sAPPα release, IP experiments, GST -fusion protein production and pulldown assay, immunocytochemistry and colocalization analysis, and structural characterization.
Supplementary Material
Acknowledgments
We thank A. Longhi and E. Zianni for technical assistance. This work was supported by funding from the European Union’s Seventh Framework Program (FP7 2007-2013) under Grant Agreement no. PIAP-GA-2008-217902 to M. Di Luca, from Fondazione CARIPLO (project number 2319-2008) to M. Di Luca, from FIRB-Accordi di Programma, project code RBAP11HSZS to M. Di Luca, from PRIN 2010-2011 2010PWNJXK to M. Di Luca, from the Italian Institute of Technology SEED project to A. Tramontano.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(6):2523–2538. doi:10.1172/JCI65401.
Alerie Guzman de la Fuente’s present address is: MRC Centre for Stem Cell Biology and Regenerative Medicine, Department of Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom.
References
- 1.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Marcello E, Epis R, Saraceno C, Di Luca M. Synaptic dysfunction in Alzheimer’s disease. Adv Exp Med Biol. 2012;970:573–601. doi: 10.1007/978-3-7091-0932-8_25. [DOI] [PubMed] [Google Scholar]
- 3.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 5.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 6.Vassar R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 7.Haass C. Take five — BACE and the gamma-secretase quartet conduct Alzheimer’s amyloid beta-peptide generation. EMBO J. 2004;23(3):483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn PH, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29(17):3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lammich S, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutwein P, et al. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003;17(2):292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116(pt 16):3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 12.Marcello E, et al. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J Neurosci. 2007;27(7):1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinverno M, et al. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J Neurosci. 2010;30(48):16343–16355. doi: 10.1523/JNEUROSCI.1984-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardoni F, et al. The neuropeptide PACAP38 induces dendritic spine remodeling through ADAM10-N-cadherin signaling pathway. J Cell Sci. 2012;125(pt 6):1401–1406. doi: 10.1242/jcs.097576. [DOI] [PubMed] [Google Scholar]
- 15.Marcello E, et al. SAP97-mediated local trafficking is altered in Alzheimer disease patients’ hippocampus. Neurobiol Aging. 2012;33(2):422.e1–422.e10. doi: 10.1016/j.neurobiolaging.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Epis R, et al. Blocking ADAM10 synaptic trafficking generates a model of sporadic Alzheimer’s disease. Brain. 2010;133(11):3323–3335. doi: 10.1093/brain/awq217. [DOI] [PubMed] [Google Scholar]
- 17.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28(2):511–525. doi: 10.1016/S0896-6273(00)00129-X. [DOI] [PubMed] [Google Scholar]
- 18.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 19.Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron. 2002;36(3):435–449. doi: 10.1016/S0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 20.Racz B, Blanpied TA, Ehlers MD, Weinberg RJ. Lateral organization of endocytic machinery in dendritic spines. Nat Neurosci. 2004;7(9):917–918. doi: 10.1038/nn1303. [DOI] [PubMed] [Google Scholar]
- 21.Carroll RC, Lissin DV, von Zastrow M, Nicoll RA, Malenka RC. Rapid redistribution of glutamate receptors contributes to long-term depression in hippocampal cultures. Nat Neurosci. 1999;2(5):454–460. doi: 10.1038/8123. [DOI] [PubMed] [Google Scholar]
- 22.Beattie EC, et al. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat Neurosci. 2000;3(12):1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 23.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 25.Seshadri S, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treusch S, et al. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334(6060):1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas RS, Lelos MJ, Good MA, Kidd EJ. Clathrin-mediated endocytic proteins are upregulated in the cortex of the Tg2576 mouse model of Alzheimer’s disease-like amyloid pathology. Biochem Biophys Res Commun. 2011;415(4):656–661. doi: 10.1016/j.bbrc.2011.10.131. [DOI] [PubMed] [Google Scholar]
- 28.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 29.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 30.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282(5392):1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapoport I, Chen YC, Cupers P, Shoelson SE, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;17(8):2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haucke V, Wenk MR, Chapman ER, Farsad K, De Camilli P. Dual interaction of synaptotagmin with mu2- and alpha-adaptin facilitates clathrin-coated pit nucleation. EMBO J. 2000;19(22):6011–6019. doi: 10.1093/emboj/19.22.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diviani D, Lattion AL, Abuin L, Staub O, Cotecchia S. The adaptor complex 2 directly interacts with the alpha 1b-adrenergic receptor and plays a role in receptor endocytosis. J Biol Chem. 2003;278(21):19331–19340. doi: 10.1074/jbc.M302110200. [DOI] [PubMed] [Google Scholar]
- 34.Kastning K, et al. Molecular determinants for the interaction between AMPA receptors and the clathrin adaptor complex AP-2. Proc Natl Acad Sci U S A. 2007;104(8):2991–2996. doi: 10.1073/pnas.0611170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63(3):503–513. doi: 10.1016/0092-8674(90)90447-M. [DOI] [PubMed] [Google Scholar]
