Abstract
Current strategies to suppress graft-versus-host disease (GVHD) also compromise graft-versus-tumor (GVT) responses. Furthermore, most experimental strategies to separate GVHD and GVT responses merely spare GVT function without actually enhancing it. We have previously shown that endogenously expressed TNF-related apoptosis-inducing ligand (TRAIL) is required for optimal GVT activity against certain malignancies in recipients of allogeneic hematopoietic stem cell transplantation (allo-HSCT). In order to model a donor-derived cellular therapy, we genetically engineered T cells to overexpress TRAIL and adoptively transferred donor-type unsorted TRAIL+ T cells into mouse models of allo-HSCT. We found that murine TRAIL+ T cells induced apoptosis of alloreactive T cells, thereby reducing GVHD in a DR5-dependent manner. Furthermore, murine TRAIL+ T cells mediated enhanced in vitro and in vivo antilymphoma GVT response. Moreover, human TRAIL+ T cells mediated enhanced in vitro cytotoxicity against both human leukemia cell lines and against freshly isolated chronic lymphocytic leukemia (CLL) cells. Finally, as a model of off-the-shelf, donor-unrestricted antitumor cellular therapy, in vitro–generated TRAIL+ precursor T cells from third-party donors also mediated enhanced GVT response in the absence of GVHD. These data indicate that TRAIL-overexpressing donor T cells could potentially enhance the curative potential of allo-HSCT by increasing GVT response and suppressing GVHD.
Introduction
While the safety of clinical allogeneic hematopoietic stem cell transplantation (allo-HSCT) has improved significantly in recent years, its success is limited by disease relapse and graft-versus-host-disease (GVHD) (1). Both allo-HSCT and a variety of immunotherapeutic strategies have demonstrated that T lymphocytes can exert potent antitumor activity. Most genetic engineering strategies have involved directing T cell specificity toward tumor-associated antigens using chimeric antigen receptors (2, 3) or transgenic T cell receptors (TCRs) (4). These strategies, while promising, are limited by requirements for clearly defined tumor-associated antigens or epitopes. They may have risks in the context of allo-HSCT, potentially by exacerbating GVHD (5) or by producing the mispairing of TCRs, leading to neoreactivity (6). In contrast, currently used strategies to prevent GVHD almost uniformly impair T cell function, with deleterious effects on graft-versus-tumor (GVT) response.
Among the major cytolytic molecules, TNF-related apoptosis-inducing ligand (TRAIL) can induce apoptotic signals in target cells expressing TRAIL receptors, which in humans include death receptor (DR) 4 and 5 molecules, and in mice include only DR5. Expression of DR5 is higher in certain tumors (7, 8); furthermore, DR5 expression by tumor cells can be induced by treatment with small molecules like proteasome inhibitors (9, 10), rendering them susceptible to TRAIL-mediated killing. We have previously demonstrated that endogenous TRAIL expression in alloreactive T cells is an important mediator of GVT effects (11). TRAIL is thus an attractive candidate for genetic engineering of donor T cells to enhance their antitumor potential. Importantly, in the setting of allo-HSCT, TRAIL does not appear to mediate GVHD lethality, although we found that TRAIL can contribute to thymic GVHD (11, 12). Here, we present our studies of the effects of genetically overexpressing TRAIL in allogeneic T cells transferred to murine bone marrow transplantation (BMT) recipients. We found that these engineered T cells indeed mediated enhanced GVT activity. However, to our surprise, these TRAIL+ T cells also ameliorated GVHD through the suppression of alloreactive T cells.
Results
TRAIL+ T cells mediate strong GVT effects.
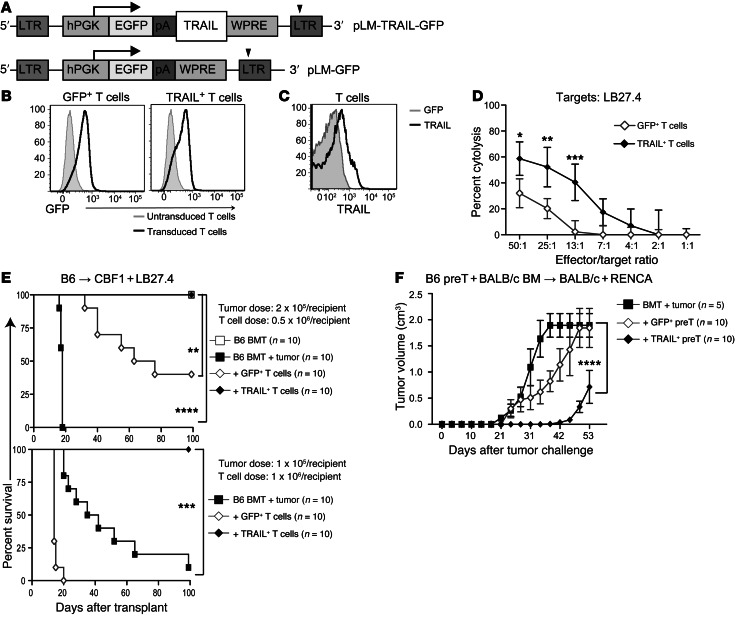
To assess the effect of constitutive TRAIL expression on donor T cells, we constructed the lentiviral vectors pLM-TRAIL-GFP to express murine TRAIL with a GFP reporter and, as a control, pLM-GFP (Figure 1A). T cells transduced with these vectors are termed TRAIL+ T cells and GFP+ T cells, respectively. We determined high transduction efficiencies measured by GFP with both vectors (Figure 1B) and also confirmed that murine T cells transduced with our pLM-TRAIL-GFP vector had increased expression of TRAIL compared with cells transduced with control vector (Figure 1C). Expression of TRAIL or GFP did not affect the expression of other cytolytic molecules, such as perforin, granzyme, or FasL (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI66301DS1).
Figure 1. TRAIL+ T cells are strong antitumor agents.
