Abstract
Because clinical trials to assess the efficacy of vaccines against anthrax are not ethical or feasible, licensure for new anthrax vaccines will likely involve the Food and Drug Administration’s “Animal Rule,” a set of regulations that allow approval of products based on efficacy data only in animals combined with immunogenicity and safety data in animals and humans. US government sponsored animal studies have shown anthrax vaccine efficacy in a variety of settings. We examined data from 21 of those studies to determine if an immunological bridge based on lethal toxin neutralization activity assay (TNA) can predict survival against an inhalation anthrax challenge within and across species and genera. The 21 studies were classified into 11 different settings, each of which had the same animal species, vaccine type and formulation, vaccination schedule, time of TNA measurement, and challenge time. Logistic regression models determined the contribution of vaccine dilution dose and TNA on prediction of survival. For most settings, logistic models using only TNA explained more than 75% of the survival effect of the models with dose additionally included. Cross species survival predictions using TNA were compared to the actual survival and shown to have good agreement (Cohen’s κ ranged from 0.55 to 0.78). In one study design, cynomolgus macaque data predicted 78.6% survival in rhesus macaques (actual survival 83.0%) and 72.6% in rabbits (actual survival, 64.6%). These data add support for the use of TNA as an immunological bridge between species to extrapolate data in animals to predict anthrax vaccine effectiveness in humans.
Keywords: animal rule, anthrax vaccine adsorbed, correlate of protection, recombinant protective antigen, toxin neutralizing activity assay
Introduction
Medical countermeasures against potential bioterrorism threats cannot be evaluated for efficacy in humans by traditional clinical trials. A set of regulations known as the Food and Drug Administration (FDA) “Animal Rule” [1, 2] may be used to approve such countermeasures by demonstration of efficacy in animals. The application of the Animal Rule requires a reasonably well-understood biological mechanism of action that includes prediction of efficacy in humans based on dose-dependent immune response and protection relationship in animals. No vaccines have been approved using the Animal Rule, but several are being developed that will require use of the Animal Rule.
Within the same study, a traditional approach for establishing a correlate of protection in vaccines is to use a cutoff of the proposed immune response, such that nearly all subjects that reach that cutoff are assumed protected [3]. Recent statistical methods have been developed that use the entire distribution of the immune response to predict protection [4, 5]. Further theoretical work has more precisely defined what we can say about the causal nature of the proposed correlate and how that correlate may or may not be used as a surrogate for protection [6–9]. In contrast to these methods developed for within-species studies, less work has been done on methods and efforts for bridging between species to predict protection in humans. Typically, once a vaccine has been shown effective in humans directly, the assumed relationship between human immunogenicity and animal models is no longer needed. For example, in the case of the whole-cell pertussis vaccines, a relationship was established between a mouse intracerebral challenge potency test and clinical efficacy [10], but the mouse potency test is not considered a model of human disease and protection and would not be assumed to predict human efficacy when testing a new vaccine. For vaccines untestable in humans there has been some discussion of bridging between efficacy studies in non-human species and immunogenicity in humans (for example for Ebola virus vaccines [11] and anthrax vaccines[12]).
Efforts by the US government to expand available medical countermeasures to anthrax considerably increased after Bacillus anthracis spores were sent via US mail in 2001. A 2002 US government-sponsored workshop [13] recommended the use of rabbits and nonhuman primates in animal models for anthrax aerosol challenge, and the use of aerosol challenge doses that could occur in an anthrax attack. The data analyzed in this paper were generated on the basis of those recommendations.
New recombinant protective antigen (rPA) vaccines and the licensed anthrax vaccine BioThrax® [anthrax vaccine adsorbed (AVA)] were studied. Both vaccines rely largely on the protection afforded by immunological responses against the PA protein [14–20]. Many studies have shown the protective efficacy of PA based anthrax vaccines in several animal genera and species including guinea pigs [21–25], rabbits [26–29] and NHPs [20, 26, 30–33]. A few studies have gone further to evaluate correlate of protection levels based on antibody to PA for AVA in rabbits [28, 29] and for rPA in rabbits [27] and guinea pigs [25]. Data sets now exist for multiple studies in multiple species using multiple vaccines. Additionally, quality assays are available to support the evaluation of meaningful endpoints across laboratories and in multiple species [34–37].
Here, we combine data from 21 US government-sponsored animal studies (15 of which are previously unpublished). The studies form an extensive series of nonclinical aerosol B. anthracis challenge experiments of AVA and different rPA candidates conducted in rabbits, rhesus macaques, and cynomolgus macaques. We assess the relationship of vaccine-induced antibody responses with survival and assess this relationship under different vaccine types, dilution doses, adjuvants, schedules, genus and species. Additionally, we use data from human immunogenicity studies to illustrate possible approaches to extrapolation from animal challenge model results to prediction of human protection [38].
Results
Analytical Approach
The goal of this paper is threefold: (i) to explore the effect of vaccine-induced antibody response on survival in different animal model settings; (ii) to assess the role of vaccine dosage (such as antigen load) and antibody level within a specific species; and (iii) to determine whether it is meaningful to extrapolate the antibody protection relationship seen in animals to infer protection in humans. To achieve this threefold goal we examined antibody-survival relationships across genera and species, PA vaccine formulation (AVA or rPA), dose, adjuvant, time of immunological measurement, and vaccination schedule. For example with rabbits receiving two injections of adjuvanted rPA at various doses, we ask whether antibody levels usefully predict survival, and whether the vaccine dose has any additional impact on protection for fixed levels of antibody. If dose has little additional effect, it suggests that antibody levels alone may allow for reliable extrapolation. The third part is the most difficult. A formal statistical approach treating the effect from each species as a random draw from an assumed distribution has difficulty with precise predictions of survival in humans because we have data from only three non-human species. Fundamentally, extrapolation from animal genera to humans is not primarily a statistical issue but relies on judgment about how well the animal model recapitulates essential features of the infection, immune response, and protection processes in humans. We can indirectly address this issue by seeing how well a given animal species predicts survival in a different animal species or genus. If these cross-species predictions are reasonably accurate, this supports the proposition that they would be relevant to humans.
Exploration of Immunological Effects on Survival under Various Animal Models
In this analysis, we combine data from US government anthrax studies in which a particular animal species was vaccinated at various dosages (different antigen levels and vaccine dilutions), measured for serum antibody response and challenged by aerosolized B. anthracis spores. Animals were monitored for survival and declared survived if they lived for at least 21 to 30 days (depending on the study) after challenge. A detailed listing of the included studies is given in Table 1. Six important aspects of the experiments change from study to study: (i) genus and species- the animal studies were conducted in two genera and three species, cynomolgus macaques (cynos, Macaca fascicularis), rhesus macaques (rhesus, Macaca mulatta), and New Zealand white rabbits (rabbits, Oryctolagus cuniculus); (ii) vaccines- studies used either the AVA vaccine or one or more of four rPA vaccines, where the rPA vaccines differ primarily by whether the PA protein was produced in Escherichia coli or B. anthracis; (iii) diluent- the rPA vaccines were diluted with either saline or adjuvant; (iv) the time at which the immune response was measured; (v) the day of challenge; and (vi) vaccine schedule. We partition the data from 21 studies listed in Table 1 into 11 different experimental settings such that each of those six important aspects is identical within a setting, but the vaccine dose varies within a setting. Note that for these data once we match on the first five aspects, we necessarily match on the sixth. This partitioning allows us to create a series of simple models, rather than creating a complicated single model, which would require choosing from among the many possible ways of controlling the effects of the first five aspects, all of which improve prediction of survival. Data from four complete studies and parts of two studies were not included in Table 1 for various reasons (the challenge happened before 4 weeks after the last vaccination; challenge day was not fixed within the study; or many different vaccine schedules were used within the study and some of those schedules did not match those of the existing settings). For tractability, differences besides those six aspects between the 21 included studies were not explicitly modeled. Animals were challenged with aerosolized B. anthracis Ames spores at target levels of 80 to 400 times the dose producing 50% death (LD50) for that genus and species. At such large challenge levels, differences between exposures are suspected to have little effect on survival[39, 40]. Settings 9 to 11 involving AVA in rhesus were focused on evaluating the duration of protection, while setting 5 was to evaluate a rabbit model for AVA. Among rabbit rPA experiments (settings 1 to 4) the role of vaccination adjuvant (1 and 3 versus 2 and 4), schedule (1 and 2 versus 3 and 4), and challenge time (1 and 2 versus 3 and 4) were all examined. Settings 6 and 7 examined the role of adjuvant for rPA vaccines in cynos, while setting 8 examined rPA vaccines in rhesus.
Table 1.
