Abstract
How cells integrate multiple patterning signals to achieve early endoderm regionalization remains largely unknown. Between gastrulation and neurulation, retinoic acid (RA) signaling is required, while Wnt/β-catenin signaling has to be repressed for the specification of the pancreas, oesophagus, stomach, and duodenum primordia in Xenopus embryos. In attempt to screen for RA regulated genes in Xenopus endoderm, we identified a direct RA target gene, N-myc downstream regulated gene 1a (ndrg1a) that showed expression early in the archenteron roof endoderm and late in the developing pancreas, oesophagus, stomach, and duodenum. Both antisense morpholino oligonucleotide mediated knockdown of ndrg1a in Xenopus laevis and the transcription activator-like effector nucleases (TALEN) mediated disruption of ndrg1 in Xenopus tropicalis demonstrate that like RA signaling, Ndrg1a is specifically required for the specification of Xenopus pancreas, oesophagus, stomach, and duodenum primordia. Immunofluorescence data suggest that RA-activated Ndrg1a suppresses Wnt/β-catenin signaling in Xenopus archenteron roof endoderm cells. Blocking Wnt/β-catenin signaling rescued Ndrg1a knockdown phenotype. Furthermore, overexpression of the putative Wnt/β-catenin target gene Atf3 phenocopied knockdown of Ndrg1a or inhibition of RA signaling, while Atf3 knockdown can rescue Ndrg1a knockdown phenotype. Lastly, the pancreas/stomach/duodenum transcription factor Pdx1 was able to rescue Atf3 overexpression or Ndrg1a knockdown phenotype. Together, we conclude that RA activated Ndrg1a represses Wnt/β-catenin signaling to allow the specification of pancreas, oesophagus, stomach, and duodenum progenitor cells in Xenopus embryos.
Introduction
The regionalization of endoderm occurs concurrently with its formation during gastrulation. By the end of gastrulation, broad antero-posterior (AP) domains within the endoderm have been established, as reflected by the anterior expression of Hhex, vpp1, Sox2, and Foxa2 and the posterior expression of the caudal type homeobox genes Cdx1, 2, and 4 [1], [2], [3]. Eventually, the endoderm gives rise to the epithelia of the respiratory and gastrointestinal tracts and their associated organs, such as the thyroid, lungs, pancreas, liver and gall bladder. The lineage tracing for vertebrate endodermal organogenesis is largely dependent on the vital dye labeling-based fate mapping studies [4], [5], [6], [7], [8]. The early expression of hhex and vpp1 in Xenopus embryos might serve to reflect the precursors for the liver and ventral pancreas, respectively [3]. So far, there are few specific marker genes reported, which can demarcate precursors for dorsal pancreas, oesophagus, stomach, and duodenum during gastrulation and neurulation.
Retinoic acid (RA) signaling plays a conserved role in the AP patterning of vertebrate endoderm as early as gastrulation [9]. Studies in frog, avian, and mice indicate that the RA signaling is specifically required for the formation of dorsal pancreas, oesophagus, stomach, and duodenum primordia [10], [11], [12], [13], [14], [15]. In zebrafish, RA signaling before the end of gastrulation is required for the initial development of both hepatic and pancreatic endoderm [16]. RA activated expression of RA-degrading enzyme cyp26a1 in the anterior trunk endoderm in turn modulates RA signaling and defines the anterior boundary of pancreatic field in zebrafish embryos [17]. mnx1 is identified as an RA downstream gene that promotes beta cell formation in the developing zebrafish endocrine pancreas [18], while RA downstream gene exdpf regulates fish exocrine pancreas development [19]. The endodermal RA target genes that mediate the early activities of RA signaling to specify the pancreas, oesophagus, stomach, and duodenum anlagen proper remain to be identified.
Canonical Wnt signaling also plays an important role in the AP patterning of vertebrate endoderm. In mice, compound knockout of Tcf1 and Tcf4 led to an anterior transformation of duodenum into stomach with little or no intestine developed [20]. The stomach mesenchymal transcription factor Barx1-mediated secretion of the Wnt antagonists SFRP1 and SFRP2 is required to inhibit endodermal Wnt/β-catenin signaling and thus to permit specification of the stomach epithelium [21]. Consistently, in Xenopus, Wnt/β-catenin signaling must be repressed in anterior endoderm between gastrula and early somite stages of development to allow the formation of the liver as well as the pancreas, stomach, and duodenum primordia [22], [23]. The secreted Wnt antagonist sfrp5 expressed in the ventral foregut endoderm coordinates the liver and ventral pancreas specification by antagonizing both canonical and noncanonical Wnt signaling [24], [25]. In addition, it has been shown that RA can repress Xenopus blastula Wnt/β-catenin signaling [26].
N-myc downstream regulated gene 1 (NDRG1) belongs to the NDRG protein family consisting of four members, NDRG1–4, which are characterized by containing a NDR domain and an α/β hydrolase-fold motif [27]. NDRG1 is an RA-inducible gene in various cell lines and human patients [28], [29], [30], [31]. Ndrg1 deficiency in mice leads to Schwann cell dysfunction, suggesting that NDRG1 is essential for maintenance of the myelin sheaths in peripheral nerves [32], [33]. Ndrg1-null mice also showed impaired mast cell maturation and degranulation and attenuated allergic responses [34]. In Xenopus, ndrg1 was identified as a gene enriched for expression in endoderm and pronephros [35], [36]. A recent study using cancer cell lines reveals that the tumor metastasis suppressor gene, NDRG1, represses Wnt/β-catenin signaling by directly interacting with the Wnt receptor, LRP6, consequently causing the suppression of Wnt/β-catenin target gene, ATF3 expression [37]. Activating transcription factor 3 (ATF3) belongs to the ATF/cyclic AMP responsive element binding family of basic leucine zipper transcription factors. It is an adaptive response gene and can act both as a transcriptional activator or repressor depending on the cell type and stimulus [38], [39], [40]. ATF3 knockout mice developed normally and did not show any discernable phenotypes under normal conditions [41]. In contrast, transgenic mice expressing ATF3 in the liver, pancreatic ductal epithelium, or pancreatic β cells led to liver dysfunction, defects in endocrine pancreas development, or islet dysfunction, respectively [41], [42], [43].
In this study, we used DNA microarray in combination with whole mount in situ hybridization to screen RA regulated genes in Xenopus endoderm. ndrg1a was identified and verified being directly activated by RA in the archenteron roof endoderm and late in the developing pancreas, oesophagus, stomach, and duodenum. We provide evidences implicating that RA-activated Ndrg1a represses Wnt/β-catenin signaling in the archenteron roof endoderm cells and consequently releases the inhibitory effect of Wnt/β-catenin signaling activities on the formation of pancreas, oesophagus, stomach, and duodenum.
Results
ndrg1a, an RA Regulated Gene, is Expressed Early in the Endodermal Archenteron Roof
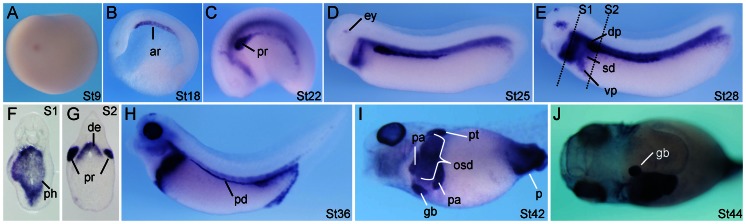
With respect to endodermal organogenesis, Xenopus embryos treated with an RA antagonist, BMS453, before the end of gastrulation displayed specific loss of the dorsal pancreas and part of ventral pancreas, stomach, and duodenum [10]. To identify RA regulated genes in endoderm, we first did microarray analysis comparing gene expression levels between wild-type and RA-deficient embryos, which were treated with BMS453 at the beginning of gastrulation and were collected at stages 12, 23, and 34, respectively. The data obtained indicate that genes down-regulated more than 2-fold at three different stages showed limited overlap (Fig. S1 and Table S1). In embryos collected at stage 23, there are 138 genes that were down-regulated more than 2-fold upon BMS453 treatment. According to the 138 genes’ structural and functional information available in the NCBI database, we gave priorities to transcription factors, kinases, RNA binding proteins, transmembrane proteins, as well as genes showing endodermal expression, and thus chose 9 genes (Table S1) to further analyze their embryonic expression patterns by whole mount in situ hybridization. One of them, ndrg1a, showed specific expression early in the archenteron roof endoderm (Fig. 1B), which was not detected by previous studies, presumably due to insufficient sensitivity of the whole mount in situ hybridization technique in previous publications [35], [36]. In agreement with the report from the Zorn laboratory [35], our data indicate that ndrg1a transcripts were detected in dorsal endoderm at early tail bud stage of development (Fig. 1C, D, E, G). At late tail bud and tadpole stages of development, in addition to its expression in eye, proctodaeum precursors, and pronephros, ndrg1a showed specific expression in the pharynx, oesophagus, pancreas, stomach, and duodenum (Fig. 1E, F, H, I, J). We were unable to detect ndrg1a expression in the liver [35]. Instead, a clear ndrg1a signal was observed in developing gall bladder (Fig. 1I, J).
