Abstract
The normal flora furnishes the host with ecological barriers that prevent pathogen attack while maintaining tissue homeostasis. Urinary tract infections (UTIs) constitute a highly relevant model of microbial adaptation in which some patients infected with Escherichia coli develop acute pyelonephritis, while other patients with bacteriuria exhibit an asymptomatic carrier state similar to bacterial commensalism. It remains unclear if the lack of destructive inflammation merely reflects low virulence or if carrier strains actively inhibit disease-associated responses in the host. Here, we identify a new mechanism of bacterial adaptation through broad suppression of RNA polymerase II–dependent (Pol II–dependent) host gene expression. Over 60% of all genes were suppressed 24 hours after human inoculation with the prototype asymptomatic bacteriuria (ABU) strain E. coli 83972, and inhibition was verified by infection of human cells. Specific repressors and activators of Pol II–dependent transcription were modified, Pol II phosphorylation was inhibited, and pathogen-specific signaling was suppressed in cell lines and inoculated patients. An increased frequency of strains inhibiting Pol II was epidemiologically verified in ABU and fecal strains compared with acute pyelonephritis, and a Pol II antagonist suppressed the disease-associated host response. These results suggest that by manipulating host gene expression, ABU strains promote tissue integrity while inhibiting pathology. Such bacterial modulation of host gene expression may be essential to sustain asymptomatic bacterial carriage by ensuring that potentially destructive immune activation will not occur.
Introduction
The skin and mucosal surfaces are sites where normal bacterial flora persists without triggering a destructive host response (1–4). Bacterial pathogens, in contrast, gain a short-term advantage at these sites by expressing virulence factors that can dysregulate a range of host signaling pathways (5). Their virulence repertoire includes a diverse array of molecules that trigger specific host cell receptors and signaling pathways or inactivate the host defense, including capsules, complement inactivators (6, 7), and secreted inhibitors of Toll-like receptor (TLR) and MyD88 signaling (8, 9). The failure of asymptomatic carrier strains to trigger disease-associated signaling pathways and pathology has generally been attributed to their lack of virulence (5, 10, 11). It is not clear if, in addition, asymptomatic carrier strains actively enhance their own persistence by modifying the host environment.
Urinary tract infections (UTIs) constitute a highly relevant model of microbial adaptation in which the contrasting effects of pathogens and commensals on host tissues are clearly displayed. While virulent Escherichia coli cause severe, potentially life-threatening disease by breaking the inertia of the mucosal barrier and infecting the kidneys, the most common outcome of bacteriuria is an asymptomatic carrier state resembling commensalism at other mucosal sites (12). Patients with asymptomatic bacteriuria (ABU) may carry the same strain for months or years without developing a destructive host response, and the bacteria co-evolve with their hosts in a niche with little microbial competition (13). The molecular determinants of long-term bacterial persistence and adaptation are still poorly understood, however. Genome sequencing has revealed that at least 50% of ABU strains have evolved from virulent uropathogenic E. coli (UPEC) strains by loss of virulence genes, suggesting a reductive evolution and an active adaptation to the host environment (14). It is unclear whether this abrogated virulence factor expression is sufficient to explain the lack of overt mucosal inflammation in asymptomatic carriers or whether a separate class of host suppressive factors enables ABU strains to modify the host response and promote their own survival.
To address this question, we examined the effects of asymptomatic bacterial carriage on host gene expression. Studying the prototype ABU strain E. coli 83972 in the human inoculation model, we observed marked suppression of host gene expression in patients who became asymptomatic carriers. ABU strains suppressed RNA polymerase II–dependent (Pol II–dependent) transcription in inoculated patients and in human kidney, bladder, and intestinal epithelial cells after in vitro infection. Signaling pathways suppressed by E. coli 83972 included TLR and type 1 IFN in contrast to the uropathogenic E. coli strain CFT073, which activated Pol II transcription and these signaling pathways. We believe that these results suggest for the first time that asymptomatic carrier strains promote their own survival by broadly manipulating host gene expression.
Results
Suppression of gene expression in patients inoculated with E. coli 83972
Therapeutic urinary tract inoculation with the prototype ABU strain E. coli 83972 is a safe alternative approach for patients with therapy-resistant, recurrent UTIs (15–17). The strain establishes persistent bacteriuria, protecting patients against superinfection with more virulent strains (17). Using this protocol, we examined whether the establishment of asymptomatic bacterial carriage alters host gene expression. Patients were inoculated with E. coli 83972 (105 CFU/ml), and in 3 individuals, significant bacteriuria was established after 24 hours (Figure 1A). Prior to inoculation, the urine was sterile and neutrophil numbers and cytokine concentrations in serum and urine were low, contradicting a strong preexisting chronic inflammatory response in the urinary tract or a preceding systemic immune response (Figure 1B).
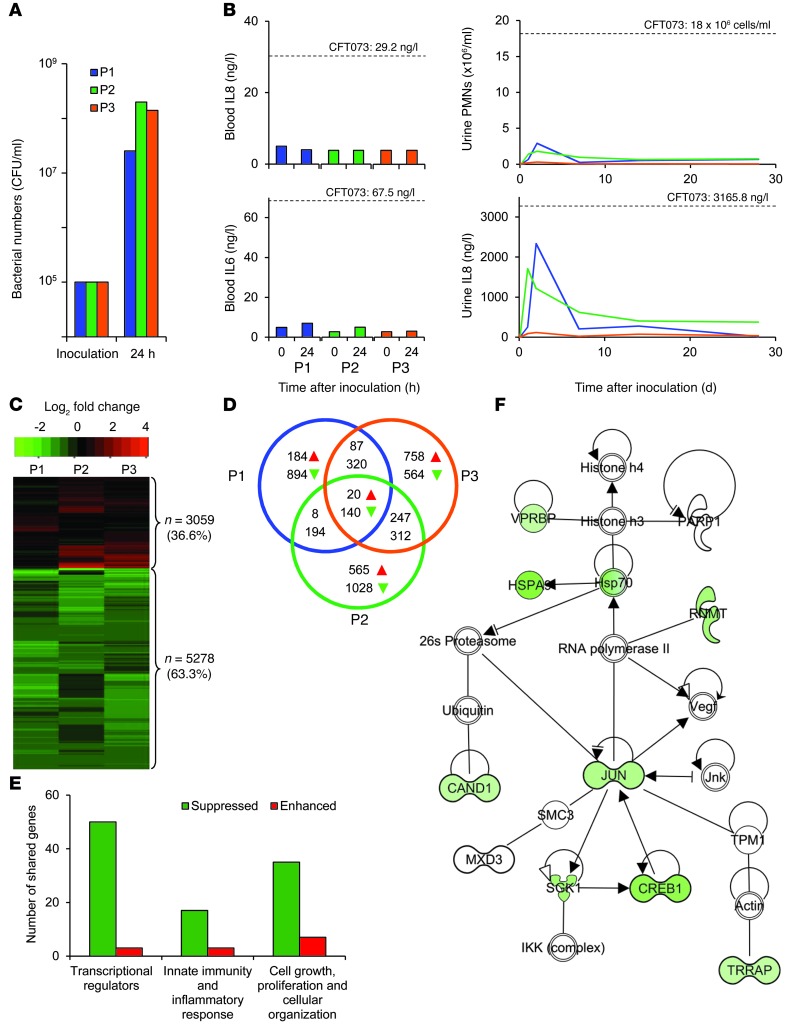
Figure 1. Inhibition of gene expression 24 hours after human inoculation with the ABU strain E. coli 83972.
(A) E. coli 83972 CFUs in urine samples obtained 24 hours after inoculation. (B) Lack of systemic innate immune response after 24 hours. Neutrophil numbers and IL8 concentrations in urine 4 weeks after inoculation. Levels in patients with symptomatic UTIs are indicated for comparison (20). (C) Heatmap illustrating the suppression of gene expression in each patient (green represents suppressed; red represents activated). (D) Venn diagram indicating the number of individually or commonly regulated genes. (E) Main functional categories among the 160 shared regulated genes (individual genes and other functional categories are shown in Supplemental Table 1). (F) IPA-generated network illustrating the in vivo suppression of genes in a Pol II network (for specific genes, see Supplemental Tables 1 and 2). PMNs, polymorphonuclear neutrophils.
The urinary tract response to E. coli 83972 inoculation in a placebo-controlled study was previously examined (18) and low, but detectable, increases in urinary neutrophils and cytokines were observed. These responses were below the levels reported during symptomatic UTIs (19, 20). Here, systemic cytokine activation (IL6 or IL8) was not detected in the inoculated patients (Figure 1B). Urinary neutrophil numbers were transiently elevated in parallel with an IL8 response (Figure 1B).