Key features of data used for analyses. Number of animals only included animals not missing TNA. Setting numbers (S#) 1–11 involved vaccination of animals followed by aerosol challenge. Additionally, 334 humans were used in Table 3. TNA values are measured 4 weeks (+/− 2 weeks) after the last vaccination, except the human TNA values which were measured 4 weeks after the second vaccination. Study name denotes the organization (B=Battelle, U= United States Army Medical Research Institute of Infectious Diseases) that implemented the study, with numbers added to differentiate studies.
| S# | Study Name [Ref.] | Spec. | Vacc. | # Animals (# Dose Groups) | Diluent | Vacc. Sch. days | Day of TNA assay (# animals) | Day of challenge | Target Chall. dose LD50s |
|---|---|---|---|---|---|---|---|---|---|
| 1 | B-1 | rabbit | rPA-A rPA-B | 134 (4) | Adjuvant | 0 | 28 (134) | 28 | 300 |
| 2 | U-1 [27] | rabbit | rPA-D | 57(6) | Saline | 0 | 21 (5) 28 (52) | 28 | 80a |
| 2 | U-2 [27] | rabbit | rPA-D | 26(2) | Saline | 0 | 21 (1) 28 (25) | 28 | 80a |
| 2 | U-3 [27] | rabbit | rPA-D | 48(3) | Saline | 0 | 28 (48) | 28 | 80a |
| 2 | U-4 [27] | rabbit | rPA-D | 58(5) | Saline | 0 | 28 (58) | 28 | 80a |
| 3 | B-2 | rabbit | rPA-A | 20(2) | Adjuvant | 0,28 | 42 (20) | 70 | 200 |
| 3 | B-3 | rabbit | rPA-B | 20(2) | Adjuvant | 0,28 | 42 (20) | 70 | 200 |
| 3 | B-4 | rabbit | rPA-E | 54(6) | Adjuvant | 0,28 | 56 (54) | 70 | 200 |
| 3 | B-5a | rabbit | rPA-E | 36(4) | Adjuvant | 0,28 | 56 (36) | 70 | 200 |
| 4 | U-5 [27] | rabbit | rPA-D | 24(4) | Saline | 0,28 | 42 (24) | 70 | 80a |
| 4 | U-6[27] | rabbit | rPA-D | 24(4) | Saline | 0,28 | 42 (24) | 70 | 80a |
| 4 | B-5s | rabbit | rPA-E | 48(5) | Saline | 0,28 | 56 (48) | 70 | 200 |
| 5 | U-7 | rabbit | AVA | 66(6) | Saline | 0,28 | 42 (66) | 70 | 100b |
| 5 | U-8 | rabbit | AVA | 90(6) | Saline | 0,28 | 42 (90) | 70 | 100b |
| 6 | B-6 | cynos | rPA-E | 29(4) | Adjuvant | 0,28 | 56 (29) | 70 | 400 |
| 7 | B-7 | cynos | rPA-E | 47(5) | Saline | 0,28 | 56 (47) | 70 | 400 |
| 7 | B-8 | cynos | rPA-E | 48(6) | Saline | 0,28 | 56 (48) | 70 | 400 |
| 8 | B-9 | rhesus | rPA-A rPA-B | 12(2) | Adjuvant | 0,28 | 56 (12) | 70 | 300 |
| 8 | B-10 | rhesus | rPA-A | 35(5) | Adjuvant | 0,28 | 56 (35) | 70 | 400 |
| 9 | B-11 | rhesus | AVA | 60(4) | Saline | 0,28, 182 | 210 (60) | 365 | 200–400 |
| 10 | B-12 | rhesus | AVA | 43(5) | Saline | 0,28, 182 | 210 (43) | 912 | 200–400 |
| 11 | B-13 | rhesus | AVA | 34(4) | Saline | 0,28, 182 | 210 (34) | 1582 | 200–400 |
| - | - | human | AVA | 334 (1) | None | 0,28c | 56 (334) | - | - |
Target dose is 80, but average inhaled doses were all over 150 times the LD50.
Target dose estimated from the mean of the delivered doses.
Other vaccines given at day 182, 365, 548, but irrelevant for our analysis.
Because immune responses change over time, measurement of immune response at a similar time point post vaccination was used for comparisons between and within studies. Here, we chose 4 weeks after the final vaccination. This is a somewhat arbitrary choice because the vaccine schedules vary among the studies (see Table 1). The 4-week time was chosen because we have data close to that time point (within 2 weeks) for all the animal settings and additionally we have immunology measurements 4 weeks after the second vaccination for 334 humans vaccinated with AVA in the Centers for Disease Control and Prevention (CDC) Anthrax Vaccine Research Program clinical trial [38].
We begin by postulating that one particular immune response explains a substantial proportion of the survival of the animals in all the studies. If such an immunological response exists we will call it a correlate of protection. Because most if not all of the known correlates of protection for existing vaccines are related to antibody measurements [3], we postulate an antibody measurement for our correlate. In particular, we study antibodies to PA as measured by either a binding enzyme-linked immunosorbent assay (ELISA) [35] or an in vitro anthrax lethal toxin neutralization activity assay (TNA) [36, 37, 41]. For some vaccines the ELISA and TNA are not necessarily highly correlated [42], so it is necessary to study both measurements unless there is a very high correlation between them.
Figure S1 plots TNA versus ELISA in humans, and similar plots are given for NHPs (fig. S2) and rabbits (fig. S3). The assay results are highly correlated with correlation coefficient values of 0.94 for humans, 0.94 for rabbits, and 0.97 for NHPs. This level of correlation indicates that ELISA and TNA would work similarly in the models. In general, ELISA can detect measurements below the limit of detection of the TNA, and is less variable than the TNA. However, as a function-based assay, the TNA is considered to be species neutral, allowing direct comparison of neutralizing activity across species [34]. Consequently only TNA responses will be evaluated here. This paper will focus on the ability of TNA to predict survival, and will not delve into the more nuanced issues of surrogacy and causality with respect to the TNA which are discussed in the Supplemental Appendix. Once an animal survives to 3 to 4 weeks after challenge, that animal is very unlikely to die from the challenge at a later time; therefore, the binary survival endpoint (that is, the animal either lived or died) we use is more appropriate than a time to death endpoint because the latter endpoint can emphasize unimportant differences in time to death during the first few days after challenge. We first examine the effect of TNA on survival using a simple logistic regression model:
where x is a log-transformed antibody response, and a and b are parameters to be estimated. If b > 0, then antibodies are positively related to protection, sometimes called a correlate of protection.
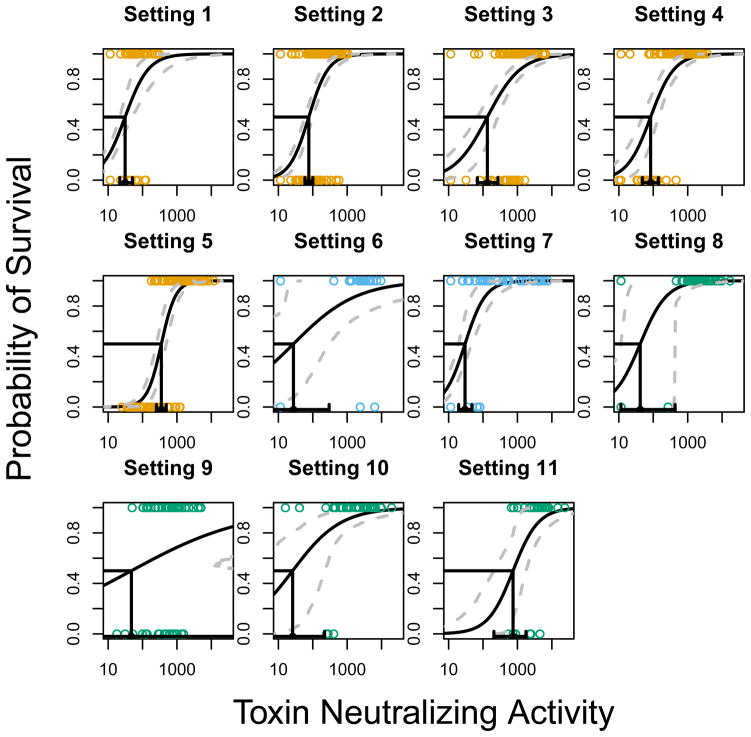
Figure 1 displays a curve with 95% confidence intervals (CIs) for each of the 11 settings under model 1 based on the estimated parameters a and b given in Table 2. The antibody level that results in 50% predicted survival (PA50) is also provided as a useful comparator, not to indicate a threshold upon which to base decisions (Figure 2). For almost all settings, there is a statistically significant relationship between antibody levels and survival [P<0.001 for all settings except setting 6 (P=0.011) and setting 9 (P=0.067)], indicating that increases in antibody levels at 4 weeks after the last vaccination increase the probability of survival.
Figure 1.
Plots of the predicted probability of survival for the 11 settings as a function of antibody (black curve) along with 95% confidence bands (dashed grey curves). The estimated antibody that provides 50% protection is given in black on the x-axis, along with a 95% confidence interval. Each point is an animal (rabbits=orange, cynos=sky blue, rhesus=blue-green).
