Dissecting the spatio-temporal interaction of biomolecules inside cells at the subcellular level is an important facet of molecular cell biology and chemical biology. Over the last few decades, a variety of fluorescent molecular probes have been developed to investigate these complex bio-interactions for both in vitro and in vivo cellular imaging and subcellular detection. In particular, semiconductor nanoparticles like quantum dots (QDs) have shown great potential as nanoparticle fluorescent probesdue to their excellent physiochemical properties, which allow them to overcome the limitations of conventional fluorescent probes such as organic dyes and fluorescent proteins.[1] These unique attributes of QDs have proven to be crucial in elucidating the intricate interactions through which small molecules and biomolecules (e.g. proteins, peptides and nucleic acids) bind to their targets in specific signaling cascades.[2] While QDs are excellent molecular probes, they also can be used as effective delivery vehicles for several therapeutic biomolecules. For example, the use of QDs for the simulataneous imaging and delivery of small interfering RNA (siRNA) for selectively knocking-down target oncogenes in tumor cells has been successfully demonstrated.[2c, 3]
However, the major limiting factor in harnessing the maximum potential of QDs as multifunctional imaging probes and delivery systemsis their inherent cytotoxicity; most of the well-established QDs are composed of highly toxic elements, such as cadmium (Cd), selenium (Se) or tellurium (Te).[2b, 4] Owing to this obstacle, the wide applications of QDs are currently delayed and the main focus of QD imaging has been limited to the cell and small animal studies. In response to the above issues, the recent development of I-III-VI2 type QDs[5] like AgInS2,[5c] CuInS2[5b, 5d] and ZnS-AgInS2[5e] offers better control of band-gap energies and demonstrates the great potential of these QDs as non-toxic molecular probes. For instance, several research groups have successfully synthesized ZnS-AgInS2 (ZAIS) QDs through the decomposition of single source precursors using thermal,[5e] hydrothermal,[6] photothermal[7] and microwave-assisted methods.[8] Yet, these conventional synthetic methodologies for preparing these I-III-VI2 type QDs have several shortcomings such as high reaction temperatures, poor control of growth rates, long reaction times, difficulty of high throughput synthesis, and the need for complicated synthetic procedures to prepare QDs with different emissions profiles, all of which would be critical in investigating the diversity and dynamic processes of multiple biomarkers in cancer and stem cells.Thus, in order to harness the full potential of QDs as biological imaging probes as well as drug delivery platforms for clinical and translational researche, there is an urgent need to develop a simple and straightforward methodology that affords both the synthesis of non-toxic QDs and the versatility of generating a library of QDs with tunable properties favoring their use as imaging agents in biology.
To address the aforementioned issues, we developed a novel sonochemical approach for the high throughput synthesis of a library of biocompatible ZnxS − AgyIn1−yS2 (ZAIS) quantum dots with tunable physical (photoluminescence, PL) properties, thereby allowing them to be used as multifunctional nanoparticles for the simultaneous imaging and effective delivery of siRNA to brain tumor cells with negligible cytotoxicity. These ZAIS QDs also proved to be useful for imaging stem cells, which are otherwise quite senstive towards nanomaterial-based molecular imaging probes (Figure 1). A sonochemical synthetic method uses ultrasound irradiation to drive the main synthetic reaction process. With the rapid growth of its use for applications in material science, the sonochemical synthetic approach is particularly attractive for the preparation of novel nanomaterials. Its advantages include a fast reaction rate (for e.g., its possible to generate a whole library of nanoparticles of several compositions in a span of few hours), controllable reaction conditions and the ability to form nanoparticles with uniform shapes, narrow size-distributions and high purity in relatively less time and at ambient conditions. Specifically, the synthetic methodology used for the preparation of our ZAIS QDs involved the sonochemical decomposition of the organometallic precursor (AgyIn1−y)Znx(S2CN(C2H5)2)4 at ambient conditions. Compared to the traditional synthetic methods for preparing QDs, this approach does not require high-temperature and high-pressure conditions (See Experimental Section for the detailed synthetic information). The resulting ZAIS QDs exhibited intense emission at room temperature, regardless of the size of the particles (Figure 2). More interestingly, the energy gap of the ZAIS QDs and their emission wavelength can be controlled by varying the concentration of each precursor element. For instance, the emission of the ZAIS QDs could be tuned from 480 nm (Blue QD) to 700 nm (Red QD) by just varying the mole ratios of zinc (Zn), silver (Ag), and indium (In) in the precursor solutions (Figure 2A). It is worthwile to note that as compared to conventional CdSe or CdTe QDs, which show size-dependent fluorescence properties, our composition-dependent tunable fluorescence of the ZAIS QDs presentsa unique advantage for obtaining QDs with emission in the near-UV range (blue or blue-green emission). This observation can be explained by the fact that a blue-colored CdSe QD needs to be less than 2.0 nm in size, which is practically impossible to obtain using traditional thermal decoposition techniques. After assessing the optimum conditions for obtaining the desired emission profiles, we observed that for a given concentration of precursor ions, ultrasonication for 5 minutes at 20 kHz and 200W output power, gave the highest emission intensity (data not shown). Figure 2B depicts a 3D heat map which summarizes the physical and chemical properties of the partial ZAIS QD library. For the different combinations of the precursor elements (i.e. Zn, Ag, and In), the 3D heat map shows the emission wavelength and the corresponding photoluminescence (PL) peak intensities of selected ZAIS QDs under UV irradiation (λex = 365nm). This data analysis not only provides the key characteristic properties for the various compositions, but it can also facilitate the selection of ZAIS QDs with appropriate physicochemical properties for further studies and applications. For example, examining the 3D heat map, it is evident that ZAIS QDs without zinc (i.e. AgyIn1−yS2) had the longest emission wavelengths (Figure 2B). We also observed that the PL peak wavelength was blue-shifted on increasing the Zn concentration. Furthermore, QDs obtained with 0.5 mol Ag and 0.5 mol In, had the longest emission wavelength (λem = 697nm), with an observed blue-shift on increasing or decreasing the Ag concentration. In conclusion, the emission profiles of the ZAIS QDs could be easily tuned by varying the Zn or Ag (In = 1 − Ag) concentrations and hence the reported method used to synthesize our QD library can allow scientists to investigate the composition-dependent physicochemical properties of QDs of interest. (See Supporting Information, Figure S1 for the detailed information about absorption and PL spectra).
Figure 1. Synthesis of a library of ZnS-AgInS2QDs for simultaneous imaging and delivery of siRNA.
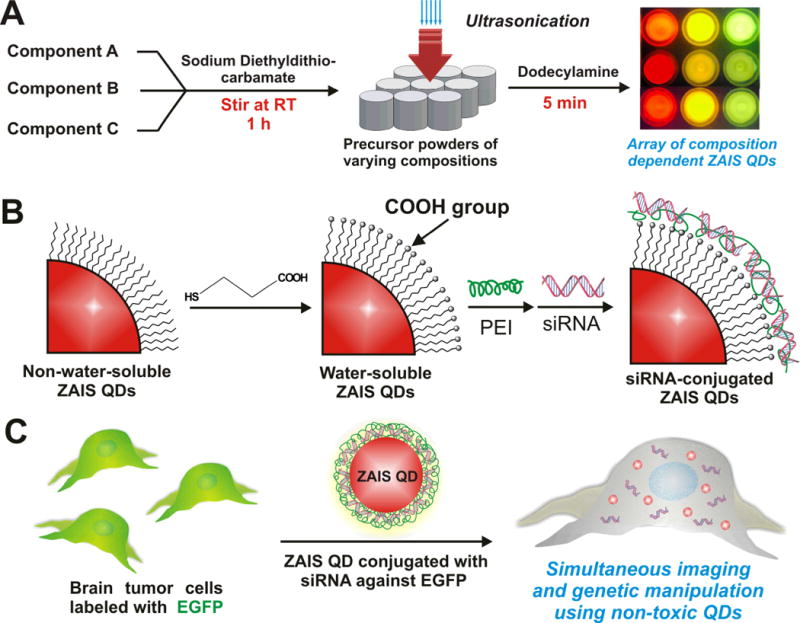
(A) Synthetic procedure to obtain a library of dodecylamine-capped hydrophobic ZAIS QDs. (B) Conversion of the the hydrophobic ZAIS QDs into water-soluble ones via ligand exchange with 3-mercaptopropionic acid (MPA) followed by attachment of siRNA using a layer-by-layer approach. (C) Delivery of the siRNA against EGFP using ZAIS QD-siRNA conjugates into brain cancer cells overexpressing EGFP.
Figure 2. Photoluminescent properties of the ZAIS QD library.