We examined the effects of the ABU strain on host gene expression using peripheral blood leucocyte RNA isolated before and 24 hours after intravesical inoculation. Substantially altered genes identified by whole genome transcriptomic profiling were sorted according to their relative expression levels (unadjusted P values less than 0.05 and absolute log2 fold change greater than 0.5).
Gene expression was markedly reduced in the 3 patients 24 hours after E. coli 83972 inoculation (63.3%, 5,278 of 8,337 probe sets) (Figure 1C). Shared genes, showing a similar change in expression in the 3 patients were mostly suppressed (87.5%, 140 of 160 probe sets, Venn diagram) (Figure 1D). Regulators of gene expression were most abundant in this group, including transcriptional repressors, transcriptional activators, and regulators of translation, chromatin, or DNA organization (Figure 1E and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI66451DS1). We further analyzed the commonly regulated genes for network interactions. Based on established molecular pathways, we identified a network of genes directly interacting with Pol II, associating Pol II with transcription factors JUN and CREB1 (Figure 1F). In total, we found that 9 of 9 regulated genes in this network were suppressed. For other strongly regulated networks, see Supplemental Table 2.
Global changes in gene expression after in vitro infection of human uroepithelial cells
To address the mechanism of transcriptional inhibition, we infected human kidney epithelial A498 cells with the ABU strain or the virulent CFT073 (APN) strain. This cell line has been validated as a model of uropathogenic E. coli infection; the cells express functional receptors for bacterial virulence ligands, and the response to virulent strains reflects human UTI (21). RNA harvested after 4 hours was subjected to whole genome transcriptomic analysis. Raw data, normalized using cross-correlation (22), were of high quality with a replicate correlation greater than 0.98 and no systematic bias (MA plots, Supplemental Figure 1). Genes with empirical Bayes-adjusted P values less than 0.05 and log2 fold changes greater than 0.585 were considered differentially expressed and were functionally characterized using the Database for Annotation, Visualization and Integrated Discovery (DAVID), Blast AceView, and Ingenuity Pathway Analysis (IPA) software.
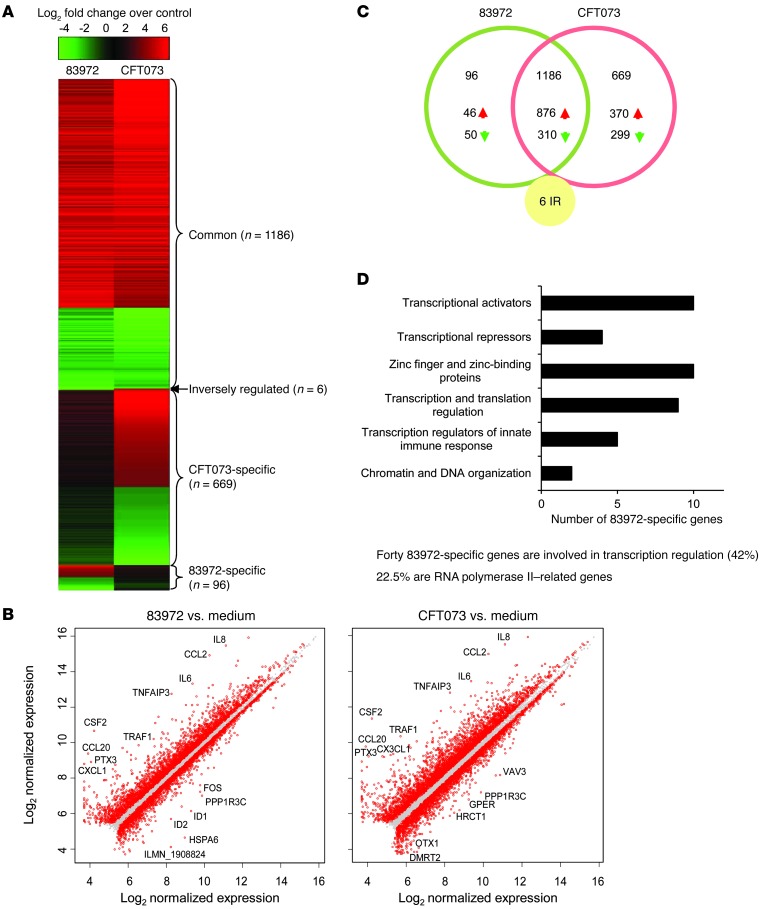
In total, 1,957 cellular genes were substantially altered by the ABU and/or APN strains compared with uninfected cells (Figure 2A). After sorting according to relative expression levels, we identified 4 patterns; 96 genes were only regulated by the ABU strain and were designated as ABU specific (Figure 2D and Supplemental Tables 3 and 4). Six genes were inversely regulated between the ABU and APN strains (Supplemental Table 3 and Supplemental Figure 2), 1,186 genes were similarly regulated by both strains (Supplemental Table 5), and 669 genes were APN specific (Figure 2A). Overall, fewer genes were transcribed in response to the ABU strain compared with the APN strain (1,288 versus 1,861; P < 0.05) (Figure 2, B and C), suggesting either active suppression of gene expression or a lack of activation.
Figure 2. Gene expression in human uroepithelial cells infected with E. coli 83972 or CFT073.
(A) Heatmap of genes sorted according to expression levels. (B) Scatter plot of log2 normalized expression illustrating a higher number of regulated genes after CFT073 stimulation than after 83972 stimulation. (C) Venn diagram specifying the number of 83972-specific, CFT073-specific, or commonly or inversely regulated genes. (D) Main categories of deregulated 83972-specific genes. Transcriptional regulators comprised 42% of the 83972-specific genes, and 22.5% were Pol II related (for 83972-specific genes, see Supplemental Tables 3 and 4).
ABU-specific changes in gene expression
To identify the effects on host gene expression unique to the ABU strain, we sorted the 96 ABU-specific and 6 inversely regulated genes according to their known or suggested functions. Substantially regulated genes involved in transcriptional regulation are listed in Supplemental Table 3 according to fold change, and the most regulated gene categories are indicated in Figure 2D. These categories correspond to those found among the shared regulated genes in the patients (Figure 1F). The regulators of gene expression were most abundant, including transcriptional repressors, transcriptional activators, zinc finger proteins, regulators of translation, and transcriptional regulators of the innate immune response, confirming the in vivo observations (Figure 2D). Genes directly involved in Pol II transcription or in regulating Pol II–dependent pathways were strongly affected by the ABU strain (22.5% of the ABU-specific genes) (Figure 2D and Supplemental Table 3). Other regulated gene categories include innate immune receptors like NOD1, TLR5, and TNFRSF10D, as well as other innate immunity and inflammatory response genes, all of which were suppressed (Supplemental Table 4). IPA identified 1 network formed by the inversely regulated genes (Supplemental Figure 2), including FOSB, HSPA6, RN7SK, RGS4, and IFIT1. Three of these genes regulate Pol II through TATA box–binding protein (TBP) (23–25). We also identified networks involved in infection and immunity (Supplemental Figure 3). The transcriptomic profile and IPA did not identify oxidative stress or ER stress response pathways in infected cells.