Table 2.
Summary of logistic models 1, 2 and 3. Using log10 TNA as the correlate of protection, we fit either a full model with separate logistic parameters for each dose group (Model 3), a reduced model that assumes dose has an additional smooth effect (Model 2) or a simple model with only log10 TNA and no effect of dose (Model 1). Parameters a and b and the PA50 values are from Model 1.
| Setting | a (std. err.) | b (std. err.) | PA50 (95% CI) | CoDa Model 3 | Model 3 vs Model 1 PPE (95%CI) | CoDa Model 2 | Model 2 vs Model 1 PPE (95%CI) |
|---|---|---|---|---|---|---|---|
| 1 | −4.4 (.82) | 2.9 (.64) | 31 (22,51) | 0.26 | 84.6 (47.6,97.6) | 0.28 | 78.3 (48.2,95.8) |
| 2 | −6.7 (.93) | 3.5 (.47) | 77 (59, 101) | 0.52 | 98.0 (82.4,100) | 0.52 | 98.6 (90.7, 100) |
| 3 | −4.4 (.85) | 2.1 (.33) | 134 (69,271) | 0.53 | 87.9 (59.1,94.4) | 0.47 | 98.9 (88.0,100) |
| 4 | −5.6 (1.09) | 2.9 (.52) | 84 (48, 142) | 0.61 | 96.8 (76.1,100) | 0.60 | 99.2 (90.0,100) |
| 5 | −12.4 (2.25) | 4.9 (.83) | 353 (254,490) | 0.76 | 96.7 (84.4,100) | 0.74 | 99.4 (96.3,100) |
| 6 | −1.6 (1.08) | 1.1 (.43) | 28 (0, 299) | 0.18 | 100 (0,100) | 0.23 | 100 (0,100) |
| 7 | −4.9 (.98) | 3.3 (.74) | 30 (20,48) | 0.53 | 85.0 (53.3,95.1) | 0.56 | 80.6 (59.4,95.0) |
| 8 | −4.2 (1.45) | 2.6 (.74) | 42 (11,431) | 0.72 | 91.9 (60.6,100) | 0.76 | 87.2 (53.6,100) |
| 9 | −1.0 (.84) | 0.6 (.34) | 47 (0,222696) | 0.11 | 34.5 (0,100) | 0.02 | 100 (0,100) |
| 10 | −2.0 (1.03) | 1.4 (.44) | 26 (2, 221) | 0.37 | 89.2 (15.6,100) | 0.33 | 100 (55.6,100) |
| 11 | −7.9 (4.09) | 2.7 (1.25) | 746 (207,1773) | 0.51 | 88.3 (38.9, 100) | 0.47 | 96.5 (50.2,100) |
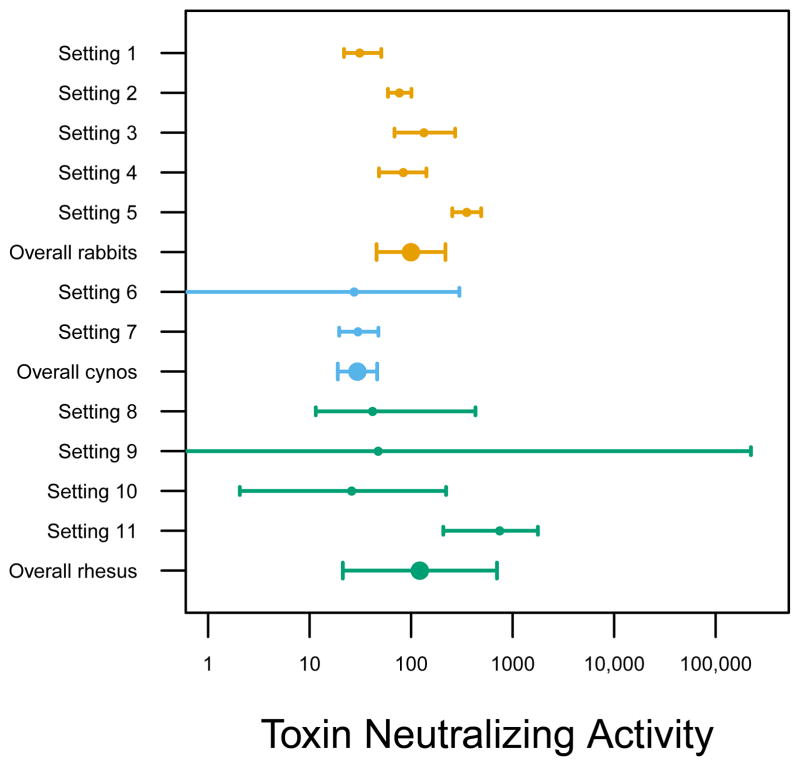
Figure 2.
Estimates of PA50, the TNA value that provides 50% protection, together with 95% confidence intervals. Each point is either a setting or the overall meta-analysis estimate (double size point) for that species (rabbits=orange, cynos=sky blue, rhesus=blue-green). Meta-analytic overall estimates use random effects models (rabbits: 100 [95% CI 46, 218], cynos: 30 [95% CI 19,46], rhesus: 122 [95% CI 21, 701]). Note the confidence interval for the overall cynos may be too small since there are only 2 settings used to estimate the random effects and it was estimated as 0.
The number of animals and doses, or more generally, the information content varies quite widely from setting to setting. Broadly speaking, settings 1 to 5 and 7 have narrower CIs for the prediction curve and the PA50 than the other settings. Tighter CIs are due primarily to large sample sizes and more complete coverage of the range of the curve for those settings. Setting 9 with 60 rhesus monkeys and four doses has the most uncertainty about the estimated slope and the antibody level that achieves 50% protection. The large uncertainty in setting 9 is due perhaps to the emphasis on highly diluted vaccine doses so that the antibody effect is harder to detect. Additionally, settings 9 to 11 completed a three dose vaccine priming series (0,1, and 6 months) and have a much longer time between our selected time of antibody measurement level and the challenge (12, 30 and 52 months).
The settings were not designed for direct comparisons, but they do demonstrate that TNA PA50 estimates vary across settings (Figure 2). We can use the conservative test that the PA50 values for two settings significantly differ if the associated 95% CIs fail to overlap. For example, settings 4 and 5 are both two-injection regimens in rabbits with a challenge 70 days after the first vaccination. The PA50 values were 84 and 353 respectively with non-overlapping 95% CIs. These settings differ in terms of the vaccine administered (rPA versus AVA) and were done by different organizations at different times. Thus, different methods of attaining a fixed level of TNA can result in different PA50 values, although precisely what is causing this difference in protection is uncertain. Settings 1 and 3 evaluate rPA with adjuvant diluent in rabbits under one and two vaccinations with a challenge at 28 and 70 days, respectively. The PA50 values are 31 and 134, respectively, with non-overlapping CIs indicating that the settings differ. This might be due to the number of vaccinations, time of measurement, later challenge time, or something else. Thus, a fixed amount of TNA can have a different impact on survival depending on how and when it was achieved. Species-level overall estimates of PA50 are given in Figure 2. The overall rabbit and rhesus PA50 values are similar, while the overall cynos PA50 is smaller but is estimated from only two settings.
Each dose group and the common control within each setting can be viewed as a very small trial. Thus, we can estimate a vaccine efficacy (VE) for each dose group with substantial uncertainty. VE is estimated in the usual way as follows:
VE can be estimated directly from the proportion of deaths in the vaccinated and control. When all placebo animals die, then the VE is simply the percent survival in the group given vaccine.
Figure 3 plots the mean TNA and estimated VE, with a 95% CI, for each dose group within each setting. For example, at the left part of the display, there are three orange circles bisecting solid vertical lines corresponding to the three (non-placebo) dose groups of setting 1. We see that as the mean TNA increases, the VE also increases. In many of the dose groups the lower bound of the 95% CI is larger than 0 indicating a significant VE.
Figure 3.
Vaccine Efficacy in the 11 experimental settings by dose group. Each point is VE estimated from one dose group compared to all control animals in that setting, vertical lines are 95% confidence intervals on the VE. The value of the point on the horizontal axis is the geometric mean of the TNA measured about 4 weeks after the last vaccination for the animals from that dose group. Orange is rabbits, sky blue is cynos and blue-green is rhesus. Logistic lines are from linear regression of empirical logits on log10(GMT) and only extend over the range for which there are geometric mean TNA values. Orange dashed is rabbits, sky blue double-dashed is cynos, and blue-green solid is rhesus.
