Abstract
Small intestine neuroendocrine tumors (SI-NETs) are the most common malignancy of the small bowel. Several clinical trials target PI3K/Akt/mTOR signaling; however, it is unknown whether these or other genes are genetically altered in these tumors. To address the underlying genetics, we analyzed 48 SI-NETs by massively parallel exome sequencing. We detected an average of 0.1 somatic single nucleotide variants (SNVs) per 106 nucleotides (range, 0–0.59), mostly transitions (C>T and A>G), which suggests that SI-NETs are stable cancers. 197 protein-altering somatic SNVs affected a preponderance of cancer genes, including FGFR2, MEN1, HOOK3, EZH2, MLF1, CARD11, VHL, NONO, and SMAD1. Integrative analysis of SNVs and somatic copy number variations identified recurrently altered mechanisms of carcinogenesis: chromatin remodeling, DNA damage, apoptosis, RAS signaling, and axon guidance. Candidate therapeutically relevant alterations were found in 35 patients, including SRC, SMAD family genes, AURKA, EGFR, HSP90, and PDGFR. Mutually exclusive amplification of AKT1 or AKT2 was the most common event in the 16 patients with alterations of PI3K/Akt/mTOR signaling. We conclude that sequencing-based analysis may provide provisional grouping of SI-NETs by therapeutic targets or deregulated pathways.
Introduction
Small intestine neuroendocrine neoplasms (SI-NENs) are the most common malignancy of the small bowel, represent the largest group of NENs by organ site, and are studied in clinical treatment trials targeting PI3K/Akt/mTOR signaling. Whether this or other canonical cancer pathways is recurrently mutated, however, is uncertain, because a genome-wide, unbiased sequence analysis of cancer genes has not been performed to date in SI-NENs.
Massively parallel, or “nextgen,” DNA sequencing is currently advancing research in other human malignancies by facilitating the collection of comprehensive, genome-wide, unbiased datasets providing a common data framework for comparing results across different tumor types and gene sets. It provides the most comprehensive technology to date to explore the potential of genomics for individualizing cancer treatment within a tumor type. To unlock and explore the potential of the technology for translational research in SI-NEN, we sequenced 48 such tumors.
Results
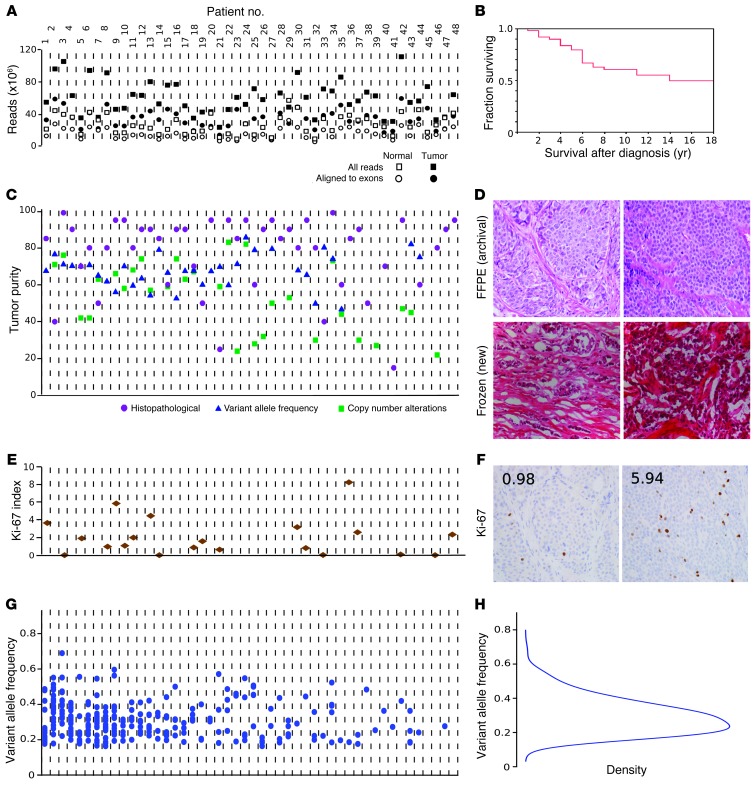
Genomic DNA libraries were exome enriched and sequenced to an average 110-fold target region of tumor coverage, supporting best practice bioinformatics analyses to detect tumor-associated point mutations, termed somatic single nucleotide variations (SNVs). For details, see Supplemental Methods (supplemental material available online with this article; doi: 10.1172/JCI67963DS1). Figure 1A shows the resulting quality control measures by sample (see also Supplemental Table 1 for summary statistics). Figure 1B highlights that the cohort was clinically typical of patients with well-differentiated SI-NENs, whose survival was measured in years. See Supplemental Table 2 for demographic and medical details for all cases. Figure 1C shows tumor purity as determined by 3 methods (see also Supplemental Table 3), which was favorably high for nextgen sequencing. All tumors were well differentiated according to the original pathology report, as confirmed by review of newly prepared frozen sections (Figure 1D). We observed 2 or fewer mitoses per 10 high-power fields in 97% of tumors, categorizing these as well-differentiated (G1) neuroendocrine tumors (NETs). Ki-67 immunohistochemistry performed in a representative subset categorized 70% of tumors as G1, and the remaining as intermediate grade (G2). Ki-67 labeling results and sample slides are shown in Figure 1, E and F. Therefore, the present study was not contaminated by high-grade NENs, and all tumors studied could be classified as G1 or G2 NETs, according to WHO 2010 classification (1). Statistical assessment and orthogonal validation by Sanger sequencing (confirmation rate, >90%; Supplemental Table 13) supported the accuracy of our somatic SNV calls. Variant allele frequency (VAF) results are summarized in Figure 1, G and H.