Figure 1. Xenopus ndrg1a is expressed early in the archenteron roof endoderm and late in pancreas, oesophagus, stomach, and duodenum primordia, as revealed by whole mount in situ hybridization.
(A) Lateral view of a blastula. (B) Whole mount in situ hybridization on bisected stage 18 neurula, anterior toward the left. (C–E) Lateral view, head toward the left. Note that the foregut ndrg1a signal in E covers pharynx, oesophagus, pancreas, stomach, and duodenum. (F, G) Transversal sections of a stage 28 embryo at the levels illustrated by the dashed lines in E. (H, I) Lateral view, head toward the left. (J) Ventral view, head toward the left. Abbreviations: ar, archenteron roof endoderm; de, dorsal endoderm; dp, dorsal pancreatic bud; ey, eye; gb, gall bladder; osd, oesophagus, stomach and duodenum anlagen; p, proctodaeum; pa, pancreas; pd, pronephric duct; ph, pharynx; pr, pronephric anlage; pt, pronephric proximal tubules; vp, ventral pancreatic bud.
ndrg1a Expression in Archenteron Roof Endoderm Cells is Directly Activated by RA
To further verify the microarray data, we analyzed ndrg1a expression in BMS453 and RA treated embryos by whole mount in situ hybridization. Upon BMS453 treatment, no ndrg1a transcripts were detected in archenteron roof endoderm cells (Fig. 2E) and only traces of ndrg1a mRNA remained in dorsal endoderm of stage 22 embryos (Fig. 2H). Later on, ndrg1a expression was completely repressed in dorsal pancreas, oesophagus, stomach, and duodenum and was partially inhibited in ventral pancreas (Fig. 2K, N). In contrast, another RA antagonist BMS493 can only partially inhibit ndrg1a expression in pancreas, oesophagus, stomach, and duodenum [11]. In addition, ndrg1a expression in pronephric proximal tubules was also severely inhibited by BMS453 (Fig. 2N). It should be noted that ndrg1a expression in eye, brain, and gall bladder was less affected upon BMS453 treatment (Fig. 2K, N). It is a common phenomenon that for genes expressing in pancreas as well as in eye and central nervous system, such as ndrg1a, esr10, neurod, and ptf1a/p48, only their pancreatic expression is inhibited upon BMS453 treatment (Fig. 2K, N and [10]). Conversely, application of RA to Xenopus embryos induced significant expansion of ndrg1a expression domains in archenteron roof endoderm (Fig. 2F), dorsal endoderm (Fig. 2I), pancreas, oesophagus, stomach, and duodenum (Fig. 2L, O). Thus, RA signaling is necessary and sufficient to activate ndrg1a expression in the archenteron roof endoderm cells.
Figure 2. RA directly activates ndrg1a expression in Xenopus archenteron roof endoderm cells.
(A–O) RA signaling is necessary and sufficient to activate ndrg1a expression in the archenteron roof endoderm cells as well as in the pancreas, oesophagus, stomach, and duodenum primordia. Embryos were treated with 0.25 µM BMS453 or 5 µM RA at stage 10 for one hour, collected at stages indicated in the control panels, and subjected to whole mount in situ hybridization analyses of ndrg1a expression. (A–I) Whole mount in situ hybridization on bisected embryos, anterior toward the left. (J–O) Lateral view, head toward the left. Red arrows in panels K and N and red triangles in panels L and O point to the loss or expansion of ndrg1a expression in pancreas, oesophagus, stomach, and duodenum upon BMS453 or RA treatment, respectively. (P–S) RA directly activates ndrg1a expression in Xenopus archenteron roof endoderm cells. Embryos were treated with cycloheximide (CHX, 10 µg/ml), 5µM RA, or both at stage 15 for one hour, fixed at stage 20, and bisected for whole mount in situ hybridization analyses of ndrg1a expression. For the combined treatment, CHX was added 15 min before the application of RA. Anterior is toward the left. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images.
To address if RA can directly regulate ndrg1a expression, we treated Xenopus embryos with cycloheximide (CHX), a protein synthesis inhibitor, 15 minutes before the application of RA. Our data indicate that RA induced expansion of ndrg1a expression in archenteron roof endoderm was not affected by CHX (Fig. 2S), supporting the notion that RA directly activates ndrg1a expression in archenteron roof endoderm cells. Indeed, an RA response element was identified in Xenopus tropicalis ndrg1 promoter region (AGTTCAacAGTTCA −1496bp to −1509bp). Unlike Xenopus laevis, the diploid Xenopus tropicalis has one ndrg1 gene.
ndrg1a Knockdown in Xenopus Embryos Phenocopies BMS453 Treatment
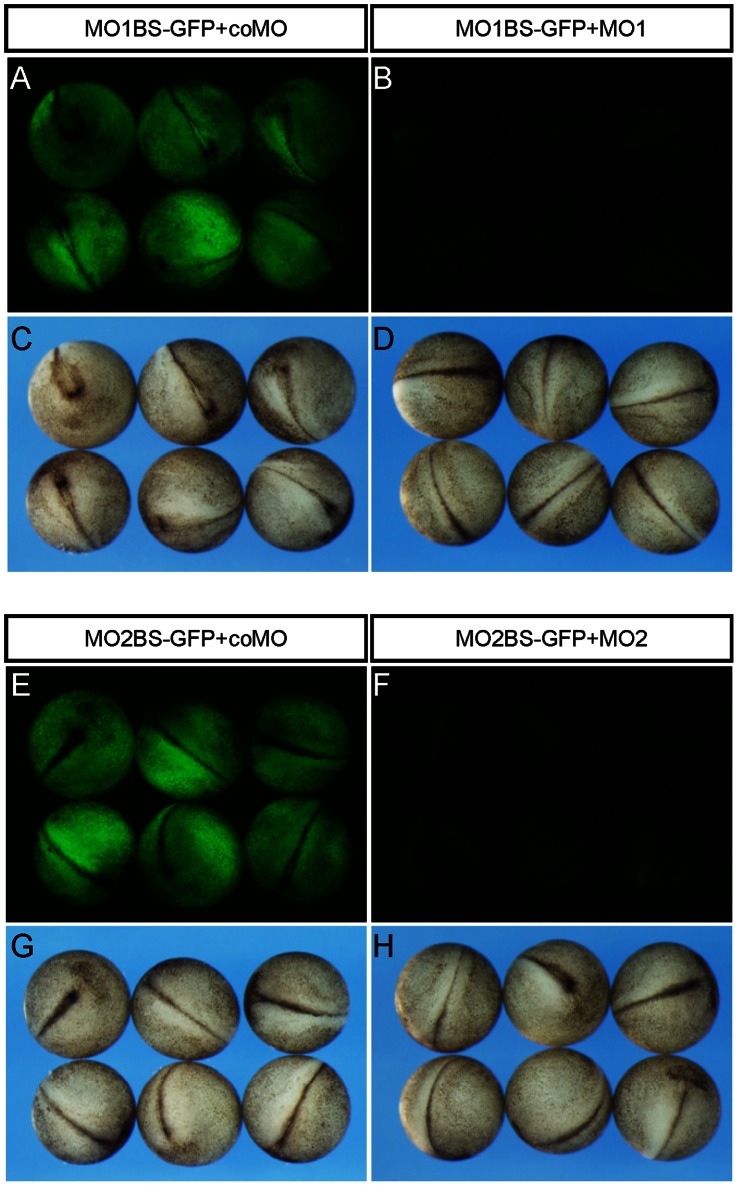
To generate ndrg1a morphants, we purchased two ndrg1a antisense morpholino oligos (MO) from Gene Tools. One covers the ATG start codon (MO1) and the other (MO2) locates in the 5′ untranslated region of the ndrg1a cDNA. Their efficiency was tested by an in vivo assay, in which GFP coding sequence was fused to a MO’s binding site and the resultant mRNA was co-injected with the MO into Xenopus fertilized eggs to evaluate live GFP translation in embryos. Both MO1 (Fig. 3B) and MO2 (Fig. 3F) were able to efficiently inhibit the translation of GFP open reading frame following the corresponding MO1 or MO2 binding sequences, respectively, while control MO (coMO) failed to do so (Fig. 3A, E).