(A) Representation of pLM-TRAIL-GFP construct: pLM-GFP-2A-TRAIL. (B) Prestimulated B6-derived T cells were transduced and transduction was measured by the expression of GFP. (C) TRAIL overexpression on transduced T cells was determined by flow cytometry. (D) TRAIL+ T cells mediate stronger killing against labeled LB27.4 targets in a 51Cr release cytolysis assay. Graphs representing 3 independent experiments are shown. (E) Lethally irradiated CBF1 recipients were reconstituted with 5 × 106 cells per recipient of WT B6 TCD BM and inoculated with 2.5 × 105 cells per recipient (upper panel) or 1 × 105 cells per recipient of LB27.4 lymphoma cells (lower panel). Designated groups were treated with 0.5 × 106 cells per recipient (upper panel) or 1 × 106 cells per recipient (lower panel) of GFP+ or TRAIL+ T cells. (F) Transduced allogeneic GFP+ or TRAIL+ pre–T cells adoptively transferred into a syngeneic BMT model. RENCA tumor cells were inoculated s.c. 2 weeks after BMT. Tumor volume is expressed in centimeters cubed measured as 1/2 × length × (width)2. Pooled data from 2 independent experiments are depicted. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. hPGK, human phosphoglycerate kinase promoter; pA, “self-cleaving” 2A peptides; WPRE, woodchuck hepatitis posttranscriptional regulatory element.
We next sought to determine the effect of TRAIL overexpression in T cells on antitumor activity. In vitro, unsorted TRAIL+ T cells mediated significantly stronger cytotoxicity against LB27.4 lymphoma targets compared with GFP+ T cells (Figure 1D). We also evaluated TRAIL+ T cell activity in vivo using the haploidentical B6→CBF1 model inoculated with 2 or 1 × 105 cells per recipient of LB27.4 lymphoma cells. We found that TRAIL+ T cells (at 0.5 or 1 × 106 cells per recipient) mediated strong antitumor activity, with 100% survival of mice inoculated with lymphoma, while mice that received control GFP+ T cells succumbed to lymphoma and GVHD (Figure 1E and Supplemental Figure 1B). Interestingly, mice that received TRAIL+ T cells had tumor-free survival and developed minimal signs of GVHD (Supplemental Figure 1C).
We next examined the effects of TRAIL overexpression in a clinically relevant model in the absence GVHD by transferring ex vivo–generated allogeneic precursor T cells (pre–T cells) in a syngeneic transplant. We previously reported that adoptively transferred pre–T cells, generated ex vivo using the OP9-DL1 coculture system, undergo maturation and selection in the recipient thymus and mediate antitumor effects across MHC barriers without alloreactivity (13). We adoptively cotransferred GFP+ and TRAIL+ B6 pre–T cells into syngeneic BMT recipients (BALB/c→BALB/c), where they comparably reconstituted the recipient thymus (Supplemental Figure 2A) and spleen at days 14 and 28 (Supplemental Figure 2, B and C). After subcutaneous challenge with a renal cell carcinoma cell line (RENCA), syngeneic (BALB/c→BALB/c) BMT recipients of TRAIL+ B6 pre–T cells had significantly reduced tumor burden, with delayed tumor growth compared with bone marrow (BM) alone and GFP+ B6 pre-T cell–treated tumor recipients (Figure 1F). We did not observe GVHD in any of the pre–T cell experiments. We conclude that TRAIL overexpression can enhance the antitumor activity of both mature and pre–T cells in BMT recipients in allogeneic and syngeneic settings.
TRAIL+ T cells suppress GVHD.
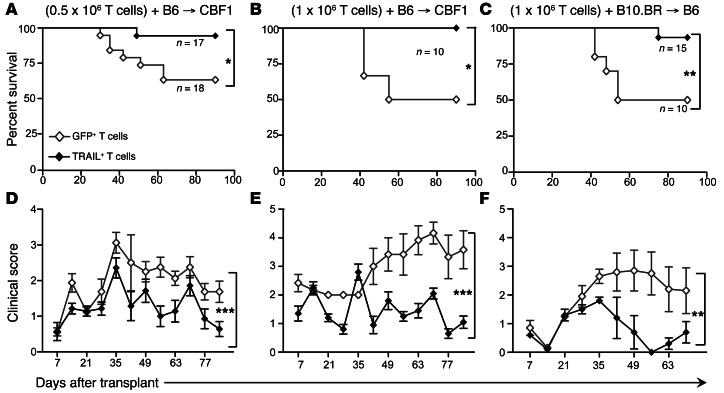
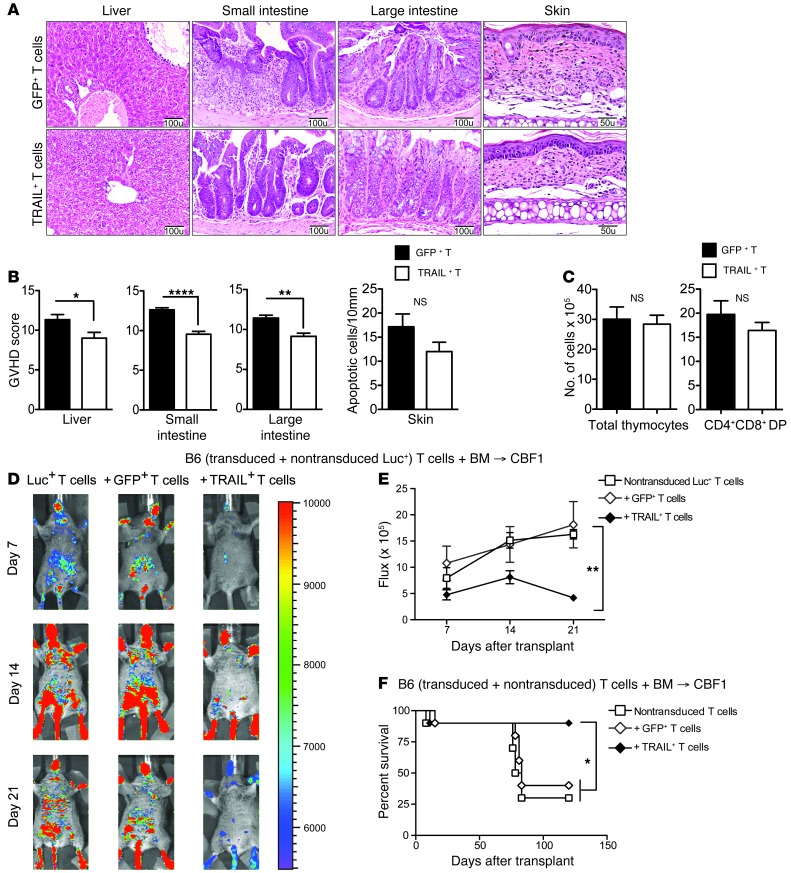
Having observed that TRAIL+ T cells produce a surprisingly mild degree of GVHD, we further investigated this in experiments with transplanted mice in the B6→CBF1 model in the absence of tumor. We again found that mice receiving TRAIL+ T cells had significantly reduced GVHD mortality at T cell doses of 0.5 × 106 (Figure 2A) and 1 × 106 (Figure 2B). Corresponding with improved survival, recipients of TRAIL+ T cells also showed significantly reduced GVHD-associated weight loss (Supplemental Figure 3, A and B) and lower clinical GVHD scores (Figure 2, D and E). We then evaluated TRAIL+ T cells in an additional MHC-disparate BMT model (B10.BR→B6) and again found that TRAIL+ T cells similarly protected BMT recipients from GVHD mortality and morbidity (Figure 2, C and F). Histologically, we found that recipients of TRAIL+ T cells had significantly less GVHD in target organs, including liver and small and large intestines (Figure 3, A and B). Skin GVHD, measured by apoptotic scores, was not significantly increased in allo-BMT recipients of TRAIL+ T cells. Thymic GVHD manifests as decreased thymic cellularity and reduced numbers of CD4+CD8+ double-positive (DP) T cells (12). Thymic GVHD was also not significantly increased in recipients of TRAIL+ T cells compared with GFP+ T cells as shown by statistically similar cellularity and DP T cell numbers (Figure 3C). Overall, these findings in mice demonstrate that overexpression of TRAIL by donor T cells diminishes GVHD morbidity, reduces organ damage in the liver and intestines, and improves survival in clinically relevant BMT models.