Figure 1. Tumor and sequencing characteristics.
Tumor-associated (somatic) mutations in the exome were determined by nextgen sequencing. (A) Sequencing quality control. Total raw sequencing reads and enrichment for the targeted exome regions were similar across samples. Quality control measures were uncorrelated with downstream results, supporting that observed disparities among tumors represent biological diversity of SI-NETs. x axes in A, C, E, and G correspond to individual tumors. (B) Survival of 48 patients. See also Supplemental Figure 2. (C) Tumor cell purity was determined by histopathology, VAF (fraction of mutated sequencing reads), and copy number alteration estimates (based on the ratio of sequence read counts for a tumor-deleted chromosome compared with germline). The 3 methods cross-validated the observation that mean tumor content was high across the dataset and uncorrelated with biological results. (D) Formalin-fixed, paraffin-embedded (FFPE) and frozen sections of 2 cases, demonstrating tumor purity and well-differentiated histology. (E) Ki-67 labeling index, demonstrating that the majority of cases were WHO 2010 classification low grade (≤2%), and a minority were intermediate grade (>3%). See also Supplemental Figure 3. (F) Ki-67 immunohistochemistry for cases in D. Numerals indicate the Ki-67 index. (G) Mutation counts and VAF for individual tumors. The number of mutations is shown for each patient as a scatter plot of the VAF for each event. While the sensitivity of sequencing technology detecting alterations in subsets of tumor cells is limited by sequence read depth, the distribution of VAF suggests that the reported mutations are dominant throughout the entirety of each tumor. (H) Unimodal distribution of VAF among mutation calls. Original magnification, ×40.
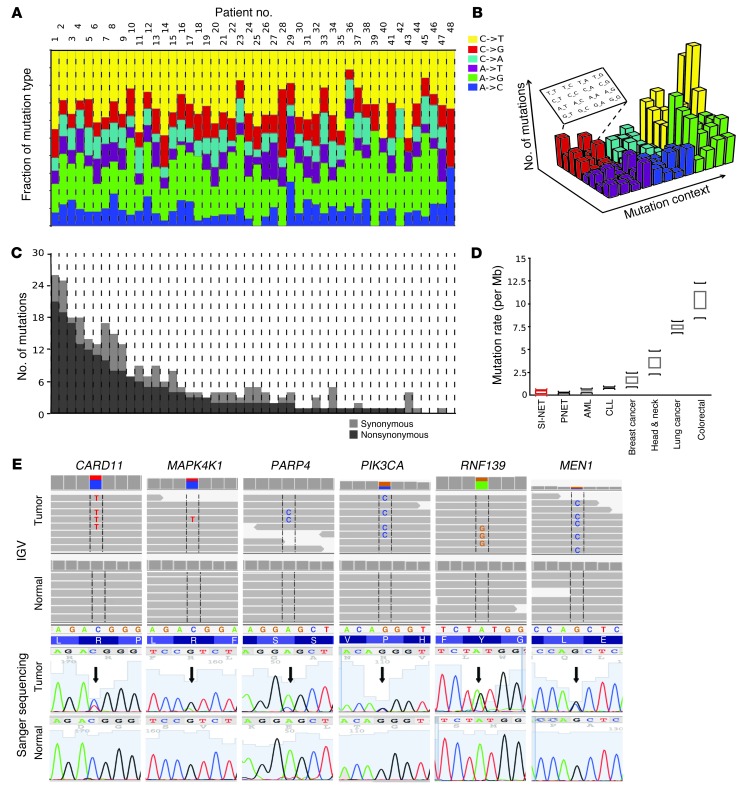
Genomic highlights of SI-NETs are detailed in Figure 2. The mutation pattern was dominated by transitions (C>T and A>G; Figure 2A). Transversions — including C>A, the hallmark of tobacco-related cancers, and C>G, dominant in viral oncogenesis — were uncommon in SI-NETs, as expected. The mutation sequence context (i.e., the nucleotides preceding and following a SNV) was also discrete (Figure 2B and Supplemental Table 4). The mutation rate of SI-NET genomes was low, at an average of 0.1 somatic SNV/106 bp in the exome (range, 0–0.59; Figure 2C). Thus, the SI-NET mutation rate was similar to lung carcinoids (2), pancreatic NET (PNET) (3), CLL (4, 5), and AML (6) and was lower than mutation rates of breast (7, 8), prostate (9), head and neck (10), colorectal (11), and lung (12) cancers (Figure 2D). Higher mutation rates in the primary SI-NETs were associated with an increased likelihood of concurrent liver metastases, which was supported by an exploratory analysis that was significant at P < 0.04 (slope of the logistic regression), hinting at a link between somatic SNV and clinical course.
