Abstract
NF-κB is constitutively activated in many cancer types and is a potential key mediator of tumor-associated inflammation, tumor growth, and metastasis. We investigated the role of cancer cell NF-κB activity in T cell–mediated antitumor responses. In tumors rendered immunogenic by model antigen expression or following administration of antitumor vaccines, we found that high NF-κB activity leads to tumor rejection and/or growth suppression in mice. Using a global RNA expression microarray, we demonstrated that NF-κB enhanced expression of several T cell chemokines, including Ccl2, and decreased CCL2 expression was associated with enhanced tumor growth in a mouse lung cancer model. To investigate NF-κB function in human lung tumors, we identified a gene expression signature in human lung adenocarcinoma cell lines that was associated with NF-κB activity level. In patient tumor samples, overall lung tumor NF-κB activity was strongly associated with T cell infiltration but not with cancer cell proliferation. These results therefore indicate that NF-κB activity mediates immune surveillance and promotes antitumor T cell responses in both murine and human lung cancer.
Introduction
Lung cancer is a leading cause of cancer-related deaths around the world, and its 5-year survival rate has remained essentially unchanged over many decades. While targeted therapies against driver oncoproteins such as EGFR and ALK have shown considerable promise, there are few therapeutic options against the vast majority of KRAS mutant tumors and tumors without known driver mutations (1–4). Thus, novel approaches and new targets are required for treating lung cancer. Among the new modalities being tested is immunotherapy, which has shown considerable promise in recent studies (5–12). T cell presence in tumors is typically associated with immune surveillance and improved patient survival (13–19). Consequently, immunotherapy using blockade of negative regulators of T cell function is an especially attractive approach (5, 11). Unlike genetic lesion-specific therapies, immunotherapy has potential for targeting tumors irrespective of driver oncogene mutation status.
Among the frequently activated pathways in cancer is the NF-κB transcription factor pathway (20–23). The NF-κB family plays a crucial role in regulating inflammatory and immune responses as well as in regulating cell proliferation and cell survival (24–27). NF-κB activation typically occurs by nuclear translocation following inducible phosphorylation of inhibitory IκB proteins by the IKKα/β/γ (IκB kinase) complex (24–27). In addition, the innate immunity-regulating IKK-like kinases IKKε (also known as IKBKE or IKKi) and TBK1 can also activate NF-κB (28–30). Importantly, NF-κB is constitutively activated in many cancers and known to directly promote tumor growth (20–23). Interestingly, a recent study of 26 different tumor types showed that genes in the NF-κB pathway are frequently impacted by copy number alterations (31). In particular, regions encoding IKKβ and IKKγ undergo frequent amplification (31). NF-κB also enhances tumor-associated inflammation, leading to increased angiogenesis and metastasis (32–35). An additional key role of the IKKβ/NF-κB pathway in inflammation-promoting myeloid cell types has also been shown to be critical in solid malignancies (36–39). Importantly, recent studies in mice have defined a key role for NF-κB in KRAS-induced lung cancer (22, 23, 40). NF-κB activation by oncogenic KRAS can be mediated by IKKβ and/or TBK1 kinase (22, 23, 40, 41). Together, these studies indicate that NF-κB promotes tumor growth and progression through diverse mechanisms.
As mentioned above, T cell presence in tumors can be associated with immune surveillance and improved patient survival (13–19). The demonstrated role of cancer “immunoediting” in selecting tumor cells best suited to escape immune attack provides additional evidence of a key function of the immune system in controlling cancer (18, 19). While recent immunotherapy strategies to boost T cell–induced responses against solid tumors have shown considerable promise (5–10), tolerance-inducing mechanisms and presence of suppressive cell types, such as Tregs and myeloid-derived suppressor cells, in the tumor microenvironment can dampen the response to immunotherapy (34, 42). It is not known whether tumor NF-κB regulates T cell–mediated antitumor responses and immune surveillance. Nonetheless, consistent with known protumor functions, it is possible that NF-κB also impairs antitumor T cell responses through cancer cell–intrinsic and/or microenvironment effects. However, we show here that in immunogenic tumors in mice NF-κB induces T cell–mediated tumor rejection. Enhanced T cell recruitment was found to be a key NF-κB–dependent mechanism for tumor rejection. Little is known about NF-κB function in human cancer. To investigate potential protumor and antitumor NF-κB functions, we developed a novel human lung cancer NF-κB gene expression signature. While we found evidence of both inflammatory and immune response functions, overall NF-κB activity was strongly associated with T cell presence in human lung cancer. These findings in both murine and human lung cancer indicate that a crucial and previously unappreciated function of tumor NF-κB is to promote T cell–mediated immune surveillance responses.
Results
Critical role of NF-κB in immunogenic tumor rejection in mice.
The presence of tumor-infiltrating T cells, which likely recognize tumor-expressed antigens, is associated with improved patient survival (13–15). To induce de novo antitumor T cell responses in mice, we expressed Kb-OVA (a single polypeptide encoding H-2Kb, β2-M, and the OVA SIINFEKL peptide recognized by CD8 T cells) (43) in poorly immunogenic Lewis lung carcinoma (LLC) to generate LLC-OVA. s.c. inoculation with LLC-OVA was sufficient to induce an OVA-specific CD8 T cell response (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI67250DS1). We next determined how tumor NF-κB activity impacts antitumor CD8 T cell responses. To selectively activate NF-κB in tumor cells, we used tumor cell–specific expression of constitutively activated–IKKβ (CA-IKKβ). IKKβ mediates NF-κB activation in response to multiple stimuli and pathways, including those activated by oncogenes such as KRAS (40). Furthermore, IKKβ is potentially amplified in human cancer (31). CA-IKKβ expression in LLC (LLC-IKK) led to increased NF-κB, but not AP-1, nuclear translocation compared with that in control MSCV-IRES-GFP–transduced (MiG-transduced) LLC (Supplemental Figure 2). Similarly, CA-IKKβ expression in LLC-OVA (LLC-OVA-IKK) enhanced nuclear NF-κB, comprising primarily RELA/p65-containing complexes (see below), and target gene Cxcl1 (also known as Kc) expression (Figure 1, A and B). However, both were induced substantially less compared with that after TNF-α treatment (Figure 1, A and B). Therefore, CA-IKKβ induces modest activation of NF-κB relative to TNF-α.
Figure 1. Impact of IKKβ-induced NF-κB on tumor rejection.
(A) EMSA showing NF-κB nuclear levels and (B) RT-PCR showing Cxcl1 expression in LLC-OVA transduced with control MiG, IKK, and MiG treated with TNF-α for 1 and 2 hours. Samples were run in triplicate (mean ± SEM). Tumor growth in C57BL/6 mice inoculated s.c. with (C) nonimmunogenic LLC-MiG and LLC-IKK and (D) immunogenic LLC-OVA-MiG and LLC-OVA-IKK. (E) Impact of immunogenic LLC tumors on peripheral T cells. Tetramer analysis of day-10 OVA-specific CD8 T cells in peripheral blood from naive mice or mice receiving LLC-OVA-MiG or LLC-OVA-IKK cells s.c. Each point represents a single mouse. *P = 0.5485, Student’s t test comparing tetramer+ CD8 T cells between mice receiving LLC-OVA-MiG and LLC-OVA-IKK tumors. (F) C57BL/6 mice received s.c. LLC-OVA-MiG or LLC-OVA-IKK, and tumor growth was monitored. Relative fold increase in tumor volume in mice at day 21 after inoculation compared with day 4 after inoculation. Combined results from 3 independent experiments are shown (n = 11 for both groups). (G) Tumor growth in Rag2–/– mice inoculated s.c. with LLC-OVA-MiG or LLC-OVA-IKK and (H) 129S4/SvJaeJ mice inoculated s.c. with immunogenic LKR-OVA-MiG and LKR-OVA-IKK. (I) C57BL/6 BALB/c F1 mice received s.c. TUBO-MiG or TUBO-IKK. After 5 days, half of the mice in each group received HER2 TriVax. Tumor growth of all mice was calculated at day 21 relative to day 5. Relative growth in vaccinated mice was compared with unvaccinated counterparts. *P = 0.0256, t test with Welch’s correction. Graph shows combined results of 2 independent experiments. (C, D, G, and H) Each line represents a single mouse. (F and I) Each point represents tumor growth from a single mouse.