- 36.Marcello E, Gardoni F, Di Luca M, Perez-Otano I. An arginine stretch limits ADAM10 exit from the endoplasmic reticulum. J Biol Chem. 2010;285(14):10376–10384. doi: 10.1074/jbc.M109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carey RM, Blusztajn JK, Slack BE. Surface expression and limited proteolysis of ADAM10 are increased by a dominant negative inhibitor of dynamin. BMC Cell Biol. 2011;12(1):20. doi: 10.1186/1471-2121-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tousseyn T, et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284(17):11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkin E, Harris B. A disintegrin and metalloproteinase (ADAM)-mediated ectodomain shedding of ADAM10. J Neurochem. 2009;108(6):1464–1479. doi: 10.1111/j.1471-4159.2009.05907.x. [DOI] [PubMed] [Google Scholar]
- 40.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng S, Kasahara C, Rickles RJ, Schreiber SL. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci U S A. 1995;92(26):12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gherardini PF, Ausiello G, Helmer-Citterich M. Superpose3D: a local structural comparison program that allows for user-defined structure representations. PLoS One. 2010;5(8):e11988. doi: 10.1371/journal.pone.0011988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newpher TM, Ehlers MD. Glutamate receptor dynamics in dendritic microdomains. Neuron. 2008;58(4):472–497. doi: 10.1016/j.neuron.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardoni F, Marcello E, Di Luca M. Postsynaptic density-membrane associated guanylate kinase proteins (PSD-MAGUKs) and their role in CNS disorders. Neuroscience. 2009;158(1):324–333. doi: 10.1016/j.neuroscience.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 45.Howard MA, Elias GM, Elias LA, Swat W, Nicoll RA. The role of SAP97 in synaptic glutamate receptor dynamics. Proc Natl Acad Sci U S A. 2010;107(8):3805–3810. doi: 10.1073/pnas.0914422107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5(12):952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- 47.Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron. 2004;44(1):181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Oh MC, Derkach VA, Guire ES, Soderling TR. Extrasynaptic membrane trafficking regulated by GluR1 serine 845 phosphorylation primes AMPA receptors for long-term potentiation. J Biol Chem. 2006;281(2):752–758. doi: 10.1074/jbc.M509677200. [DOI] [PubMed] [Google Scholar]
- 49.Otmakhov N, et al. Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J Neurophysiol. 2004;91(5):1955–1962. doi: 10.1152/jn.00941.2003. [DOI] [PubMed] [Google Scholar]
- 50.Lin JW, et al. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat Neurosci. 2000;3(12):1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Otano I, et al. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9(5):611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall RA, Soderling TR. Quantitation of AMPA receptor surface expression in cultured hippocampal neurons. Neuroscience. 1997;78(2):361–371. doi: 10.1016/S0306-4522(96)00525-8. [DOI] [PubMed] [Google Scholar]
- 53.Roche KW, O’Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16(6):1179–1188. doi: 10.1016/S0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 54.Gardoni F, Bellone C, Cattabeni F, Di Luca M. Protein kinase C activation modulates alpha-calmodulin kinase II binding to NR2A subunit of N-methyl-D-aspartate receptor complex. J Biol Chem. 2001;276(10):7609–7613. doi: 10.1074/jbc.M009922200. [DOI] [PubMed] [Google Scholar]
- 55.Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313(5794):1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- 56.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33(3):121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44(5):749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 58.Kamenetz F, et al. APP processing and synaptic function. Neuron. 2003;37(6):925–937. doi: 10.1016/S0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 59.Tampellini D, et al. Synaptic activity reduces intraneuronal Abeta, promotes APP transport to synapses, and protects against Abeta-related synaptic alterations. J Neurosci. 2009;29(31):9704–9713. doi: 10.1523/JNEUROSCI.2292-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20(2):164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6(1):32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 62.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 63.Lichtenthaler SF. Alpha-secretase cleavage of the amyloid precursor protein: proteolysis regulated by signaling pathways and protein trafficking. Curr Alzheimer Res. 2011;9(2):165–177. doi: 10.2174/156720512799361655. [DOI] [PubMed] [Google Scholar]
- 64.Prox J, et al. Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell Mol Life Sci. 2012;69(17):2919–2932. doi: 10.1007/s00018-012-0960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cirrito JR, et al. Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron. 2005;48(6):913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 67.Mauceri D, Cattabeni F, Di Luca M, Gardoni F. Calcium/calmodulin-dependent protein kinase II phosphorylation drives synapse-associated protein 97 into spines. J Biol Chem. 2004;279(22):23813–23821. doi: 10.1074/jbc.M402796200. [DOI] [PubMed] [Google Scholar]
- 68.Mammen AL, Huganir RL, O’Brien RJ. Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J Neurosci. 1997;17(19):7351–7358. doi: 10.1523/JNEUROSCI.17-19-07351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.