Effects on RNA Pol II–dependent transcription
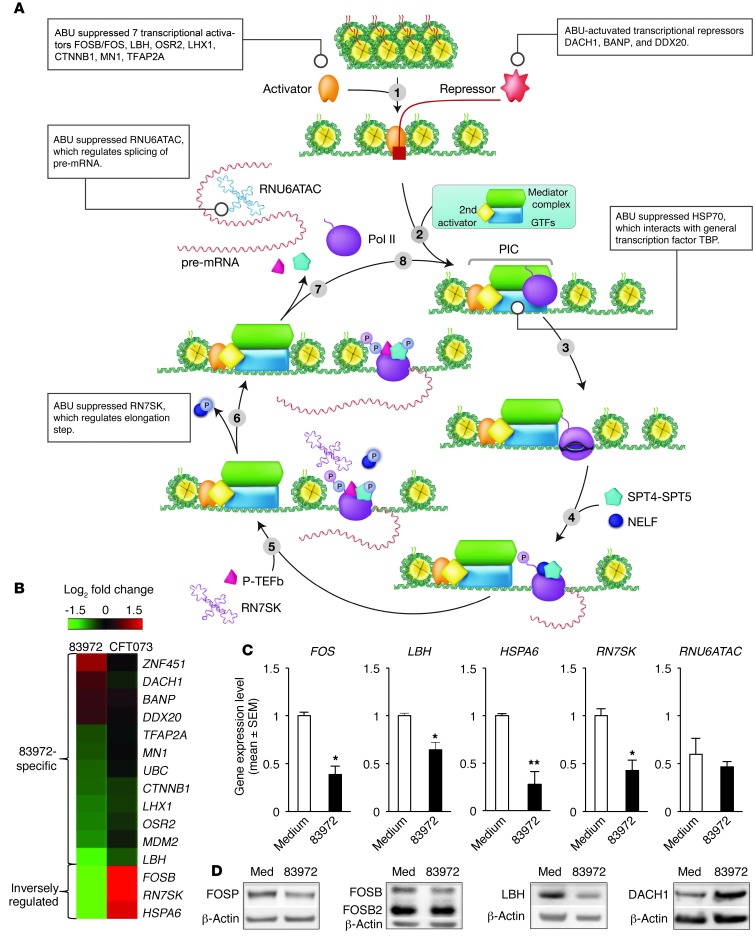
RNA Pol II controls eukaryotic gene expression through mRNA precursors, most small nuclear RNAs (snRNAs), and microRNAs (miRNAs) (26). The effects of the ABU strain on critical steps involved in the Pol II transcription cycle are shown in Figure 3A, and molecules whose expression was modified in an ABU-specific manner are listed in Supplemental Table 3 according to their role in Pol II–dependent gene expression (see legend, Figure 3A). In addition, genes inversely regulated by APN and ABU were included. A heatmap of genes modified by the ABU strain is shown in Figure 3B, and the corresponding effects of infection with the APN strain are shown for comparison.
Figure 3. Suppression of RNA Pol II–dependent transcription by E. coli 83972.
(A) Overview of the Pol II–dependent transcription cycle, with genes modified by E. coli 83972 indicated in boxes adjacent to each step (modified from Fuda et al., ref. 38). For log2 fold changes of individual genes, see Supplemental Table 3. Step 1: Chromatin opening and binding of the Pol II transcriptional complex to DNA is modified by specific activators or repressors. E. coli 83972 suppressed 6 transcriptional activators and activated 3 repressors. Step 2: Preinitiation complex formation. HSP70 interacting with GTFs was inhibited. Genes involved in steps 3 (initiation of transcription), 4 (promoter escape/clearance), and 5 (escape from pausing) were not affected. Step 6: Message elongation. Suppressed snRNA RN7SK, which binds to the transcription elongation factor P-TEFb. Step 7: Termination. Suppressed snRNA RNU6ATAC, involved in pre-mRNA splicing. See also Supplemental Figure 2 and Supplemental Table 3. (B) Heatmap of 83972-specific genes interacting with Pol II and regulating transcription. (C) Verification of the transcriptomic analysis. RT-PCR confirmed a reduction in transcript levels for genes in the Pol II cycle (FOS, LBH, HSPA6, RN7SK, and RNU6ATAC) in A498 cells infected with E. coli 83972. Data represent the means ± SEM. *P < 0.05; **P < 0.01. (D) Western blot analysis confirming suppression by E. coli 83972. Reduced levels of FOS, FOSB, LBH, and increased levels of DACH1 were detected.
Transcriptional activators and repressors (step 1).
The ABU strain suppressed 7 activators of Pol II–dependent transcription (FOSB, LBH, OSR2, LHX1, CTNNB1, MN1, and TFAP2A) (Figure 3A and Supplemental Table 3). The FOS transcription factor family has pleiotropic cellular effects. LBH recruits Pol II to specific promoter sites and may regulate FOS through the serum-responsive element (SRE) in the FOS promoter (27). OSR2 regulates genes involved in proliferation and development (28). LHX1 is highly expressed in the urinary tract and is a potential transcription factor. CTNNB1 is a pleiotropic regulator that recruits Pol II to promoters for cell growth and adhesion, and recruits the transcriptional activators LBH (through WNT3A), MN1, SOX4, and BRIX1, which are necessary for ribosome biogenesis (29). MN1 is a transcriptional coregulator of genes induced by retinoic acid (30). TFAP2A belongs to the transcription factor AP2 superfamily, which is inhibited by viral infection (31).
Remarkably, we found that the ABU strain also activated several repressors of Pol II–dependent transcription, including DACH1, BANP, and DDX20, which prevent Pol II from accessing multiple promoters through local or global chromatin alterations. DACH1 regulates FOS by binding SRE in the FOS promoter (32), potentially providing a mechanism for suppression of the innate immune response by the ABU strain (Supplemental Table 3). BANP represses CCND1 transcription and promotes TP53 Ser15 phosphorylation, causing cell cycle arrest (33). DDX20 interacts directly with the RNA Pol II complex (34), associates with histone deacetylases, and enhances NFKB suppression by miRNAs (35).
Pol II complex formation and initiation of transcription (steps 2 and 3).
The ABU strain suppressed 5 genes recognizing TBP (MDM2, CTNNB1, FOSB, RN7SK, and HSPA6). In contrast, the APN strain increased the expression of HSPA6, a member of the HSP70 family that has been suggested to facilitate transcription when TBP is inhibited (23). In addition, the APN strain upregulated HSPA9, another member of the HSP70 family that interacts with GTF2A1 (a TFIIA component) (24) and also upregulated 2 genes that interact with TFIIB (NR4A2 and GMNN, a repressor of transcription factor binding). The APN strain stimulated the expression of 3 mediator complex components (MED10, MED13, MED15) but the ABU strain had only a weak effect on MED13 (log fold change 0.65).
Genes regulating preinitiation complex release and productive elongation (steps 4–6).
The ABU strain suppressed the snRNA RN7SK, a regulator of the Pol II transcription cycle that reversibly sequesters transcription elongation factor b (P-TEFb) into a large kinase-inactive complex (25). As RN7SK also directly interacts with TBP, this snRNA has great potential as a broad transcriptional regulator in ABU. RNU6ATAC, which regulates splicing of pre-mRNA (36), was the most strongly suppressed gene in ABU-stimulated cells (log fold change –4.14). In addition, the ABU strain inhibited the transcription of RNA28S5, which is Pol I dependent. The ABU strain increased the expression of PDCD4, which inhibits the helicase activity of translation initiation factors EIF4A and EIF4F and blocks their binding to RNA (37).
RT-PCR and Western blot validation
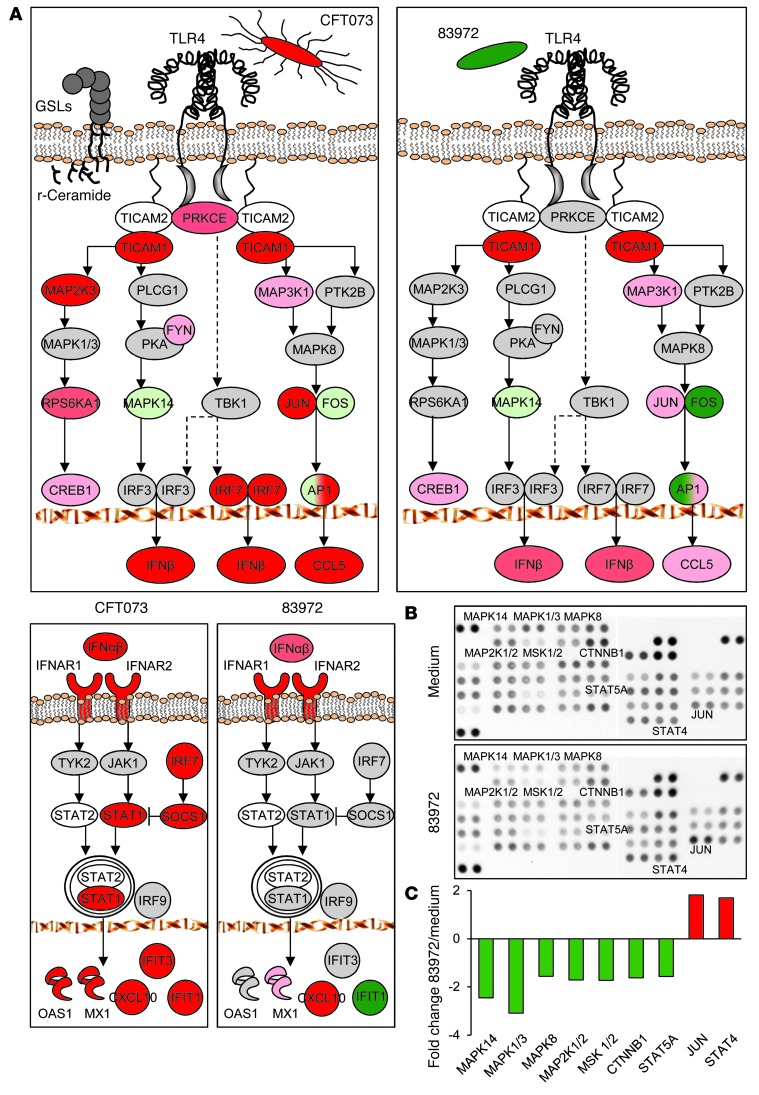
We confirmed the effects of the ABU strain on gene expression by RT-PCR of mRNA and snRNA obtained after stimulation of A498 cells (Figure 3C). FOS and LBH expression was reduced compared with uninfected cells, as was heat shock protein HSPA6 and snRNAs RN7SK and RNU6ATAC. Western blot analysis (Figure 3D) detected a reduction in FOS and LBH proteins, whereas the transcriptional repressor DACH1 was increased, consistent with the gene expression data. These genes were also suppressed in the patients (for a comparison with the in vitro data, see Supplemental Figure 4A). mRNA levels for genes involved in Pol II regulation and transcription were perturbed, including LBH and FOS, as well as transcription factors IFIT1, EGR1, and ID2, which were suppressed, and DACH1, which was increased (Supplemental Figure 4, A and B).