Compared with control LLC-MiG, LLC-IKK had no significant effect on s.c. growth of nonimmunogenic LLC in syngeneic C57BL/6 mice (Figure 1C). Interestingly, LLC-OVA-IKK tumors grew initially but were rejected subsequently while LLC-OVA-MiG tumors grew unrestrained (Figure 1D). Importantly, a similar number of activated OVA-specific CD8 T cells was detected in peripheral blood of LLC-OVA-MiG and LLC-OVA-IKK mice (Figure 1E), suggesting that reduced growth of LLC-OVA-IKK tumors was not due to enhanced T cell priming. Combined results from 3 experiments indicated that once tumors were perceptible (day 4), 10 out of 11 LLC-OVA-MiG mice showed 2-fold or greater tumor growth, while only 3 out of 11 LLC-OVA-IKK mice showed similar growth (Figure 1F). The difference in the number of tumors that showed growth in the 2 groups was significant (P = 0.008, Fisher’s exact test). Analysis of LLC-OVA-IKK tumors undergoing rejection showed the presence of numerous lymphocytic infiltrates, which were substantially less plentiful in LLC-OVA-MiG tumors (Supplemental Figure 3). In addition, extensive fibrosis was evident in LLC-OVA-IKK tumors, likely resulting from tumor cell loss (Supplemental Figure 3). Importantly, LLC-OVA-IKK tumors grew robustly in lymphocyte-deficient recombination activating gene 2 (Rag2–/–) mice (Figure 1G), demonstrating a role for lymphocytes in rejection of LLC-OVA-IKK tumors. To extend these studies to a different lung tumor model, we used KRAS mutant LKR-13 cells (44). As in LLC, CA-IKKβ expression in LKR-13 cells also resulted in NF-κB activation (Supplemental Figure 4). Importantly, while LKR-OVA-MiG tumors showed robust growth, LKR-OVA-IKK tumor growth was drastically reduced (Figure 1H). These results therefore indicate that NF-κB activation induces rejection and/or growth suppression of immunogenic lung tumors in mice.
We then determined the effect of NF-κB activation in vaccine-induced responses against the breast carcinoma TUBO line, which expresses HER2/NEU (45). Notably, HER2/NEU amplification is also frequent in human lung cancer (46). TUBO-MiG– and TUBO-IKK–injected mice were randomly split into control (no vaccine) and TriVax (47, 48) vaccine groups, which were immunized using a synthetic peptide from HER2/NEU (49). The TriVax-induced HER2-specific T cell increase in peripheral blood was similar in the 2 groups (Supplemental Figure 5). Tumor growth in vaccinated TUBO-MiG and TUBO-IKK mice was then determined relative to their unvaccinated counterparts. While vaccination reduced growth of both TUBO-MiG and TUBO-IKK tumors, the reduction was significantly more pronounced in TUBO-IKK tumors (P = 0.025) (Figure 1I). We conclude that NF-κB activation restrains tumor growth in de novo and vaccine-induced T cell models.
We next used a metastatic model in which i.v. injected LLC forms tumor foci in lungs, the natural environment of LLC. Importantly, large tumor foci were evident in LLC-OVA-MiG–injected mice but not in LLC-OVA-IKK–injected mice (Figure 2A). However, both nonimmunogenic LLC-MiG and LLC-IKK mice showed multiple large foci (Figure 2A). Further examination of lungs of LLC-OVA-IKK–injected mice showed the presence of microscopic foci together with lymphocytic infiltrates (Figure 2, B–D). Larger lesions showed lymphocytic infiltrates peripherally and within tumors (Figure 2B), while smaller foci showed numerous lymphocytes with few remaining tumor cells (Figure 2, C and D). Together with results of LLC-OVA-IKK s.c. tumors, these results suggest that lymphocytic infiltrates may be limiting the growth of LLC-OVA-IKK lung tumor foci.
Figure 2. LLC growth in a metastatic model of lung cancer.
(A) Growth of LLC in a metastatic model of lung cancer. H&E staining of lungs from mice 24 days after receiving i.v. nonimmunogenic LLC-MiG or LLC-IKK or immunogenic LLC-OVA-MiG or LLC-OVA-IKK. Scale bars: 4 mm. (B–D) LLC-OVA-IKK–injected mice showing the presence of microscopic foci together with lymphocytic infiltrates. (B) Larger lesions showed lymphocytic infiltrates peripherally and within tumors, (C and D) while smaller foci showed numerous lymphocytes with few remaining tumor cells. Tumor cells (T) and lymphocytic infiltrates (L) are indicated. Scale bars: 50 μM.
NF-κB–induced T cell chemokine expression is crucial for tumor rejection.
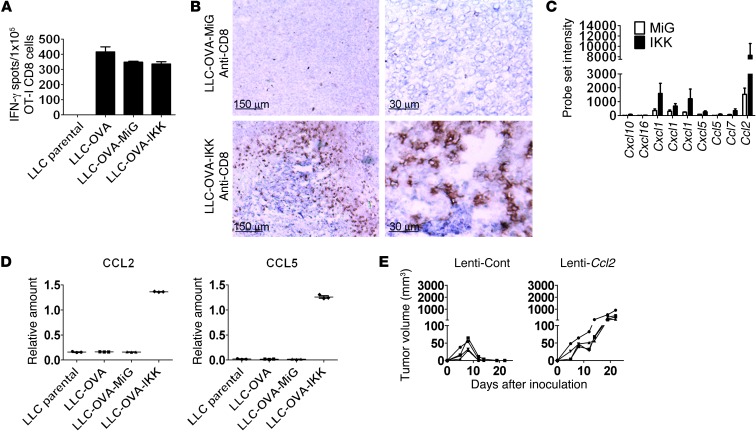
We next investigated possible mechanisms involved in rejection of LLC-OVA-IKK tumors. Rejection was not likely mediated by increased numbers of OVA-specific CD8 T cells (Figure 1E). In addition, LLC-OVA-IKK cells were not superior stimulators of CD8 T cell IFN-γ expression (Figure 3A). Importantly, while LLC-OVA-MiG tumors had a small number of infiltrating CD8 T cells, LLC-OVA-IKK tumors showed greatly increased CD8 T cell presence (Figure 3B). These results suggest that NF-κB–regulated expression of T cell chemokines may be responsible for increased T cell recruitment. To identify T cell chemokines involved, global RNA expression studies using microarray analysis were performed on LLC-OVA-MiG and LLC-OVA-IKK cells. We used a 2-fold cutoff to identify genes upregulated or downregulated by IKKβ in 2 independent experiments (Supplemental Table 1). In total, 88 genes were upregulated and 83 genes were downregulated in both experiments (Supplemental Table 1). These included multiple chemokines involved in both T cell and neutrophil chemotaxis (Figure 3C and Supplemental Figure 6). Among T cell chemokines identified, CCL2, CCL5, and CXCL10 are known to mediate activated T cell chemotaxis (50). RT-PCR confirmed upregulation of Ccl2 and Ccl5 in LLC-OVA-IKK cells (Figure 3D). Similarly, Ccl2 and Ccl5 expression was also substantially enhanced in LKR-OVA-IKK cells (data not shown).
Figure 3. Impact of T cell chemokines on tumor rejection.