Figure 2. Somatic mutation landscape across the coding genes (exome) in SI-NETs.
(A) Mutation pattern. A similar characteristic nucleotide mutation pattern was observed in all tumors. Somatic SNVs were most commonly the result of either of the 2 possible “transitions”: C>T (pyrimidine-to-pyrimidine) or A>G (purine-to-purine). The 4 possible transversions (pyrimidine-to-purine or reverse) were uncommon. x axes in A and C correspond to individual tumors. (B) Sequence context of SNVs. Extending the analysis of mutation patterns to the 16 possible motifs of nucleotides preceding and following an SNV, a pattern corresponding to known mechanisms of mutagenesis was observed. Most prominent was the CpG context, rendering the base C susceptible to deamination resulting in uracil and subsequent replacement by thymidine. Figure adapted from Journal of Clinical Investigation (29). (C) Mutation rate. SI-NET exonic mutation counts were uniform among tumors varying within only 1 order of log10 magnitude, suggesting absence of a hypermutator subtype (usually seen with mismatch repair deficiencies). (D) Comparison of mutation rate with well-characterized human cancers. Shown is the median mutation rate and range reported for the indicated cancer types. (E) Somatic SNVs representative of the preponderance of mutations in cancer genes in SI-NETs. Top: Primary sequence data showed each respective mutation in a fraction of the sequence reads consistent with an alteration in only 1 gene allele in the tumor or contamination of the tumor sample with normal tissue. No mutation was seen in any sequence read of the germline. SNVs are framed and shown as letters within the reads. IGV, Integrative Genomics Viewer. Bottom: Sanger sequencing. Arrows denote SNVs. PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α.
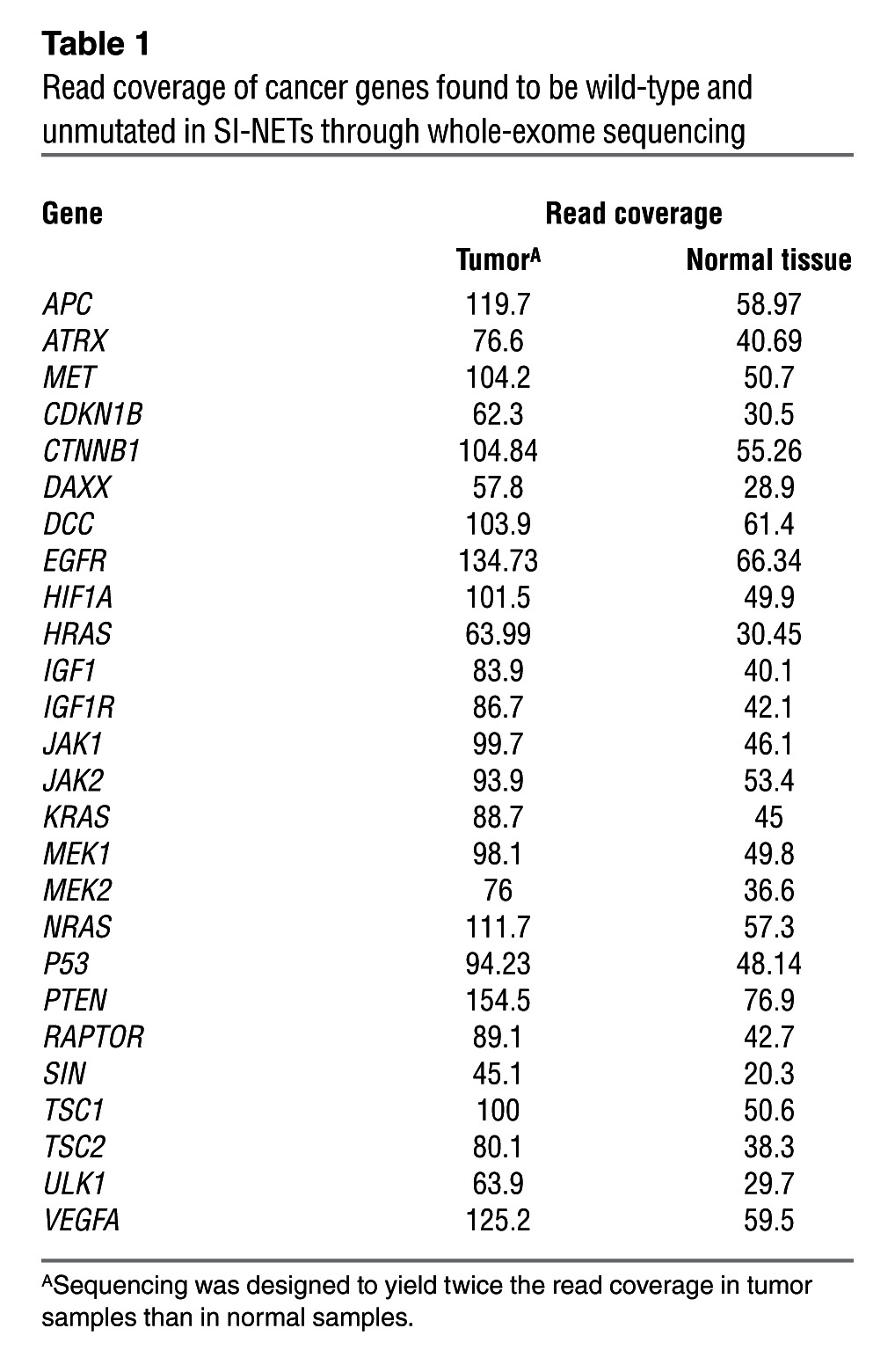
Review of somatic SNVs demonstrated SI-NET genetic diversity among an overall marked preponderance of cancer genes. 