Figure 2. Adoptive transfer of TRAIL+ T cells does not cause lethal GVHD.
(A, B, D, and E) Lethally irradiated CBF1 recipients were reconstituted with 5 × 106 cells per recipient of WT B6 TCD BM. Designated groups were treated with (A and D) 0.5 × 106 cells per recipient or (B and E) 1 × 106 cells per recipient of B6-derived GFP+ or TRAIL+ T cells. Survival was monitored and clinical GVHD scores were recorded weekly in a blinded fashion. (C and F) Lethally irradiated B6 recipients were reconstituted with 5 × 106 cells per recipient of WT B10.BR TCD BM. Designated groups were treated with 1 × 106 cells per recipient of B10.BR GFP+ or TRAIL+ T cells. Survival was monitored (A–C) and clinical GVHD scores were recorded weekly (D–F) in a blinded fashion. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 3. TRAIL+ T cells suppress GVHD.
(A and B) Livers and small and large intestines from B6→CBF1 mice treated with GFP+ or TRAIL+ T cells were harvested on day 14 after BMT and skin was harvested on day 21 and scored for GVHD pathology. Representative micrographs are shown (original magnification, ×200 for liver, small and large intestines; and ×400 for skin). GVHD scores pooled from 2 independent experiments are shown (n = 8–10 per group). (C) Thymi from B6→CBF1 mice treated with GFP+ or TRAIL+ T cells were harvested on day 21 after BMT. Total cellularity was obtained from counts of live thymocyte suspension and numbers of DP T cells were derived from flow cytometric determination of CD4+CD8+ T cell proportions. Data pooled from 2 independent experiments are shown (n = 8–10 per group). (D and E) Lethally irradiated CBF1 recipients were reconstituted with 5 × 106 cells per recipient of B6 TCD BM. Designated groups were treated with 0.5 × 106 cells per recipient of GFP+ or TRAIL+ T cells mixed with nontransduced Luc+ T cells. Bioluminescence imaging of the transplanted mice was performed weekly (D) and flux was measured (E). Animals representative of 1 experiment (n = 7 per group) and flux pooled from 3 independent experiments are shown. (F) Lethally irradiated CBF1 recipients were reconstituted with 5 × 106 cells per recipient of B6 TCD BM. Designated groups were treated with 0.5 × 106 cells per recipient of GFP+ or TRAIL+ T cells mixed with nontransduced T cells (n = 10 per group). Survival was monitored daily. *P < 0.05; **P < 0.01; ****P < 0.0001. NS, not significant.
Recent studies have indicated that the endogenous induction of TRAIL has suppressive effects on other T cells (14, 15). To investigate whether TRAIL+ T cells can suppress alloreactive T cells, we adoptively transferred luciferase-expressing (Luc+) nontransduced T cells at a 1:1 ratio with either GFP+ or TRAIL+ T cells (all B6 background) into CBF1 allo-BMT recipients and measured the expansion of nontransduced Luc+ T cells after transfer. We found that Luc+ T cells underwent alloreactive expansion when cotransferred with GFP+ T cells, which was significantly attenuated upon transfer with TRAIL+ T cells (Figure 3, D and E). In addition, CBF1 recipients of B6 BM were adoptively transferred with (a) nontransduced T cells, (b) nontransduced T cells and GFP+ T cells, (c) nontransduced T cells and TRAIL+ T cells. Recipients of TRAIL+ T cells had significantly improved survival compared with either those that received nontransduced T cells cotransferred with GFP+ T cells or nontransduced T cells alone (Figure 3F). These findings suggest that TRAIL+ T cells suppress GVHD by limiting alloreactive T cell expansion.
We next examined whether TRAIL-mediated suppression of alloreactive T cell expansion can inhibit antiviral responses. We assessed viral burden after lymphocytic choriomeningitis virus (LCMV) infection in CBF1 recipients of allografts treated with GFP+ or TRAIL+ T cells (Supplemental Figure 3C). Recipients of GFP+ or TRAIL+ T cells had statistically similar viral burden, which was significantly lower than the viral load of allo-BMT recipients without adoptive transfer of T cells. These results indicate that TRAIL+ T cells do not significantly impair antiviral responses.
TRAIL receptors are induced on host APCs following radiation.