Inhibition of Pol II Ser2 phosphorylation by the ABU strain
Productive elongation of the mRNA requires Pol II phosphorylation (38). Carboxy-terminal domain (CTD) repeats on Ser5 are phosphorylated by the kinase subunit of the general transcription factor TFIIH, followed by the phosphorylation of Pol II CTD repeats on Ser2 by the kinase complex positive transcription elongation factor b (P-TEFb). Pol II hyperphosphorylation by TFIIH is necessary for breaking Pol II contacts with some promoter-bound factors and for its release from the core promoter region.
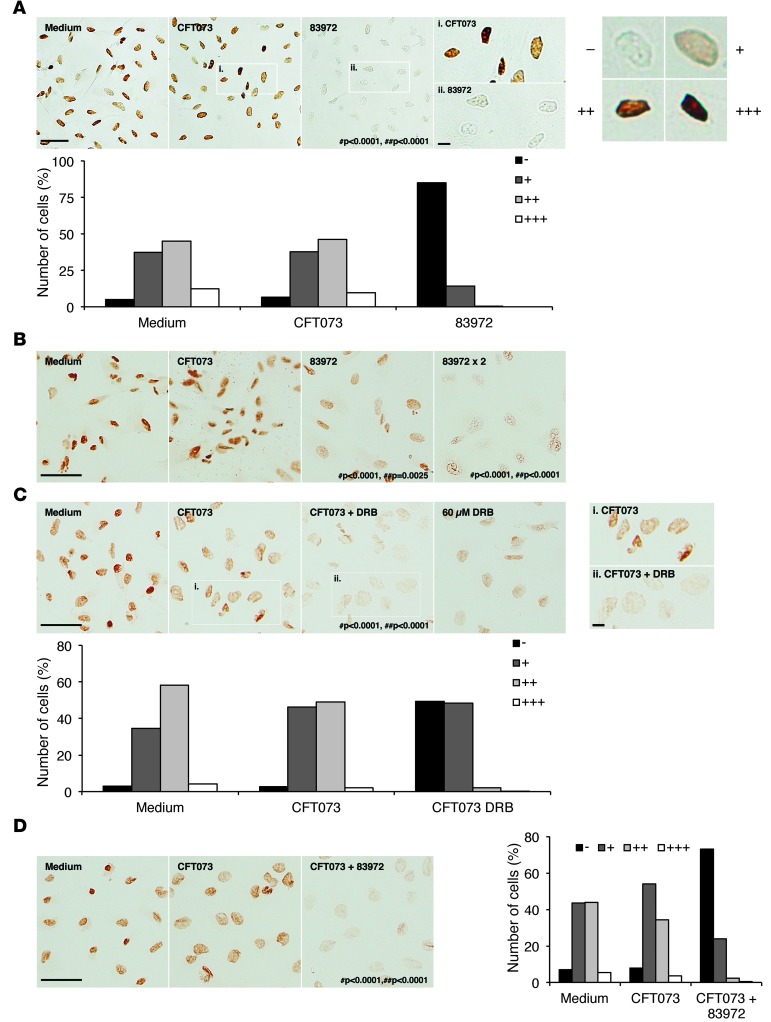
We observed changes in Pol II phosphorylation after infection with the ABU strain (for 4 hours) (Figure 4). We infected the A498 human kidney epithelial cell line or pediatric kidney cells in primary culture with the ABU strain, and detected phosphorylated Pol II by immuno-peroxidase staining using anti–phospho Ser2 antibodies. Cells infected with the APN strain were included as controls (Figure 4, A and B). For cell morphology, see Supplemental Figure 5A. A marked reduction in nuclear phosphorylated Pol II staining was detected in A498 cells infected with the ABU strain compared with uninfected cells (Figure 4A). The frequency of cells with negative staining (–) increased from 5% to 85% (P < 0.0001, χ2 test for independence). This effect was confirmed by Western blot analysis of phosphorylated Pol II in whole-cell extracts (Supplemental Figure 5B). Four additional ABU strains inhibited Pol II phosphorylation (Supplemental Figure 5C), but infection with the APN strain had no significant effect (P = 0.742) (Figure 4A).
Figure 4. Suppression of Pol II phosphorylation by E. coli 83972.
(A) Active Ser2 phosphorylated Pol II was stained using monoclonal primary human phospho CTD Ser2 antibodies and peroxidase-labeled secondary antibodies (brown). E. coli 83972 markedly suppressed Pol II phosphorylation compared with uninfected or CFT073-infected human kidney cells (A498). E. coli CFT073 had a nonsuppressive effect. (B) Inhibition by E. coli 83972 of Pol II phosphorylation in primary human kidney cells (HRTEC) compared with uninfected or CFT073-infected cells. (C) The specific Pol II inhibitor DRB (60 μM) abrogated Pol II phosphorylation in response to E. coli CFT073 compared with A498 cells without an inhibitor. For dose-dependent inhibition of Pol II phosphorylation in uninfected A498 cells, see Supplemental Figure 7A. (D) Competition between E. coli CFT073 and 83972 in A498 cells. E. coli 83972 inhibited Pol II phosphorylation in the presence of E. coli CFT073. (A and C) Scale bars: 50 μm and 10 μm (insets). #P = compared with medium control; ##P = compared with CFT073-infected cells; χ2 test for independence.
We observed a parallel reduction in Pol II phosphorylation in primary human renal tubular cells. A dose-dependent reduction in Ser2 phosphorylation was detected in ABU-infected cells (P < 0.0001 compared with uninfected control cells) (Figure 4B). Furthermore, in the human bladder cell lines J82 and HTB-9, we observed a reduction in phosphorylation after exposure to the ABU strain (Supplemental Figure 6).
Effects of Pol II inhibition on the cellular response to infection
To specifically address whether Pol II inhibition alters the response to infection, we pretreated human kidney cells with 5,6-dichloro-1-b-D-ribofuranosylbenzimidazole (DRB). This adenosine analog has been proposed to specifically and reversibly inhibit Pol II transcription without directly affecting other cellular functions (39). We quantified the dose-dependent effect of DRB as the reduction in Ser2 staining in A498 cells (Supplemental Figure 7A). At these concentrations, no reduction in cell viability was observed (Supplemental Figure 8A). Pol II phosphorylation in APN-infected cells was specific (Supplemental Figure 8B) and was reduced by DRB (60 μM; P < 0.0001) (Figure 4C). We observed a similar reduction in Pol II phosphorylation in cells cochallenged with the ABU and APN strains compared with the APN strain alone, suggesting that the ABU strain inhibits the response to the APN strain (P < 0.0001) (Figure 4D).
We confirmed the broad suppression of gene expression by transcriptomic analysis in DRB-treated (60 μM) human kidney cells (Supplemental Figure 7, B and C) and found that the effect of the ABU strain was more restricted (1,577 versus 155 inhibited genes; absolute fold change greater than 2). Genes regulated by both DRB and the ABU strain included FOS, LBH, DACH1, and RNU6ATAC, as defined by RT-PCR. We found that HSPA6 was suppressed only by the ABU strain, and RN7SK was not regulated in this experiment (Supplemental Figure 7D). Several studies have proposed that DRB targets casein kinase II and that inhibition of casein kinase II may block Pol II carboxy-terminal domain phosphorylation (39–41). Furthermore, casein kinase II inhibits CREB1 and ATF1, which regulate the MAPK1 pathway (42, 43). These effects are consistent with the profile of suppression by the ABU strain, but the molecular basis for Pol II inhibition and the narrower spectrum of inhibited genes compared with DRB remain to be identified.