197 SNVs were nonsynonymous mutations/disruptions of a protein-encoding gene region, and 14 were mutations of splice sites. Mutations in canonical COSMIC genes were detected in FGF receptor 2 (FGFR2), MEN1, HOOK3, EZH2, MLF1, CARD11, VHL, NONO, FANCD2, and BRAF. Among the remaining genes, many had obvious roles in carcinogenesis, such as BIRC5 and RNF139. See Supplemental Results for a gene-centered discussion and Supplemental Table 5 for an annotated list of mutated genes. Examples of mutation discovery and validation data are depicted in Figure 2E. A hallmark of the by-gene mutation analysis was the wild-type sequence status of genes known to be mutated in other malignancies. Genes reported as being recurrently mutated in other malignancies, including other types of neuroendocrine tumors, were found to be normal in SI-NETs. Table 1 shows that read coverage for these genes was adequate to designate these genes wild-type in our dataset. In order to confirm that Sanger sequencing as executed was capable of detecting the recurrence of mutations in different tumor samples, we resequenced somatic SNVs in metastases from the same patients and found the same mutation in 83% of instances, as detected in the corresponding primary tumors (Supplemental Table 6). To also confirm the absence of mutations by a traditional technology, 372 Sanger sequencing reactions were performed on both strands covering unaffected sites in tumor and germline samples, which confirmed wild-type status in 100% of the cases (Supplemental Table 7). Assuming that the stochastic variation resulting from random sampling of patients (some with and some without a certain mutation) is the main source of variation, the present study with 48 samples could be estimated to have a power greater than 90% to detect a gene as being mutated if the true mutation rate in SI-NETs were greater than 5%; thus, it is unlikely that the size of the study cohort limited its statistical power to detect whether SNVs in any of the genes listed here are responsible for a large proportion of SI-NETs. To address other possible pitfalls limiting the detection of mutations in common cancer genes, such as a low mutation VAF, additional experiments were performed using a commercial nextgen cancer gene panel test (Illumina) that is designed for optimal sensitivity for a small panel of cancer-relevant genes in a subset of patients. No additional mutation was found among this panel of cancer genes (Supplemental Results and Supplemental Table 8), further supporting the relative sensitivity of the original exome sequencing procedure.
Table 1.