Figure 3. The efficiency of ndgr1a MO1 and MO2 was verified in vivo.
(A–D) Fertilized Xenopus eggs were co-injected with 3.5 picomoles of coMO or ndrg1a MO1 in combination with an mRNA containing MO1 binding site followed by GFP coding sequence (MO1BS-GFP) and evaluated for live GFP translation at stage 18 with an Olympus SZX16 fluorescence microscope. (A, B) GFP. (C, D) Bright field view. (E–H) ndrg1a MO2 specifically inhibited the translation of GFP following its binding site (MO2BS-GFP), as revealed by the in vivo assay as above. (E, F) GFP. (G, H) Bright field view.
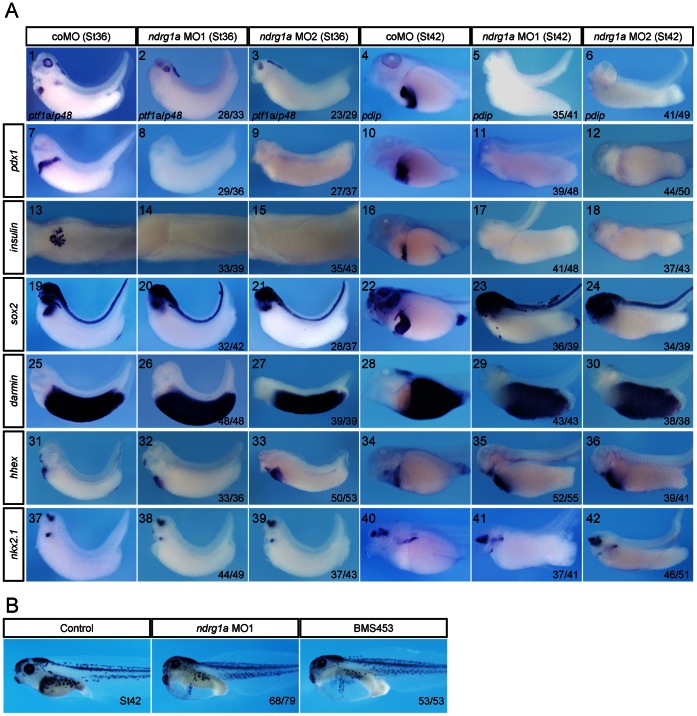
To test if Ndrg1a can functionally mediate RA signaling to specify foregut endoderm, we injected MO1, MO2, or coMO separately into the vegetal part of all four blastomeres of 4-cell stage embryos, collected the embryos at stages 36 and 42, and analyzed with a panel of endodermal marker genes. ptf1a/p48 is specific for pancreatic precursor cells during early embryogenesis and later becomes restricted to the exocrine pancreas [10], [44], [45]. pdx1 specifically demarcates developing pancreas, stomach, and duodenum [46]. insulin, an endocrine marker gene, is exclusively expressed in the dorsal pancreatic bud of Xenopus tail bud stage embryos [47], [48]. pdip is an exocrine pancreas specific marker gene [49]. sox2 marks the oesophagus, stomach and an anterior portion of the duodenum [50]. darmin is an intestine specific marker gene [35], [51]. hhex serves as a liver and thyroid marker gene [52]. nkx2.1 marks developing lung and thyroid [53].
The data obtained indicate that MO1 and MO2 equally efficiently abolished insulin, ptf1a/p48, pdip, pdx1, and sox2 expression in pancreas, oesophagus, stomach, and duodenum, but had minor influence on darmin, hhex, and nkx2.1 expression in intestine, liver, thyroid, and lung, which is very similar to the effect of BMS453 (Fig. 4A and [10]). The only difference is that no ventral pancreatic expression of ptf1a/p48, pdip, or pdx1 was observed in ndrg1a morphants, but traces of ptf1a/p48, pdip, and pdx1 expression in the ventral pancreatic buds were maintained in BMS453 treated embryos (Fig. 4A and [10]). The morphological phenotype of ndrg1a knockdown also resembles that of BMS453 treatment. Embryos injected with MO1 or MO2 appeared normal before stage 40 and subsequently displayed severe gut malformations with a loss of gut coiling and formation of edema (Fig. 4B and [10]). As MO1 and MO2 caused identical phenotypes, we used MO1 to carry out the rest studies. Together, these data suggest that RA and Ndrg1a are in the same genetic hierarchy in controlling pancreas, oesophagus, stomach, and duodenum development.
Figure 4. ndrg1a knockdown in Xenopus embryos specifically disturbed pancreas, oesophagus, stomach, and duodenum formation, which phenocopied BMS435 treatment.
(A) 3.5 picomoles of coMO, ndrg1a MO1 or ndrg1a MO2 was vegetally injected into all four blastomeres at four cell stage of development and collected at stages 36 and 42 for whole mount staining with marker genes indicated on the left side or in the images 1–6. (A1–12 and A16–42) Lateral view, head toward the left. (A13–15) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. (B) Morphology of ndrg1a MO1 injected and BMS453 treated embryos collected when control siblings developed to stage 42. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images.
Injection of even 4 ng of ndrg1a mRNA into Xenopus embryos caused neither morphological abnormality nor expression alterations of the marker genes analyzed (Fig. S2A, B). Co-injection of ndrg1a mRNA with its MOs could not rescue the MO phenotype either (data not shown). It seems that we were unable to get functional Ndrg1a protein in Xenopus embryos through the routine mRNA injection protocol that works for most if not all other genes analyzed. An Ndrg1a-GFP construct demonstrated that the protein was produced (Fig. S3).We have recently established TALEN mediated gene targeting in Xenopus [54]. To further verify the MO phenotype, we designed ndrg1 TALENs within the third exon of Xenopus tropicalis ndrg1 gene (Fig. 5A), which efficiently caused somatic mutations at the targeted loci (Fig. 5B). Consistent with the MO phenotype obtained in Xenopus laevis embryos, insulin and pdx1 expression was severely inhibited in ndrg1 TALEN mRNA injected Xenopus tropicalis embryos (Fig. 5C), albeit with a lower frequency in comparison to the MO injected ones, which happened also to the application of ptf1a/p48 TALENs in our previous study [54]. A stable ndrg1 knockout frog line is yet to be established. We dissected 25 froglets that showed pancreas, stomach, and duodenum hypoplasia or even aplasia but with normal liver and gall bladder developed (Fig. 5D). Thus, these data confirmed the ndrg1a MO phenotype obtained in Xenopus laevis.
Figure 5. TALEN mediated disruption of ndrg1 in Xenopus tropicalis confirmed MO mediated ndrg1a knockdown phenotype in Xenopus laevis.
