Abstract
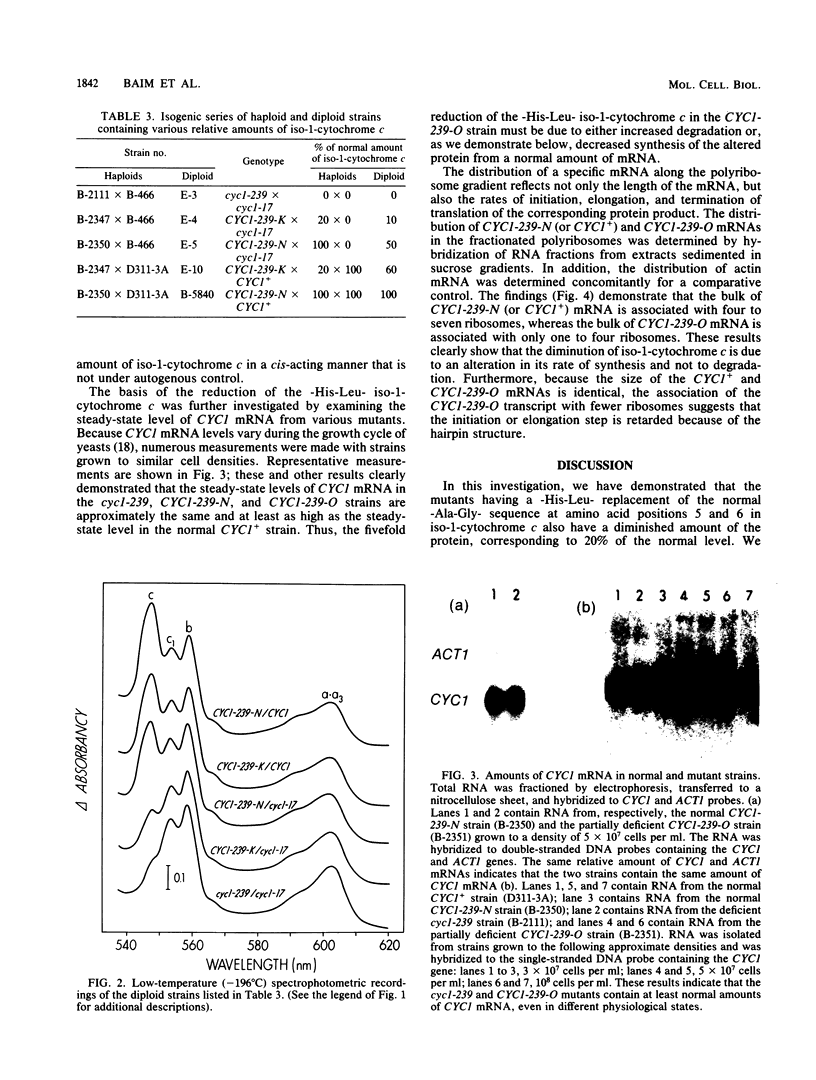
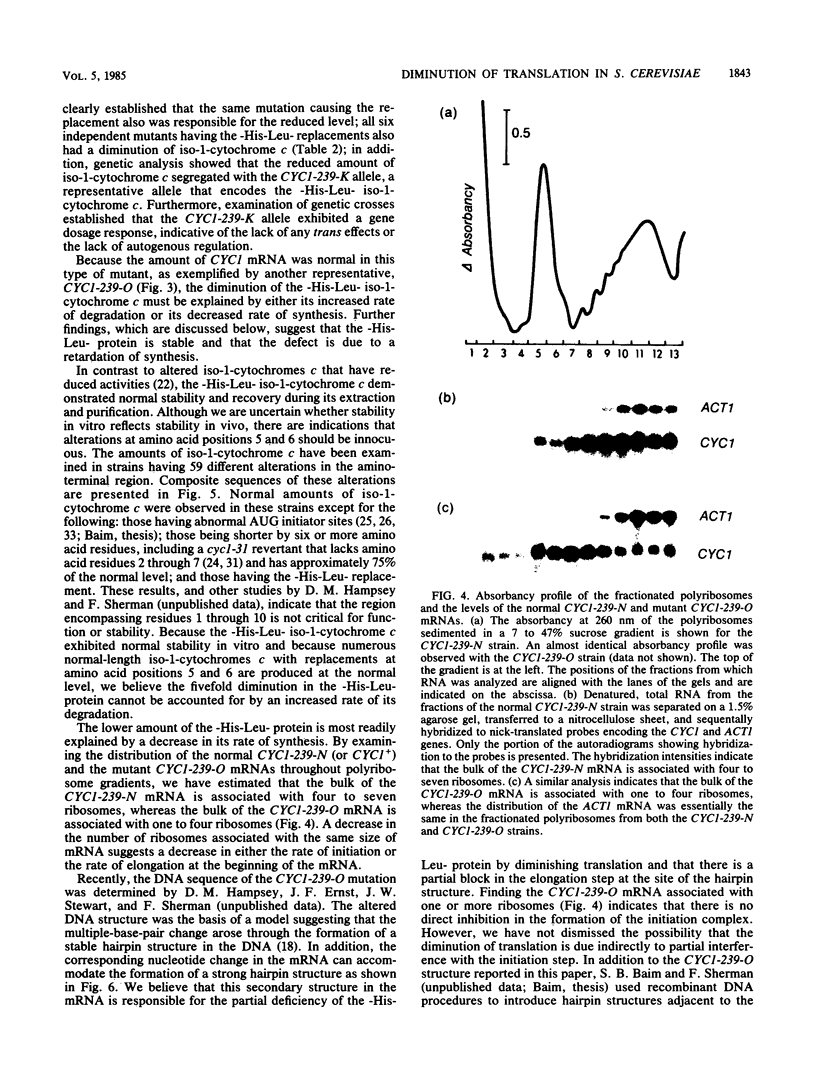
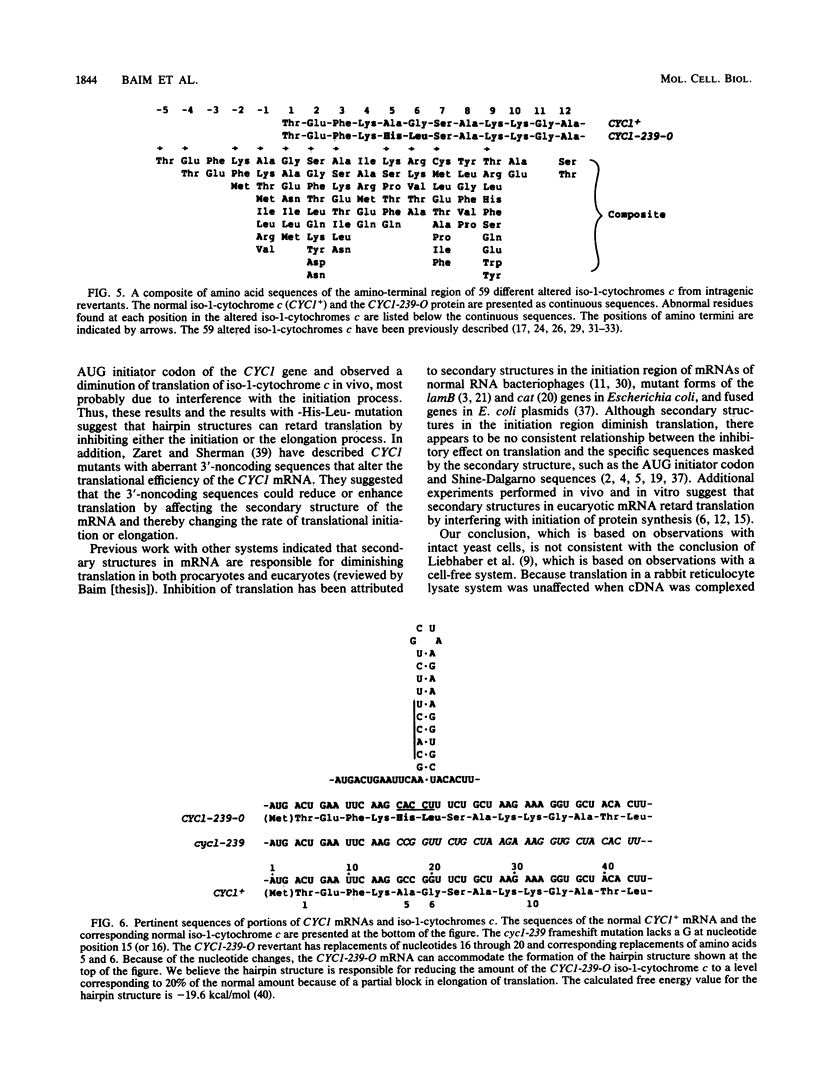
The CYC1-239-O mutation in the yeast Saccharomyces cerevisiae produces a -His-Leu- replacement of the normal -Ala-Gly- sequence at amino acid positions 5 and 6, which lie within a dispensable region of iso-1-cytochrome c; this mutation can accommodate the formation of a hairpin structure at the corresponding site in the mRNA. The amount of the altered protein was diminished to 20% of the wild-type level, whereas the amount of the mRNA remained normal. However, in contrast to the normal CYC1+ mRNA that is associated mainly with four to seven ribosomes, the bulk of the CYC1-239-O mRNA is associated with one to four ribosomes. These results suggest that the stable secondary structure within the translated region of the CYC1 mRNA diminishes translation by inhibiting elongation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballinger D. G., Pardue M. L. The control of protein synthesis during heat shock in Drosophila cells involves altered polypeptide elongation rates. Cell. 1983 May;33(1):103–113. doi: 10.1016/0092-8674(83)90339-2. [DOI] [PubMed] [Google Scholar]
- Gheysen D., Iserentant D., Derom C., Fiers W. Systematic alteration of the nucleotide sequence preceding the translation initiation codon and the effects on bacterial expression of the cloned SV40 small-t antigen gene. Gene. 1982 Jan;17(1):55–63. doi: 10.1016/0378-1119(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Kastelein R. A., Berkhout B., Overbeek G. P., van Duin J. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983 Sep;23(3):245–254. doi: 10.1016/0378-1119(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980 Jan;19(1):79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Sherman F., Jackson M., Gilmore R. A. Mapping and gene conversion studies with the structural gene for iso-1-cytochrome C in yeast. Genetics. 1975 Dec;81(4):615–629. doi: 10.1093/genetics/81.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laz T. M., Pietras D. F., Sherman F. Differential regulation of the duplicated isocytochrome c genes in yeast. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4475–4479. doi: 10.1073/pnas.81.14.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhaber S. A., Cash F. E., Shakin S. H. Translationally associated helix-destabilizing activity in rabbit reticulocyte lysate. J Biol Chem. 1984 Dec 25;259(24):15597–15602. [PubMed] [Google Scholar]
- Lizardi P. M., Mahdavi V., Shields D., Candelas G. Discontinuous translation of silk fibroin in a reticulocyte cell-free system and in intact silk gland cells. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6211–6215. doi: 10.1073/pnas.76.12.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J Biol Chem. 1971 Dec 10;246(23):7131–7138. [PubMed] [Google Scholar]
- Lodish H. F. Specificity in bacterial protein synthesis: role of initiation factors and ribosomal subunits. Nature. 1970 May 23;226(5247):705–707. doi: 10.1038/226705a0. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payvar F., Schimke R. T. Methylmercury hydroxide enhancement of translation and transcription of ovalbumin and conalbumin mRNA's. J Biol Chem. 1979 Aug 25;254(16):7636–7642. [PubMed] [Google Scholar]