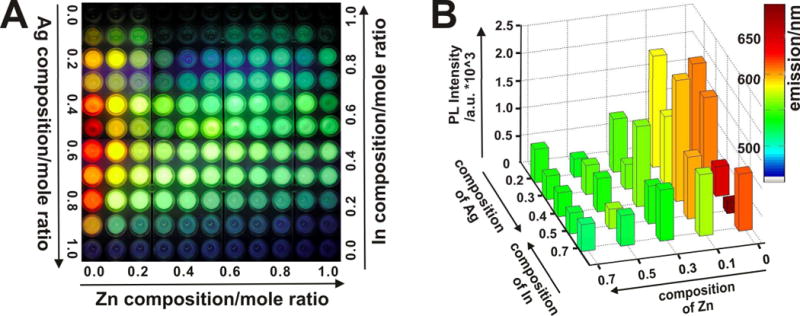
(A) Representative fluorescent image of the entire library of ZAIS QDs with varying compositions (ZnxS − AgyIn1−yS2)synthesized via the sonochemical approach. (B) Heat map depicting the PL intensity (z axis) vs. Zn and Ag/In concentrations (x and y axis) for select compositions of the ZAIS nanoparticles library. Column color indicates the maximum emission wavelength.
Comprehensive characterization of the resulting ZAIS QDs was performed using several different methods. The elemental composition of one representative ZAIS QD (Zn0S − Ag0.2In0.8S2) was analyzed using X-Ray fluorescence spectroscopy (XRF). It was observed that the relative atomic ratios of the constituent elements was consistent with the calculated mole ratios (Figure 3A), thus confirming the efficiency of the sonochemical synthetic methodology in obtaining QDs with desired chemical compositions. We also analyzed the crystal structures of several compositions of the ZAIS QDs using powder X-ray diffraction (XRD). The particles prepared with varying mole ratios of Zn, Ag and In exhibited three broad peaks which lie in between the diffraction patterns of the tetragonal AgInS2 and bulk cubic ZnS crystals (Figure 3B). Furthermore, there was a clear peak shift towards a higher angle with an increase in the amount of Zn2+, thus indicating that the QDs obtained were a not a just mixture of bulk ZnS and AgInS2 but a ZnS-AgInS2 solid solution.[5e] Transmission electron microscopy (TEM) analysis clearly revealed the spherical shape and monodispersity of our ZAIS QDs (Figure 3C). The average size of the QDs as determined by TEM was found to be 12 ± 1.3 nm. This was further confirmed by measuring the hydrodynamic size and the polydispersity index (PDI) of the ZAIS QDs using dynamic light scattering (Figure S3). Additionally, the size of the QDs did not significantly change when their composition was altered (data not shown). We also determined the PL quantum yields of the ZAIS QDs and compared them to the quantum yield of the CdSe/ZnS QDs (~ 0.4). It was found that the relative quantum yields of the ZAIS QDs depended on their composition and were comparable or in some cases higher than that of the CdSe/ZnS QDs (Figure S4). Finally, in terms of stability, the physical properties of the ZAIS QDs such as absorption and photoluminescence remained unchanged for two months when stored at ambient conditions (see Figure S2). Hence, these ZAIS QDs could be potentially used for long term cellular labeling without any loss of photoluminescence.
Figure 3. Physical characterization of the ZnxS − AgyIn(1−y)S2QDs.
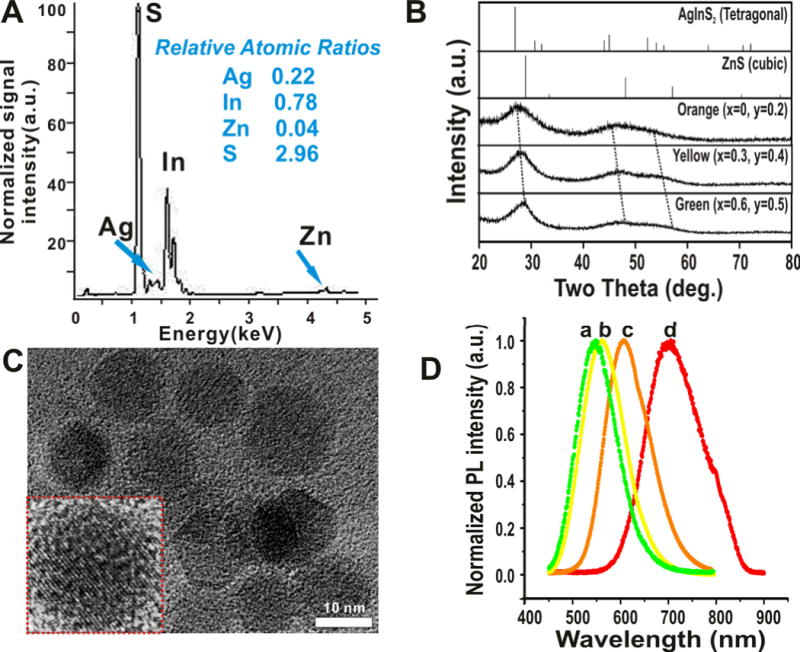
(A) X-ray Fluorescence analysis of one of the ZAIS QD (x = 0, y = 0.2) showing the elemental composition and relative atomic mole ratios of the constituent elements. The composition agrees to that calculated theoretically. (B) X-ray diffraction patterns of the ZAIS QDs prepared using sonochemistry. The values of x and y are indicated in the figure. Reference patterns of bulk ZnS and AgInS2 are also shown. Broad peak width is attributed to the amorphous nature of the ZAIS QDs. (C) TEM images of one of the ZAIS QDs (x = 0, y = 0.2) clearly shows the monodispersity and narrow size distribution (n = 100). Inset shows the high resolution image of a single nanoparticle (Scale bar = 10 nm). (D) Photoluminescence spectra of select water-soluble ZAIS QDs (a: x = 0.6, y = 0.5.; b: x = 0.3, y = 0.4; c: x = 0, y = 0.2; d: x = 0, y = 0.5)