Inhibition of innate immune signaling defined by pathway analysis and phosphorylation arrays
Uropathogenic E. coli activate a pathogen-specific, TLR4/CREB/IRF3/7-dependent innate immune response, which is crucial for host resistance to acute pyelonephritis (9, 44). P fimbriae bind to glycosphingolipids, and ceramide release activates TLR4 signaling (44). Irf3–/– mice develop acute pyelonephritis with urosepsis, followed by tissue damage within 1 week of infection (9). Uropathogens also stimulate IFNβ expression and infected Ifnb mice develop kidney pathology, suggesting that type 1 IFNs are essential for a functional antibacterial defense in the kidneys (9).
These pathways were less strongly activated by the ABU strain than by the APN strain, as shown by transcriptomic analysis of infected A498 kidney cells (at 4 hours) (Figure 5A and Supplemental Figure 9) TLR4, MAPK14, FOS, JUN, and CCL5. In addition, IL1B expression was reduced in ABU-infected cells, and MAP2K3, MAP3K1, PRKCE, and IRF7 were not activated. IFNB was less strongly induced by the ABU strain, and IFIT1, downstream of IFNB, was suppressed. Importantly, the ABU strain still activated the NFKB-dependent pathway and TNF, IL6, and IL8 responses to the ABU and APN strains were similar in vitro (Supplemental Figure 10).
Figure 5. Pathogen-specific signaling in infected A498 cells.
(A) Suppression by E. coli 83972 (right panels) of genes in the TLR4/CREB/IRF3/7 and type 1 IFN pathways. For log2 fold changes of individual genes, see Supplemental Table 3. Activation by E. coli CFT073 is shown for comparison. Increased expression (red), reduced expression (green), and no change (gray). Color intensity reflects the fold change. (B) Phosphorylation of molecules in the TLR4/CREB/IRF3/7 signaling pathway examined by phosphoarray analysis of whole-cell extracts from E. coli 83972-infected A498 cells. (C) The ABU strain suppressed the phosphorylation of 7 targets compared with uninfected cells. Quantification based on pixel intensities of phosphorylated targets. Cutoff is set at a fold change less than –1.5 or greater than 1.5.
The ABU strain also reduced the phosphorylation of signaling molecules downstream of TLR4 (MAP2K1/2 and MAPK1, as well as MAPK14 and MAPK8), as determined by phosphoarray analysis of whole-cell extracts of ABU-infected cells (Figure 5, B and C). A corresponding increase in phosphorylation in response to virulent strains has been reported (9). MAPKs, and especially MAPK14, were previously shown to regulate the phosphorylation of CREB1 and the nuclear translocation of IRF3 (9), and MAPKs regulate AP1 formation from FOS and JUN.
Long-term suppression of pathogen-specific signaling in inoculated patients
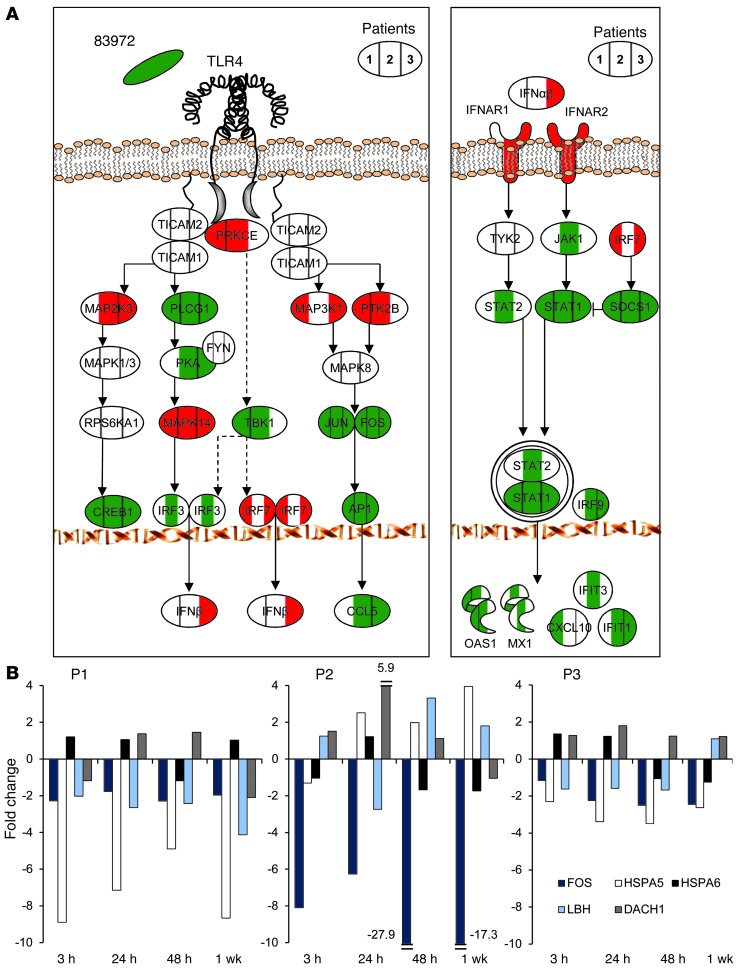
We also detected the inhibition of TLR4/CREB/IRF3/7 and type 1 IFN signaling in the inoculated patients (Figure 6, Supplemental Figure 11, and Supplemental Table 1). After 24 hours, both pathways were suppressed compared with the preinoculation samples in each patient (reduced JUN, FOS, CREB, and PLCG1 expression) (Figure 6A). Type I IFN pathway genes were either suppressed or unchanged, including SOCS1, STAT1-, and IFN-stimulated genes (IFIT1, IFIT3, OAS1, MX1, CXCL10) (Figure 6A).
Figure 6. Suppression by E. coli 83972 of pathogen-specific signaling in inoculated patients.
(A) Inhibition in inoculated patients of the TLR4/CREB/IRF3/7 pathway and suppression of type I IFN signaling. Each symbol is divided into 3 parts and colored according to individual gene expression levels in P1, P2, and P3. Log2 fold change greater than 0.4 (red); log2 fold change less than –0.4 (green); and no change (white). For log fold change of individual genes, see Supplemental Table 1. (B) Transcriptomic analysis of RNA samples obtained from each patient after 3, 24, 48 hours, and 7 days. Pol II cycle–related genes remained suppressed for at least 1 week after inoculation. DACH1 expression was increased, except after 1 week in P1 and P2.
To address whether the ABU strain continued to suppress host gene expression during in vivo carriage, we obtained repeated RNA samples from each patient (at 3, 24, 48 hours and 7 days) (Figure 6B). Suppression of Pol II cycle–related genes was detected up to 1 week after inoculation (FOS, HSPA5, functional analogs of HSPA6 and LBH). DACH1 expression was increased at all time points except after the first week in patient no. 1 (P1) and patient no. 2 (P2). The results show that the ABU strain actively suppresses essential regulators of gene expression and pathogen-specific innate immune signaling and that the suppressive effect is maintained in inoculated patients for at least 1 week.
Frequency of Pol II suppression among E. coli strains from APN, ABU, or the fecal flora
We examined Pol II suppression in an epidemiologically defined collection of ABU isolates (n = 75) from which E. coli 83972 was derived (10, 45, 46). APN strains (n = 88) were obtained from children with febrile UTIs and fecal strains (n = 46) from children in the same area who had no history of UTI and negative urine cultures at screening.