(A) The TALEN targeting site in Xenopus tropicalis ndrg1 locus (GenBank accession no. NM_001008145.1) was designed in the third exon highlighted in gray with the flanking introns in plain text. The TALEN recognition sequences are highlighted in yellow. The underlined sequences are PCR primers (P1 and P2) used for the evaluation of the gene targeting efficiency. (B) Somatic mutations induced by ndrg1 TALENs in Xenopus tropicalis embryos. Deletions (Δ) are indicated by red dashes and insertions (+) by lowercase red letters against a gray background. The numbers in parentheses show the number of deleted or inserted base pairs. The frequency of the mutation in the sequenced samples is shown in the square brackets. Up to 90% (28/31) of ndrg1 loci sequenced were disrupted. (C) Whole mount in situ hybridization analysis of pdx1 and insulin expression in stage 40 Xenopus tropicalis embryos with or without injection of ndrg1 TALEN mRNAs. All images are lateral view with head toward the left. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images. (D) Xenopus tropicalis froglets once subjected to the injection of ndrg1 TALEN mRNAs showed pancreas, stomach, and duodenum hypoplasia (upper right image), while their liver and gall bladder developed normally (lower right image). Among 25 froglets sacrificed and dissected, 6 showed pancreas aplasia as illustrated in D (upper right image) and the rest showed severe pancreas hypoplasia, while all of them showed stomach and duodenum hypoplasia. The pancreas in control froglet is outlined by white dashed lines. Abbreviations: du, duodenum; gb, gall bladder; he, heart; li, liver; st, stomach.
RA Activated Ndrg1a Represses Wnt/β-catenin Signaling in Archenteron Roof Endoderm Cells to Allow Pancreas, Oesophagus, Stomach, and Duodenum Specification
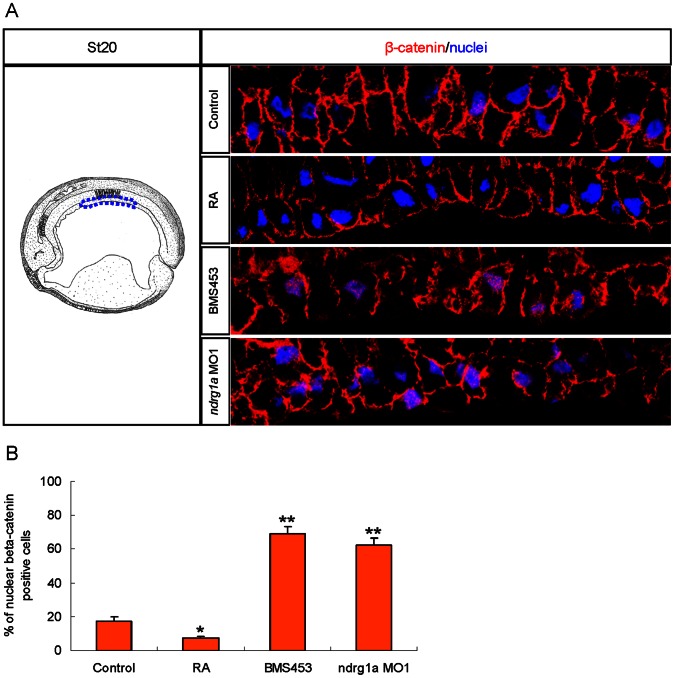
Mechanistically, NDRG1 can interact with Wnt receptor LRP6 and block Wnt/β-catenin signaling in cancer cell lines [37]. To ask if RA activated Ndrg1a can repress Wnt/β-catenin signaling in Xenopus embryos, we compared nuclear β-catenin levels in ndrg1a positive archenteron roof endoderm cells of stage 20 embryos that were subjected to RA, BMS453 treatment, or MO1 injection. Immunolocalization of nuclear β-catenin is a reliable method to characterize Wnt/β-catenin signaling activity in Xenopus embryos [55]. In the selected area of archenteron roof endoderm, a few cells showed nuclear β-catenin staining under normal condition, which became further less upon RA treatment. In contrast, the number of nuclear β-catenin positive cells in the archenteron roof endoderm significantly increased upon BMS453 treatment, or MO1 injection (Fig. 6A, B). Together, these data suggest that RA and Ndrg1a repress Wnt/β-catenin signaling in Xenopus archenteron roof endoderm cells.
Figure 6. Nuclear β-catenin localization in archenteron roof endoderm cells appears to be suppressed by RA activated Ndrg1a in Xenopus laevis.
Embryos were treated with 5 µM RA, 0.25 µM BMS453 for one hour at stage 10, or vegetally injected with 3.5 picomoles of ndrg1a MO1 at four cell stage and collected at stage 20 for immunofluorescence. (A) Left panel is a schematic drawing illustrating a midsagittal section of stage 20 embryos (after Hausen and Riebesell [71]). The dashed blue lines outline the archenteron roof endoderm where ndrg1a is expressed. Right panels are representative immunofluorescence images showing β-catenin signals (red channel) and DAPI staining (blue channel) in the outlined archenteron roof endoderm cells. (B) Quantification data obtained from three independent experiments. Nine embryos in total (three for every experiment) from each group were sectioned to evaluate the mean percentage of β-catenin positive cells in the outlined archenteron roof endoderm illustrated in the left panel of A. For each embryo, the outlined archenteron roof endoderm cells in the 30 consecutive parasagittal sections central to the median plane were scanned for nuclear β-catenin signals. *, p<0.05. **, p<0.01 (Student’s t-test, two-tailed distribution).
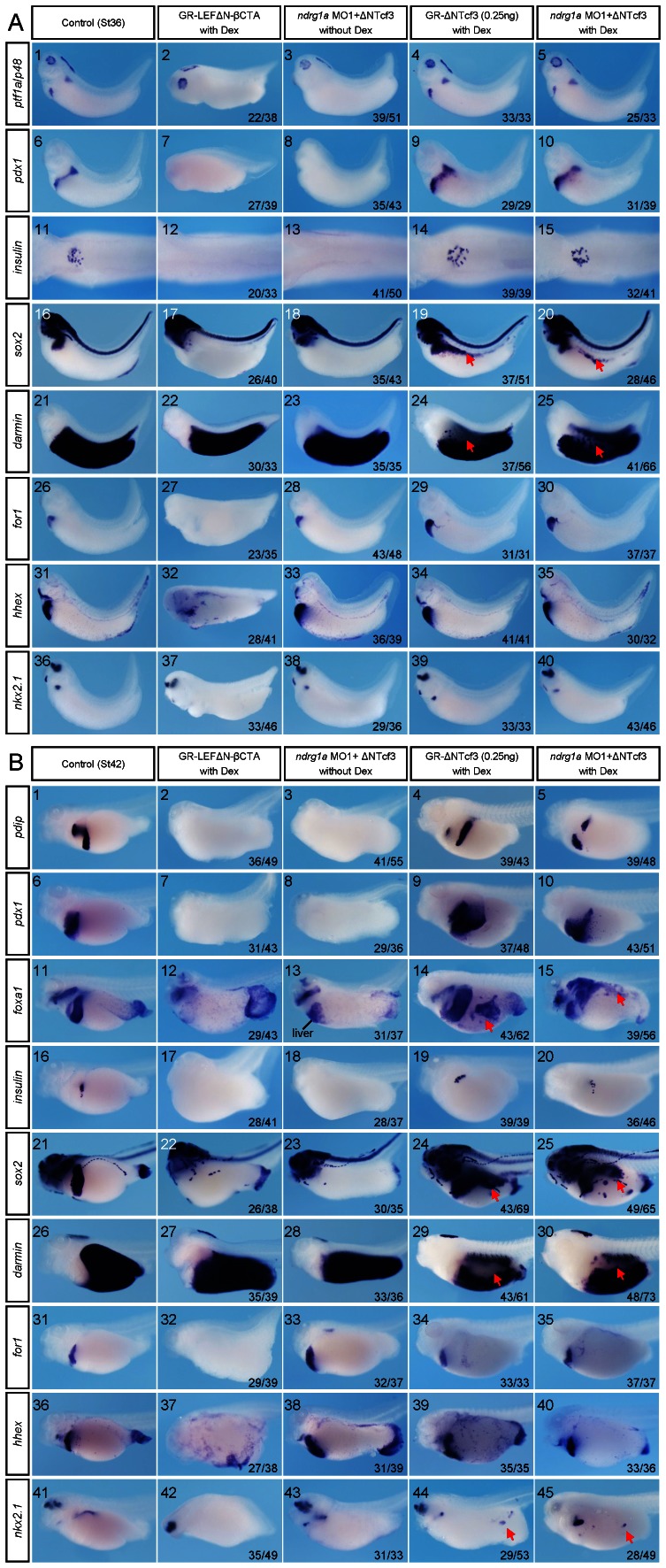
To test if RA, Ndrg1a, and Wnt/β-catenin signaling are functionally linked in the same cascade in controlling pancreas, oesophagus, stomach, and duodenum specification, we co-injected ndrg1a MO1 with GR-ΔNTcf3, a dominant negative form of Tcf3 fused with the hormone-binding domain of the glucocorticoid receptor serving to repress Wnt/β-catenin signaling upon dexamethasone induction [56]. Our data indicate that repressing Wnt/β-catenin signaling rescued ndrg1a knockdown phenotype (Fig. 7A, B). Ectopic activation of nkx2.1 (Fig. 7B44, 45), sox2 (Fig. 7A19, 20 and B24, 25), and foxa1 (Fig. 7B14, 15), a forkhead transcription factor marking stomach, duodenum, and proctodaeum [44], is correlated with the loss of darmin expression (Fig. 7A24, 25, 7B29, 30) upon injection of 0.25 ng of GR-ΔNTcf3 mRNA. In line with the previous study [22], higher dose (0.8 ng) of GR-ΔNTcf3 mRNA is required to induce ectopic expression of pdx1 and for1,a liver specific marker gene [22], [57], in Xenopus embryos (data not shown).