Read coverage of cancer genes found to be wild-type and unmutated in SI-NETs through whole-exome sequencing
Large and small (focal) somatic copy number alterations (SCNAs) were determined from exome read counts (13). Setting a significance level of P < 0.02 by permutation testing, we found 1,013 SCNAs in 48 SI-NETs, with an average of 12.6 amplifications and 8.7 deletions per tumor (Supplemental Tables 9–12). The amplitude of focal amplifications was regularly much higher, with normalized read count ratios corresponding to up to 16-fold amplification. We also found recurrent loss of chromosomes 11 and 18 and gains of chromosomes 4, 5, 19, and 20, as previously shown by array-based methods in SI-NETs (14–19), providing external validation. See Supplemental Figure 4 for array-comparative genomic hybridization (aCGH) validation of SCNAs detected by sequencing. Direct comparison of all SCNAs found in the present study with previous results by Kulke et al. (14) is shown as Supplemental Figure 5. SCNAs in other cancers vary, from less than 5 per tumor in myeloproliferative disorders to greater than 120 in dedifferentiated liposarcoma (20), placing SI-NETs — with their estimated 21.1 SCNAs per tumor — among the more stable tumor types.
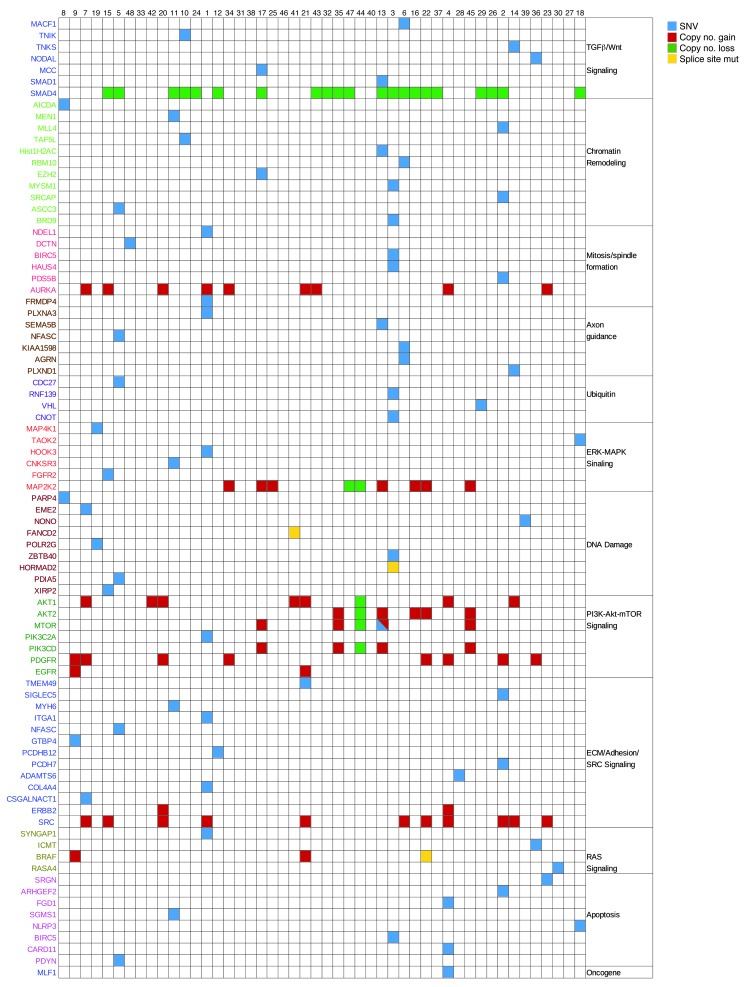
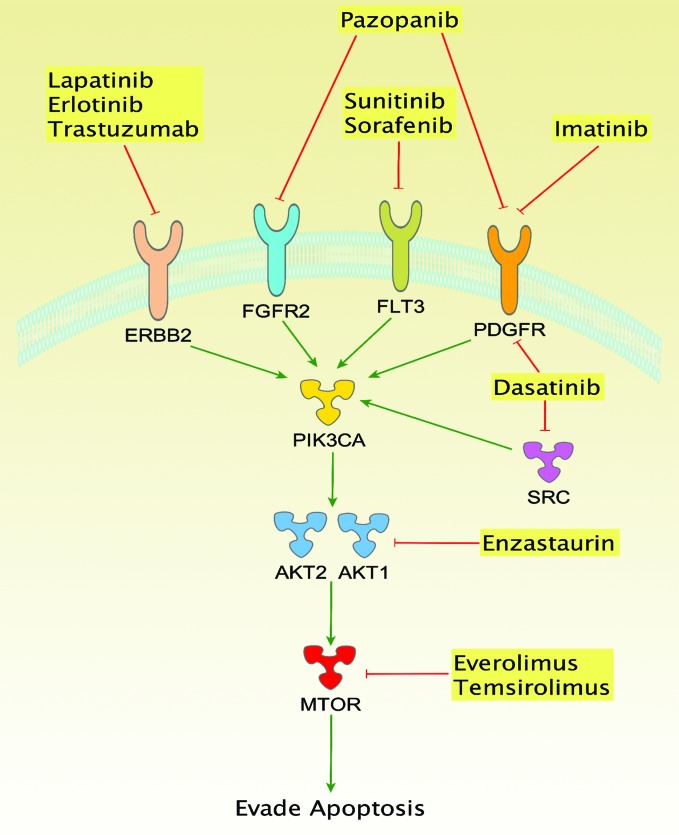
An integrative analysis was performed with a dual approach. First, canonical mechanisms of cancer were found to be recurrently mutated (Figure 3). Second, an individualized genome analysis for candidate “actionable” alterations was performed (Supplemental Table 14). SI-NET genomes have not been systematically searched for candidate therapeutic targets to date. We adopted an “n = 1” knowledge-mining approach, searching each of the 48 SI-NET exomes individually for candidate therapeutic targets. Somatic genome alterations were mapped to cancer-related molecular concepts and prioritized according to mutations and signaling pathways targeted by approved or clinically available experimental therapeutics. Genetic alterations in the PI3K/Akt/mTOR pathway were noted as a top therapeutic priority in 14 patients (29%). Amplification of RAC-α serine-threonine protein kinase 1 and 2 (AKT1/2) was found in 13 cases. Mammalian target of rapamycin (MTOR) was altered by amplification with or without mutation in 4 patients. A member of the PIK3 family of genes was altered in 5 of the above patients (along with MTOR or AKT1). The convergence of these multiple genomic alterations on the PI3K/Akt/mTOR pathway is consistent with the observation that MTOR inhibitors are active in a number of SI-NET patients (Figure 4). Furthermore, direct targeting of PI3K or Akt may be promising alternatives in patients in which these genes appear to be the deregulating pathway component.