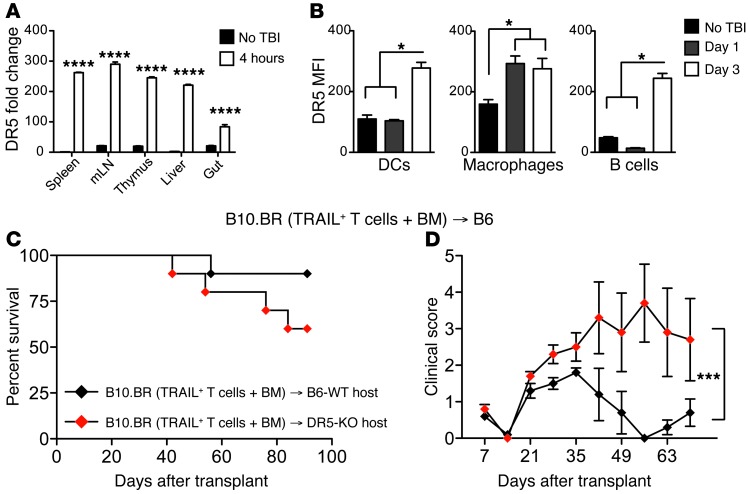
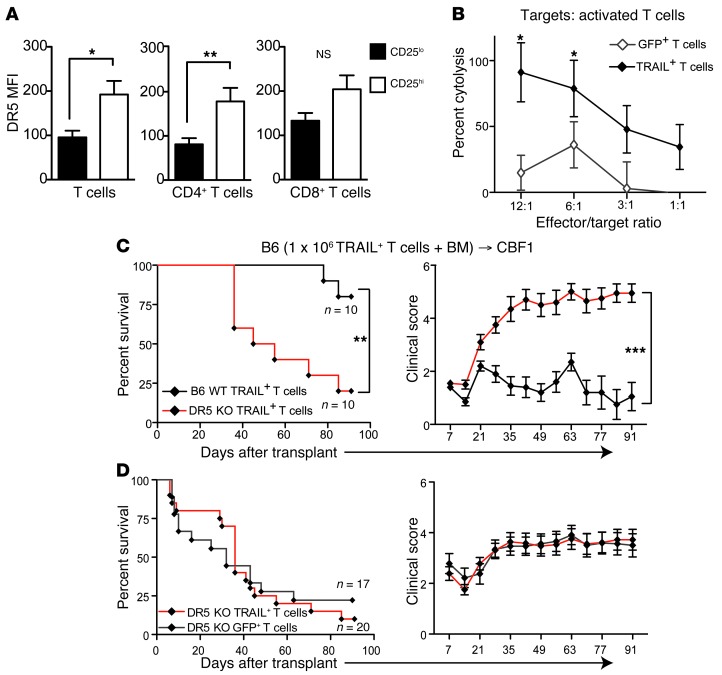
To explore the potential mechanisms by which TRAIL+ T cells suppress the expansion of alloreactive T cells, we first examined the effects of TRAIL+ T cells on host APCs. In mice, TRAIL exerts cytolysis through its receptor, DR5, which has been reported to be upregulated after irradiation (16). We indeed found increased expression of DR5 transcripts in lymphoid organs, liver, and intestine within 4 hours after irradiation (Figure 4A) and significantly increased DR5 cell surface expression by splenic APCs, including DCs, macrophages, and B cells within 1 to 4 days after irradiation (Figure 4B). To assess whether negative regulation by TRAIL+ T cells interacting with DR5+ host APCs contributed to the suppression of GVHD, we performed experiments with B10.BR TRAIL+ T cells in B6 DR5 KO or B6 WT recipients (Figure 4, C and D). DR5 KO recipients had significantly increased GVHD morbidity and somewhat greater GVHD mortality, although this did not reach statistical significance. Taken together, our results demonstrate that DR5 expression is upregulated in host tissues following irradiation, and that host DR5 expression plays a role in the suppressive effect of TRAIL+ T cells on GVHD.
Figure 4. TRAIL+ T cells can eliminate residual host APCs.
(A) Tissues from lethally irradiated CBF1 mice were harvested at designated time points and qPCR was performed for DR5. (B) DR5 was assessed by flow cytometry on splenocytes gated on CD11c+ DCs, CD11b+ macrophages, and B220+ B cells. Representative data from 2 independent experiments are shown (n = 3 per group). *P < 0.05. (C and D) Lethally irradiated WT or DR5 KO B6 recipients were reconstituted with 5 × 106 cells per recipient of WT B10.BR TCD BM. Designated groups were treated with 1 × 106 cells per recipient of B10.BR TRAIL+ T cells. Survival was monitored (C) and clinical GVHD scores were recorded weekly (D) in a blinded fashion. Graph representative of 2 independent experiments is shown. *P < 0.05; ***P < 0.001; ****P < 0.0001. mLN, mesenteric lymph node.
Fratricidal TRAIL+ T cells suppress GVHD.
We next investigated whether TRAIL+ T cells suppressed GVHD by directly inhibiting other alloreactive donor T cells. Since naive human T cells express DR5 on their surface following activation (17), we assessed DR5 expression on alloactivated murine T cells. Four days after adoptive transfer into lethally irradiated CBF1 recipients, the CD25hi alloactivated donor T cells expressed significantly higher levels of cell surface DR5 than the CD25lo T cells (Figure 5A). CD4+ T cells in particular expressed significantly higher levels of DR5, with a similar trend in donor CD8+ T cells. Since activation of DR5 can induce cell death, we next studied whether TRAIL+ T cells could cause fratricide of alloactivated T cells. We found that TRAIL+ T cells mediated significantly enhanced cytotoxicity against activated T cells in vitro compared with GFP+ T cell controls (Figure 5B). To investigate the role of DR5 signaling in the suppression of alloreactive T cells by TRAIL+ T in vivo, we adoptively transferred donor B6 WT TRAIL+ T cells or DR5 KO TRAIL+ T cells into our B6→CF1 haploidentical model. DR5 KO TRAIL+ T cells, which are impervious to TRAIL-mediated fratricide, caused significantly greater GVHD mortality and morbidity compared with B6 WT TRAIL+ T cells (Figure 5C). Similar GVHD mortality and morbidity were observed in groups treated with DR5 KO TRAIL+ T and DR5 KO GFP+ T cells (Figure 5D). Together these findings suggest that TRAIL+ T cells can suppress GVHD by fratricide of alloactivated T cells.