Figure 7. Ndrg1a represses Wnt/β-catenin signaling, thus specially allowing pancreas, oesophagus, stomach, and duodenum specification.
(A, B) Xenopus laevis embryos were vegetally injected with the reagents indicated on the top, treated with 10 µM dexamethasone (Dex) at stage 15, collected at stages 36 (A) and 42 (B), and subjected to whole mount staining with probes indicated on the left side. Doses of the reagents injected are as follows: ndrg1a MO1, 3.5 picomoles; GR-ΔNTcf3 mRNA, 0.25 ng; GR-LEFΔN-βCTA mRNA, 0.5 ng. Red arrows in images A19, 20, 24, 25, B14, 15, 24, 25, 29, 30, 44, and 45 point to either ectopic or loss of expression of marker genes indicated upon inhibition of Wnt/β-catenin signaling. (A11–15) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. All the rest images in A and B are lateral view with head toward the left. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images.
In full agreement with the earlier study [22], activation of canonical Wnt signaling in Xenopus endoderm by vegetal injection of 0.5 ng of GR-LEFΔN-βCTA mRNA containing Lef1 DNA-binding domain and the β-catenin transactivation domain [58] into 4 blastomeres of the 4-cell stage embryos resulted in identical phenotype caused by Ndrg1a knockdown in the context of pancreas, oesophagus, stomach, and duodenum development (Fig. 7A, B). The suppression effect of Wnt/β-catenin signaling on the formation of liver (Fig. 7A27, 32, B32, 37) and lung (Fig. 7A37, B42), was not seen in Ndrg1a morphants, suggesting that the early repression of Wnt/β-catenin signaling in the territory of liver and lung forming endoderm is executed by factors other than RA activated Ndrg1a. Taken together, these data suggest that Wnt/β-catenin signaling is downstream of Ndrg1a and is repressed by Ndrg1a to allow the specification of pancreas, oesophagus, stomach, and duodenum.
Atf3, a Putative Wnt/β-catenin Target Gene in Xenopus Embryos, Specifically Inhibits Xenopus Pancreas, Oesophagus, Stomach, and Aduodenum Specification
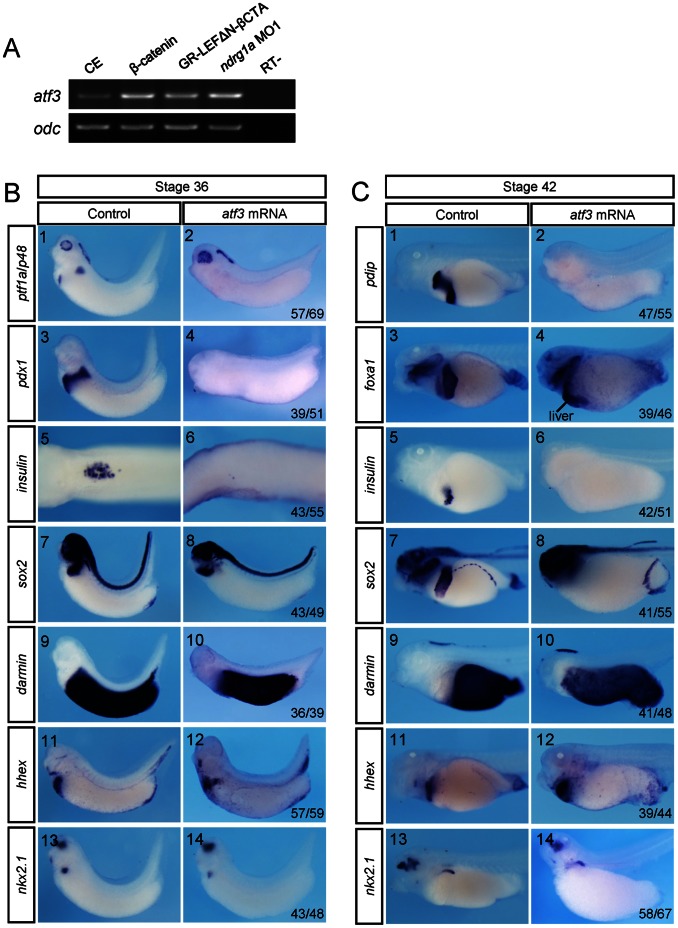
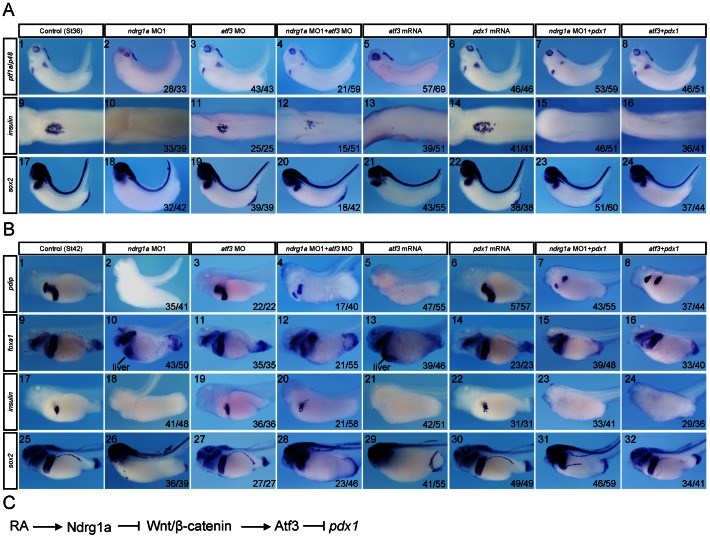
It was shown that ATF3, a Wnt/β-catenin target gene in cancer cell lines [37], can either transcriptionally repress Pdx1 expression [59] or physically interact with PDX1 to block PDX1 mediated transactivation in a murine β-cell line [60]. During Xenopus embryogenesis, atf3 is hardly detected by whole mount in situ hybridization (Fig. S4). RT-PCR analysis indicated that atf3 expression was up-regulated upon ndrg1a knockdown or β-catenin overexpression in Xenopus embryos (Fig. 8A). We found three core Tcf/Lef binding sites (5′-A/T A/T CAAAG-3′) in Xenopus tropicalis atf3 promoter region, which locate at −1467 bp (TACAAAG), −5591 bp (AACAAAG) and −8889 bp (AACAAAG), respectively. Together, these data suggest that atf3 is also a Wnt/β-catenin target gene in Xenopus embryos. Strikingly, overexpression of Atf3 in Xenopus endoderm led to complete loss of insulin, ptf1a/p48, pdip, pdx1, foxa1, and sox2 expression in the pancreas, oesophagus, stomach, and duodenum, but had no obvious effect on darmin, hhex, and nkx2.1 expression (Fig. 8B, C), which is just identical to ndrg1a knockdown phenotype (Fig. 4A). More importantly, atf3 MO, which alone caused minor effects on the expression of marker genes analyzed, was able to rescue insulin, ptf1a/p48, pdip, foxa1, and sox2 expression inhibited by ndrg1a MO (Fig. 9A, B). Furthermore, Pdx1 was able to rescue Atf3 overexpression or Ndrg1a knockdown phenotype with respect to ptf1a/p48, pdip, foxa1, and sox2 expression except for insulin (Fig. 9A, B). It is known that Pdx1 is neither necessary nor sufficient for the activation of early insulin expression in Xenopus embryos [44]. Altogether, our data suggest an epistasis that RA activated Ndrg1a represses Wnt/β-catenin signaling and consequently releases the suppression activity of Wnt/β-catenin signaling activities, which may be partially mediated by Atf3, on pancreas, oesophagus, stomach, and duodenum formation.