Figure 3. Integrative analysis of genomic alterations in SI-NET.
SNVs and SCNAs are clustered into pathways of carcinogenesis and tumor maintenance. Canonical concepts and signaling models relevant to molecular carcinogenesis were found to be recurrently altered by nonsynonymous, potentially deleterious somatic SNVs and SCNAs. Chromatin remodeling, mitosis/spindle formation, the DNA damage response, and the regulation of apoptosis are concepts controlling basic mechanisms of cell growth and have established roles in cancer. RAS, ERK/MAPK, and Wnt signaling are classic pathways of cancer. Axon guidance genes have recently been recognized as tumor suppressors (30). PI3K/Akt/mTOR signaling is a mechanism of tumorigenesis. Blue, nonsynonymous exonic point mutation; yellow, splice donor or splice acceptor site mutation; red, amplification; green, deletion.
Figure 4. Signaling connected with the PI3K/Akt/mTOR pathway and altered in SI-NETs.
Cancer therapeutics target multiple proteins in connected signaling cascades. Candidate therapeutically actionable genome alterations were noted in 72% of SI-NET patients. FLT3, fms-like tyrosine kinase receptor 3.
The most commonly amplified oncogene was SRC (11 of 48 tumors; Figure 3). SMAD genes were recurrently mutated or deleted in 22 patients. The most common event was deletion of SMAD2 and SMAD4; 1 patient had a predicted damaging mutation in SMAD1. Additional candidate therapeutic targets were identified in smaller patient subsets. The HSP90 gene was amplified in 5 patients, consistent with our previous observation of HSP90 overexpression in NETs, increased transcript and protein levels in SI-NET cell lines, and sensitivity of SI-NET cells in culture to nanomolar concentrations of 17AAG, an inhibitor of HSP90 (21). Extending knowledge mining to all somatic SNVs and SCNAs in each SI-NET genome, spanning numerous altered genes in most patients, implicated a wide spectrum of additional candidate therapeutic targets. Some were notable because corresponding drugs are available, such as imatinib for PDGF receptor (PDGFR; 10 patients), herceptin for ERBB2 (2 patients), and erlotinib for EGFR (2 patients). For other repeatedly amplified molecular targets, including SRC (11 patients) and AURKA (9 patients), experimental inhibitory drugs are available.
Discussion
This study of whole-exome sequencing of 48 SI-NETs and their normal tissue counterparts represents the first genome-wide sequencing study for this tumor type. While our results suggest a paucity of somatic mutations, we found several recurrent SCNAs among the 48 tumors. Integrative bioinformatic analysis of our data implicated several cancer-related pathways, including PI3K/Akt/mTOR signaling, the TGF-β pathway (through alterations in SMAD genes), and the SRC oncogene.