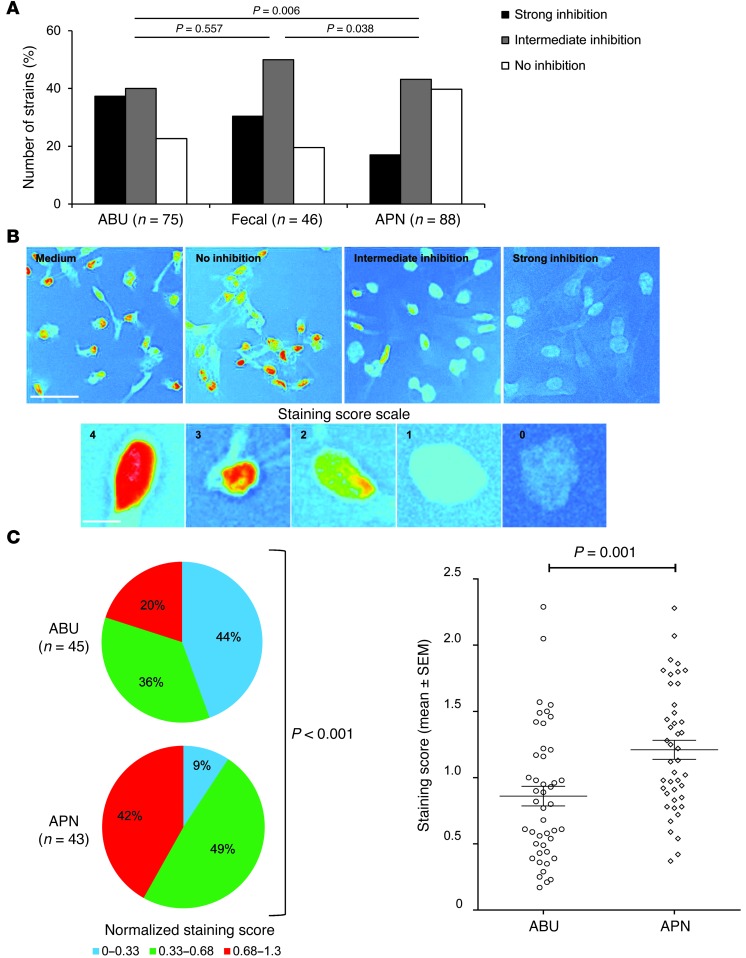
We infected human kidney cells with each strain, and the inhibition of Pol II phosphorylation was graded according to the overall staining intensity as strong, intermediate, or negative relative to uninfected control cells (Figure 7, A–C). Seventy-seven percent of the ABU strains inhibited phosphorylation and 37% were strongly inhibitory compared with 17% for the strongly inhibitory APN strains (P = 0.006, X2 test for independence) (Figure 7A). The fecal strains resembled the ABU group, but with the majority of the strains being intermediately inhibitory (P = 0.038 compared with the APN group).
Figure 7. Epidemiologic analysis of Pol II suppression.
(A) Higher frequency of Pol II suppression in A498 cells infected with ABU or fecal strains than with APN strains. (B) Light microscopic images of Pol II phosphorylation at Ser2 in A498 cells, analyzed by ImageJ, using changed lookup tables and inverted colors. Scale bar: 50 μm (upper panel). Each strain was assigned a staining score based on the scale and is illustrated in the lower panel (scale bar: 10 μm). (C) Strongly inhibitory strains were more abundant in the ABU (44%) than in the APN group (9%, P < 0.001), and the staining score was lower (P = 0.001).
To further characterize the difference in Pol II phosphorylation, we quantified the staining in a subset of the ABU (n = 45) or APN (n = 43) strains (Figure 7B). The Pol II phosphorylation score was substantially lower in the ABU group than in the APN group (P < 0.001, χ2 test for independence) (Figure 7C). The results suggest that ABU strains inhibit Pol II activation more often than virulent strains and that this property is shared with certain E. coli strains from the intestinal flora.
To address whether the inhibitory effect might be relevant in the intestinal environment, we exposed 2 colon cancer cell lines (HT29 and DLD-1) to E. coli 83972 and stained for Pol II phosphorylation. The strain inhibited phosphorylation in both cell lines (Supplemental Figure 12, A and B). The cells were subsequently exposed to E. coli strains from the fecal flora, one defined as inhibitory and one as noninhibitory in the epidemiologic survey based on uroepithelial cells (Figure 7A and Supplemental Figure 12C). The pattern of inhibition was reproduced in the colonic epithelial cells. In addition, we exposed the 2 bladder epithelial cell lines, J82 and HTB-9, to two acute cystitis strains, one with inhibitory activity in A498 human kidney cells and one lacking such activity. The pattern of Pol II inhibition resembled that observed in A498 cells (Supplemental Figure 6, C and D).
Discussion
As pathogens express new tissue attack mechanisms, the host defense retaliates by increasing the complexity of innate and acquired immunity. While this positive and competitive mode of evolution is well established, we believe this to be a new mechanism that shifts the balance in favor of commensals or symbionts and away from disease. We propose that certain nonvirulent bacteria may act as guardians of the host environment by controlling the quality and quantity of host gene expression. Such suppressive strains may add essential negotiating power to the host, aiming to ensure that constant, potentially destructive immune activation will not occur. The inhibitory effect retained specificity, however, as only certain facets of the host response repertoire were perturbed. For example, the suppressive effect of the ABU strain did not include all innate immune signaling pathways, since type 1 IFN signaling was inhibited, but NFKB was not. Through this new mechanism of transcriptional modulation, bacteria may establish a host-beneficial, pathology-averse symbiotic state.
DNA is transcribed by RNA polymerases, and the large RNA Pol II complex is essential for protein expression (38). For specificity, the assembly of Pol II is tightly controlled, and the efficiency of transcription is modified at individual promoters by activators and repressors, general or specific transcription factors, and mRNA-modifying steps (47, 48). Approximately 10% of all expressed genes have been shown to be involved in transcriptional regulation, and the assembly and control of the Pol II complex and transcription cycle have been extensively characterized (38). Targeting of the host Pol II transcription machinery by bacteria has, to our knowledge, not been investigated, however. We show here that E. coli strains from asymptomatic carriers directly inhibit Pol II–dependent transcription in vitro in human kidney cells and in vivo during asymptomatic bacterial carriage in the human urinary tract. A marked reduction in overall gene expression was achieved by the combined stimulation of critical Pol II transcriptional repressors and by suppression of transcriptional activators and transcription factors. Further network analysis showed that critical innate immune response pathways were suppressed, especially the pathology-generating pathways activated by uropathogenic E. coli. The majority of ABU strains from a population-based screen shared the ability to suppress Pol II activation, as did many E. coli strains from the fecal flora, suggesting relevance of this mechanism for asymptomatic carriage in the urinary and gastrointestinal tracts. Suppressive activity was also seen among the APN strains, though less frequently. The expression by the APN strains of virulence factors that activate the host response might also provide a mechanism to counter suppression of Pol II.
Normal gut flora has mainly been studied as a positive force in tissue homeostasis and immune maturation (2, 3, 49). Bacterial colonization stimulates the formation of Peyer’s patches and mucosal lymphocyte maturation in germ-free mice (50), and symbiotic flora regulates postnatal gut development and host physiology, including the degradation of otherwise indigestible plant polysaccharides and dietary oxalates, the biotransformation of conjugated bile acids, or vitamin synthesis (51). Bacteroides infection modulated the expression of genes involved in nutrient absorption, sodium transport, host lipid absorption, metabolism, and postnatal immune maturation (4). Commensals were also proposed to enhance influenza virus immunity in the lung (52). Here, we add a mechanism for broad modulation of the host environment by bacteria, as the ABU strain suppressed key transcription factors involved in innate immunity. Selectivity was indicated by the inactivation of key signaling pathways generating mortality and pathology, and by the activation of these pathways by the virulent APN strain under the same experimental conditions (9). By suppressing host gene expression, the asymptomatic carrier strains may succeed in tethering the host defense, thereby generating a well-balanced, rather than an overreactive, immune environment.
These findings may appear paradoxical, as immune avoidance is an established survival strategy for pathogens rather than commensals. However, commensals have also been proposed to suppress the innate immune response. Using gene chip analysis, a reduction in IFN-α signaling was detected in specific pathogen-free (SPF) mice compared with germ-free mice (53). Intestinal colonization with bifidobacteria restored oral tolerance in neonatal mice, and an intact flora reduced allergy-type hypersensitivity reactions to food allergens (54). The normal flora has also been proposed to suppress immune activation through NFKβ-dependent pathways (49). Deleterious effects of TNF or IFNγ were reversed by “probiotics’’ (55), and some commensal bacteria inhibited TLR4 signaling specifically by elevating PPARγ expression and uncoupling NFKβ-dependent target genes in a negative feedback loop (56). Recently, normal gut microbiota were shown to modulate brain development by reducing motor neuron excitability and anxiety in SPF mice (57), suggesting effects on tissues distant from the site of colonization. The inhibition of Pol II–dependent gene expression observed in the present study may provide a mechanistic framework to further explain some of the previously observed inhibitory effects.