Ideally, we want to combine the VE estimates to examine the relationship of TNA to survival within species as suggested by Daniels and Hughes [43], to see whether achieved antibody as measured by TNA is a substitute for survival. To do this we regress the empirical logit of the estimated VE on the average immune response for a single dose group using least squares regression, that is, using the following model:
where a and b are parameters to be estimated, and gmean (TNA) is the geometric mean TNA immune response for a specific dose group within a specific setting. Separate curves are estimated separately for each species. These logistic curves are given in Figure 3. Although all three curves show an increasing survival with increasing mean TNA, when we test for statistical significance for predicting survival by this method we have low power because all effects of TNA within a (noncontrol) dose group are modeled to act only through the geometric mean for the animals in that dose group. Additionally, this is a crude approach because it mixes data from different settings within a species. Nevertheless, we find a significant protective TNA effect on survival for rabbits by this method (rabbits two-sided p=0.014) and nonsignificant trends toward protective effects for TNA for the monkeys (cynos p=0.14, rhesus p=0.28). The rest of this paper will focus on the more sophisticated approach that divides the studies into settings based on the study designs.
Effect of Dose Beyond its Effects through Antibody as Measured by TNA
In the previous section, we showed that TNA can predict survival (Fig. 1), and that most positive doses have significant vaccine effects which appear to be related to average TNA value within a dose group (Figure 3). In this section, we examine whether dose has any effects on survival beyond its effects on TNA. To isolate the combined effects of antibody and dose on protection, we expand model 1 to allow increasing flexibility for the effect of dose in each of the 11 settings:
where x and d are the log10 antibody response and log10 dose for a selected animal [for dose=0 we used log10 of one half of the smallest dose, that is, log10(0.005) for rPA and log10(l/512) for AVA], and i indexes dose group within a setting. In model 1 animals that achieve an immune response x have the same probability of survival no matter what dose was used. Model 2 allows for an additional smooth effect of dose so that an animal that achieves x with a high dose could have a somewhat greater probability of survival than an animal who achieves x through a low dose. Model 3 allows each dose to have its own relationship of immune response to survival, a very flexible approach. Visually oriented readers can see examples of models 1 and 2 in Fig. 4. Model 3 is the same as repeatedly using model 1 on each dose group.
Figure 4.
The effect of TNA on survival for the 130 rabbits of setting 3. Each point is an animal and random noise was added to the points to avoid overlap. Colored curves are from logistic model with a continuous effect for log10dose from Model 2, where log10(0) is set to log10(0.005). The curves only cover the range for which there are observed TNA values in that dose group, so no lines are drawn for dose=0 and dose=0.01 since all TNA values are below the limit of detection. The black line is Model 1 where dose is not included.
Within a setting, the main question of interest is the extent to which the probability of survival differs for an antibody level of x achieved from a dose d compared to an antibody level of x achieved from a dose d′. Figure 4 displays model 2 for setting 3 where 130 rabbits in nine dose groups received two injections of an rPA vaccine with saline diluent. Recall that in model 2, we require dose to have a smooth effect on survival beyond the effect of antibody. Here we have a strong effect of TNA on the probability of survival with the additional effect of dose (apart from its effect via TNA) being insignificant (P=.18), where higher (lower) doses have estimated survival slightly higher (lower) than predicted from model 1 which solely uses TNA. For example, the very top dark orange curve gives the estimated probability of survival as a function of TNA achieved by dose=10. Visually, this curve gives only slightly higher probability of survival compared to the overall black curve that ignores dose, and most of the effect of the large dose is explained by the resulting increase in TNA. Other dose-specific curves are close to the overall black curve as well reflecting the irrelevance of dose at a fixed level of antibody.
Although the difference between model 1 and model 2 for setting 3 is not significant, with increasing numbers of animals we could potentially achieve statistical significance for an effect that is perhaps unimportant. Thus, we need some statistic to quantify the extent to which dose has an additional predictive effect beyond antibody within settings. To do this we introduce the statistic called percent of prediction explained (PPE). The coefficient of discrimination (CoD) is an R squared measure for logistic regression, defined as the difference in average survival probabilities for those who live minus the average survival probabilities for those who die [44]. This ranges from 0 (when the model is useless) to 1 (for example, when all animals with x > some threshold T live, and all those with x < T die). Because we are comparing CoD from different models and we do not want the models with more parameters to have an unfair advantage, we use an adjusted CoD (CoDa), which we define analogously to the adjusted R squared value: CoDa = 1−[(n−1)/(n−p)](1−CoD), where n is the number of animals and p is the number of estimable parameters. To describe the improvement in prediction with the addition of dose to immune response, we form the percent prediction (of model 3) explained (by model 1) as follows:
The PPE attempts to describe the relative impact of the addition of dose to a model using antibody alone to predict survival. We bound the PPE so that it ranges from 0 (if antibody has no effect) to 100 (if dose has no effect given antibody). We can test whether PPE differs significantly from 100 by seeing if the upper 95% confidence limit excludes 100, and this is equivalent to testing whether model 1 differs from model 3. This testing strategy is consistent with that of Freedman et al [45] and Buyse and Molenberghs [46] for examination of the surrogacy of a candidate x. Similar analyses were done comparing model 1 to model 2.
We explain the PPE for setting 3 in detail. In this setting, model 3 fits model 1 within each of the nine dose groups thus allows for 9 completely independent curves. It is thus geared to predict quite well. In setting 3, 84 of the rabbits survived and more than 50% of these survivors had a predicted probability of survival greater than 0.90 using model 3. Conversely 46 of the rabbits died and more than 30% of them had predicted probabilities of survival less than 0.10 using model 3. Overall we see that the rabbits that survived generally had quite high probabilities of survival and that animals that perished had quite low probabilities of survival, reflecting the strong predictive ability of antibody for model 3. However, model 1, although much simpler, does fairly well in predicting survival. For model 1 (3), the mean probability of survival is 0.812 (0.853) for rabbits that ultimately lived. For model 1 (3), the mean probability of death is 0.657 (0.731) for rabbits that ultimately died. The CoDs are 0.469=[0.812 − (1−0.657)] and 0.584=[0.853 −(1−0.731)] for models 1 and 3, respectively. However, model 1 only has only 2 estimable parameters while model 3 has 16 [2 for each dose group, except 1 for the 2 smallest dose groups where each animal has the same level (undetectable) of TNA], so the CoDa values are as follows: 0.465 = 1− (129/128)(1−0.469) and 0.529 = 1−(129/114)(1−0.584). Thus the proportion of the model 3 prediction explained by model 1, or PPE is 100(0.465/0.529) which gives 87.9% in Table 2. The 95% CI for the PPE is 59.1 to 94.4%. Thus, 87.9% significantly differs from 100%, and dose provides a statistically significant though modest additional benefit beyond antibody alone in terms of predicting survival using the very flexible model 3. The important point for this analysis is not the statistically significant additional benefit, but the fact that 87.9% of the possible predictability with dose included was already explained by the TNA before dose is added to the model.
Table 2 evaluates the additional benefit of incorporating dose in addition to TNA within each setting by comparing the simple model 1 to more complex models using PPEs. When comparing model 3 to model 1,10 of 11 settings have PPE estimates more than 75%. Looking at the lower 95% confidence limits, we see that for most of the settings (6 of 11), we are 95% sure that the PPE is greater than 50%. When we repeat this exercise using model 1 versus model 2 the PPEs are similar with all estimated PPEs greater than 75%, with only three settings with lower 95% limits less than 50%. In two settings the upper limit is different from 100%, suggesting that dose, as used in model 2, is not always useless in predicting response once antibody is known; however, in general the magnitude of the improvement from model 1 to model 2 is quite modest with 8 of 11 of the estimated PPEs exceeding 95%.
Comparing both models 3 and 2 to model 1 via the PPEs provides complementary information. Model 3 is extremely flexible but because of this flexibility may over fit the data, leading to PPEs that are less reliably estimated. Model 2 is quite parsimonious and stringent, requiring only one parameter to explain the additional smooth effect of dose. This leads to PPEs that are more reliably estimated. In either comparison, the PPEs suggest that a substantial fraction of the effect of the dose of vaccine is captured by the immune response. This suggests that within a specific setting of species and formulation, model 1 may accurately predict survival for a new dose.
Cross Species Prediction
Because humans cannot be challenged with inhalation anthrax, we cannot directly evaluate how an animal-derived statistical model applies to humans. As a proxy, we evaluate how a statistical model derived from one species predicts survival in another species for a specific vaccine regimen. If these cross-species predictions are accurate, we have more confidence in extrapolating from the animal models to humans.
Two groups of settings have similar designs except species: Settings 3, 6, and 8 all have a day 0 and day 28 vaccine schedule with rPA vaccines, diluted with adjuvant, and challenge at day 70, while settings 4 and 7 have similar designs except they are diluted with saline. We compare the effect of TNA on survival between the species in two different ways.