Figure 5. Adoptive transfer of TRAIL+ T cells leads to suppression of alloactivated T cells.
(A) Purified T cells from B6 mice were injected into lethally irradiated CBF1 hosts. Single-cell suspension of splenocytes was analyzed by flow cytometry on day 4. DR5 MFI in CD25hi and CD25lo cells was analyzed in total donor T cells, CD4+ T cells, and CD8+ T cells. Representative data (n = 4 per group) from 2 independent experiments are shown. (B) Activated B6 T cells were used as targets of GFP+ and TRAIL+ T cells in a CTL assay. Graph representative of 2 independent experiments is shown. (C and D) Lethally irradiated CBF1 recipients were reconstituted with 5 × 106 cells per recipient of WT B6 TCD BM and 1 × 106 cells per recipient of B6 WT or DR5 KO B6 TRAIL+ T cells. Survival was monitored (C) and clinical GVHD scores were recorded weekly (D) in a blinded fashion. Graph representative of 2 independent experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
TRAIL+ human T cells can eliminate tumors and alloreactive T cells.
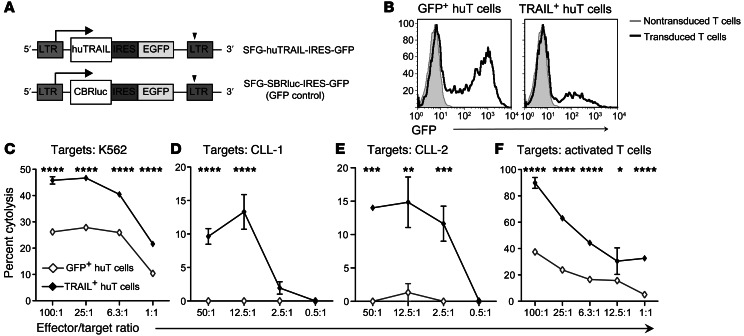
Given our promising findings in murine BMT models, we investigated the antitumor and alloactivated T cell–killing potential of human T (huT) cells engineered to express TRAIL. We designed a human (hu)TRAIL-expressing retroviral vector, SFG-huTRAIL-IRES-GFP, and its control, SFG-CBRLuc-IRES-GFP (Figure 6A). We observed efficient transduction of activated huT cells by the expression of GFP using flow cytometry (Figure 6B). We then evaluated the ability of TRAIL+ huT cells to mediate cytotoxicity. TRAIL+ huT cells mediated significantly stronger killing of the human erythroleukemia cell line K562 than did GFP+ huT cells (Figure 6C). We then further tested the antitumor potential of TRAIL+ huT cells against PBMCs obtained from 2 chronic lymphocytic leukemia (CLL) patients. Fifty-six percent of PBMCs from patient CLL-1 and 60% of PBMCs from patient CLL-2 were CD19+CD5+ cells (putative malignant cells) and had high expression of huTRAIL receptors DR4 and DR5 (Supplemental Figure 4). TRAIL+ huT cells mediated stronger cytolytic activity against PBMCs obtained from the 2 CLL patients compared with the GFP+ huT cells, demonstrating enhanced antitumor effects against primary tumors (Figure 6, D and E). As we had observed in our murine models, TRAIL+ huT cells also mediated significantly enhanced cytotoxicity against activated huT cells (Figure 6F). Altogether, these data show that TRAIL overexpression in huT cells can enhance cytolysis of both tumor cells and activated T cells, similar to our findings in murine models.
Figure 6. Genetically engineered TRAIL+ huT cells have increased GVT potential and can suppress activated T cells.
(A) Representation of SFG-huTRAIL-GFP construct: SFG-huTRAIL-IRES-GFP. SFG-CBRLuc-IRES-GFP was used as a GFP control. (B) Prestimulated huT cells were transduced and transduction was measured by the expression of GFP. (C–E) 51Cr release assays comparing tumor cytolysis between GFP+ and TRAIL+ huT cells against the (C) K562 cell line and PBMCs derived from CLL patients (D) CLL-1 and (E) CLL-2. (F) 51Cr release assay comparing cytolysis mediated by GFP+ and TRAIL+ huT cells against activated T cells. Representative graphs of at least 3 assays are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. IRES, internal ribosome entry site.
Discussion
TRAIL is a potent inducer of apoptosis through either the DR4 or DR5 receptors (18). These receptors are often expressed on cells upon malignant transformation. Systemic administration of recombinant TRAIL has had limited success in clinical trials, possibly due to the rapid clearance of recombinant TRAIL protein, as well as the presence of decoy receptors (19). The relative absence of toxicities observed in trials of TRAIL therapies, however, argues that TRAIL remains a safe therapeutic option for cancer immunotherapy.
We genetically engineered T cells with a TRAIL overexpression vector and found that murine and human TRAIL+ T cells can cause strong antitumor effects in vitro and in vivo. Antitumor activity mediated by TRAIL+ T cells is antigen independent and directed against malignant cells expressing the TRAIL receptors. While this strategy could easily be applied in syngeneic settings for tumor immunotherapy, we focused on using TRAIL-overexpressing T cells across MHC barriers to increase the feasibility and abundance of T cell availability. We used donor-derived T cells in the context of allogeneic BMT and demonstrated strong antitumor response across MHC barriers. In this setting, TRAIL+ T cells have an added benefit of suppressing GVHD, leading to improved outcomes. In an additional clinically applicable strategy, the adoptive transfer of allogeneic ex vivo–generated pre–T cells expressing TRAIL mediated significant antitumor activity in syngeneic BMT recipients. The development and persistence of GFP+ and TRAIL+ B6 pre-T–derived T cells indicate a relative tolerance toward the adoptively transferred allogeneic pre–T cells, consistent with our previous studies (13). TRAIL+ pre–T cells can therefore be used as an “off-the-shelf” cell therapy in allogeneic and autologous settings.