Interestingly, a number of fecal strains inhibited Pol II phosphorylation, raising the question about the relevance of transcriptional regulation for the commensal flora in the large intestine. Prior to causing UTI, uropathogenic E. coli reside in the large intestine, which serves as a reservoir for such strains, and virulence factors associated with UTI facilitate their persistence and spread to the urinary tract (58). ABU strains are also successful colonizers of the gut, but lack the adaptive advantages of the virulent strains and so far, their molecular adaptation strategies have not been addressed. This study implicates Pol II regulation as a mechanism of adaptation also in the gut environment. The ABU strain was shown to suppress Pol II phosphorylation in 2 human colonic epithelial cell lines, as did a strain from the fecal flora previously shown to be inhibitory for urinary tract cells. Further in vivo studies are required to evaluate the contribution of this mechanism in the intestinal environment, however.
It may appear paradoxical that suppression of innate immunity by ABU strains would benefit a host, who frequently encounters uropathogenic E. coli. Innate immunity is essential for the antibacterial defense of the urinary tract and exaggerated or dysregulated innate immunity causes symptoms and tissue damage. The ABU strain did not entirely stop transcription, however, but reduced the magnitude of gene expression. Based on human genetic variation affecting innate immunity, we have previously identified several common polymorphisms that reduce the level of gene expression rather than inactivate the gene. For example, TLR4 promoter polymorphisms suppressing TLR4 expression have been identified in patients with ABU, suggesting that being a low responder is advantageous during mucosal infection. The present study adds active suppression of innate immunity by ABU bacteria as a mechanism of “taming’’ the host environment rather than eliminating the defense entirely. Control of gene expression and the mucosal environment might thus be achieved by the combined effect of host genetics and bacteria that actively modify the host transcriptional machinery.
Methods
Bacterial strains and stimulation of human epithelial cells.
The prototype ABU or APN strains E. coli 83972 (OR:K5:H–) (13, 15–17) and E. coli CFT073 (O6:K2:H1) (59) were cultured on tryptic soy agar (16 hours, 37°C), harvested in PBS (pH 7.2, 1010 CFU/ml), and diluted as appropriate. Human kidney carcinoma (A498, ATCC HTB44), human colon cancer (HT29, DLD-1, ATCC), and bladder carcinoma cells (J82, ATCC HTB-1 and ATCC HTB-9) were cultured in RPMI-1640 supplemented with 1 mM sodium pyruvate, 1 mM nonessential amino acids, gentamycin (50 μg/ml–1), and 5% FCS (PAA). The A498 cell line is an established model to study UTI pathogenesis (21). Primary human renal tubular epithelial cells (9), provided by Diana Karpman (Department of Pediatrics, Lund University, Lund, Sweden), were cultured in supplemented DMEM/F12 1:1 (HyClone, Thermo Scientific) with 15% FCS. Confluent cells in 6-well plates (Thermo Fisher Scientific) were exposed to bacteria in fresh, serum-free supplemented RPMI (109 or 2 × 109 CFU/ml).
Human therapeutic inoculation.
The protocol for therapeutic bladder inoculation of patients with E. coli 83972 (15–17) has been used by investigators in Sweden and the US (15–17). Briefly, after antibiotic treatment to remove prior infection, patients are inoculated with E. coli 83972 through a catheter (30 ml, 105 CFU/ml in saline). Blood and urine samples were obtained before and repeatedly after inoculation. Viable counts in urine were determined, and IL6 and IL8 concentrations were quantified by Immulite (Siemens).
Global gene expression.
Total RNA was extracted from kidney cells with Trizol (Invitrogen) followed by RNeasy Cleanup (QIAGEN). RNA was reverse-transcribed and converted to biotin-labeled cRNA using a TargetAmp Nano-g Biotin-aRNA Labeling Kit (EPICENTRE Biotechnologies), hybridized onto an Illumina Human HT-12 Expression Beadchip for 16 hours at 58°C, then washed, stained (Illumina Wash Protocol), and scanned (BeadArray Scanner 500GX). Patient RNA was extracted from PBLs before and at various times after inoculation with E. coli 83972 using a QIAamp RNA Blood Mini Kit (QIAGEN) and was then amplified and hybridized onto Affymetrix Human Genome U219 arrays (AROS Applied Biotechnology).
Illumina data were preprocessed and normalized using cross-correlation (22). Normalized human kidney cell data were of high quality, with a replicate correlation greater than 0.98 and no systematic bias (microarray plots, Supplemental Figure 1). Affymetrix data were normalized using robust multi-array average (RMA) implemented in the free software packages R and Bioconductor ( http://www.r-project.org). Differentially expressed genes were functionally categorized using DAVID (60) and IPA software (Ingenuity Systems). The microarray data are available in the NCBI’s Gene Expression Omnibus database (accession number GSE43887).
Real-time PCR.
Total RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) and quantified in real time using QuantiTect SYBR Green–based PCR Kits for mRNA (QIAGEN) or QiantiFast Probe Assay (QIAGEN) for snRNA on a RotorGene Q cycler (QIAGEN). Quantitative RT-PCR (qRT-PCR) reactions were run in biological duplicates and triplicates, and gene expression was done based on a standard curve method.
Western blot analysis.
Cell lysates from A498 cells (RIPA lysis buffer supplemented with protease and phosphatase inhibitors; Roche Diagnostics) were separated by SDS-PAGE (4%–12% Bis-Tris gels; Invitrogen), and blotted PVDF membranes were incubated with antibodies against RNA polymerase II subunit B1 (phospho CTD Ser2), clone 3E10 (Millipore), or antibodies against FOSB (Cell Signaling Technology), cFOS-P (Abcam), LBH (Biosite), and DACH1 (Abcam), followed by the appropriate anti–IgG-HRP with ECL Plus Western Blotting Reagent (GE Healthcare). Restore Western Blot Stripping Buffer (Pierce Biotechnology) was used as indicated. Band intensities were quantified using ImageJ software 1.44p (NIH).
Pol II staining and microscopy.
Infected and control cells cultured on Nunc Lab-Tek 8-well chamber glass slides (Thermo Fisher Scientific) (2.5 × 104 cells per well) were infected for 4 hours, washed, permeabilized (0.25% Triton X 100), and stained (rat anti-human polymerase II Ser2 and goat anti–rat-HRP antibodies, Millipore; DAB kit, Invitrogen). Endogenous peroxidase was quenched (0.3 % H2O2 in PBS). Duplicates were counterstained with H&E. The cells were sequentially dehydrated with alcohol and xylene, mounted (Histomount; Invitrogen) and inspected by Microphot-FX Microscopy (LRI Instrument AB). The Pol II inhibitor 5,6-dichlorobenzimidazole DRB (Sigma-Aldrich) was first solubilized in DMSO and subsequently diluted in cell culture media to a final concentration of 60 μM. DMSO itself did not affect Pol II staining (Supplemental Figure 8B).
Phosphoarray.
Protein phosphorylation was quantified using the Human Phospho-Kinase Array Kit (R&D Systems). Protein extracts were prepared from 100% confluent A498 cells infected with 109 CFU/ml of E. coli 83972 with untreated cells as controls. Nitrocellulose membranes were exposed to 170 μg of protein extract, blocked, incubated with a detection antibody cocktail and streptavidin-HRP according to the manufacturer’s instructions. The signals were detected using the ECL Plus Western Blotting Detection System (GE Healthcare).
Statistics.
For gene expression analysis, genes with empirical Bayes-adjusted P values less than 0.05 and log2 fold changes greater than 0.585 were considered differentially expressed and were functionally characterized using DAVID, Blast AceView, and IPA. An unpaired 2-tailed Student’s t test for homoscedastic variances was used to evaluate RT-PCR data and staining score values. The χ2 test for independence was used to compare Pol II phosphorylation (Figures 4 and 7) with InStat software, version 3.06 (GraphPad). In all cases, P values less than 0.05 were considered significant.
Study approval.
The human inoculation study was approved by the human ethics committee at Lund University, and patients provided signed informed consent forms (Dnr 298/2006).
Supplementary Material
Acknowledgments
We thank Wen Huang, Henry Yang, and Fredrik Alsin for experimental help. We gratefully acknowledge the support of the Swedish Medical Research Council, Medical Faculty (Lund University); the Swedish Cancer Society, Söderberg and Österlund; and the Sharon D. Lund, Anna-Lisa, and Sven-Erik Lundgren, Maggie Stephens, Inga-Britt and Arne Lundberg, and HJ Forssman Foundations; and the Royal Physiographic Society and Network of Excellence: EuroPathoGenomics.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(6):2366–2379. doi:10.1172/JCI66451.
Jingyao Zhang’s present address is: Genome Institute, Agency for Science, Technology, and Research (A*STAR), BIOPOLIS, Singapore.
See the related Attending physician beginning on page 2348.