The solubility and stability of QDs in an aqueous solution is essential for their wide application as molecular probes in molecular cell biology. The ZAIS QDs reported here were functionalized with 3-mercaptopropionic acid (MPA) to render them soluble in physiological conditions (See Experimental section for the details regarding the surface modification).[9] These water-soluble ZAIS QDs were found to be extremely stable at physiological conditions without any signs of aggregation, even when stored for several months (Figure S5). This was further confirmed by monitoring the PL intensity of one of the ZAIS QDs (x = 0, y = 0.2) in phosphate buffered saline (PBS, pH = 7.4) at 37 ºC over a period of 6 days. The ZAIS QDs were found to be quite stable without any significant loss of photoluminescence over the test period (Figure S6). Another critical factor limiting the use of conventional QDs for various biological applications is their inherent cellular toxicity.[10] This could be partially attributed to the lack of suitable methods to generate QDs with different compositions in a high throughput manner for subsequent toxicological screening. Hence, the generation of a QD library and assaying their cytotoxity in a simple and quick way can facilitate the selection of appropriate QDs eliciting minimum toxic effects and thus enabling the long term cellular imaging of cancer and stem cells in vitro and in vivo. To test the biocompatibility of our ZAIS QDs for use as imaging probes in vitro, a cytotoxicity assay was carried out in both cancer and stem cells (Figure 4A). The concentration-dependent cytotoxicity of the water-soluble ZAIS QDs was assessed in human bone marrow-derived mesenchymal stem cells (hMSCs) and human brain tumor cells (U87 glioblastoma cell line) for two days using a cell proliferation assay (MTS). The toxicity of the ZAIS QDs was also compared to the water-soluble CdSe/ZnS QDs (control sample). The ZAIS QDs showed significantly improved biocompatibility (less cytotoxic, 95 % cell viability) in both U87 cells and hMSCs even at high concentrations (upto 100 μg/mL) in comparison to the control sample (CdSe/ZnS QDs), which were found to noticeably cytotoxic at even low concentrations. The cytotoxicity of other compositions of the ZAIS QDsis presented in the supporting information (Figures S7 and S8). In addition, we carried out the cytotoxicity assay of the ZAIS QDs in normal healthy cells (NIH-3T3 mouse fibroblasts). The ZAIS QDs were found to be extremely biocompatible as compared to the CdSe/ZnS QDs (Figure S9). The prolonged exposure of nanoparticles, especially QDs, to an oxidative environment (aerial oxidation or UV-induced oxidation) has been known to catalyse their decomposition, thereby leading to the leaching of the metal ions, which is known to induce cytotoxic effects in cells. In order to test the cytotoxicity of our ZAIS QDs under an oxidative environment, we subjected the ZAIS QDs to high-intensity UV light to catalyze the oxidation process. The water-soluble ZAIS QD solutions were exposed to a UV-light source (λem = 365 nm, power density of 12 mW/cm2) for 1 – 4 hours after which they were incubated with U87 cells. It was found that even after 4h of UV-induced photo-oxidation, the ZAIS QDs were not toxic to the cells (> 85% viability, Figure S10) which is in stark contrast to the photo-toxicity of CdSe/ZnS QDs reported previously. For our cellular imaging experiment, Figure 4B shows the internalization of the water-soluble ZAIS QDs (with 606 nm emission) in human MSCs. The QDs were primarily found to be present in the cytoplasm of the hMSCs with a few QDs being found in the nucleus of the cells. Furthermore, fluorescent microscopy images of U87 cells (Figure 4C) and hMSCs (Figure 4D) incubated with the water soluble ZAIS-QDs proved that the QDs showed strong fluorescence even when inside the cells. Collectively, our ZAIS-QDs showed excellent biocompatibility (non-toxic) and these results can be extended to their wide use in cellular imaging and delivery applications, especially for stem cells, which are known to be extremely sensitive towards nanomaterials.