TRAIL+ T cells were also found to suppress GVHD. We found a marked improvement in GVHD survival with reduced clinical and pathological scores with a variety of T cell doses in 2 different GVHD models. Mechanistically, TRAIL+ T cells appear to inhibit the proliferation of other alloreactive T cells. While a body of evidence suggests a role for the TRAIL/DR5 pathway in immune regulation (15, 20), its physiological role in suppressing GVHD has not been clear (11). We found that TRAIL receptor DR5 expression is induced in APCs postirradiation and that GVHD suppression by TRAIL+ T cells is less effective in DR5 KO recipients, suggesting that DR5 expression (possibly on host APCs) could play a role in GVHD suppression by TRAIL+ T cells. In addition, we found that TRAIL-mediated suppression of GVHD was critically dependent on the elimination of other alloactivated T cells expressing DR5. The GVHD potential of DR5 KO TRAIL+ T cells was similar to DR5 KO GFP+ T cells, further suggesting that TRAIL+ T cells suppress GVHD by directly inducing apoptosis of alloreactive T cells.
Considering that TRAIL+ T cells suppress alloreactive expansion independent of antigen specificity, it is possible that TRAIL+ T cells can suppress T cells activated by pathogens. In fact, Brenden et al. showed that TRAIL can be involved in the elimination of antiviral effector T cells (21). However, in our experiments with LCMV infection, we did not find significant suppression of antiviral immunity by T cells. This could be due to increased cytotoxicity of TRAIL-overexpressing T cells, as previously shown in an influenza model (22).
We have previously shown that the TRAIL/DR5 pathway is required for thymic GVHD mediated by alloreactive T cells (12). In this study, we thus evaluated whether TRAIL overexpression could result in aggravated thymic GVHD, but did not find evidence for this. One possible explanation is that physiological expression of TRAIL by alloactivated T cells develops with the coexpression of other cytolytic ligands, and isolated overexpression of TRAIL may not be adequate to exacerbate thymic GVHD without the contribution of multiple other cytolytic mediators downstream of the alloreactive activation known to play roles in GVHD and GVT activity (23).
In summary, we demonstrate that overexpression of TRAIL by murine or human T cells enhances tumor killing while simultaneously having the unexpected benefit of reducing GVHD through fratricide of alloactivated T cells (and possibly eliminating host APCs). Genetic engineering to induce constitutive TRAIL expression on allogeneic T cells therefore represents a uniquely promising strategy to enhance GVT without a requirement for antigen specificity, while actively reducing GVHD.
Methods
Lentiviral vectors and mouse T cell transduction.
Murine TRAIL (mTRAIL) cDNA was obtained from activated T cells and cloned into the lentiviral construct pLM-GFP-2A (pLM-GFP) to create pLM-GFP-2A-TRAIL (pLM-TRAIL-GFP). Lentiviral vectors were produced by tripartite transfection of 293T cells with transfer genes (control pLM-GFP or pLM-TRAIL-GFP), pCMVΔR8.92, and pUCMD.G53 using TransIT-293 (Mirus Bio). Concentrated lentiviral vector supernatants were used for transductions.
Mouse T cells were transduced as described previously (24). Briefly, T cells derived from splenocytes were stimulated with Concanavalin A (Sigma-Aldrich) in the presence of IL-7 and IL-2 (Miltenyi Biotec). Concentrated lentiviral supernatants containing vectors for control GFP and TRAIL GFP were used to transduce T cells to obtain GFP+ and TRAIL+ T cells, respectively.
Generation of pre–T cells.
GFP+ and TRAIL+ pre–T cells were generated ex vivo for adoptive transfer (25). Lin–Sca+Kithi (LSK) cells were obtained from donor BM and prestimulated with SCF, TPO, IL-3, IL-6 (R&D Systems), and Flt3L (Miltenyi Biotec). The prestimulated cells were then transduced with concentrated lentiviral supernatants. Pre–T cells were generated from the transduced LSK cells by coculturing with OP9-DL1 cells in the presence of IL-7 and FLT3-L (Miltenyi Biotec).
Mice, syngeneic, and allo-BMT.
C57BL6 (B6-H2b), BALB/c (H2d) CBF1 ([BALB/c × B6]F1), B10.BR (H2k), and B6-expressing congeneic marker Ly5.1 (B6-Ly5.1-H2b) were obtained from The Jackson Laboratory (11). DR5 KO (B6 strain) mice and their littermate control mouse colonies were set up from cryopreserved spermatozoa (MMRRC-NIH) (16). Animals were housed in the Memorial Sloan-Kettering Cancer Center–specific pathogen-free barrier facilities. For syngeneic BMT, lethally irradiated mice (850 cGy; BALB/c) were reconstituted with lineage-depleted BALB/c BM. For allo-BMT, we used a haploidentical B6→CBF1 model or complete MHC-mismatched B10.BR→B6 combinations. Lethally irradiated (1,300 cGy for CBF1; 1,100 cGy for B6; and 850 cGy for BALB/c) mice were reconstituted with 5 × 106 cells per recipient of B6 WT T cell–depleted donor BM (TCD BM) as described previously (11, 25, 26).
Retroviral vectors and huT cell transduction.
In the TRAIL-expressing vector hTRAIL, cDNA (pORF-hTRAIL; InvivoGen) was placed under the control of the retroviral long terminal repeat (LTR) promoter followed by the IRES-GFP sequence. The final vector was labeled SFG-huTRAIL-IRES-GFP. The click beetle red luciferase encoding the SFG-CBRLuc-IRES-GFP retroviral vector was used as a GFP positive control vector (27). The PG13 viral producer cell line was transduced with the vectors mentioned above and selected based on GFP expression.
T cells from consenting healthy volunteers were isolated from buffy coat (New York Blood Center) using the density gradient separation method. Forty-eight hours after stimulation with phytohemagglutinin (2 μg/ml; Fisher Scientific), T cells were transduced using GALV-pseudotyped retroviral particles obtained from transduced PG13 cells, as described previously (27), in the presence of human recombinant (hr)IL-2 (20 U/ml; R&D Systems). During T cell expansion, the medium was supplemented with hrIL-15 at a concentration of 10 ng/ml (R&D Systems).