References
- 1.Pamer EG. Immune responses to commensal and environmental microbes. Nat Immunol. 2007;8(11):1173–1178. doi: 10.1038/ni1526. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5(7):e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rautava S, Walker WA. Commensal bacteria and epithelial cross talk in the developing intestine. Curr Gastroenterol Rep. 2007;9(5):385–392. doi: 10.1007/s11894-007-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 5.Ragnarsdottir B, Lutay N, Gronberg-Hernandez J, Koves B, Svanborg C. Genetics of innate immunity and UTI susceptibility. Nat Rev Urol. 2011;8(8):449–468. doi: 10.1038/nrurol.2011.100. [DOI] [PubMed] [Google Scholar]
- 6.Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: learning from staphylococci and meningococci. Nat Rev Microbiol. 2010;8(6):393–399. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- 7.Oelschlaeger TA, Dobrindt U, Hacker J. Virulence factors of uropathogens. Curr Opin Urol. 2002;12(1):33–38. doi: 10.1097/00042307-200201000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Cirl C, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14(4):399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 9.Fischer H, et al. Pathogen specific, IRF3-dependent signaling and innate resistance to human kidney infection. PLoS Pathog. 2010;6(9):e1001109. doi: 10.1371/journal.ppat.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svanborg-Edén C, Hanson L, Jodal U, Lindberg U, Sohl-Åkelund A. Variable adherence to normal urinary tract epithelial cells of Escherichia coli strains associated with various forms of urinary tract infection. . Lancet. 1976;1(7984):490–492. [PubMed] [Google Scholar]
- 11.Bergsten G, Wullt B, Svanborg C. Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int J Med Microbiol. 2005;295(6–7):487–502. doi: 10.1016/j.ijmm.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Kunin C. Urinary Tract Infections: Detection, Prevention and Management. Baltimore, Maryland, USA: Williams and Wilkins; 1997. [Google Scholar]
- 13.Zdziarski J, et al. Host imprints on bacterial genomes — rapid, divergent evolution in individual patients. PLoS Pathog. 2010;6(8):e1001078. doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect Immun. 2008;76(2):695–703. doi: 10.1128/IAI.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agace WW, Hedges SR, Ceska M, Svanborg C. Interleukin-8 and the neutrophil response to mucosal gram-negative infection. J Clin Invest. 1993;92(2):780–785. doi: 10.1172/JCI116650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wullt B, et al. P fimbriae enhance the early establishment of Escherichia coli in the human urinary tract. Mol Microbiol. 2000;38(3):456–464. doi: 10.1046/j.1365-2958.2000.02165.x. [DOI] [PubMed] [Google Scholar]
- 17.Sunden F, Hakansson L, Ljunggren E, Wullt B. Escherichia coli 83972 bacteriuria protects against recurrent lower urinary tract infections in patients with incomplete bladder emptying. J Urol. 2010;184(1):179–185. doi: 10.1016/j.juro.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez JG, Sunden F, Connolly J, Svanborg C, Wullt B. Genetic control of the variable innate immune response to asymptomatic bacteriuria. PLoS One. 2011;6(11):e28289. doi: 10.1371/journal.pone.0028289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson SH, Lu Y, Brauner A. Soluble interleukin-6 receptor, interleukin-10 and granulocyte colony-stimulating factor in acute pyelonephritis: relationship to markers of bacterial virulence and renal function. Nephron. 1998;80(4):401–407. doi: 10.1159/000045211. [DOI] [PubMed] [Google Scholar]
- 20.Sheu JN, et al. Serum and urine levels of interleukin-6 and interleukin-8 in children with acute pyelonephritis. Cytokine. 2006;36(5–6):276–282. doi: 10.1016/j.cyto.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Hedlund M, Svensson M, Nilsson A, Duan RD, Svanborg C. Role of the ceramide-signaling pathway in cytokine responses to P-fimbriated Escherichia coli. J Exp Med. 1996;183(3):1037–1044. doi: 10.1084/jem.183.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chua SW, Vijayakumar P, Nissom PM, Yam CY, Wong VV, Yang H. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 2006;34(5):e38. doi: 10.1093/nar/gkl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffar G, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15(1):95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 24.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol. 2006;26(2):630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ai J, et al. A human homolog of mouse Lbh gene, hLBH, expresses in heart and activates SRE and AP-1 mediated MAPK signaling pathway. Mol Biol Rep. 2008;35(2):179–187. doi: 10.1007/s11033-007-9068-4. [DOI] [PubMed] [Google Scholar]
- 28.Kawai S, Abiko Y, Amano A. Odd-skipped related 2 regulates genes related to proliferation and development. Biochem Biophys Res Commun. 2010;398(2):184–190. doi: 10.1016/j.bbrc.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto-Sato E, et al. A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS One. 2010;5(2):e9289. doi: 10.1371/journal.pone.0009289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Wely KH, et al. The MN1 oncoprotein synergizes with coactivators RAC3 and p300 in RAR-RXR-mediated transcription. Oncogene. 2003;22(5):699–709. doi: 10.1038/sj.onc.1206124. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell PJ, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 32.Wu K, et al. Cell fate determination factor DACH1 inhibits c-Jun-induced contact-independent growth. Mol Biol Cell. 2007;18(3):755–767. doi: 10.1091/mbc.E06-09-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol Cell Biol. 2005;25(19):8415–8429. doi: 10.1128/MCB.25.19.8415-8429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152(1):75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takata A, et al. A miRNA machinery component DDX20 controls NF-kappaB via microRNA-140 function. Biochem Biophys Res Commun. 2012;420(3):564–569. doi: 10.1016/j.bbrc.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 36.Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273(5283):1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 37.Yang HS, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23(1):26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandomeni RO. Kinetics of inhibition by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole on calf thymus casein kinase II. Biochem J. 1989;262(2):469–473. doi: 10.1042/bj2620469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palancade B, Dubois MF, Bensaude O. FCP1 phosphorylation by casein kinase 2 enhances binding to TFIIF and RNA polymerase II carboxyl-terminal domain phosphatase activity. J Biol Chem. 2002;277(39):36061–36067. doi: 10.1074/jbc.M205192200. [DOI] [PubMed] [Google Scholar]
- 41.Yankulov K, Yamashita K, Roy R, Egly JM, Bentley DL. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270(41):23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 42.Shanware NP, Trinh AT, Williams LM, Tibbetts RS. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J Biol Chem. 2007;282(9):6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- 43.Gupta P, Prywes R. ATF1 phosphorylation by the ERK MAPK pathway is required for epidermal growth factor-induced c-jun expression. J Biol Chem. 2002;277(52):50550–50556. doi: 10.1074/jbc.M209799200. [DOI] [PubMed] [Google Scholar]
- 44.Fischer H, Ellström P, Ekström K, Gustafsson L, Gustafsson M, Svanborg C. Ceramide as a TLR4 agonist; a putative signalling intermediate between sphingolipid receptors for microbial ligands and TLR4. Cell Microbiol. 2007;9(5):1239–1251. doi: 10.1111/j.1462-5822.2006.00867.x. [DOI] [PubMed] [Google Scholar]
- 45.Lindberg U, Claesson I, Hanson LA, Jodal U. Asymptomatic bacteriuria in schoolgirls. VIII. Clinical course during a 3-year follow-up. J Pediatr. 1978;92(2):194–199. doi: 10.1016/s0022-3476(78)80003-1. [DOI] [PubMed] [Google Scholar]
- 46.Salvador E, et al. Asymptomatic bacteriuria E. coli isolates from healthy individuals versus hospital patients: long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun. 2012;80(2):668–678. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21(2):225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 49.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker WA. Mechanisms of action of probiotics. Clin Infect Dis. 2008;46(suppl 2):S87–S91. doi: 10.1086/523335. [DOI] [PubMed] [Google Scholar]
- 51.Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munakata K, et al. Importance of the interferon-alpha system in murine large intestine indicated by microarray analysis of commensal bacteria-induced immunological changes. BMC Genomics. 2008;9:192. doi: 10.1186/1471-2164-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159(4):1739–1745. [PubMed] [Google Scholar]
- 55.Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-α- and IFN-γ-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130(3):731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Dubuquoy L, et al. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology. 2003;124(5):1265–1276. doi: 10.1016/S0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 57.Heijtz RD, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108(7):3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plos K, et al. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. J Infect Dis. 1995;171(3):625–631. doi: 10.1093/infdis/171.3.625. [DOI] [PubMed] [Google Scholar]
- 59.Nielubowicz GR, Mobley HL. Host-pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7(8):430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 60.Huang da W, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.