Figure 8. Overexpression of Atf3 phenocopied ndrg1a knockdown.
(A) Xenopus laevis embryos were vegetally injected with 0.25 ng of β-catenin mRNA, 0.5 ng of GR-LEFΔN-βCTA mRNA or 3.5 picomoles of ndrg1a MO1 at 4-cell stage, treated with 10 µM Dex at stage 11, and subjected to RT-PCR analysis of atf3 expression at stage 30. Ornithine decarboxylase (odc) was used as the RNA loading control. (B, C) Xenopus laevis embryos were vegetally injected with 0.3 ng of atf3 mRNA at 4-cell stage and collected at stages 36 (B) and 42 (C) for whole mount staining with probes indicated on the left side. (B5, 6) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. All the rest images in B and C are lateral view with head toward the left. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images. Abbreviation: CE, control embryos.
Figure 9. atf3 MO was able to rescue ndrg1a knockdown phenotype.
Pdx1 partially rescued ndrg1a knockdown or Atf3 overexpression phenotypes. (A, B) Xenopus laevis embryos were vegetally injected with the reagents indicated on the top at 4-cell stage and collected at stages 36 (A) and 42 (B) for whole mount staining with probes indicated on the left side. The doses of the reagents injected are as follows: atf3 MO, 2 picomoles; pdx1 mRNA, 75 pg; ndrg1a MO1, 3.5 picomoles; atf3 mRNA, 300 pg. (A9–16) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. All the rest images in A and B are lateral view with head toward the left. The numbers of embryos showing the illustrated phenotypes are given in the corresponding images. (C) The data obtained suggest an epistasis that RA activated Ndrg1a represses Wnt/β-catenin signaling and consequently releases the inhibitory effect of Wnt/β-catenin, which may be partially mediated by Atf3, on pancreas, oesophagus, stomach, and duodenum formation.
Discussion
ndrg1a is directly activated by RA in the archenteron roof endoderm cells of Xenopus neurulae. Nuclear β-catenin level is very low in the archenteron roof endoderm cells and it appears that RA activated Ndrg1a represses Wnt/β-catenin signaling in these cells. Knockdown of ndrg1a mimics inhibition of RA signaling or activation of Wnt/β-catenin signaling in Xenopus embryos with respect to the pancreas, oesophagus, stomach, and duodenum development, which leads to an almost complete loss of the specification of those organ primordia. Blocking Wnt/β-catenin signaling can rescue ndrg1a knockdown phenotype. Thus, Ndrg1a coordinates RA and Wnt/β-catenin signaling in the specification of Xenopus pancreas, oesophagus, stomach, and duodenum.
ndrg1a might be a Specific Marker Gene for Dorsal Pancreas, Oesophagus, Stomach, and Duodenum Precursors in Xenopus Neurulae
Several studies have described microarray based screening of RA responsive genes in Xenopus [61] or zebrafish [17], [18], [62] embryos. Using microarray in combination with whole mount in situ hybridization, we were able to identify archenteron roof endoderm specific gene ndrg1a. A vital dye staining based fate mapping study has shown that archenteron roof endoderm cells later give rise to dorsal pancreas, oesophagus, stomach, and duodenum [5]. shirin appears to be expressed in Xenopus archenteron roof endoderm, but meanwhile it is also expressed in mesoderm and ectoderm [63]. fetuinish specifically marks the archenteron roof endoderm in Xenopus early neurulae, but later it displays expression in liver and intestine with no expression in pancreas, oesophagus, stomach, and duodenum [35]. Only ndrg1a shows consistent expression early in the archenteron roof endoderm and later in pancreas, oesophagus, stomach, and duodenum. We speculate that ndrg1a can serve as a specific dorsal pancreas, oesophagus, stomach, and duodenum precursor marker gene in Xenopus, which remains to be verified by genetic lineage tracing studies.
Ndrg1a is Indispensable for Xenopus Pancreas, Oesophagus, Stomach, and Duodenum Specification
Early endodermal expression of cyp26a1 and cdx4 in zebrafish embryos appears to counteract RA signaling from mesoderm to set anterior and posterior boundaries of pancreatic territory respectively [17], [64]. ndrg1a is the first gene identified that is directly activated by RA early in Xenopus archenteron roof endoderm cells and it mediates RA signaling to positively pattern endoderm cells into pancreas, oesophagus, stomach, and duodenum. Both MO based knockdown of ndrg1a and TALEN mediated disruption of ndrg1a demonstrate that the Ndrg1a activity in Xenopus endoderm patterning is indispensable, which was not observed in Ndrg1 knockout mice [32], [33], [34]. One explanation for this discrepancy is that the essential role of Ndrg1a in Xenopus endodermal patterning is not conserved in mammals. Alternatively, no endodermal organ defects reported in mouse Ndrg1 gene targeting studies might be due to leaky expression of NDRG1 in Ndrg1 deficient mice [32], [33] or due to functional redundancy among NDRG family members 1–4 in mice.
A number of studies indicate that overexpression of NDRG1 in cancer cell lines generates conspicuous phenotypes [27]. It is also reported that overexpression of Ndrg1a in Xenopus embryos results in a reduced pronephros and disorganized somites [36]. For reasons unknown, we could not get ectopic Ndrg1a in function in Xenopus embryos. It should be pointed out that a truncated version of Ndrg1a with the first 48 amino acids in the N-terminal missing was used for both mRNA injection and MO design in the earlier study [36]. We also tried mRNA injection with this short version and the injected embryos were healthy even at high dose (4 ng of mRNA). We could not rescue our ndrg1a MO phenotype with this truncated version of Ndrg1a either. Lastly, the strong endodermal expression of ndrg1a illustrated in our study and described by Costa et al. [35] was not detected in that study [36].
Ndrg1a Mediated Crosstalk between RA and Wnt/β-catenin Signaling is Restricted to Xenopus Pancreas, Oesophagus, Stomach, and Duodenum Forming Cells
During embryogenesis, cells must constantly integrate multiple signaling pathways to achieve their distinct fate at the right time and place. The integrated role of FGF, RA, and Wnt signaling pathways in specifying lung primordium and controlling lung bud growth defined in mice [65] is conserved in Xenopus [66]. In mice, it seems that RA represses Wnt antagonist Dkk1 expression in the prospective lung endoderm of E8.5–E9.5 embryos, thus allows canonical Wnt signaling mediated pulmonary specification [65]. RA and Wnt pathways are linked by RA downstream target genes osr1 and osr2 to maintain pectoral fin development in zebrafish [67]. Together, these cases reveal a circumstance that RA promotes Wnt/β-catenin signaling.