Figure 4. Biocompatibility and cellular imaging studies of ZAIS-QDs in mammalian cells.
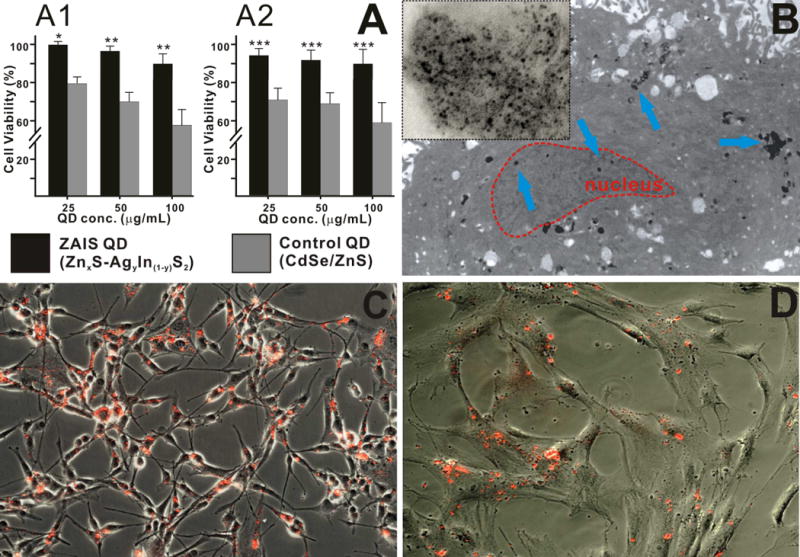
(A) Comparison of the cellular cytotoxicity of the water soluble ZAIS QD (Zn0S − Ag0.2In0.8S2) and CdSe/ZnS QDs at different concentrations in U87 cells (A1) and hMSCs (A2). The results are presented as means ± SD from three independent experiments. Student’s unpaired t-test was used for evaluating the statistical significance of the cytotoxicity of ZAIS QDs (* = P < 0.001, ** = P < 0.01, *** = P < 0.05) as compared to the CdSe/ZnS QDs (B) Transmission electron microscopy of the ZAIS QD in hMSCs. The image clearly shows the presence of the QDs (marked by blue arrows) in the cytoplasm and the nucleus (marked in red). The inset depicting the magnified image of the QD cluster, confirms the monodisperse nature of the QDs inside the cell. (C) and (D) Fluorescence microscopy imaging demonstrating the uptake of the water soluble ZAIS-QD (λem = 606 nm) in U87 cells (C) and hMSCs (D).