(A) IFN-γ production by OT-1 CD8 T cells. ELISpot of OT-I T cells cultured with LLC parental, LLC-OVA, LLC-OVA-MiG, or LLC-OVA-IKK tumor cells (1 × 105 LLC cells per well; 1 × 105 T cells per well). Samples were run in triplicate (mean ± SEM). (B) C57BL/6 mice received s.c. LLC-OVA-MiG or LLC-OVA-IKK. Tumors were excised at day 9, and CD8 expression was determined using IHC. Typical results from 1 mouse per group out of 4 mice is shown. Scale bars: 150 μm (left); 30 μm (right). (C) Affymetrix probe set signal intensity of indicated chemokines in LLC-OVA-IKK compared with that in LLC-OVA-MiG. Genes identified in 2 separate microarray experiments are shown (mean ± SEM). (D) RT-PCR showing CCL2 and CCL5 expression in LLC parental, LLC transduced with OVA, and LLC transduced with OVA and MiG or OVA and IKK. Samples were run in triplicate (average ± SEM). (E) Lentivirus-expressing scrambled shRNA (Lenti-Cont) and LLC-OVA-IKK Ccl2 shRNA (Lenti-Ccl2) cells were injected s.c. in C57BL/6 mice, and tumor growth was monitored. Each line represents tumor growth in a single mouse. All results are representative of at least 2 independent experiments.
We next determined cytoplasmic and nuclear levels of NF-κB subunits in LLC-OVA-MiG and LLC-OVA-IKK cells (Supplemental Figure 7). Nuclear levels of RELA and RELB, but not cREL, p50, or p52, were increased in LLC-OVA-IKK cells compared with those in LLC-OVA-MiG cells (Supplemental Figure 7, A–C). Interestingly, increased cytoplasmic levels of p52 precursor NFKB2/p100 (Supplemental Figure 7C) and RELB (Supplemental Figure 7B) in LLC-OVA-IKK cells are consistent with their positive regulation by canonical NF-κB pathway activation (51). However, lack of increase in p52 cytoplasmic or nuclear levels suggests that IKKβ does not lead to increased processing of NFKB2/p100 (Supplemental Figure 7C). Furthermore, these results also suggest that increased nuclear RELB likely results from increased expression rather than increased formation of p52/RELB complexes from NFKB2/p100 processing. Importantly, enhanced phosphorylation of RELA at S536, a site known to be important for transcriptional activation (52), was also detected in LLC-OVA-IKK cells (Supplemental Figure 7A). In contrast, no increase in phospho-ERK was seen in LLC-OVA-IKK cells (Supplemental Figure 7D). The role of the 2 subunits (RELA and RELB), showing increased nuclear translocation in LLC-OVA-IKK cells, was also determined following shRNA-mediated knockdown. These studies showed that RELA plays a more critical role than RELB in expression of T cell chemokines in LLC-OVA-IKK cells (Supplemental Figure 8). Thus, increased RELA nuclear translocation and phosphorylation likely play a crucial role in T cell chemokine expression in LLC-OVA-IKK cells.
Ccl2 in particular exhibited a dramatically higher microarray probe set signal compared with that of T cell chemokines Ccl5 and Cxcl10 (Figure 3C), suggesting it may dominate T cell recruitment responses by LLC-OVA-IKK. In addition, CCL2 protein expression was greatly increased in LLC-OVA-IKK cells compared with that in LLC-OVA-MiG cells (Supplemental Figure 9). Previous studies have indicated an important role for CCL2 in breast cancer metastasis through monocyte recruitment (53) but also in T cell recruitment (54). Given high expression and potentially diverse functions, we determined whether CCL2 was specifically required for rejection of LLC-OVA-IKK tumors by knockdown of CCL2 expression (Supplemental Figure 10A). Ccl2 shRNA did not impact LLC-OVA-IKK growth in vitro or OVA-specific T cell expansion in vivo (Supplemental Figure 10B). However, while control LLC-OVA-IKK tumors were readily rejected, expression of Ccl2 shRNA resulted in robust tumor growth (Figure 3E and Supplemental Figure 10C). These results therefore suggest that Ccl2 is a potentially crucial immune surveillance–regulating NF-κB target gene that is required for LLC-OVA-IKK rejection.
Generation of a gene expression signature to predict NF-κB activity in human lung cancer.
Intrigued by the above findings in mice, we determined whether NF-κB activity is also associated with T cell presence in human lung cancer. To determine NF-κB activity, we reasoned that an NF-κB–driven gene expression signature will provide a superior indication of activity than the activation state of individual subunits or NF-κB pathway kinases. Furthermore, previous studies have shown the predictive potential of gene expression signatures in determining pathway activation state (55, 56). To our knowledge, no such gene expression signature to predict NF-κB activity in human lung cancer exists. We used 5 human lung adenocarcinoma cell lines (A549, H23, H358, PC9, and HCC827) to generate such a signature. In each cell line, we identified genes that were impacted by NF-κB inhibition (using retrovirus-expressed “superrepressor” of NF-κB [IκBαSR]) (57) and/or NF-κB activation (using CA-IKKβ described above) (58). As shown in Figure 4A, IκBαSR reduced and CA-IKKβ increased NF-κB heterodimer nuclear activity compared with that in control MiG retrovirus–infected cells. Although NF-κB is widely considered to be a tumor growth-promoting transcription factor, we found surprisingly little effect on survival or proliferation of these 5 lung cancer cell lines following NF-κB inhibition by IκBαSR expression.
Figure 4. Generation and validation of a lung cancer NF-κB signature.
(A) EMSA analysis of sorted H23, PC9, and HCC827 cells infected with GFP, IκBαSR, or CA-IKKβ retroviruses. Parental (Par) cells were not infected. The major NF-κB complexes are indicated with arrows. The upper complex corresponds to NF-κB subunit heterodimers, while the lower complex corresponds to homodimers. (B) NF-κB signature scores were determined by building a classifier to determine relative signature activity in different T cell lines (Supplemental Methods and Supplemental Table 3). Each “x” represents an individual microarray experiment. The median of the relative signature activity in all cell lines used for this analysis is also indicated. (C) H1299, H157, and H226 cells were transduced with MiG or IκBαSR retroviruses, following which TNFAIP3 and BIRC3 mRNA was determined by RT-PCR (mean ± SEM). (D) Expression of CCL2, CCL5, CXCL1–CXCL3, and IL8 was determined in H157 cells transduced with MiG and IκBαSR (mean ± SEM). (E) NF-κB activity determined by EMSA in NF-κB signature low and high cell lines. Major NF-κB complexes are indicated with arrows.
RNA isolated from these 5 cell lines was used to perform gene expression studies using Affymetrix U133 Plus 2.0 microarrays. Initially, all probe sets impacted 1.4-fold or more by IκBα or IKKβ relative to the control vector were identified in each cell line. This low stringency cutoff was used to accommodate a second selection criteria. Next, the 5 cell line signatures were used to identify probe sets similarly regulated by IκBα and IKKβ (with IκBα and IKKβ causing opposite effects on gene expression) in the different cell lines in ≥60% of the experimental conditions (i.e., coregulation score of ≥0.6). A total of 240 probe sets were identified using this approach (Supplemental Table 2). The rationale for this was that the identification of genes similarly regulated by NF-κB in multiple cell lines will not only eliminate false positives but also generate a signature that is broadly applicable. As examples, 2 probe sets for the well-known NF-κB target gene BIRC3 (also known as CIAP2) (59, 60) had coregulation scores of 1.0 and 0.8, and both probe sets of another known NF-κB target gene TNFAIP3 (also known as A20) (61) had coregulation scores of 1.0. In addition, IκBα increased expression of certain genes, while IKKβ repressed their expression; these likely represent genes negatively impacted by NF-κB activity. In the 240 probe sets (Supplemental Table 2), approximately 200 individual genes were present that were either upregulated or downregulated by NF-κB. A substantial number of NF-κB signature genes regulate immune cell chemotaxis (including T cell chemokines CCL2 and CCL5), inflammation, and immune regulation (Table 1).