First, we test for differences in the PA50 values. Consider the group with vaccine diluted with adjuvant first. The PA50 for setting 3 (rabbits) is 134, that for setting 6 (cynos) is 28, and that for setting 8 (rhesus) is 42. The ratio of the PA50 for setting 3 over the PA50 for setting 6 is 4.86 (95% CI, 0.28 to 3785; P=0.22). The corresponding ratios for settings 3 to 8 and 6 to 8 are 3.20 (95% CI, 0.28 to 17.3; P=0.22) and 0.66 (95% CI, 0.0003 to 15.6; P=0.79), respectively. The group with vaccine diluted with saline has a PA50 ratio (setting 4/setting 7) of 2.79 (95% CI, 1.35 to 5.80; P=0.01). Only the last ratio denotes species having PA50 values that are statistically significantly different from each other. For these analyses, the important issue is not if there are statistically significant differences between species, but how large that difference is. If two species are statistically significantly different, but the difference is small, then useful cross-species predictions can still be made. The wide CIs for the other ratios reflect the relatively poor statistical power in these data.
A way to focus on the practical differences between species is via prediction. We thus plug in the cyno immune responses into model 1 with a and b estimated from rabbits, and average the associated probabilities of survival. This rabbit-to-cyno predicted average can be compared to the proportion of cynos who survived challenge. Additionally, we can see how well the binary predicted survival (yes/no) for each cyno agrees with the actual survival using Cohen’s κ, an agreement coefficient that corrects for chance agreement [47]. Cohen’s κ ranges from −1 to 1, with 0 indicating agreement is no different from chance, and 1 indicating perfect agreement. For determining agreement, animals with a predicted probability of survival greater than 0.5 were considered to predict survival. In general, κ values can be classified as fair (0.21 to 0.40), moderate (0.41 to 0.60), or substantial (0.61 to 0.80) [48].
Table 3 gives the results of the second type of cross-species protection estimates. Cross-species estimates were performed on all cases where there were two or more settings that differ only by species. In each of these cases, protection for human was also estimated, although in the human case there are other setting differences in addition to species. Figure 5 illustrates the cross-species protection calculation of the first row of Table 3 where a model estimated in the 130 rabbits of setting 3 is applied to the 29 cynos of setting 6. The actual TNAs achieved by the cynos are transformed into estimated probabilities of survival using the rabbit prediction equation and averaged to 70.1%.
Table 3.
Using TNA to predict survival in one species from another. Model data is the setting and species used to estimate the logistic model, and predicting data is the setting and species whose TNA values are inserted into that model to obtain a predicted survival. Values in parentheses are 95% CIs (using exact methods for the percent observed survival and using the percentile bootstrap method with 2000 replications for the others).
| Model Data | N (model) | Predicting Data | N (pred) | Percent Predict Survival | Percent Observed Survival | Cohen’s Kappa |
|---|---|---|---|---|---|---|
| 3. Rabbit | 130 | 6. Cynos | 29 | 70.1 (55.1,83.5) | 75.9 (56.5,89.7) | 0.55 (0.14,0.85) |
| 8. Rhesus | 47 | 74.8 (65.0,85.0) | 83.0 (69.2,92.4) | 0.78 (0.50,1.00) | ||
| Human | 334 | 53.7 (39.7,64.6) | NA | |||
| 6. Cynos | 29 | 3. Rabbits | 130 | 72.6 (54.0,90.6) | 64.6 (55.8,72.8) | 0.58 (0.00,0.73) |
| 8. Rhesus | 47 | 78.6 (62.8,93.0) | 83.0 (69.2,92.4) | 0.78 (0.00,1.00) | ||
| Human | 334 | 68.2 (42.3,95.0) | NA | |||
| 8. Rhesus | 47 | 3. Rabbits | 130 | 76.5 (61.2,88.5) | 64.6 (55.8,72.8) | 0.60 (0.00,0.73) |
| 6. Cynos | 29 | 76.8 (58.9,90.8) | 75.9 (56.5,89.7) | 0.55 (0.00,0.85) | ||
| Human | 334 | 75.8 (20.5,95.4) | NA | |||
| 4. Rabbit | 96 | 7. Cynos | 95 | 31.4 (20.3,42.3) | 46.3 (36.0,56.8) | 0.61 (0.36,0.74) |
| Human | 334 | 65.0 (53.3,75.1) | NA | |||
| 7. Cynos | 95 | 4. Rabbit | 96 | 71.6 (62.5,79.7) | 59.4 (48.9,69.3) | 0.59 (0.44,0.76) |
| Human | 334 | 83.5 (75.0,91.3) | NA |
Figure 5.
Graphical representation of first row in Table 3. Orange logistic line is the predicted survival based on Setting 3 (rabbits). The lines going from the horizontal axis to the logistic curve, then to the vertical axis represent the TNA values for the 29 cyno monkeys in setting 6. Random noise was added to the lines close to TNA=11.5 (half the limit of detection), and those lines represent 8 monkeys, 5 that died and 3 that survived. The sky blue tick on the vertical axis represents the mean predicted survival (70.1) for cyno macaques based on rabbit efficacy data.
Consider settings 4 and 7. In setting 4, rabbits predict a low percent of cynos surviving (31.4%) and the actual surviving percent is 46.3%. The agreement between the observed and the binary predicted survival in the cynos is substantial (κ=0.61) and significantly different from chance. The cynos in setting 7 predict 71.6% of rabbits surviving, and the actual surviving percent is 59.4% with moderate agreement (κ=0.59). Some of the particular rows of Table 3 may not have sufficient power to show statistically significant agreement (some CIs for κ have the lower 95% limit equal to 0), but when we take the average κ coefficient for all of Table 3, we get substantial agreement (average κ=0.63; 95% CI by nonparametric bootstrap 0.32 to 0.74).
Although humans received AVA vaccines and the animal settings in Table 3 are for rPA vaccines, we also perform the calculation to predict human protection based on the animal models. In practice we would want animals and humans to use the same vaccine formulation, so these calculations are more for illustration. In these predictions for humans we use TNA 4 weeks after the second vaccination because this matches the timing and schedules in the animals. The predicted survivals range from 54 to 84% with higher predictions from the NHP models.
Discussion
This paper has analyzed an extensive and heterogeneous suite of inhalation anthrax challenge experiments to determine if an immune measure could be correlated with protection in the varied experiments, and to ascertain if the models could predict protection across species. We find in the final models for all three animal species that increases in vaccine-induced TNA 4 weeks after the last vaccination were strongly associated with increased survival. Because of differences in study designs, 4 weeks after vaccination time has a different meaning in different studies (for example, 4 weeks after the last vaccination of a 0–1 month schedule and challenge at 10 weeks is different from 4 weeks after the last vaccination of a 0-1-6 month schedule and challenge at 1 year or later), and we emphasize that the 4 weeks is arbitrary and other times and other immune measures (such as antibodies as measured by ELISA) may work similarly. We have studied 11 different settings, where within each setting the species, vaccine type (rPA or AVA), vaccine schedule, vaccine formulation, time of TNA measurement, and challenge time are the same, and we have found that the TNA measurement can be used to predict survival in each setting. When we hold constant all factors except vaccine dose, we find that immune response explains a substantial fraction of the combined effect of dose and immune response. This lends support to the idea that immune response alone can be used to predict survival outcomes between different doses of vaccine. The fundamental question, however, is the extent to which this supports extrapolation to humans, which is much more ambitious than extrapolating to a different dose. We approached this indirectly by looking at cross-species extrapolation while holding other factors constant. For nonhumans the extrapolation appeared reasonably accurate (Cohen’s κ 0.55 to 0.78; Table 3), which supports the idea that such extrapolation from animal to human data may also be informative.
How might this information be used for dose selection in humans? Knowing that the TNA endpoint is a reasonable correlate and that results of the animal challenge studies should be predictive of efficacy in humans, one can assess that the likely protection afforded a human population from a safe and logistically sound vaccination regimen.
Extrapolation from animals to humans is fundamentally not a statistical issue because humans cannot be intentionally challenged with inhalation anthrax. The estimate of what might happen to humans relies on a holistic judgment of many sorts of evidence, only some of which can be statistically manipulated as done in this paper. Knowledge of the pathogenicity of the bacterium and the likely protective mechanism of the vaccine form the basis of extrapolating from animals to humans. Anthrax is primarily a toxin mediated disease and as such, should be ameliorated by anti-toxin antibodies. In the 1950s an AVA-like vaccine was evaluated in a series of trials that randomized textile mill workers who worked with raw goat hair to vaccine or placebo. The overall reported VE for cutaneous anthrax was 0.925 [49]. This directly demonstrated the ability of PA based vaccines to protect humans from B. anthracis infection. Additionally, passive transfer experiments convincingly demonstrate that infusing sufficient antibodies early in the infection process can reliably ensure survival in animals [50]. Nonetheless, in passive immunization, the amount of circulating antibodies required for protection is much larger than that required in active vaccination. Thus, other players in the vaccine induced acquired immune response, such as memory B cells, T cells, and/or unidentified processes must have a causative role in protection. Implicitly, we accept for the vaccine formulations evaluated in this analysis that these other unmeasured players are likely associated with antibody measured by TNA 4 weeks after the last injection. When taken in total, the evidence supports that PA protein based vaccines protect humans and also that the TNA measures a very important, although not exclusive, mechanism of protection.