Here, we provide convincing evidence showing for the first time, to our knowledge, that RA activated endodermal expression of Ndrg1a represses Wnt/β-catenin signaling and consequently releases the suppression activity of Wnt/β-catenin signaling on Xenopus pancreas, oesophagus, stomach, and duodenum formation, which does not apply to the liver and lung specification. In support of our finding, overexpression of a secreted Wnt antagonist Sfrp5 can substitute for RA in respect to the induction of exocrine pancreas differentiation in VegT injected animal caps in Xenopus [24]. The detrimental effects of early ectopic Wnt/β-catenin signaling activation on the liver and lung bud formation seen in this study as well as in an earlier study [22] is not contradictory to the positive role of canonical Wnt signaling in promoting liver and lung development, which was observed when Wnt/β-catenin signaling was activated in Xenopus embryos from stages 30 to 42 for liver and from stages 28 to 35 for lung, respectively [22], [66]. The repression effect of RA on Xenopus blastula Wnt/β-catenin signaling [26] is unlikely mediated by Ndrg1a, since ndrg1a is not maternally provided and its zygotic expression is not activated before neurulation, as revealed by both whole mount in situ hybridization and RT-PCR analyses (Fig. 1A, [36] and data not shown). It remains to be investigated if Ndrg1a can interact with Lrp6 and how Ndrg1a represses Wnt/β-catenin signaling in Xenopus embryos.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol (2010052) was approved by the Institutional Animal Care and Use Committee (IACUC) of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences. All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Embryo Cultivation and Microarray Analysis
Wild type Xenopus laevis or Xenopus tropicalis embryos were obtained by in vitro fertilization and staged according to the normal table of Xenopus development [68]. Xenopus laevis embryos were treated with 0.25 µM BMS453 (a gift from Bristol Myers Squibb) for one hour at stage 10. Corresponding amount DMSO was added to control embryos. Embryos were collected at stages 12, 23, and 34. Total RNA was extracted using TRIZOL reagent (Invitrogen), purified using Qiagen RNAeasy kit, and subjected to microarray analyses using Xenopus laevis Genome Arrays (Affymetrix, version 2.0). The array was not repeated. We randomly chose 15 down-regulated genes (Table S1) and validated their expression by RT-PCR. Indeed, all the tested genes showed down-regulation in the BMS453 treated embryos. The raw and normalized data were stored in the ArrayExpress database (accession no. E-MTAB-1419).
Whole Mount in situ Hybridization and Immunofluorescence
Whole mount in situ hybridization was performed as described [69]. The antisense probes for darmin, for1, foxa1, insulin, hhex, pdip, pdx1, and ptf1a/p48 were prepared as described previously [10], [22], [44], [49]. ndrg1a (GenBank accession no. NM_001094390), atf3 (GenBank accession no. NM_001094018), sox2 (GenBank accession no. NM_213704), and nkx2.1 (GenBank accession no. AF281080) were cloned into pGEM-T Easy vector by RT-PCR. The resultant constructs were cut with SalI and transcribed with T7 RNA polymerase to get their antisense probes.
Immunofluorescence was performed as described [55] with following modifications. Stage 20 embryos were fixed in MEMFA (0.1 M MOPS, pH 7.4; 2 mM EGTA; 1 mM MgSO4 and 4% formaldehyde) for two hours at room temperature, embedded in paraffin, and sectioned sagittally at a thickness of 5 µm. Serial sections were incubated with rabbit anti-β-catenin antibody (1∶250, H-102, Santa Cruz Biotechnology, #sc-7199) and followed by goat anti-rabbit Cy3 (1∶200, Bi Yuntian, #A0516) as a secondary antibody. The nuclei were counterstained with DAPI (1 µg/ml, Roche, #10-236-276-001). Fluorescent images were captured using a Zeiss LSM 710 confocal microscope with 40× objectives.
MO and mRNA Injection in Xenopus laevis Embryos
ndrg1a MO1 (5′-ATAGCCGTTTGCCTGTGTAAGAAGA-3′), MO2 (5′-ATACCCTGGTGTCCTGATGCTGCGC-3′), atf3 MO (5′-AGCATCATTTTCGTGCTGTGTCGGT-3′), and standard control morpholino oligonucleotide (5′-CCTCTTACCTCAGTTACAATTTATA -3′) were purchased from GeneTools, LLC. The open reading frames of Xenopus laevis ndrg1a, atf3 and pdx1 were obtained by RT-PCR and cloned into the pCS2+ vector. pCS2+HA-GR-ΔNLEF-βCTA [58] and pT7TSHA-GR-ΔNTcf3 [56] were obtained from Zorn laboratory. Capped mRNA was generated with SP6 or T7 mMessage mMechine Kit (Ambion) and purified using Qiagen RNAeasy kit. All the reagents were injected into four blastomeres of the 4-cell stage embryos from the vegetal pole.
Construction and Application of Xenopus tropicalis ndrg1 TALENs
A pair of Xenopus tropicalis ndrg1 TALENs was constructed through Golden Gate TALEN Assembly method [54], [70]. The resultant ndrg1 TALEN mRNAs were injected into animal pole of fertilized Xenopus tropicalis eggs. Five injected embryos were collected at stage 40 for somatic gene targeting analysis. The rest were either collected for marker gene expression analysis, or raised for late phenotyping and establishing of stable knockout lines. Primer 1 (5′-GTGCTGCAAGTTGGAGTGAT-3′) and primer 2 (5′-ACTCTAGGTGGCATGACAGC-3′), bridging the right and left binding sites of ndrg1 TALENs in wild type Xenopus tropicalis genomic DNA, were used to amplify the targeting region of ndrg1. The obtained PCR fragments were subcloned to pGEM-T Easy vector and single colonies were picked for sequencing.
Chemical Treatment
RA (all-trans-RA, Sigma) and cycloheximide (CHX, Sigma) were first prepared as 10 mM and 10 mg/ml stock solutions in 100% ethanol and then diluted into desired concentrations with 0.1×MBS (at least 1∶1000 dilution). Carrier controls were performed at the highest solvent concentration that the experimental embryos received in each set. For the activation of GR fusion proteins, dexamethasone (Dex, Sigma) was prepared as 5 mM stock solution in 100% ethanol and applied to the control and mRNA-injected embryos at stage 11 in a final concentration of 10 µM in 0.1× MBS.
RT-PCR
RT-PCR was carried out as previously described [10]. The following primers and cycle numbers were used: atf3 (forward 5′-TTTAGATTCGGTGGTGGTGTCC-3′ and reverse 5′-ATCTGCTGGATGAAGAGGTTGC-3′, 28cycles), and ornithine decarboxylase (forward 5′-TGAATTGATGAAAGTGGCAAGG-3′ and reverse 5′-CAGGGCTGGGTTTATCACAGAT-3′, 23cycles).
Supporting Information
The down-regulated genes upon BMS453 treatment appear dynamic during early development. A Venn diagram indicates that only a few genes showing overlapping at different stages of development. The 9 genes selected for further whole mount in situ analysis are listed in the diagram.
(TIF)
Injection of ndrg1a mRNA into Xenopus embryos generated no phenotype. Xenopus laevis embryos were vegetally injected with 4 ng of ndrg1a mRNA at 4-cell stage and collected at stages 36 (A) and 42 (B) for whole mount staining with probes indicated on the left side. (A5, 6) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. All the rest images in A and B are lateral view with head toward the left. The numbers of embryos manipulated are given in the individual images.
(TIF)
Ndrg1a-GFP fusion protein was properly synthesized in Xenopus embryos. 1 ng of ndrg1a-GFP mRNA was injected into the vegetal part of all four blastomeres of 4-cell stage Xenopus laevis embryos. (A) Live GFP signals were observed under an Olympus SZX16 fluorescence microscope when the injected embryos developed to stage 35. (B) Western blot analysis with an anti-GFP antibody confirmed that fusion protein was properly generated in stage 35 embryos. For Western blot analysis, in addition to the control uninjected embryos, we also injected GFP mRNA as a control. CE, control embryos.
(TIF)
atf3 expression is nearly undetectable in developing Xenopus embryos by whole mount in situ hybridization. All images are lateral view. (C–G) Head toward the left.
(TIF)
List of genes down regulated more than 2-fold in BMS453 treated Xenopus laevis embryos collected at stages 12, 23, and 34 based on one Affymetrix Genome Array. The overlapping ones among three stages analyzed are crossed in additional columns. The 9 genes that were chosen for further in situ hybridization analysis are highlighted in bold.
(XLS)
Acknowledgments
We are grateful to Dr. A. Zorn for generously providing constructs used in this study. We thank members of the Chen laboratory for their help during the study.