Table 1.
NF-κB signature genes divided in broad categories based on known functions

Using publically available microarray data from 126 human lung cancer cell lines (Supplemental Methods), we next identified cell lines with high or low NF-κB signature activity. NF-κB signature scores were determined by building a classifier, as previously described (62), that allowed determination of relative signature activity in the different cell lines (Supplemental Methods and Supplemental Table 3). Based on this analysis, we selected (Figure 4B) 4 cell lines with high NF-κB signature (H226, H157, H1299, and H650) and 4 cell lines with low NF-κB signature (H322, H1395, H522, and H1437), which were used for additional studies. These non-small-cell lung cancer cell lines include adenocarcinoma (H650, H322, H1395, H522, and H1437), squamous (H226 and H157), and large cell (H1299) cell lines. When expression of individual genes was determined, the highest expression was invariably noted in one or more of the NF-κB high cell lines, but with substantial variation among these cell lines (Supplemental Figure 11). These results indicate, as expected, that overall signature activity will likely be a more reliable indicator of NF-κB activity than expression of individual genes.
Genes present in the NF-κB signature may also be regulated by non–NF-κB pathways. Thus, while H226, H157, H1299, and H650 cell lines have high expression of NF-κB signature genes, it is unclear whether this is indeed due to NF-κB activity. To determine NF-κB involvement, we transduced H226, H157, and H1299 cells with MiG and IκBαSR retroviruses (H650 cells showed little or no retrovirus infection). Importantly, expression of both genes was markedly reduced by IκBαSR expression (Figure 4C). In addition, expression of genes relevant to this study (i.e., T cell and neutrophil chemokines) were similarly reduced by IκBαSR expression (Figure 4D and data not shown). These results therefore indicate that high expression of NF-κB signature genes in these cell lines is dependent on NF-κB activity. We next determined the association between the NF-κB signature and NF-κB DNA binding activity. Importantly, EMSA showed that the 4 lines with high NF-κB signature indeed had high NF-κB site binding activity (Figure 4E). However, the low NF-κB signature H322 cell line also showed high NF-κB activity (Figure 4E). Therefore, NF-κB DNA binding activity alone does not provide an unambiguous indication of NF-κB transcriptional activity. To seek a better understanding of NF-κB function in human lung cancer, we next used the signature for studies in human lung adenocarcinoma. Specifically, we determined correlation between NF-κB–regulated chemokine gene expression and T cell presence as well as the association of these genes with patient survival.
Association of T cell chemokines, but not neutrophil chemokines, with presence of T cells in human lung cancer.
We first determined the association of different chemokines present in the NF-κB signature with T cell presence. For these studies, we used the Consortium for the Molecular Classification of Lung Adenocarcinoma (CMCLA) survival prediction study (62). CMCLA is the largest and most comprehensive study in which microarray-based gene expression data from tumors were used to predict survival of 442 patients with early-stage lung adenocarcinoma (62). Tumor samples used were from 4 different institutions, including the Moffitt Cancer Center (62). Specifically, we determined whether neutrophils (CXCL1–CXCL3 and IL8) and T cell chemokines (CCL2, CCL5, and CXCL10) were differentially associated with T cell presence (although CXCL10 did not achieve the criteria for inclusion in the NF-κB signature, we included it in this analysis because it was identified as a target gene in LLC and in some human cell lines). To detect T cells, we used T cell receptor α (TRAC) and T cell receptor β chain (TRBC1) gene expression as a marker for T cell presence. As expected, TRAC expression was highly correlated with TRBC1 expression (Figure 5A). Importantly, we found that expression of CCL2, CCL5, and CXCL10, but not neutrophil chemokine expression (CXCL1–CXCL3 and IL8), was positively correlated with these T cell markers (Figure 5A). These results therefore indicate a functional link between T cell chemokine expression and T cell presence in tumors. Interestingly, expression of neutrophil chemokine genes was highly correlated (Figure 5A), suggesting that NF-κB–regulated inflammatory and immune response functions predominate in different tumors. To help understand how genes associated with different NF-κB functions may be differentially regulated, we next determined their association with key NF-κB activators.
Figure 5. Association of T cell chemokines, but not neutrophil chemokines, with T cell presence in human lung adenocarcinoma.
(A) Correlation plot based on Spearman correlation r values of CMCLA gene expression data in human adenocarcinomas (n = 442) for T cell chemokines (CCL2, CCL5, and CXCL10), neutrophil chemokines (CXCL1–CXCL3 and IL8), and T cell receptor (TRAC and TRBC1) genes. Gene names and Affymetrix probe set ID numbers are shown. (B) Correlation r values of LTβ expression with neutrophil chemokines, T cell chemokines, and T cell presence in CMCLA data set (n = 442). Gene name and Affymetrix probe set ID numbers for genes with 2 probe sets are shown. (C) mRNA expression of indicated genes normalized to 18s rRNA in HCC827 lung cancer cells determined by RT-PCR. Fold difference in expression of genes after LTα1/β2 treatment compared with untreated cells is shown. Samples were run in triplicate (mean ± SEM). (D) mRNA expression of indicated genes normalized to 18s rRNA in HCC827 lung cancer cells determined by RT-PCR. Fold difference in expression of genes after TNF-α treatment compared with untreated cells is shown. Samples were run in triplicate (mean ± SEM).
LTβ expression is differentially associated with T cell chemokines in human lung cancer.
Multiple NF-κB–activating cytokines may be present in the lung tumor microenvironment (33). To identify cytokines potentially involved in differential expression of T cell and neutrophil chemokines, we tested association with expression of TNF-α, IL-1α, IL-1β, LTα (also known as TNF-β), and LTβ. These cytokines not only induce NF-κB activation but can also be transcriptionally regulated by NF-κB. However, only LTβ was identified as an NF-κB target gene in lung cancer cells (Table 1), and, interestingly, only LTβ showed a greater correlation with expression of T cell chemokines compared with that of neutrophil chemokines (Figure 5B and Supplemental Figure 12). LTβ receptor (LTβR) engagement by heterodimers of LTα and LTβ (LTα1/β2) activates NF-κB RELA and RELB, typically in association with p52 (63–66). Mirroring correlation in tumors, soluble LTα1/β2 induced expression of T cell chemokines but not neutrophil chemokines in human lung cancer cells (Figure 5C). In contrast, TNF-α strongly induced both neutrophil and T cell chemokines (Figure 5D). These results suggest that differential expression of NF-κB–regulated T cell chemokines, compared with that of neutrophil chemokines, in human lung cancer could be achieved through agents that selectively induce these different chemokine subsets, such as the LTβR ligand.
NF-κB signature genes are associated with distinct overall survival of patients with lung cancer.
The above results indicate that NF-κB–regulated inflammatory and immune response genes are differentially expressed in lung cancer. We next determined whether expression of these genes was also associated with distinct overall survival (OS) of patients. Using the CMCLA data set, we determined 5-year OS of patients exhibiting high versus low expression of these genes using a median cutoff. A striking effect on survival was seen for NF-κB target genes involved in neutrophil chemotaxis and inflammation (IL8, CXCL1, CXCL3, and IL6), all of which were associated with significantly poor OS (Figure 6). Poor survival in human cancer is associated with increased metastasis. Although direct clinical evidence is lacking, both IL-8 and CXCL1 have been linked to increased metastasis through effects on infiltrating myeloid cells in preclinical studies (67, 68), suggesting association with poor survival can be through increased metastatic dissemination. On the other hand, high expression of a subset of genes involved in T cell chemotaxis and T cell responses, including CCL2, LTB, ICAM1, and CD83, was associated with significantly improved OS (Figure 7, A–D). These results suggest that, in addition to promoting T cell chemotaxis, NF-κB–regulated expression of multiple key genes may promote T cell responses against tumors. LTβ association with improved OS was especially pronounced, perhaps because LTβ can enhance expression of multiple genes involved in T cell responses. Importantly, high expression of T cell receptor α and T cell receptor β chain genes was also associated with significantly improved OS in patients with lung cancer (Figure 7E). Therefore, while both T cell presence and expression of NF-κB signature genes associated with T cell responses are associated with improved OS, the high expression of inflammatory genes is associated with poor OS. Hence, distinct functions of NF-κB in human lung cancer are associated with potentially different survival outcomes.