Having demonstrated the potential of our ZAIS QDs as non-toxic fluorescent imaging probes, we evaluated their ability to be used as vehicles for the efficient delivery and tracking of siRNA in vitro. In the past decade, there has been a considerable interest in the development of nanomaterial-based siRNA and gene delivery methods for controlling cell fate and behavior.[11] RNA interference (RNAi) involves the use of small interfering RNAs (siRNAs) to selectively mediate the cleavage of complementary mRNA sequences and thus regulate target gene expression. In combination with other modalities like small molecules and peptides, RNAi could prove to be a powerful tool to manipulate the cellular microenvironments.[11b] In this context, the use of QDs as a multimodal delivery vehicle becomes a promising choice, thereby allowing for the efficient delivery of siRNA and real-time tracking of the siRNA-mediated gene knockdown.[2c] As a proof-of-concept experiment, we used a brain tumor cell line (U87) which was genetically labeled to express the green fluorescent protein (GFP). In order to deliver the siRNA to target brain tumor cellsexpressing EGFP (U87-EGFP), the MPA-coated ZAIS QDs were conjugated to the siRNA (against the EGFP gene) using polyethyleneimine (PEI) as a cationic polymer via a layer-by-layer approach (Figure 1B).[12] The efficiency of the PEI coating and siRNA conjugation was monitored using zeta potential (Figure S11). The decrease in the green fluorescence due to the siRNA-mediated knockdown of the EGFP gene using our ZAIS QD-siRNA constructs was then monitored (Figure 5) in order to to assess the transfection efficiency and RNA interference (RNAi) activity. It was found that the ZAIS QD-siRNA constructs were efficiently taken up by cells as evident by the intracellular red fluorescence of the QDs. In addition, the QD localization correlated well with the decrease of the green fluorescence (~ 80 % after 3 days of transfection) resulting from the siRNA-mediated knockdown of the EGFP mRNA. These results show the ability of our ZAIS QDs to efficiently translocate siRNA into the cells and achieve gene knockdown in vitro. With further surface modifications using targeting ligands (RGD or TAT peptides and antibodies) and bifunctional linkers (cleavable linkers or covalent linkers), the ZAIS QDs could be used for the targeted delivery of siRNA to cancer/stem cells and for the real-time monitoring of its delivery in an efficient manner. In addition, the presence of a library of multicolored QDs obtained via our sonochemical approach, would allow the for multiplexed imaging of transplanted cell populations in vivo (i.e., tracking different cell populations with different QDs using different emission wavelengths at the same time).
Figure 5. In vitro testing of the ZAIS-QD-siEGFP cell uptake and silencing efficiency in stably transfected U87-EGFP glioblastoma cells.
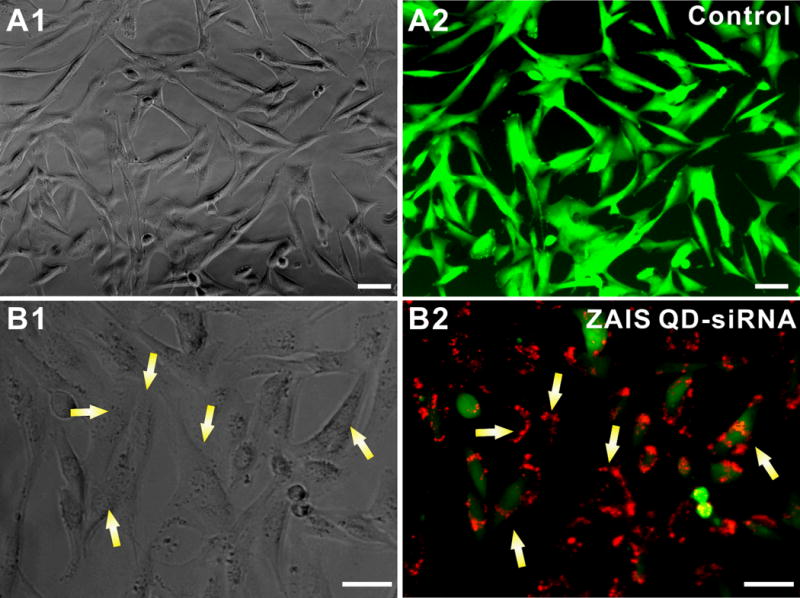
(A) Control U87-EGFP cells with PEI-coated ZAIS-QD; (A1) represents the phase contrast image and (A2) is the corresponding fluorescence image. (B): EGFP knockdown using the ZAIS QD-siRNA constructs; (B1) Phase contrast image showing the the viability of U87-EGFP cells has not changed appreciably after the transfection of the ZAIS QD-siRNA constructs as compared to the control cells in (A). (B2) Fluorescence image clearly shows the knockdown of EGFP in cells which have internalized the siRNA-QDs (red) after 72 hrs. The red fluorescence from the ZAIS QDs correlates well with the loss of the green fluorescence in cells (indicated by yellow arrows). Scale bar is 50 μm
In summary, we successfully demonstrated the preparation of ZnxS − AgyIn1−yS2 (ZAIS) QDs using a facile sonochemical synthetic method. The physicochemical and bio-relevant properties of the resulting QDs can be easily tuned over the entire visible spectrum by varying the chemical composition of the precursors. We also demonstrated that our ZAIS QDs can exhibit excellent biocompatibility for the efficient delivery of siRNA and simultaneous imaging/tracking of the same in cancer cells (and stem cells) with negligible QD-induced cytotoxicity. While the ZAIS QDs show great potential for the imaging and delivery of siRNA in vitro, a more thorough investigation of their long-term cytotoxicity is needed before they can be used in vivo. Efforts in this direction are underway. Overall, the ease of the synthesis of the ZAIS QDs, their excellent cyto-compatibility and their versatility as multiplexed imaging agents provides an attractive alternative over conventional QD-based molecular imaging probes and siRNA delivery vehicles. The above methodology could be potentially extended to synthesize libraries of various types of nanoparticles (magnetic nanoparticles and upconverting near-IR fluorescent nanoparticles), thereby allowing for rapid screening of the nanomaterials for biomedical applications such as drug delivery and cellular labeling.