- Protzel A., Morris A. J. Gel chromatographic analysis of nascent globin chains. Evidence of nonuniform size distribution. J Biol Chem. 1974 Jul 25;249(14):4594–4600. [PubMed] [Google Scholar]
- Putterman G. J., Margoliash E., Sherman F. Identification of missense mutants by amino acid replacements in iso-1-cytochrome c from yeast. J Biol Chem. 1974 Jul 10;249(13):4006–4015. [PubMed] [Google Scholar]
- Ripley L. S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. D. Structure analysis of three T7 late mRNA ribosome binding sites. J Mol Biol. 1981 Mar 25;147(1):55–71. doi: 10.1016/0022-2836(81)90079-6. [DOI] [PubMed] [Google Scholar]
- Schottel J. L., Sninsky J. J., Cohen S. N. Effects of alterations in the translation control region on bacterial gene expression: use of cat gene constructs transcribed from the lac promoter as a model system. Gene. 1984 May;28(2):177–193. doi: 10.1016/0378-1119(84)90255-5. [DOI] [PubMed] [Google Scholar]
- Schwartz M., Roa M., Débarbouillé M. Mutations that affect lamB gene expression at a posttranscriptional level. Proc Natl Acad Sci U S A. 1981 May;78(5):2937–2941. doi: 10.1073/pnas.78.5.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber M. E., Stewart J. W., Sherman F. Primary site and second site revertants of missense mutants of the evolutionarily invariant tryptophan 64 in iso-1-cytochrome c from yeast. J Biol Chem. 1979 May 25;254(10):4132–4143. [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Parker