Figure 6. Association of NF-κB signature inflammatory genes with patient survival.
Association of mRNA expression of indicated inflammatory genes (Affymetrix probe set IDs are shown) with OS in CMCLA data set (n = 442). 5-year OS of patients exhibiting high versus low expression of indicated genes using a median cutoff. Kaplan-Meier method was used to generate survival curves, and the log-rank test was used to test survival difference between the low and high expression groups by median cutoff for each gene. The P values shown were adjusted by false discovery rate for multiple testing. Patients alive after different time periods in high and low expression (Exprs) groups (each starting with n = 221) are indicated above the x axis.
Figure 7. Association of NF-κB signature immune response genes and T cell presence with patient survival.
(A–D) Association of mRNA expression of indicated immune response genes (Affymetrix probe set IDs are shown) with OS in CMCLA (n = 442). 5-year OS of patients exhibiting high versus low expression of indicated genes using a median cutoff. Kaplan-Meier method was used to generate survival curves, and the log-rank test was used to test survival difference between the low and high expression groups by median cutoff for each gene. The P values shown were adjusted by false discovery rate for multiple testing. (E) Association of T cell presence detected by expression of T cell receptor genes (TRAC and TRBC1) on patient survival was determined as with above genes. Patients alive after different time periods in high and low expression (Exprs) groups (each starting with n = 221) are indicated above the x axis.
NF-κB activity in human lung cancer is strongly associated with T cell presence.
The above findings indicate differential expression of distinct NF-κB target genes in tumors. By allowing investigation of combined expression of signature genes, principal component analysis (PCA) can simplify evaluation of pathway-dependent gene expression activity and has been widely used to derive and validate gene signatures in various cancer studies (69–71). Of the 240 NF-κB signature probe sets present in Affymetrix U133 Plus 2.0 microarrays, 159 were present (Supplemental Table 4) in the CMCLA data set that used Affymetrix 133A (62). We first identified GBP1, PSMB9, IRF1, TAP1, TNFAIP3, CCL5, PSMB8, IL32, SH2B3, and NFKBIE as NF-κB signature-driver genes (i.e., the top 10 genes with the highest PCA weight). The first principal component (PC1) is associated with the largest variance of the data (e.g., variance of NF-κB signature activity in the CMCLA data set), followed by each succeeding PC. Using association with expression of driver genes, we found that PC1 but not PC2 is associated with NF-κB activity (Supplemental Figures 13 and 14). Using the NF-κB signature PC1, we next determined whether NF-κB activity was associated with T cell presence. For these studies, we used the PC1 of T cell receptor α and β chain gene expression (Supplemental Figure 15). Importantly, a strong association was seen between NF-κB activity and T cell presence (r = 0.68) (Figure 8A). Therefore, while NF-κB target genes are associated with different functions in tumors, the overall NF-κB activity, as determined by PCA, is strongly associated with T cell presence.
Figure 8. Association of T cell presence with NF-κB activity in human lung cancer.
(A) Spearman correlation plot with r value of NF-κB signature (159 probe sets) PC1 with T cell PC1 are shown for CMCLA data (n = 442). (B) Correlation plot of NF-κB signature (159 probe sets) PC1 with MR signature PC1. (C) Correlation plot of NF-κB signature PC1 (159 probe sets) with 10-gene NF-κB signature PC1. (D) Correlation plot of T cell PC1 with 10-gene NF-κB signature PC1. (E) Correlation plot of MR signature PC1 with 10-gene NF-κB signature PC1.
In addition to inflammation, NF-κB also exerts protumor effects through cancer cell–intrinsic regulation of cell survival and proliferation (22, 23, 36–40). In previous studies, we described a breast and non-small-cell lung cancer malignancy-risk (MR) signature that is rich in proliferation and cell cycle genes (69–71). Using the MR signature PC1, we next determined whether NF-κB activity was also associated with cancer cell proliferation. However, little correlation between the NF-κB and MR signatures was noticed (r = 0.21) (Figure 8B). Thus, NF-κB activity is associated with T cell presence and potential immune surveillance functions but not with cancer cell proliferation.
We next determined whether expression of the 10 driver genes mentioned above was associated with overall NF-κB activity and T cell presence. Indeed, the PC1 of these genes (Supplemental Figure 16) was highly correlated with the NF-κB signature PC1 (r = 0.92) (Figure 8C). Therefore, this smaller 10-gene signature can be used in lieu of the NF-κB signature to determine NF-κB activity. Importantly, the 10-gene PC1 was also strongly correlated with the T cell PC1 (r = 0.79) (Figure 8D) but not with the MR signature PC1 (r = 0.19) (Figure 8E). Using an additional lung cancer microarray data set (n = 133) (72), we found a similarly high correlation of the 10-gene signature with T cell presence (r = 0.8) but not with the MR signature (r = –0.16) (Supplemental Figure 17). In conclusion, our findings from both mouse and human studies indicate that tumor NF-κB activity is strongly associated with T cell presence and immune surveillance in lung cancer.
Discussion
While previous studies have extensively documented protumor functions of NF-κB (22, 23, 36–40), our results indicate a potential tumor suppressor function of NF-κB that is mediated by enhancement of T cell–induced antitumor responses. In mouse studies, we found that NF-κB activation renders immunogenic tumors susceptible to T cell rejection. A key mechanism of rejection is increased infiltration of T cells into tumors through NF-κB–induced expression of T cell chemokines. We found an especially important role for the T cell chemokine CCL2 in tumor rejection in mice, high expression of which was also associated with improved survival in human lung cancer. However, it is likely that other NF-κB–induced T cell chemokines, such as CCL5, also play a crucial role in T cell recruitment and tumor rejection in other tumor models. Studies by Schreiber and colleagues have defined distinct phases of the immune response against tumors (18). In light of these studies, we believe the impact of NF-κB–mediated immune surveillance may be especially important in the “elimination” phase, wherein tumors with high NF-κB could undergo spontaneous rejection.
NF-κB function in lung cancer has been extensively studied in mice (22, 23, 40), yet little is known about NF-κB function in human lung cancer. A major reason for this is the lack of an appropriate functional readout of NF-κB activity in human lung cancer. One of our main findings is the description a gene expression signature that can be used to define NF-κB transcriptional activity. Using this signature, we investigated both protumor and immune surveillance functions of NF-κB in human lung cancer. Importantly, as in mouse studies, we found that NF-κB activity is strongly associated with T cell infiltration in human lung cancer. Previous studies have defined the presence of both cytotoxic (CD8) and helper (CD4) T cells in human tumors (13–15). Indeed, our results suggest association of both CD8 and CD4 subsets with tumor NF-κB activity (Supplemental Figure 18). Thus, NF-κB may lead to recruitment of different effector T cell subsets and thus facilitate antitumor responses through distinct cellular mechanisms. In addition to effector T cells, antitumor responses are also controlled by Tregs. While previous studies have focused on CD4 Tregs, CD8 Tregs expressing FoxP3 have also been identified (73). In LLC-OVA-IKK tumors, we determined whether CD8+FoxP3+ T cells were also present. However, as shown in Supplemental Figure 19B, virtually no CD8+FoxP3+ T cells were detected in LLC-OVA-IKK tumors. While not excluding tumor IKK/NF-κB involvement in regulatory CD8 (or CD4) T cell responses, these results nonetheless suggest that in the LLC model, activation of NF-κB primarily recruits cytotoxic CD8 T cells that can mediate tumor rejection.