There are subtle aspects to the statistical reasoning that go into these experiments. The extrapolation is built up within different nonhuman species and then bridged from nonhumans to humans. The amount of data and resources that are devoted to estimating the effect of antibody on protection, exploring the impact of dose, and the similarity of models across species needs to be judged with this in mind. For example, the ratio of PA50 values from rabbits (in setting 3) to cynos (in setting 6) was 2.79 (95% CI, 1.35 to 5.80). There is a statistically significant difference between species here, and we can estimate this ratio more precisely by studying more animals. Nevertheless, one needs to question whether increasing that precision will substantially fortify the extrapolation from animal models to human protection models. Clearly, there is a difference between rabbits and cynos. While greater precision about that difference may not tell us much about the difference between cynos and humans it may at least reduce the uncertainty of extrapolation.
For anthrax, we have shown that TNA measured at a specific time after vaccination can obtain good agreement when predicting survival between species. We have also shown that the actual TNA values needed to predict at least 50% survival can vary between settings and genera.
Although other strategies could be used to try to more precisely isolate a causative mechanism, the analyses presented here support a rational approach for bridging VE in animals to vaccine effectiveness in humans. Detailed knowledge of the infectious process, how the immune system successfully defeats infection, and how the vaccine successfully enhances the immune response contributes to the ability to extrapolate from animal models to human use. We believe that the approaches described here are relevant to the requirements codified in the FDA’s Animal Rule.
Materials and Methods
Description of Animal Studies
The animal studies included were designed in a data-driven, iterative manner to develop and refine animal models that would support licensure of new anthrax vaccines using the Animal Rule. Data were provided by Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases (MAID); National Center for Immunization and Respiratory Diseases, CDC; and the United States Army Medical Research Institute of Infectious Diseases (USAMRIID), Department of Defense. Each institution performed various studies to assess the effect of vaccine dilution dose on survival, the immediacy and duration of vaccine-induced protection, the immunological responses to vaccination or a combination of aspects. Study designs varied depending on the primary purpose of the study. Although not all studies were specifically designed to determine immunologic correlates of protection, the basic approach of vaccination with a range of vaccine dilutions to modulate the immune response, humoral immune response assessment, and then challenge with high doses of aerosolized virulent B. anthracis spores allowed rational combination of data.
Diluents
For the AVA vaccine, dose was varied by diluting the standard human dose with saline so that a fractional dose was obtained. Doses are expressed as the fraction of the full dose, that is, 1 is equal to a full dose, 0.5 is equal to a half dose and so forth. For the rPA vaccines, dose was expressed by micrograms of rPA protein and dose was varied by diluting the vaccine to desired protein concentration. The rPA vaccines were diluted either with buffer containing aluminum hydroxide adjuvant or with saline. Thus, when diluted with buffer containing adjuvant, the adjuvant concentration was held constant for all protein doses. When diluted with saline, the ratio of adjuvant to protein remained constant because the two components were diluted together. Control groups (placebo injections) were also included, some of which were injected with saline and others with buffer containing adjuvant.
Immunological Assays
ELISAs to detect antibody to PA were based on the CDC methods [35] with minor variations among the laboratories. In general, recombinant PA was passively adsorbed to the wells of a 96 well plate overnight at about 4°C. Samples and reference sera were diluted in phosphate buffered saline (PBS) (pH 7.4)/5% skim milk/0.5% Tween 20. Plates were washed with PBS/0.1 % Tween 20, and samples, reference sera and controls were plated and incubated for 60 min at 37°C. Plates were then washed and horseradish peroxidase conjugated immunoglobulin G-specific antibody appropriate for the species was diluted in PBS/5% skim milk/0.5% Tween 20 and added to the plates. After incubation at 37°C for 60 min, plates were washed and the appropriate substrate was added and the plates were developed. Plates were generally read at dual wavelength and the samples were quantified against the reference material calibrated in microgram per milliliter [51] and analyzed with a four parameter logistic regression.
The laboratories performed the TNA essentially as described [36, 37, 41]. Briefly, serum samples were titrated by with twofold serial dilutions in a 96-well plate, followed by the addition of a constant amount of lethal toxin (LT) to each dilution. The concentration of LT added was that needed to kill about 95% of the cells in the absence of any neutralization. After pre-incubation of the test serum with the LT, the mixtures were transferred to another 96-well plate that had been seeded with J774A.1 cells in late log phase. LT that was not neutralized by anti-LT antibodies in the serum would intoxicate and kill the cells. Following intoxication, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added to the plates, followed by the addition of a solubilization buffer to lyse the cells and solubilize the MTT. The cell plates were then incubated, and the optical density (OD) values were read with a microplate reader to determine cell viability [52]. All incubations were carried out at 37°C in about 5% C02. Neutralization of anthrax LT was manifest as a suppression of cytotoxicity and, hence, the preservation of cell viability. A four-parameter logistic regression model was used to analyze the OD versus the reciprocal of the serum dilution. The inflection point was reported as the effective dilution at 50% inhibition (ED50).
These assays were conducted for all studies at the study site. An interlaboratory comparison was conducted that included most of the laboratories contributing to the data set for this study and TNA data from all laboratories were found to be similar [34]. A long-term assay performance study of the TNA estimated that the SE of log10(TNA) for replications was 0.11[53]. Values below the limit of detection were set at one half the limit of detection.
Other assays that examined multiple aspects of humoral and cell-mediated immunity were used in settings 9 to 11. For these settings, statistical analyses indicated that antibody measured with either TNA or ELISA at different time points provided the best prediction of survival with limited improvement with the addition of other assays.
Statistical Methods
Although the logistic regression models posit a specific parametric form for the model, we use nonparametric bootstrap methods with percentile CIs which give asymptotically correct coverage even if the parametric models do not fit the data [54]. For example, in Fig. 1, we refit the logistic model 1 for each of 2000 bootstrap samples, calculate the predicted survival at a fine grid of points, and then take the middle 95% of those 2000 replicates at each point in the grid. The PA50 values and the CIs (Fig. 2 and Table 2) are just one of those points in the grid. Statistical significance for each setting in Figure 1 was determined by permutation test on the TNA values, that is, seeing if the observed slope of the logistic model was large compared to 2000 slopes calculated after randomly permuting the TNA values 2000 times. In Figs. 1 and 2, percentile bootstrap CIs were used for the settings, and for overall PA50, we used a random effects weighted mean model with the Paule-Mandel estimator and using within setting variances estimated by bootstrap [55]. A similar permutation test to that in Fig. 1 was done to test if the slopes are significantly different from 0 in Fig. 3, and in that case we permute only within setting and the permutation test automatically adjusts for the fact that the same controls are used within each setting to calculate the VEs for that setting. For the empirical logits, the VEs were estimated using the ratio of proportion deaths of the vaccinated to control groups, with the proportions adjusted to ensure that VE values were less than 100% using two sequential adjustments: First, the vaccinated proportion was estimated by adding 1/2 to the numerator and 1 to the denominator, and second any control sample proportion less than the adjusted vaccine proportion was replaced with the unadjusted vaccinated proportion. Confidence limits for VE in Figure 3 were obtained with an asymptotic method [56]. Nonparametric bootstrap percentile method was used in the cross species comparisons for CIs on predicted survival, ratios of PA50 values and Cohen’s κ. This approach bootstraps both data from animals used in the creation of the logistic model and data from animals whose immune responses are predicted from the model [4]. The CIs for the observed survival (Table 3) are exact [57]. All P-values are two-sided. Calculations were done using R 2.15.0.
Ethics
The animal procedures done by Battelle were approved by Battelle’s Institutional Animal Care and Use Committee. The research was conducted in compliance with the Animal Welfare Act and followed the principles in the Guide for the Care and Use of Laboratory Animals [58]. Similar ethics were followed for the animal studies at USAMRIID (see [27]). The human study was approved by several human investigations committees and was registered at clinicaltrials.gov (identifier: NCT00119067), and all subjects provided informed consent [38].
Supplementary Material
Acknowledgments
We acknowledge the Battelle Biomedical Research Center (BBRC) (NIAID Contracts N01-AI-25494 and N01-AI-30061, and CDC Contract 200-2000-100065) and the USAMRIID for conduct of studies that generated data analyzed in this paper. We are also grateful to Dr Judy Hewitt (NIAID), Dr. Louise Pitt (USAMRIID) and Dr. Jim Estep (Battelle) for assistance in study planning, design and implementation. We are grateful to Larry Moulton who suggested the strategy of cross-species extrapolation to examine surrogacy.
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/4/151/151ra126/DC1
Appendix: Correlation, surrogacy, and causation.
Fig. S1. Human data.