Cell lines and in vitro cytotoxicity assays.
The cell lines K562, LB27.4, and RENCA have been described (11, 28). In vitro cytotoxicity assays were performed by measuring chromium-51 (51Cr) release from labeled target cells as described previously (11). In some experiments, activated murine T cells were generated by incubating splenic T cells in wells coated with anti-CD3 and anti-CD28 in the presence of IL-2 and IL-7 for 24 hours before the assay. For activated human T cells, T cells derived from buffy coat were stimulated for 48 hours in the presence of hrIL-2 and hrIL15.
Primary CLL cells.
PBMCs from the CLL patients (CLL-1 and CLL-2) were isolated using density gradient separation (Ficoll-Paque PLUS; GE Healthcare). No further manipulation was performed. PBMCs were pelleted and resuspended in freezing medium composed of 90% FBS (Omega Scientific) plus 10% DMSO (Sigma-Aldrich), then frozen and stored in liquid nitrogen. The cells were thawed and then washed thrice prior to use.
GVHD and GVT.
To model GVHD, GFP+ or TRAIL+ T cells were adoptively transferred into allo-BMT recipients as described above. In some experiments MACS-purified (Miltenyi Biotec) splenic T cells were admixed with the transduced T cells at a 1:1 ratio and adoptively transferred. All BMT recipients were monitored daily for survival and scored weekly on a 10-point scale in a blinded fashion for signs of clinical GVHD (11, 29). Animals that scored greater than 5 were immediately euthanized. For histopathological assessment of GVHD, liver, small intestine, and large intestine were harvested 14 days after BMT, and skin was harvested 21 days after BMT. The organs were formalin preserved, paraffin embedded, sectioned, and stained with H&E. Blinded scoring was performed at Massachusetts General Hospital (Boston, Massachusetts, USA), and University of Florida (Gainesville, Florida, USA), as previously described (29).
To study GVT effects, LB27.4 (H2b/d) lymphoma cells were i.v. inoculated separately at the time of allo-BMT and adoptive transfer (11). Survival was monitored daily and animals found dead were necropsied to determine the presence or absence of tumor. RENCA (H2d) cells were s.c. inoculated in recipients 14 days after BMT. Tumors were measured with calipers and the volumes are expressed in centimeters cubed measured as 1/2 × length × (width)2.
Flow cytometry.
Flow cytometric analyses of cells were performed with fluorochrome-labeled antibodies (all antibodies were purchased from BD Biosciences and eBioscience) or TRAIL-R2-FcIg fusion protein (Alexis Biochemicals) and used as described previously (11). The cells were acquired on an LSR II flow cytometer (BD Biosciences) and analyzed with FlowJo software (Tree Star).
LCMV challenge.
On day 14 after BMT, mice were challenged i.p. with 2 × 105 LCMV Armstrong PFUs. Splenocytes were harvested 8 days after challenge and weighed. PFU assays were performed as previously described (30). The plaques were read the next day.
In vivo bioluminescence imaging.
T cells from luciferase-expressing mice (a gift from Robert Negrin, Stanford University, Palo Alto, California, USA) were admixed with TRAIL+ or GFP+ T cells and visualized using in vivo bioluminescence imaging systems (Caliper Life Sciences) (31). The bioluminescent flux was analyzed using Living Image software, version 4.3 (Caliper Life Sciences).
Quantitative PCR.
Tissue was harvested, mRNA was extracted, and cDNA were prepared at the Memorial Sloan-Kettering Cancer Center Genomics Core Facility. Quantitative real-time PCR (qRT-PCR) was performed for DR5 expression using a Taqman assay according to the manufacturer’s protocols (Applied Biosystems).
Statistics.
Calculations were performed using Excel (Microsoft) and graphed using Prism V5.0 software (GraphPad Software). Survival curves were analyzed with a Mantel-Cox test, and other comparisons were made using a Mann-Whitney U test or 2-way ANOVA. Data represent the means ± SEM. P values less than 0.05 were considered statistically significant.
Study approval.
All animal protocols were approved by the IACUC of Memorial Sloan-Kettering Cancer Center. We used samples from 2 CLL patients and volunteers who provided signed, informed consent to have their samples used for research purposes, in accordance with the Declaration of Helsinki and approved by the IRB of Memorial Sloan-Kettering Cancer Center.
Supplementary Material
Acknowledgments
This research was supported by grants from the NIH (R01-HL069929, R01-CA107096, R01-AI080455, P01-CA33049, and R01-HL095075, all to M. van den Brink). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Support was also received from the US Department of Defense (USAMRAA Award W81XWH-09-1-0294, to M. van den Brink), the Radiation Effects Research Foundation (RERF-NIAID, to M. van den Brink), the Ryan Gibson Foundation, The Lymphoma Foundation, and Alex’s Lemonade Stand. A. Ghosh has been a recipient of a Dr. Mildred Scheel fellowship of Deutsche Krebshilfe and the Judah Folkman Fellowship from the American Association for Cancer Research (AACR). A. Ghosh is a fellow of the Lymphoma Research Foundation. M.G. Sauer was supported by the Deutsche Forschungsgemeinschaft (SFB 738, TP A3). E. Velardi was supported by a fellowship from the Italian Foundation for Cancer Research (FIRC). M. Sadelain received support from the Major Family Foundation and the Lewis Sander Fund.
We appreciate the help of Julie White, LCP, RARC, of the Genomics Core Facility and the Flow Cytometry Core Facility of Memorial Sloan-Kettering Cancer Center. We thank Robert Negrin of Stanford University for his gift of the luciferase mice. We also appreciate the help of Kate Takvorian, Lauren Dimenna, Susan Puckett, Lao-Tzu Allan-Blitz, Anna Maria Mertelsmann, and Kristina Carrigan for their contributions to this project.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(6):2654–2662. doi:10.1172/JCI66301.
See the related Commentary beginning on page 2362.