Of the 48 tumors in our dataset, 22 had mutated or deleted SMAD genes. Multiple studies have suggested that the TGF-β pathway might be an important regulator of growth in SI-NET. Gilbert et al. examined formalin-fixed, paraffin-embedded tissue from 104 neuroendocrine cancers with immunohistochemistry, and enhanced expression (3+/2+) of TGF-β was found in all but 1 tumor (21). Moreover, preclinical data from studies in cell lines suggest that the TGF-β pathway might be a therapeutic target in small bowel NENs (22, 23). The SI-NET cell line KRJ was induced to proliferate by TGF-β1 (24), increasing c-myc expression, downregulating p21, and cross-activating ERK1/2 in KRJ SI-NET cells, but not in normal small intestinal enterochromaffin cells. Furthermore, SMAD alterations might adversely affect responses to fluorouracil-based therapy, as previously seen in colon cancer (25), and suggest the TGF-β pathway as a tentative indirect therapeutic corollary (26). The implications for the treatment of SI-NET could therefore be 2-fold. First, patients with unresectable and otherwise therapy-resistant disease and TGF-β alterations might be considered for enrollment into phase I clinical trials investigating the targeting of the TGF-β pathway. Second, it could also be tested whether patients with intact SMAD4 might be more likely to respond to capecitabine or 5FU than patients with SMAD4 deletions. Another common alteration was amplification of the SRC gene. A role for SRC in the carcinogenesis of neuroendocrine tumors has recently been suggested, as activated SRC stimulated MTOR activity in neuroendocrine cells (27). Furthermore, the combination of a SRC inhibitor and an MTOR inhibitor acted synergistically to inhibit neuroendocrine tumor cell growth in vitro (28). While the combination of SRC inhibitors (e.g., dasatinib) and MTOR inhibitors has not yet been studied in phase I clinical trials, this combination would seem especially attractive for patients with neuroendocrine carcinomas that show evidence of dysregulation of either pathway.
The exome sequencing technology used in this study did not cover regulatory elements or epigenomic alterations, leaving such potential mechanisms of carcinogenesis unaddressed for SI-NETs. Another limitation was the reliance on a reference genome, which reflects the current best practice in cancer bioinformatics, because alternatives such as de novo assembly of cancer genomes remain challenging with the sequencing read length provided by nextgen technology and available computational tools.
Candidate therapeutically actionable genomic alterations were seen in the majority of patients (72%). Thus, the therapeutic roadmap for SI-NETs might feature few or no broad avenues, but rather many related but distinct paths definable by personalized genomics.
Methods
Patients and tissue samples.
Primary SI-NETs and adjacent nonmalignant tissue were collected during surgical resection of primary tumors. Tissues were immediately frozen in liquid nitrogen and stored at –80°C. Surgical specimens were assessed by microscopic pathology for the presence of tumor and for well-differentiated type histology of SI-NET (see also Supplemental Figure 3). Formalin-fixed, paraffin-embedded sections were stained with H&E and reviewed by a board-certified pathologist. Only tumors with well-differentiated histology were selected for our study.
DNA isolation and nextgen whole-exome sequencing.
2 μg DNA was sheared by sonication to a fragment length of 200–300 bp (peak on electrophoresis). Illumina DNA sequencing libraries were constructed from each sample using adapters suitable for “indexing,” a method for identifying samples during a multiplexed sequencing run with several samples per lane of the sequencing flow cell. Exome enrichment was performed using the Illumina TruSeq exome enrichment kit. Sequencing was performed by the Mayo Clinic Genomics Core on an Illumina Hiseq2000 instrument to 100 bp in length in paired-end mode (each read representing 100 bp × 2).
aCGH.
See Supplemental Methods. Array data have been deposited at GEO (accession no. GSE45190).
Validation by capillary (Sanger) sequencing.
Sanger sequencing was performed on high-value SNVs. Custom PCR primers were designed for each SNV, and amplicons of 300–400 nucleotides in length were generated using tumor and germline genomic DNA as template. Capillary sequencing was performed bidirectionally.
Data processing.
See Supplemental Methods and Supplemental Figure 1 for computational processing of primary sequencing reads, somatic SNV calling, and SCNA calling.
Statistics.
Statistical testing was 2-sided in all instances, with differences considered significant only when the estimated type I error was P < 0.05.
Study approval.