Fig. S2. Nonhuman primate data.
Fig. S3. Rabbit data.
Data.
Program. Includes description of data and instructions on using R functions.
Author contributions: Drafted sections of paper [MPF, DAF, FL, EON], guidance in interpreting results and editing of paper [MPF, DAF, FL, JS, GS, RK, CPQ, EON], data analysis [MPF, DAF], analysis advice [RK, JS, FL, CPQ, EON], database assembly [JS, FL, MPF, GS], study design and coordination [FL, CPQ, EON, GS]. None of the authors declare any competing interests.
References and Notes
- 1.Food and Drug Administration. Subpart H--Approval of Biological Products When Human Efficacy Studies Are Not Ethical or Feasible. Food and Drug Administration; Silver Spring, MD: 2009. [Google Scholar]
- 2.Gronvall GK, Trent D, Borio L, Brey R, Nagao L. The FDA animal efficacy rule and biodefense. Nat Biotechnol. 2007;25(10):1084–7. doi: 10.1038/nbt1007-1084. [DOI] [PubMed] [Google Scholar]
- 3.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401–9. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 4.Chan IS, Li S, Matthews H, Chan C, Vessey R, Sadoff J, Heyse J. Use of statistical models for evaluating antibody response as a correlate of protection against varicella. Stat Med. 2002;21(22):3411–30. doi: 10.1002/sim.1268. [DOI] [PubMed] [Google Scholar]
- 5.Kohberger RC, Jemiolo D, Noriega F. Prediction of pertussis vaccine efficacy using a correlates of protection model. Vaccine. 2008;26(27–28):3516–21. doi: 10.1016/j.vaccine.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 6.De Gruttola VG, Clax P, DeMets DL, Downing GJ, Ellenberg SS, Friedman L, Gail MH, Prentice R, Wittes J, Zeger SL. Considerations in the evaluation of surrogate endpoints in clinical trials. Summary of a National Institutes of Health workshop. Control Clin Trials. 2001;22(5):485–502. doi: 10.1016/s0197-2456(01)00153-2. [DOI] [PubMed] [Google Scholar]
- 7.Fleming TR. Surrogate endpoints and FDA’s accelerated approval process. Health Aff (Millwood) 2005;24(1):67–78. doi: 10.1377/hlthaff.24.1.67. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert PB, Qin L, Self SG. Evaluating a surrogate endpoint at three levels, with application to vaccine development. Stat Med. 2008;27(23):4758–78. doi: 10.1002/sim.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin L, Gilbert PB, Corey L, McElrath MJ, Self SG. A framework for assessing immunological correlates of protection in vaccine trials. Journal of Infectious Diseases. 2007;196(9):1304–12. doi: 10.1086/522428. [DOI] [PubMed] [Google Scholar]
- 10.Council MR. Vaccination Against Whooping-Cough: Relation Between Protection in Children and Results of Laboratory Tests. British Medical Journal. 1956;2(4990):454–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat Rev Microbiol. 2009;7(5):393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joellenbeck LM, Zwanzinger LL, Durch JS, Strom BL. In: The Antrhrax Vaccine: Is It Safe? Does It Work? Strom BL, Barlow WE, Blazer DG, Cowan LD, Edwards KM, Faustman DL, et al., editors. The National Academies Press; 2002. [PubMed] [Google Scholar]
- 13.Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Anthrax Vaccines: Efficacy Testing and Surrogate Markers of Immunity Workshop (4/23/2002) 2002 [cited. Available from: http://www.fda.gov/BiologicsBloodVaccines/NewsEvents/WorkshopsMeetingsConferences/TranscriptsMinutes/default.htm.
- 14.Ivins BE, Ezzell JW, Jr, Jemski J, Hedlund KW, Ristroph JD, Leppla SH. Immunization studies with attenuated strains of Bacillus anthracis. Infect Immun. 1986;52(2):454–8. doi: 10.1128/iai.52.2.454-458.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivins BE, Ristroph JD, Nelson GO. Influence of body weight on response of Fischer 344 rats to anthrax lethal toxin. Appl Environ Microbiol. 1989;55(8):2098–100. doi: 10.1128/aem.55.8.2098-2100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivins BE, Welkos SL. Cloning and expression of the Bacillus anthracis protective antigen gene in Bacillus subtilis. Infect Immun. 1986;54(2):537–42. doi: 10.1128/iai.54.2.537-542.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivins BE, Fellows PF, Nelson GO. Efficacy of a standard human anthrax vaccine against Bacillus anthracis spore challenge in guinea-pigs. Vaccine. 1994;12(10):872–4. doi: 10.1016/0264-410x(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 18.Boor AK, Tresselt HB. An antigen prepared in vitro effective for immunization against anthrax. II. Concentration of the protective antigen by chemical fractionation. J Infect Dis. 1955;97(2):203–6. doi: 10.1093/infdis/97.2.203. [DOI] [PubMed] [Google Scholar]
- 19.Boor AK, Tresselt HB. Protection of sheep against anthrax by immunization with an antigen prepared in vitro. Am J Vet Res. 1955;16(60):425–8. [PubMed] [Google Scholar]
- 20.Tresselt HB, Boor AK. An antigen prepared in vitro effective for immunization against anthrax. III. Immunization of monkeys against anthrax. J Infect Dis. 1955;97(2):207–10. doi: 10.1093/infdis/97.2.207. [DOI] [PubMed] [Google Scholar]
- 21.Ivins BE, Welkos SL, Little SF, Crumrine MH, Nelson GO. Immunization against anthrax with Bacillus anthracis protective antigen combined with adjuvants. Infect Immun. 1992;60(2):662–8. doi: 10.1128/iai.60.2.662-668.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivins BE, Welkos SL, Knudson GB, Little SF. Immunization against anthrax with aromatic compound-dependent (Aro-) mutants of Bacillus anthracis and with recombinant strains of Bacillus subtilis that produce anthrax protective antigen. Infect Immun. 1990;58(2):303–8. doi: 10.1128/iai.58.2.303-308.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride BW, Mogg A, Telfer JL, Lever MS, Miller J, Turnbull PC, Baillie L. Protective efficacy of a recombinant protective antigen against Bacillus anthracis challenge and assessment of immunological markers. Vaccine. 1998;16(8):810–7. doi: 10.1016/s0264-410x(97)00268-5. [DOI] [PubMed] [Google Scholar]
- 24.Singh Y, Ivins BE, Leppla SH. Study of immunization against anthrax with the purified recombinant protective antigen of Bacillus anthracis. Infect Immun. 1998;66(7):3447–8. doi: 10.1128/iai.66.7.3447-3448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuveny S, White MD, Adar YY, Kafri Y, Altboum Z, Gozes Y, Kobiler D, Shafferman A, Velan B. Search for correlates of protective immunity conferred by anthrax vaccine. Infect Immun. 2001;69(5):2888–93. doi: 10.1128/IAI.69.5.2888-2893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawla A, Midha S, Bhatnagar R. Efficacy of recombinant anthrax vaccine against Bacillus anthracis aerosol spore challenge: preclinical evaluation in rabbits and Rhesus monkeys. Biotechnol J. 2009;4(3):391–9. doi: 10.1002/biot.200800213. [DOI] [PubMed] [Google Scholar]
- 27.Little SF, Ivins BE, Fellows PF, Pitt ML, Norris SL, Andrews GP. Defining a serological correlate of protection in rabbits for a recombinant anthrax vaccine. Vaccine. 2004;22(3–4):422–30. doi: 10.1016/j.vaccine.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Pitt ML, Little S, Ivins BE, Fellows P, Boles J, Barth J, Hewetson J, Friedlander AM. In vitro correlate of immunity in an animal model of inhalational anthrax. J Appl Microbiol. 1999;87(2):304. doi: 10.1046/j.1365-2672.1999.00897.x. [DOI] [PubMed] [Google Scholar]
- 29.Pitt MLM, Little SF, Ivins BE, Fellows P, Barth J, Hewetson J, Gibbs P, Dertzbaugh M, Friedlander AM. In vitro correlate of immunity in a rabbit model of inhalational anthrax. Vaccine. 2001;19(32):4768–73. doi: 10.1016/s0264-410x(01)00234-1. [DOI] [PubMed] [Google Scholar]
- 30.Ivins BE, Pitt ML, Fellows PF, Farchaus JW, Benner GE, Waag DM, Little SF, Anderson GW, Jr, Gibbs PH, Friedlander AM. Comparative efficacy of experimental anthrax vaccine candidates against inhalation anthrax in rhesus macaques. Vaccine. 1998;16(11–12):1141–8. doi: 10.1016/s0264-410x(98)80112-6. [DOI] [PubMed] [Google Scholar]