Experimental
Synthesis of Precursor Complexes
An aqueous solution of sodium diethyldithiocarbamate (0.05 M, 5.0 mL) was mixed with an aqueous solution containing appropriate amounts of AgNO3, In(NO3)3.xH2O and Zn(NO3)2.6H2O in order to get the required mole ratios (Total concentration of the metal ions was 0.025M). The solution was allowed to stir for 5 minutes after which it was filtered using a buchhner funnel, washed several times with distilled water and MeOH and finally dried in a convection oven at 60 deg C overnight to obtain the precursor as a dried powder. Several precursor powders were synthesized by varying the mole ratios of the metal salts.
Sonochemical synthesis of dodecylamine-capped ZnS-AgInS2 quantum dots
The precursor complex (0.1 g) and dodecylamine (10.0 mL) were put into a 20 mL vial and sonicated using a tip probe-based high frequency sonicator (Branson) for 5 minutes in an air-atmosphere. The resulting suspension was allowed to sit at room temperature for two minutes after which 5.0 mL of chloroform (5.0 mL) and MeOH (5.0 mL) were added to it and centrifuged at 4000 rpm. The supernatant containing the ZAIS QDs was collected and equal amount of MeOH was added to it in order to isolate the nanoparticles. The obtained ZAIS QDs were then resuspended in chloroform for absorbance and photoluminescence measurements.
Surface modification of ZAIS-QDs with 3-mercaptopropionic acid (MPA)
The dodecylamine-capped ZAIS QDs were subjected to a ligand exchange reaction using 3-mercaptopropionic acid (MPA) according to a previously reported protocol.[9] Briefly, a 3.0 mL ethanolic solution of MPA (0.2 M) and KOH (0.3 M) was added dropwise to an equal amount of the dodecylamine-capped ZAIS QD solution in chloroform. The turbid solution was stirred for 3h at room temperature followed by centrifugation at 4000 rpm. The wet precipitate of the MPA-coated ZAIS QDs was washed with EtOH and redissolved in phosphate-buffered saline (PBS). The water soluble MPA-coated ZAIS QDs were stable in buffer solution with no significant change in absorption and photoluminescence for upto 2 months, when stored at ambient conditions.
Culture of human U87 glioblastoma cells, NIH-3T3 mouse fibroblasts and human mesenchymal stem cells
The EGFRvIII overexpressed U87 glioblastoma cells (U87) and human mesenchymal stem cells (hMSCs) were cultured using previously reported methods. For U87-EGFRvIII cells, DMEM with high glucose, 10% fetal bovine serum (FBS, Gemini Bioproducts), 1% Streptomycin-penicillin and 1% Glutamax (Invitrogen, Carlsbad, CA) were used as basic components of growth media including Hygromycin (100 μg/ml, Invitrogen) as a selection marker. Human bone marrow-derived MSCs (Lonza, Walkerville) were cultured in the conditioned media (Lonza, Walkerville) according to manufacturer’s recommendations. For the NIH-3T3 mouse fibroblasts, DMEM (with high glucose) supplemented with 10% fetal calfserum (FCS, Gemini Bioproducts), 1% Streptomycin-penicillin and 1% Glutamax was used as the growth medium. All cells were maintained at 37 o C in humidified 5% CO2 atmosphere.
Supplementary Material
Acknowledgments
We thank Prof. Gene Hall for help with the XRF measurements, Dr. Tom Emge and Vaishali Thakral for their support with the XRD and the IAMDN (Rutgers) for allowing us to use their high resolution TEM facility. We are also grateful to Drs. Joung Kyu Park and Jeong Je Cho for their scientific input and the KBLee group members for their valuable suggestions for the manuscript. KBLee acknowledges the NIH Director’s Innovator Award (1DP20D006462-01) and is also grateful to the N.J. Commission on Spinal Cord grant (09-3085-SCR-E-0). This study is also funded in part by the US EPA (Grant# 83469302) and the UK NERC (Grant# NE/H012893).
Footnotes
Supporting Information is available from the Wiley Online Library orfrom the author.