Collection and testing of all samples was approved by the Mayo Clinic IRB (approval no. 1683-01); oral or written informed consent was not required, because the research involved no more than minimal risk to the subjects, the waiver did not adversely affect the rights and welfare of the subjects, the research could not practicably be carried out without the waiver, and, whenever appropriate, the subjects were to be provided with additional pertinent information after participation.
Supplementary Material
Acknowledgments
The authors thank Gary Schroth and Illumina for access to TruSeq exome enrichment reagents. The study was supported by grants from the Center for Individualized Medicine (Biomarker Program), Mayo Clinic (to M.S. Banck and M.M. Ames), and the Schulze Family Foundation (to A.S. Beutler).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(6):2502–2508. doi:10.1172/JCI67963.
References
- 1.Rindi G, et al. WHO Classification Of Tumours Of The Digestive System. 2010. Nomenclature and classification of neuroendocrine neoplasms of the digestive system. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, eds. pp. 13–14. Lyon, France: IARC Press; [Google Scholar]
- 2.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonigro RJ, et al. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia. 2011;13(11):1019–1025. doi: 10.1593/neo.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulke MH, et al. High-resolution analysis of genetic alterations in small bowel carcinoid tumors reveals areas of recurrent amplification and loss. Genes Chromosomes Cancer. 2008;47(7):591–603. doi: 10.1002/gcc.20561. [DOI] [PubMed] [Google Scholar]
- 15.Kim do H, Nagano Y, Choi IS, White JA, Yao JC, Rashid A. Allelic alterations in well-differentiated neuroendocrine tumors (carcinoid tumors) identified by genome-wide single nucleotide polymorphism analysis and comparison with pancreatic endocrine tumors. Genes Chromosomes Cancer. 2008;47(1):84–92. doi: 10.1002/gcc.20510. [DOI] [PubMed] [Google Scholar]
- 16.Tönnies H, et al. Analysis of sporadic neuroendocrine tumours of the enteropancreatic system by comparative genomic hybridisation. Gut. 2001;48(4):536–541. doi: 10.1136/gut.48.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson E, Swärd C, Stenman G, Ahlman H, Nilsson O. High-resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocr Relat Cancer. 2009;16(3):953–966. doi: 10.1677/ERC-09-0052. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham JL, Díaz de Ståhl T, Sjöblom T, Westin G, Dumanski JP, Janson ET. Common pathogenetic mechanism involving human chromosome 18 in familial and sporadic ileal carcinoid tumors. Genes Chromosomes Cancer. 2011;50(2):82–94. doi: 10.1002/gcc.20834. [DOI] [PubMed] [Google Scholar]
- 19.Zikusoka MN, Kidd M, Eick G, Latich I, Modlin IM. The molecular genetics of gastroenteropancreatic neuroendocrine tumors. Cancer. 2005;104(11):2292–2309. doi: 10.1002/cncr.21451. [DOI] [PubMed] [Google Scholar]
- 20.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert JA, et al. Molecular markers for novel therapies in neuroendocrine (carcinoid) tumors. Endocr Relat Cancer. 2010;17(3):623–636. doi: 10.1677/ERC-09-0318. [DOI] [PubMed] [Google Scholar]
- 22.Kidd M, Schimmack S, Lawrence B, Alaimo D, Modlin IM. EGFR/TGFα and TGFβ/CTGF signaling in neuroendocrine neoplasia: theoretical therapeutic targets. Neuroendocrinology. 2013;97(1):35–44. doi: 10.1159/000334891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wimmel A, Wiedenmann B, Rosewicz S. Autocrine growth inhibition by transforming growth factor beta-1 (TGFβ-1) in human neuroendocrine tumour cells. Gut. 2003;52(9):1308–1316. doi: 10.1136/gut.52.9.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd M, et al. Small bowel carcinoid (enterochromaffin cell) neoplasia exhibits transforming growth factor-beta1-mediated regulatory abnormalities including up-regulation of C-Myc and MTA1. Cancer. 2007;109(12):2420–2431. doi: 10.1002/cncr.22725. [DOI] [PubMed] [Google Scholar]
- 25.Papageorgis P, et al. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-β targeted cancer therapy. Int J Biol Sci. 2012;8(7):964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaur P, et al. Identification of cancer stem cells in human gastrointestinal carcinoid and neuroendocrine tumors. Gastroenterology. 2011;141(5):1728–1737. doi: 10.1053/j.gastro.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Florio A, et al. Src kinase activity coordinates cell adhesion and spreading with activation of mammalian target of rapamycin in pancreatic endocrine tumour cells. Endocr Relat Cancer. 2011;18(5):541–554. doi: 10.1530/ERC-10-0153. [DOI] [PubMed] [Google Scholar]
- 29.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.