Our results indicate that only OVA-expressing tumors are rejected by IKK expression. Thus, levels of T cell chemokines induced by NF-κB may not be sufficient to recruit low numbers of T cells and/or low-affinity T cells capable of recognizing tumor antigens. We hypothesized that greater NF-κB activation may override strong antigen (e.g., OVA) requirement in tumor immune surveillance. To this end, we isolated an LLC-IKK subline with greater IKK-induced NF-κB activation (LLC-IKKhi) (Supplemental Figure 20A) as well as CCL2 and CCL5 expression (Supplemental Figure 20B). Interestingly, compared with LLC-IKK tumors, s.c. LLC-IKKhi tumors showed significantly reduced early tumor growth (e.g., day 10; Supplemental Figure 20C). Notably, a striking difference was noticed in the LLC metastatic model, wherein numerous tumor foci were evident in LLC-IKK–injected mice but not in LLC-IKKhi–injected mice (Supplemental Figure 21A). Furthermore, LLC-IKKhi tumors had substantially greater lymphocyte infiltration in tumors compared with LLC-IKK tumors (Supplemental Figure 21, B and C). While the precise mechanisms and cell types involved are unclear, these results nonetheless suggest that high NF-κB activity may override requirement for strong antigens in controlling tumor growth.
Multiple genes capable of enhancing T cell responses were found in the NF-κB signature. Thus, while NF-κB–regulated expression of T cell chemokines is likely crucial for initial T cell recruitment, the expression of HLA genes, genes important for antigen-processing (e.g., TAP1, PSMB8, and PSMB9) and cell-cell interactions (e.g., ICAM1 and CD83) (Table 1), may help sustain and amplify antitumor T cell responses. Therefore, the association between lung tumor NF-κB activity and T cell presence is likely the result of multifaceted enhancement of T cell responses. Although it remains to be tested, we suspect an immune surveillance function of NF-κB will also be evident in other cancer types. Indeed, a recent study showed that enhancing NF-κB activation by IFN-α+ TLR ligand treatment leads to enhanced T cell recruitment in colorectal tumors (74).
We generated the NF-κB signature to specifically identify genes regulated by NF-κB in lung cancer cells. Consequently, this signature lacks typical NF-κB–regulated genes in lymphocytes (e.g., BCLXL, BCL2, and IL2), antigen-presenting cells (e.g., CD80, CD86, and CD40), and monocyte/macrophage lineages (e.g., IL12A/B, TNFA, IL1A, and IL1B). Nonetheless, it is likely that at least some of the signature genes are also regulated by NF-κB in nonmalignant cell types present in tumors. The 10-gene signature may be especially useful for detecting NF-κB activity associated with immune surveillance functions. These 10 genes were also expressed at high levels in lung cancer cell lines, suggesting a strong contribution of malignant cells in tumor activity of this signature. It is also interesting to note that expression of at least 5 out of these 10 genes in malignant cells is expected to have a stimulatory effect on T cell responses (CCL5, TAP1, PSMB8, PSMB9, and IRF1). Thus, association of the 10-gene signature with T cell presence could be mediated through both recruitment and stimulatory functional effects. However, the use of a smaller signature may also limit the ability to reliably detect NF-κB activity. Thus, it is possible that intertumoral and intratumoral heterogeneity will confound use of the 10-gene signature more significantly than the full NF-κB signature.
Using the NF-κB signature, we also evaluated tumor-promoting NF-κB functions in human lung cancer. Previous studies indicate that the 2 main protumor NF-κB functions are (a) induction of inflammation and (b) cancer cell–intrinsic regulation of survival and tumor growth (22, 23, 36–40). Indeed, we found that inflammatory genes, such as neutrophil chemokines, regulated by NF-κB are associated with significantly poor patient survival. Based on previous studies, it is likely that these chemokines impact survival through increased metastasis mediated by myeloid cell recruitment (67, 68). This effect on survival was in marked contrast with that of NF-κB genes involved in immune response regulation, which were associated with improved survival. Interestingly, these functionally distinct genes appear to be differentially expressed in tumors. While underlying mechanisms are unclear, we suggest that this can be accomplished through agents capable of selectively regulating these functionally distinct genes. Thus, we found that expression of the NF-κB activator and target gene LTβ is associated with T cells but not neutrophil chemokines. Importantly, LTβR engagement in human lung cancer cells induced T cell but not neutrophil chemokine expression. Thus, NF-κB target genes involved in immune surveillance versus those involved in inflammatory functions may be differentially expressed through presence of agents (such as LTβ) that can selectively regulate their expression. Nonetheless, it is likely that immune surveillance functions of NF-κB are not absolutely dependent on the LTβR pathway. For example, unlike that in human cell lines, LTβ expression in LLC was not induced by IKKβ; in addition, the lack of both NFKB1/p100 processing and RELB requirement for T cell chemokine induction suggests that the LTβR-induced noncanonical NF-κB pathway is not functionally critical in LLC-OVA-IKK cells (Supplemental Figure 7). Thus, activation of NF-κB through multiple pathways may lead to enhancement of immune surveillance functions. Importantly, key oncogenes such as KRAS can also activate NF-κB in lung cancer (22, 23, 40). Furthermore, recent studies indicate increased NF-κB activity in cells harboring both KRAS and p53 mutations (23). It is therefore possible that in addition to their known effects on tumor cell proliferation and survival, occurrence of sequential mutations in cancer will also modulate NF-κB activity and immune surveillance functions.
In addition to inflammation, NF-κB also exerts protumor effects through cancer cell–intrinsic regulation of cell survival and tumor growth (22, 23, 40). Mouse studies have shown that NF-κB is important for development and progression of KRAS mutant tumors, especially in a p53-null background (23). NF-κB requirement was strongly associated with NF-κB activation state (23, 41). In addition, the RELA NF-κB subunit was specifically associated with generation of high-grade KRAS mutant tumors (40). In view of these findings, we hypothesized that greater NF-κB activity may be associated with highly proliferative tumors; however, no such association was seen. It is possible that, with multiple driver mutations, human tumors do not require high NF-κB to the same degree as genetically defined KRAS mutant mouse tumors. This finding is also consistent with the lack of effect of NF-κB inhibition by IκBSR expression on human lung cancer cell survival and proliferation and the lack of proliferation-regulating genes in the NF-κB signature. NF-κB activation in nonimmunogenic LLC also did not enhance tumor growth in syngeneic mice. While we cannot overlook existence of potentially crucial cancer cell–intrinsic roles of NF-κB, our results nonetheless suggest that induction of inflammation may be a more important protumor function of NF-κB than regulation of cancer cell survival and proliferation. While protumor NF-κB functions have been extensively documented, a tumor suppressor function has been specifically identified in skin cancer (75, 76). Other studies suggest that in a lymphoma model, NF-κB can enhance the response to chemotherapy by promoting senescence (77). Unlike the tumor suppressor functions of NF-κB identified in these studies, we believe that NF-κB–regulated immune surveillance is likely to be important across multiple cancer types.
Recent clinical trials have shown that immunotherapy can enhance survival of patients with advanced cancer (5, 12). However, this beneficial effect is limited to a small percentage of patients, indicating the need to identify specific biomarkers to predict response. While there are few, if any, reliable biomarkers available to predict the response to immunotherapy, recent studies suggest that an immune-active tumor microenvironment is associated with response (78). T cell chemokine expression, in particular, is associated with T cell presence and survival benefit in multiple tumors types (54, 79, 80). Based on our findings, indicating that NF-κB activity is associated with expression of T cell chemokines and T cell presence, we believe that high NF-κB activity may signal the presence of a tumor microenvironment favorable to immunotherapy.