- 31.Ivins BE, Fellows PF, Pitt ML, Estep JE, Welkos SL, Worsham PL, Friedlander AM. Efficacy of a standard human anthrax vaccine against Bacillus anthracis aerosol spore challenge in rhesus monkeys. Salisbury Medical Bulletin. Special Supplement No. 1996;87:125–6. [Google Scholar]
- 32.Pitt MLM, Ivins BE, Estep JE, Farchaus J, Friedlander AM. Comparison of the efficacy of purified protective antigen and MDPH to protect non-human primates from inhalation anthrax. Salisbury Medical Bulletin. Special Supplement No. 1996;87:130. [Google Scholar]
- 33.Williamson ED, Hodgson I, Walker NJ, Topping AW, Duchars MG, Mott JM, Estep J, Lebutt C, Flick-Smith HC, Jones HE, Li H, Quinn CP. Immunogenicity of recombinant protective antigen and efficacy against aerosol challenge with anthrax. Infect Immun. 2005;73(9):5978–87. doi: 10.1128/IAI.73.9.5978-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omland KS, Brys A, Lansky D, Clement K, Lynn F. Interlaboratory comparison of results of an anthrax lethal toxin neutralization assay for assessment of functional antibodies in multiple species. Clin Vaccine Immunol. 2008;15(6):946–53. doi: 10.1128/CVI.00003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn CP, Semenova VA, Elie CM, Romero-Steiner S, Greene C, Li H, Stamey K, Steward-Clark E, Schmidt DS, Mothershed E, Pruckler J, Schwartz S, Benson RF, Helsel LO, Holder PF, Johnson SE, Kellum M, Messmer T, Thacker WL, Besser L, Plikaytis BD, Taylor TH, Freeman AE, Wallace KJ, Dull P, Sejvar J, Bruce E, Moreno R, Schuchat A, Lingappa JR, Marano N, Martin SK, Walls J, Bronsdon M, Carlone GM, Bajani-Ari M, Ashford DA, Stephens DS, Perkins BA. Specific, sensitive, and quantitative enzyme-linked Immunosorbent assay for human immunoglobulin G antibodies to anthrax toxin protective antigen. Emerg Infect Dis. 2002;8(10):1103–10. doi: 10.3201/eid0810.020380. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hering D, Thompson W, Hewetson J, Little S, Norris S, Pace-Templeton J. Validation of the anthrax lethal toxin neutralization assay. Biologicals. 2004;32(1):17–27. doi: 10.1016/j.biologicals.2003.09.003. [Article] [DOI] [PubMed] [Google Scholar]
- 37.Li H, Soroka SD, Taylor TH, Jr, Stamey KL, Stinson KW, Freeman AE, Abramson DR, Desai R, Cronin LX, Oxford JW, Caba J, Pleatman C, Pathak S, Schmidt DS, Semenova VA, Martin SK, Wilkins PP, Quinn CP. Standardized, mathematical model-based and validated in vitro analysis of anthrax lethal toxin neutralization. Journal of Immunological Methods. 2008;333(1–2):89–106. doi: 10.1016/j.jim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Marano N, Plikaytis BD, Martin SW, Rose C, Semenova VA, Martin SK, Freeman AE, Li H, Mulligan MJ, Parker SD, Babcock J, Keitel W, El Sahly H, Poland GA, Jacobson RM, Keyserling HL, Soroka SD, Fox SP, Stamper JL, McNeil MM, Perkins BA, Messonnier N, Quinn CP, Anthrax W. Vaccine Res Program, Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months - A randomized trial. Jama-Journal of the American Medical Association. 2008;300(13):1532–43. doi: 10.1001/jama.300.13.1532. [DOI] [PubMed] [Google Scholar]
- 39.Coleman ME, Thran B, Morse SS, Hugh-Jones M, Massulik S. Inhalation anthrax: Dose response and risk analysis. Biosecurity and Bioterrorism-Biodefense Strategy Practice and Science. 2008;6(2):147–59. doi: 10.1089/bsp.2007.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Anthrax Vaccines: Bridging Correlates of Protection In Animals to Immunogenicity In Humans (11/09/2007) 2007 http://www.fda.gov/BiologicsBloodVaccines/NewsEvents/WorkshopsMeetingsConferences/TranscriptsMinutes/default.htm.
- 41.Friedlander AM, Bhatnagar R, Leppla SH, Johnson L, Singh Y. Characterization of Macrophage Sensitivity and Resistance to Anthrax Lethal Toxin. Infect Immun. 1993;61(1):245–52. doi: 10.1128/iai.61.1.245-252.1993. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes GK, Naess LM, Rosenqvist E, Guerin PJ, Caugant DA G Fractional Doses Vaccine Study. Avidity of Serogroup A Meningococcal IgG Antibodies after Immunization with Different Doses of a Tetravalent A/C/Y/W135 Polysaccharide Vaccine. Scandinavian Journal of Immunology. 2011;74(1):87–94. doi: 10.1111/j.1365-3083.2011.02535.x. [DOI] [PubMed] [Google Scholar]
- 43.Daniels MJ, Hughes MD. Meta-analysis for the evaluation of potential surrogate markers. Statistics in Medicine. 1997;16(17):1965–82. doi: 10.1002/(sici)1097-0258(19970915)16:17<1965::aid-sim630>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 44.Tjur T. Coefficients of Determination in Logistic Regression Models-A New Proposal: The Coefficient of Discrimination. American Statistician. 2009;63(4):366–72. [Google Scholar]
- 45.Freedman LS, Graubard BI, Schatzkin A. Statistical Validation of Intermediate End-points for Chroinc Diseases. Statistics in Medicine. 1992;11(2):167–78. doi: 10.1002/sim.4780110204. [DOI] [PubMed] [Google Scholar]
- 46.Buyse M, Molenberghs G. Criteria for the validation of surrogate endpoints in randomized experiments. Biometrics. 1998;54(3):1014–29. [PubMed] [Google Scholar]
- 47.Cohen J. A Coefficient of Agreement for Nominal Scales. Educational and Psychological Measurement. 1960;20(1):37–46. [Google Scholar]
- 48.Landis JR, Koch GG. Measurement of Observer Agreeement for Categorical Data. Biometrics. 1977;33(1):159–74. [PubMed] [Google Scholar]
- 49.Brachman PS, Fekety FR, Gold H, Werrin M, Ingraham NR, Plotkin SA. Field Evaluation of Human Anthrax Vaccine. American Journal of Public Health and the Nations Health. 1962;52(4):632. doi: 10.2105/ajph.52.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.I. Human Genome Sciences. Anti-Infectives Advisory Committee Breifing Document: Raxibacumab, Treatment of Inhalation Anthrax, BLA 125349, September 21, 2009. 2009. [Google Scholar]
- 51.Semenova VA, Steward-Clark E, Stamey KL, Taylor TH, Schmidt DS, Martin SK, Marano N, Quinn CP. Mass value assignment of total and subclass immunoglobulin G in a human standard anthrax reference serum. Clin Diagn Lab Immunol. 2004;11(5):919–23. doi: 10.1128/CDLI.11.5.919-923.2004. [Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosmann T. Rapid Colorimetric Assay for Cellular Growth and Survival- Application to Proliferation and Cyto-toxicity Assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [Article] [DOI] [PubMed] [Google Scholar]
- 53.Soroka SD, Schiffer JM, Semenova VA, Li H, Foster L, Quinn CP. A two-stage, multilevel quality control system for serological assays in anthrax vaccine clinical trials. Biologicals. 2010;38(6):675–83. doi: 10.1016/j.biologicals.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman and Hall; 1993. [Google Scholar]
- 55.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: An update. Contemporary Clinical Trials. 2007;28:105–14. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 56.Farrington CP, Manning G. Test Statistics and Sample-Size Formulas for Comparative Binomial Trials with Null Hypothesis of Nonzero Risk Difference or Non-unity Relative Risk. Statistics in Medicine. 1990;9(12):1447–54. doi: 10.1002/sim.4780091208. [DOI] [PubMed] [Google Scholar]
- 57.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–13. [Google Scholar]
- 58.National Research Council. The Care and Use of Laboratory Animals. National Academy of Sciences; 2011. [cited. Available from: http://oacu.od.nih.gov/regs/guide/guide.pdf. [PubMed] [Google Scholar]
- 59.Joffe MM, Greene T. Related Causal Frameworks for Surrogate Outcomes. Biometrics. 2009;65(2):530–8. doi: 10.1111/j.1541-0420.2008.01106.x. [DOI] [PubMed] [Google Scholar]
- 60.Prentice RL. Surrogate Endpoints in Clincial Trials - Definition and Operational Criteria. Statistics in Medicine. 1989;8(4):431–40. doi: 10.1002/sim.4780080407. [DOI] [PubMed] [Google Scholar]
- 61.Pearl J, Bareinboim E. Techincal Report. 2011. Trasportability across studies: A formal approach. [Google Scholar]
- 62.Pearl J. Causality: models, reasoning, and inference. 2. Cambridge; New York: Cambridge University Press; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.