References
- 1.a) Pons T, Mattoussi H. Annals of Biomed Eng. 2009;37:1934–1959. doi: 10.1007/s10439-009-9715-0. [DOI] [PubMed] [Google Scholar]; b) Solanki A, Kim JD, Lee KB. Nanotechnology. 2008;3:567–578. doi: 10.2217/17435889.3.4.567. [DOI] [PubMed] [Google Scholar]
- 2.a) Delehanty JB, Mattoussi H, Medintz IL. Analytical and Bioanalytical Chemistry. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]; b) Derfus AM, Chan WCW, Bhatia SN. Nano Lett. 2003;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jung J, Solanki A, Memoli KA, Kamei KI, Kim H, Drahl MA, Williams LJ, Tseng HR, Lee KB. Angew Chem Int Ed. 2010;49:103–107. doi: 10.1002/anie.200905126. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Nat Biotech. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 3.a) Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconj Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]; b) Smith AM, Duan H, Mohs AM, Nie S. Adv Drug Del Rev. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. J Am Chem Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su Y, He Y, Lu H, Sai L, Li Q, Li W, Wang L, Shen P, Huang Q, Fan C. Biomat. 2009;30:19–25. doi: 10.1016/j.biomaterials.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 5.a) Allen PM, Bawendi MG. J Am Chem Soc. 2008;130:9240–9241. doi: 10.1021/ja8036349. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cassette E, Pons T, Bouet C, Helle M, Bezdetnaya L, Marchal F, Dubertret B. Chem Mat. 2010;22:6117–6124. [Google Scholar]; c) Hamanaka Y, Ogawa T, Tsuzuki M, Kuzuya T. J Phys Chem C. 2011;115:1786–1792. [Google Scholar]; d) Tang X, Cheng W, Choo ESG, Xue J. Chem Comm. 2011;47:5217–5219. doi: 10.1039/c1cc10417a. [DOI] [PubMed] [Google Scholar]; e) Torimoto T, Adachi T, Okazaki KI, Sakuraoka M, Shibayama T, Ohtani B, Kudo A, Kuwabata S. J Am Chem Soc. 2007;129:12388–12389. doi: 10.1021/ja0750470. [DOI] [PubMed] [Google Scholar]; f) Xie R, Rutherford M, Peng X. J Am Chem Soc. 2009;131:5691–5697. doi: 10.1021/ja9005767. [DOI] [PubMed] [Google Scholar]
- 6.Zhong H, Zhou Y, Ye M, He Y, Ye J, He C, Yang C, Li Y. Chem Mat. 2008;20:6434–6443. [Google Scholar]
- 7.Nairn JJ, Shapiro PJ, Twamley B, Pounds T, von Wandruszka R, Fletcher TR, Williams M, Wang C, Norton MG. Nano Lett. 2006;6:1218–1223. doi: 10.1021/nl060661f. [DOI] [PubMed] [Google Scholar]
- 8.Gardner J, Shrudha E, Lau L, Wang C, Rodriguez R, Pak J. J Nanoparticle Research. 2008;10:633–641. [Google Scholar]
- 9.a) Chen CC, Yet CP, Wang HN, Chao CY. Langmuir. 1999;15:6845–6850. [Google Scholar]; b) Mitchell GP, Mirkin CA, Letsinger RL. J Am Chem Soc. 1999;121:8122–8123. [Google Scholar]
- 10.a) Mitchell P. Nat Biotech. 2001;19:1013–1017. doi: 10.1038/nbt1101-1013. [DOI] [PubMed] [Google Scholar]; b) Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. Current Opinion in Biotechnology. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]; c) Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Shah BS, Clark PA, Moioli EK, Stroscio MA, Mao JJ. Nano Lett. 2007;7:3071–3079. doi: 10.1021/nl071547f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Park JK, Jung J, Subramaniam P, Shah BP, Kim C, Lee JK, Cho JH, Lee C, Lee KB. Small. 2011;7:1647–1652. doi: 10.1002/smll.201100012. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shah BP, Kim C, Subramaniam P, Lee KB. Mol Pharmaceutics. 2011;8:1955–1961. doi: 10.1021/mp100460h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang H, Chen K-J, Wang S, Ohashi M, Kamei K-I, Sun J, Ha JH, Liu K, H-R T. Chem Comm. 2010;46:1851–1853. doi: 10.1039/b923711a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xue X, Wang F, Liu X. J Mater Chem. 2011;21:13107–13127. [Google Scholar]
- 12.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Nano Lett. 2009;9:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