Methods
Mice and cell culture
Mice were maintained under specific pathogen–free conditions. Mouse LLC, derived from lung carcinoma in C57BL mice, was obtained from ATCC. The Turin-Bologna (TUBO) cell line was derived from HER2/NEU tumors in BALB/c mice. LLC and TUBO cells were cultured in DMEM supplemented with 10% FBS. LKR-13 cells were originally derived from KRASLA1 mice and provided by J.M. Kurie (MD Anderson Cancer Canter, Houston, Texas, USA). Cells were stimulated with 20 ng/ml TNF-α (R&D Systems) and recombinant human lymphotoxin α1/β2 (R&D Systems) at 100 ng/ml for different time points. Human cell lines (H157 and HCC827) and LKR-13 cells were cultured in RPMI supplemented with 10% FBS.
Retroviral and lentiviral transduction
Retroviruses were prepared by transfecting HEK 293T cells with Kb-OVA, MiG, or MiG with activated IKKβ (S177, S181 to E mutations; IKKβEE) and packaging vectors, as previously described (58, 81). MiG expresses GFP, which allowed retrovirus-transduced cells to be sorted based on GFP expression using a FACSVantage sorter (BD Biosciences) (81). Lentiviruses were prepared by transfecting HEK 293T cells with a scrambled control pLKO.1 or pLKO.1 expressing shRNA specific for CCL2 or RELB (Open Biosystems), along with packaging vectors (81). Four to five different shRNA were tested, and detailed studies were performed with shRNA showing the best knockdown of protein expression. Lentivirus-transduced cells were selected for puromycin resistance. RELA and cREL shRNA constructs were obtained from Addgene, and knockdown was performed as described previously (22, 23).
Flow cytometric analysis
Retrovirus-infected LLC and human cell lines were sorted based on GFP expression to yield >95% purity. Tetramer staining was performed as described previously (48) with the following changes: cells were incubated for 5 minutes at room temperature with Fc block, and DAPI was added to cells prior to analysis for viability gating. HER2/NEU tetramer has been described previously (49), and H2-Kb OVA tetramer was purchased from Beckman Coulter. Flow cytometric analysis was performed on an LSR II cytometer (BD Biosciences). Aggregates and dead cells were excluded from analysis. Data were acquired using CellQuest software (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Tumor studies
Cells were harvested in logarithmic growth after being cultured for less than 2 weeks and washed once in injection medium (phenol-free DMEM supplemented with 2% FBS) and counted. 5 × 105 LLC cells were injected either s.c. (in a volume of 100 μl) or i.v. (in a volume of 200 μl). 3 × 105 TUBO cells were injected s.c. in a volume of 100 μl. s.c. tumors were monitored for growth and measured 2–3 times per week. Mice receiving i.v. LLC injections were monitored for morbidity. Mice were sacrificed when s.c. tumors reached a diameter of 20 mm or when they showed signs of morbidity (i.v. or s.c.). Tumor volume was calculated as previously described (82). Relative tumor growth between treatment groups was analyzed using the t test with Welch’s correction. Mice receiving TUBO cells were split into TriVax treatment and nontreatment groups on day 5. Immunization with HER2 TriVax was performed as described previously (49).
RNA analysis and microarray studies
RNA was isolated using a Qiagen RNeasy Kit and then reverse transcribed and subjected to quantitative PCR analysis, as described previously (57), in an Applied Biosystems 7900HT Sequence Detection System with SYBR Green I Master Mix (Applied Biosystems) using gene-specific primers. All samples were run in triplicate and were normalized to rRNA 18s or β-actin. Primers were obtained from RealTimePrimers.com. Microarray studies are described in the Supplemental Methods. Microarray data was submitted to the GEO repository (accession no. GSE44619).
Histology
Mouse tumors were excised and sections were embedded in Tissue Tek OCT compound (Sakura Fintek) or placed in 10% formalin and subsequently embedded in paraffin (FFPE) blocks. Frozen sections of mouse tumors were stained with anti-CD8 and the Rat-IgG HRP Detection Kit from BD Biosciences. Mouse lungs were insufflated in 10% formalin and paraffin embedded prior to H&E staining.
Statistics
Statistical analysis was performed using 2-tailed Student’s t test, Student’s t test with Welch’s correction, and Fisher’s exact test. GraphPad Prism 5 software (GraphPad Software Inc.) was used with significance determined at P < 0.05.
PCA.
PCA uses orthogonal transformation of possibly correlated variables (e.g., expression levels of genes in a signature) into uncorrelated variables called PCs. As such, the PC1 accounts for the largest variance in the data, while additional PCs (PC2, PC3, etc.) account for the next largest variance, with the limitation that each PC is orthogonal (uncorrelated) with preceding PCs. We used the PC1, as it accounts for the largest variability in the data, to represent the overall expression level for NF-κB activation. That is, the NF-κB activation score equals Σwixi, a weighted average expression among the NF-κB–activated genes, where gene i at expression level x is expressed as xi, wi is the corresponding weight (loading coefficient) with Σwi2 = 1, and the wi values maximize the variance of Σwixi. This approach has been used to derive a malignancy pathway gene signature in a breast cancer study (83). PCA provides weighted average expression of signature genes, whereby genes with high expression have correspondingly greater weight (loading coefficient). Genes with highest loading coefficients were identified as the “driver” genes: GBP1, PSMB9, IRF1, TAP1, TNFAIP3, CCL5, PSMB8, IL32, SH2B3, and NFKBIE.
Correlation and survival studies.
Human tumor studies were performed on the microarray data from the Director’s Challenge study (CMCLA), consisting of 442 stage I and stage II adenocarcinoma patient samples from 4 institutions ( https://array.nci.nih.gov/caarray/project/jacob-00182) (62). 159 NF-κB signature probe sets (Supplemental Table 4) were used for this analysis. Additional correlation studies were performed on the GSE14814 data set (n = 133) (72). Association among T cell chemokines, neutrophil chemokines, T cell presence, and NF-κB signature activity was evaluated by Spearman correlation analysis to estimate correlation coefficient values (r). Correlation plot was generated to display the correlation matrix. Kaplan-Meier method was used to generate survival curves, and the log-rank test was used to test survival difference between the low and high expression groups by median cutoff for each gene. The P value was adjusted by false discovery rate for multiple testing as described previously (69, 84).
Study approval
All animal experiments were performed with approval and following the guidelines of the University of South Florida Institutional Animal Care and Use Committee. Human studies were conducted on publically available data sets and did not require institutional approval.
Supplementary Material
Acknowledgments
We acknowledge help provided by Flow Cytometry, Molecular Genomics, Microscopy, and Pathology core facilities at the Moffitt Cancer Center. We also thank Steven Eschrich, Anders Berglund, and Eric Welsh for helpful comments on this manuscript. This work was supported by lung cancer SPORE P50 CA119997, National Functional Genomics Center, and James and Esther King Biomedical Research Program.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(6):2509–2522. doi:10.1172/JCI67250.