References
- 1.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12(6):443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brentjens RJ, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. 2011;118(18):4817–4828. doi: 10.1182/blood-2011-04-348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doubrovina E, Carpenter T, Pankov D, Selvakumar A, Hasan A, O’Reilly RJ. Mapping of novel peptides of WT-1 and presenting HLA alleles that induce epitope-specific HLA-restricted-T-cells with cytotoxic activity against WT-1+ leukemias. . Blood. 2012;120(8):1633–1646. doi: 10.1182/blood-2011-11-394619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendle GM, et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med. 2010;16(5):565–570. doi: 10.1038/nm.2128. [DOI] [PubMed] [Google Scholar]
- 6.van Loenen MM, et al. Mixed T cell receptor dimers harbor potentially harmful neoreactivity. Proc Natl Acad Sci U S A. 2010;107(24):10972–10977. doi: 10.1073/pnas.1005802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth MJ, Takeda K, Hayakawa Y, Peschon JJ, van den Brink MR, Yagita H. Nature’s TRAIL — on a path to cancer immunotherapy. Immunity. 2003;18(1):1–6. doi: 10.1016/S1074-7613(02)00502-2. [DOI] [PubMed] [Google Scholar]
- 8.Frew AJ, et al. Combination therapy of established cancer using a histone deacetylase inhibitor and a TRAIL receptor agonist. Proc Natl Acad Sci U S A. 2008;105(32):11317–11322. doi: 10.1073/pnas.0801868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers TJ, et al. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL-mediated apoptosis by reducing levels of c-FLIP. Blood. 2003;102(1):303–310. doi: 10.1182/blood-2002-09-2975. [DOI] [PubMed] [Google Scholar]
- 10.Sun K, et al. Inhibition of acute graft-versus-host disease with retention of graft-versus-tumor effects by the proteasome inhibitor bortezomib. Proc Natl Acad Sci U S A. 2004;101(21):8120–8125. doi: 10.1073/pnas.0401563101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmaltz C, et al. T cells require TRAIL for optimal graft-versus-tumor activity. Nat Med. 2002;8(12):1433–1437. doi: 10.1038/nm1202-797. [DOI] [PubMed] [Google Scholar]
- 12.Na IK, et al. The cytolytic molecules Fas ligand and TRAIL are required for murine thymic graft-versus-host disease. J Clin Invest. 2010;120(1):343–356. doi: 10.1172/JCI39395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakrzewski JL, et al. Tumor immunotherapy across MHC barriers using allogeneic T-cell precursors. Nat Biotechnol. 2008;26(4):453–461. doi: 10.1038/nbt1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen EM, et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. . Nature. 2005;434(7029):88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 15.Wolkers MC, et al. Nab2 regulates secondary CD8+ T-cell responses through control of TRAIL expression. . Blood. 2012;119(3):798–804. doi: 10.1182/blood-2011-08-373910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnberg N, et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol Cell Biol. 2005;25(5):2000–2013. doi: 10.1128/MCB.25.5.2000-2013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barblu L, Herbeuval JP. Three-dimensional microscopy characterization of death receptor 5 expression by over-activated human primary CD4+ T cells and apoptosis. . PLoS One. 2012;7(3):e32874. doi: 10.1371/journal.pone.0032874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheridan JP, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277(5327):818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Lostao L, Marzo I, Anel A, Naval J. Targeting the Apo2L/TRAIL system for the therapy of autoimmune diseases and cancer. Biochem Pharmacol. 2012;83(11):1475–1483. doi: 10.1016/j.bcp.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 20.Iyori M, Zhang T, Pantel H, Gagne BA, Sentman CL. TRAIL/DR5 plays a critical role in NK cell-mediated negative regulation of dendritic cell cross-priming of T cells. J Immunol. 2011;187(6):3087–3095. doi: 10.4049/jimmunol.1003879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brincks EL, Katewa A, Kucaba TA, Griffith TS, Legge KL. CD8 T cells utilize TRAIL to control influenza virus infection. J Immunol. 2008;181(7):4918–4925. doi: 10.4049/jimmunol.181.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamada H, et al. Multiple redundant effector mechanisms of CD8+ T cells protect against influenza infection. . J Immunol. 2013;190(1):296–306. doi: 10.4049/jimmunol.1200571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Montagut T, et al. IFN-gamma and Fas ligand are required for graft-versus-tumor activity against renal cell carcinoma in the absence of lethal graft-versus-host disease. J Immunol. 2007;179(3):1669–1680. doi: 10.4049/jimmunol.179.3.1669. [DOI] [PubMed] [Google Scholar]
- 24.Kessels HW, de Visser KE, Tirion FH, Coccoris M, Kruisbeek AM, Schumacher TN. The impact of self-tolerance on the polyclonal CD8+ T cell repertoire. . J Immunol. 2004;172(4):2324–2331. doi: 10.4049/jimmunol.172.4.2324. [DOI] [PubMed] [Google Scholar]
- 25.Zakrzewski JL, et al. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nat Med. 2006;12(9):1039–1047. doi: 10.1038/nm1463. [DOI] [PubMed] [Google Scholar]
- 26.Hanash AM, et al. Abrogation of donor T-cell IL-21 signaling leads to tissue-specific modulation of immunity and separation of GVHD from GVL. Blood. 2011;118(2):446–455. doi: 10.1182/blood-2010-07-294785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobrenkov K, et al. Monitoring the efficacy of adoptively transferred prostate cancer-targeted human T lymphocytes with PET and bioluminescence imaging. J Nucl Med. 2008;49(7):1162–1170. doi: 10.2967/jnumed.107.047324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda K, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell Immunol. 2001;214(2):194–200. doi: 10.1006/cimm.2001.1896. [DOI] [PubMed] [Google Scholar]
- 29.Waldman E, et al. Absence of beta7 integrin results in less graft-versus-host disease because of decreased homing of alloreactive T cells to intestine. Blood. 2006;107(4):1703–1711. doi: 10.1182/blood-2005-08-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland AM, et al. Extrathymic development of murine T cells after bone marrow transplantation. J Clin Invest. 2012;122(12):4716–4726. doi: 10.1172/JCI60630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edinger M, et al. CD4+CD25+ regulatory T cells preserve graft-versus-tumor activity while inhibiting graft-versus-host disease after bone marrow transplantation. . Nat Med. 2003;9(9):1144–1150. doi: 10.1038/nm915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.