J. H., Inhaber E., Shipman N. A., Putterman G. J., Gardisky R. L., Margoliash E. The mutational alteration of the primary structure of yeast iso-1-cytochrome c. J Biol Chem. 1968 Oct 25;243(20):5446–5456. [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Schweingruber A. M. Mutants of yeast initiating translation of iso-1-cytochrome c within a region spanning 37 nucleotides. Cell. 1980 May;20(1):215–222. doi: 10.1016/0092-8674(80)90249-4. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Specific recognition of non-initiator regions in RNA bacteriophage messengers by ribosomes of Bacillus stearothermophilus. J Mol Biol. 1973 Jan;73(1):1–16. doi: 10.1016/0022-2836(73)90155-1. [DOI] [PubMed] [Google Scholar]
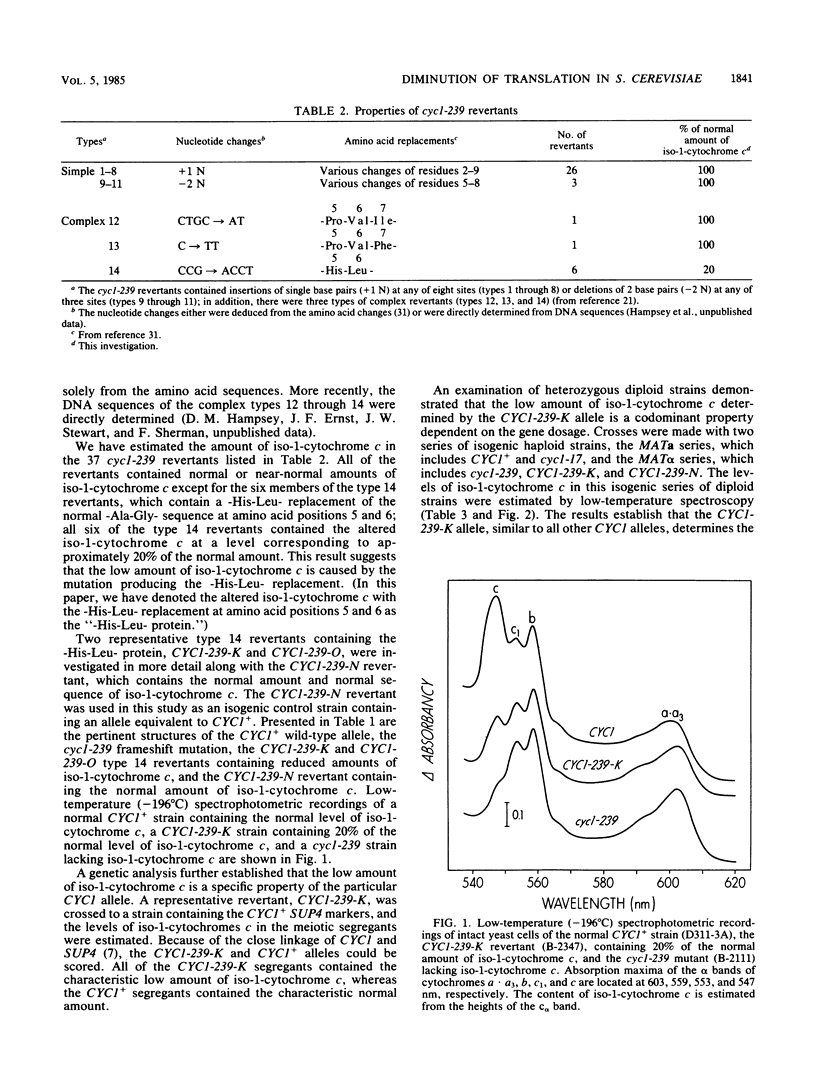
- Stewart J. W., Sherman F., Jackson M., Thomas F. L., Shipman N. Demonstration of the UAA ochre codon in bakers yeast by amino-acid replacements in iso-1-cytochrome c. J Mol Biol. 1972 Jul 14;68(1):83–96. doi: 10.1016/0022-2836(72)90264-1. [DOI] [PubMed] [Google Scholar]
- Stewart J. W., Sherman F., Shipman N. A., Jackson M. Identification and mutational relocation of the AUG codon initiating translation of iso-1-cytochrome c in yeast. J Biol Chem. 1971 Dec 25;246(24):7429–7445. [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeau M., Stanley K. K. Enhanced expression of cro-beta-galactosidase fusion proteins under the control of the PR promoter of bacteriophage lambda. EMBO J. 1982;1(10):1217–1224. doi: 10.1002/j.1460-2075.1982.tb00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]