References
- 1.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 2.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 3.Downward J. Targeting RAS and PI3K in lung cancer. Nat Med. 2008;14(12):1315–1316. doi: 10.1038/nm1208-1315. [DOI] [PubMed] [Google Scholar]
- 4.Shaw AT, Yasothan U, Kirkpatrick P. Crizotinib. Nat Rev Drug Discov. 2011;10(12):897–898. doi: 10.1038/nrd3600. [DOI] [PubMed] [Google Scholar]
- 5.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pentcheva-Hoang T, Corse E, Allison JP. Negative regulators of T-cell activation: potential targets for therapeutic intervention in cancer, autoimmune disease, and persistent infections. Immunol Rev. 2009;229(1):67–87. doi: 10.1111/j.1600-065X.2009.00763.x. [DOI] [PubMed] [Google Scholar]
- 7.Brahmer JR, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch TJ, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line treatment in stage IIIB/IV non-small-cell lung cancer: results from a randomized, double-blind, multicenter phase II study. J Clin Oncol. 2012;30(17):2046–2054. doi: 10.1200/JCO.2011.38.4032. [DOI] [PubMed] [Google Scholar]
- 9.Weber J. Immunotherapy for melanoma. Curr Opin Oncol. 2011;23(2):163–169. doi: 10.1097/CCO.0b013e3283436e79. [DOI] [PubMed] [Google Scholar]
- 10.Weber J. Immune checkpoint proteins: a new therapeutic paradigm for cancer--preclinical background: CTLA-4 and PD-1 blockade. Semin Oncol. 2010;37(5):430–439. doi: 10.1053/j.seminoncol.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higano CS, et al. Integrated data from 2 randomized, double-blind, placebo-controlled, phase 3 trials of active cellular immunotherapy with sipuleucel-T in advanced prostate cancer. Cancer. 2009;115(16):3670–3679. doi: 10.1002/cncr.24429. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 14.Fridman WH, Galon J, Pages F, Tartour E, Sautes-Fridman C, Kroemer G. Prognostic and predictive impact of intra- and peritumoral immune infiltrates. Cancer Res. 2011;71(17):5601–5605. doi: 10.1158/0008-5472.CAN-11-1316. [DOI] [PubMed] [Google Scholar]
- 15.Pages F, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353(25):2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 19.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 20.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 21.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 22.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meylan E, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 25.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 26.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 27.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 28.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 29.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Beutler B, et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7(10):753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 31.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12(8):715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 33.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 35.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 2011;25(24):2559–2572. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121(7):977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 38.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi H, Ogata H, Nishigaki R, Broide DH, Karin M. Tobacco smoke promotes lung tumorigenesis by triggering IKKbeta- and JNK1-dependent inflammation. Cancer Cell. 2010;17(1):89–97. doi: 10.1016/j.ccr.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basseres DS, Ebbs A, Levantini E, Baldwin AS. Requirement of the NF-kappaB subunit p65/RelA for K-Ras-induced lung tumorigenesis. Cancer Res. 2010;70(9):3537–3546. doi: 10.1158/0008-5472.CAN-09-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue W, et al. Response and resistance to NF-κB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1(3):236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, et al. A single peptide-MHC complex positively selects a diverse and specific CD8 T cell repertoire. Science. 2009;326(5954):871–874. doi: 10.1126/science.1177627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DuPage M, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19(1):72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S, Yagita H, Sayers TJ, Celis E. Optimized combination therapy using bortezomib, TRAIL and TLR agonists in established breast tumors. Cancer Immunol Immunother. 2010;59(7):1073–1081. doi: 10.1007/s00262-010-0834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanton C, Futreal A, Eisen T. Her2-targeted therapies in non-small cell lung cancer. Clin Cancer Res. 2006;12(14 pt 2):4377s–4383s. doi: 10.1158/1078-0432.CCR-06-0115. [DOI] [PubMed] [Google Scholar]
- 47.Assudani D, Cho HI, DeVito N, Bradley N, Celis E. In vivo expansion, persistence, and function of peptide vaccine-induced CD8 T cells occur independently of CD4 T cells. Cancer Res. 2008;68(23):9892–9899. doi: 10.1158/0008-5472.CAN-08-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho HI, Celis E. Optimized peptide vaccines eliciting extensive CD8 T-cell responses with therapeutic antitumor effects. Cancer Res. 2009;69(23):9012–9019. doi: 10.1158/0008-5472.CAN-09-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nava-Parada P, Forni G, Knutson KL, Pease LR, Celis E. Peptide vaccine given with a Toll-like receptor agonist is effective for the treatment and prevention of spontaneous breast tumors. Cancer Res. 2007;67(3):1326–1334. doi: 10.1158/0008-5472.CAN-06-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat Immunol. 2008;9(9):970–980. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- 51.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128(2):369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F, Tang E, Guan K, Wang CY. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol. 2003;170(11):5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
- 53.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 56.Chang JT, et al. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell. 2009;34(1):104–114. doi: 10.1016/j.molcel.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, et al. Lack of essential role of NF-kappa B p50, RelA, and cRel subunits in virus-induced type 1 IFN expression. J Immunol. 2007;178(11):6770–6776. doi: 10.4049/jimmunol.178.11.6770. [DOI] [PubMed] [Google Scholar]
- 58.Valenzuela JO, et al. PKCtheta is required for alloreactivity and GVHD but not for immune responses toward leukemia and infection in mice. J Clin Invest. 2009;119(12):3774–3786. doi: 10.1172/JCI39692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci U S A. 1997;94(19):10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c- IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 61.Harhaj EW, Dixit VM. Deubiquitinases in the regulation of NF-kappaB signaling. Cell Res. 2011;21(1):22–39. doi: 10.1038/cr.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shedden K, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25(6):280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Dejardin E, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17(4):525–535. doi: 10.1016/S1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 65.Gommerman JL, Browning JL. Lymphotoxin/light, lymphoid microenvironments and autoimmune disease. Nat Rev Immunol. 2003;3(8):642–655. doi: 10.1038/nri1151. [DOI] [PubMed] [Google Scholar]
- 66.Wolf MJ, Seleznik GM, Zeller N, Heikenwalder M. The unexpected role of lymphotoxin beta receptor signaling in carcinogenesis: from lymphoid tissue formation to liver and prostate cancer development. Oncogene. 2010;29(36):5006–5018. doi: 10.1038/onc.2010.260. [DOI] [PubMed] [Google Scholar]
- 67.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6(5):447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 68.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150(1):165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen DT, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst. 2011;103(24):1859–1870. doi: 10.1093/jnci/djr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen DT, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119(2):335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen DT, Nasir A, Venkataramu C, Fulp W, Gruidl M, Yeatman T. Evaluation of malignancy-risk gene signature in breast cancer patients. Breast Cancer Res Treat. 2010;120(1):25–34. doi: 10.1007/s10549-009-0357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu CQ, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28(29):4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Le DT, et al. CD8(+) Foxp3(+) tumor infiltrating lymphocytes accumulate in the context of an effective anti-tumor response. International journal of cancer. Int J Cancer. 2011;129(3):636–647. doi: 10.1002/ijc.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muthuswamy R, et al. NF-kappaB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the recruitment of cytolytic T effector cells. Cancer Res. 2012;72(15):3735–3743. doi: 10.1158/0008-5472.CAN-11-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dajee M, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421(6923):639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 76.van Hogerlinden M, Rozell BL, Ahrlund-Richter L, Toftgard R. Squamous cell carcinomas and increased apoptosis in skin with inhibited Rel/nuclear factor-kappaB signaling. Cancer Res. 1999;59(14):3299–3303. [PubMed] [Google Scholar]
- 77.Jing H, et al. Opposing roles of NF-kappaB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 2011;25(20):2137–2146. doi: 10.1101/gad.17620611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji RR, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61(7):1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coppola D, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol. 2011;179(1):37–45. doi: 10.1016/j.ajpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moran CJ, et al. RANTES expression is a predictor of survival in stage I lung adenocarcinoma. Clin Cancer Res. 2002;8(12):3803–3812. [PubMed] [Google Scholar]
- 81.Wang J, et al. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J Immunol. 2007;178(11):6777–6788. doi: 10.4049/jimmunol.178.11.6777. [DOI] [PubMed] [Google Scholar]
- 82.Torabian SZ, et al. Ribozyme-mediated targeting of IkappaBgamma inhibits melanoma invasion and metastasis. Am J Pathol. 2009;174(3):1009–1016. doi: 10.2353/ajpath.2009.080207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen DT, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119(2):335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.