Funding Statement
This work was supported by funds from the National Basic Research Program of China (2009CB941200) and the National Natural Science Foundation of China (31271554). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lewis SL, Tam PP (2006) Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn 235: 2315–2329. [DOI] [PubMed] [Google Scholar]
- 2. Zorn AM, Wells JM (2009) Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25: 221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao H, Han D, Dawid IB, Pieler T, Chen Y (2012) Homeoprotein hhex-induced conversion of intestinal to ventral pancreatic precursors results in the formation of giant pancreata in Xenopus embryos. Proc Natl Acad Sci U S A 109: 8594–8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Warga RM, Nusslein-Volhard C (1999) Origin and development of the zebrafish endoderm. Development 126: 827–838. [DOI] [PubMed] [Google Scholar]
- 5. Chalmers AD, Slack JM (2000) The Xenopus tadpole gut: fate maps and morphogenetic movements. Development 127: 381–392. [DOI] [PubMed] [Google Scholar]
- 6. Tremblay KD, Zaret KS (2005) Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Developmental Biology 280: 87–99. [DOI] [PubMed] [Google Scholar]
- 7. Franklin V, Khoo PL, Bildsoe H, Wong N, Lewis S, et al. (2008) Regionalisation of the endoderm progenitors and morphogenesis of the gut portals of the mouse embryo. Mech Dev 125: 587–600. [DOI] [PubMed] [Google Scholar]
- 8. Kimura W, Yasugi S, Stern CD, Fukuda K (2006) Fate and plasticity of the endoderm in the early chick embryo. Developmental Biology 289: 283–295. [DOI] [PubMed] [Google Scholar]
- 9. Kam RK, Deng Y, Chen Y, Zhao H (2012) Retinoic acid synthesis and functions in early embryonic development. Cell Biosci 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Pan FC, Brandes N, Afelik S, Solter M, et al. (2004) Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Developmental Biology 271: 144–160. [DOI] [PubMed] [Google Scholar]
- 11. Stafford D, Hornbruch A, Mueller PR, Prince VE (2004) A conserved role for retinoid signaling in vertebrate pancreas development. Dev Genes Evol 214: 432–441. [DOI] [PubMed] [Google Scholar]
- 12. Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, et al. (2005) Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Developmental Biology 284: 399–411. [DOI] [PubMed] [Google Scholar]
- 13. Molotkov A, Molotkova N, Duester G (2005) Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn 232: 950–957. [DOI] [PubMed] [Google Scholar]
- 14. Pan FC, Chen Y, Bayha E, Pieler T (2007) Retinoic acid-mediated patterning of the pre-pancreatic endoderm in Xenopus operates via direct and indirect mechanisms. Mech Dev 124: 518–531. [DOI] [PubMed] [Google Scholar]
- 15. Bayha E, Jorgensen MC, Serup P, Grapin-Botton A (2009) Retinoic acid signaling organizes endodermal organ specification along the entire antero-posterior axis. Plos One 4: e5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stafford D, Prince VE (2002) Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol 12: 1215–1220. [DOI] [PubMed] [Google Scholar]
- 17. Kinkel MD, Sefton EM, Kikuchi Y, Mizoguchi T, Ward AB, et al. (2009) Cyp26 enzymes function in endoderm to regulate pancreatic field size. Proc Natl Acad Sci U S A 106: 7864–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dalgin G, Ward AB, Hao le T, Beattie CE, Nechiporuk A, et al. (2011) Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development 138: 4597–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Z, Song J, Qi F, Xiao A, An X, et al. (2008) Exdpf is a key regulator of exocrine pancreas development controlled by retinoic acid and ptf1a in zebrafish. PLoS Biol 6: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gregorieff A, Grosschedl R, Clevers H (2004) Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(−/−)/Tcf1(−/−) embryos. EMBO J 23: 1825–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA (2005) The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell 8: 611–622. [DOI] [PubMed] [Google Scholar]
- 22. McLin VA, Rankin SA, Zorn AM (2007) Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134: 2207–2217. [DOI] [PubMed] [Google Scholar]
- 23. Rankin SA, Kormish J, Kofron M, Jegga A, Zorn AM (2011) A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Developmental Biology 351: 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Damianitsch K, Melchert J, Pieler T (2009) XsFRP5 modulates endodermal organogenesis in Xenopus laevis. Developmental Biology 329: 327–337. [DOI] [PubMed] [Google Scholar]
- 25. Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, et al. (2008) Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev 22: 3050–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S, Lou X, Wang J, Liu B, Ma L, et al. (2008) Retinoid signaling can repress blastula Wnt signaling and impair dorsal development in Xenopus embryo. Differentiation 76: 897–907. [DOI] [PubMed] [Google Scholar]
- 27. Melotte V, Qu X, Ongenaert M, van Criekinge W, de Bruine AP, et al. (2010) The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J 24: 4153–4166. [DOI] [PubMed] [Google Scholar]
- 28. Tschan MP, Shan D, Laedrach J, Eyholzer M, Leibundgut EO, et al. (2010) NDRG1/2 expression is inhibited in primary acute myeloid leukemia. Leuk Res 34: 393–398. [DOI] [PubMed] [Google Scholar]
- 29. Chen S, Han YH, Zheng Y, Zhao M, Yan H, et al. (2009) NDRG1 contributes to retinoic acid-induced differentiation of leukemic cells. Leuk Res 33: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 30. Jung EU, Yoon JH, Lee YJ, Lee JH, Kim BH, et al. (2010) Hypoxia and retinoic acid-inducible NDRG1 expression is responsible for doxorubicin and retinoic acid resistance in hepatocellular carcinoma cells. Cancer Lett 298: 9–15. [DOI] [PubMed] [Google Scholar]
- 31. Piquemal D, Joulia D, Balaguer P, Basset A, Marti J, et al. (1999) Differential expression of the RTP/Drg1/Ndr1 gene product in proliferating and growth arrested cells. Biochim Biophys Acta 1450: 364–373. [DOI] [PubMed] [Google Scholar]
- 32. King RH, Chandler D, Lopaticki S, Huang D, Blake J, et al. (2011) Ndrg1 in development and maintenance of the myelin sheath. Neurobiol Dis 42: 368–380. [DOI] [PubMed] [Google Scholar]
- 33. Okuda T, Higashi Y, Kokame K, Tanaka C, Kondoh H, et al. (2004) Ndrg1-deficient mice exhibit a progressive demyelinating disorder of peripheral nerves. Molecular and Cellular Biology 24: 3949–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taketomi Y, Sunaga K, Tanaka S, Nakamura M, Arata S, et al. (2007) Impaired mast cell maturation and degranulation and attenuated allergic responses in Ndrg1-deficient mice. Journal of Immunology 178: 7042–7053. [DOI] [PubMed] [Google Scholar]
- 35. Costa RM, Mason J, Lee M, Amaya E, Zorn AM (2003) Novel gene expression domains reveal early patterning of the Xenopus endoderm. Gene Expression Patterns 3: 509–519. [DOI] [PubMed] [Google Scholar]
- 36. Kyuno J, Fukui A, Michiue T, Asashima M (2003) Identification and characterization of Xenopus NDRG1. Biochem Biophys Res Commun 309: 52–57. [DOI] [PubMed] [Google Scholar]
- 37. Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, et al. (2012) N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO Mol Med 4: 93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, et al. (2006) Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441: 173–178. [DOI] [PubMed] [Google Scholar]
- 39. Hai T, Wolford CC, Chang YS (2010) ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr 15: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hunt D, Raivich G, Anderson PN (2012) Activating transcription factor 3 and the nervous system. Front Mol Neurosci 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hartman MG, Lu D, Kim ML, Kociba GJ, Shukri T, et al. (2004) Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Molecular and Cellular Biology 24: 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T (2001) The roles of ATF3 in glucose homeostasis. A transgenic mouse model with liver dysfunction and defects in endocrine pancreas. Journal of Biological Chemistry 276: 29507–29514. [DOI] [PubMed] [Google Scholar]
- 43. Allen-Jennings AE, Hartman MG, Kociba GJ, Hai T (2002) The roles of ATF3 in liver dysfunction and the regulation of phosphoenolpyruvate carboxykinase gene expression. Journal of Biological Chemistry 277: 20020–20025. [DOI] [PubMed] [Google Scholar]
- 44. Afelik S, Chen Y, Pieler T (2006) Combined ectopic expression of Pdx1 and Ptf1a/p48 results in the stable conversion of posterior endoderm into endocrine and exocrine pancreatic tissue. Genes Dev 20: 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jarikji ZH, Vanamala S, Beck CW, Wright CV, Leach SD, et al. (2007) Differential ability of Ptf1a and Ptf1a-VP16 to convert stomach, duodenum and liver to pancreas. Developmental Biology 304: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wright CV, Schnegelsberg P, De Robertis EM (1989) XlHbox 8: a novel Xenopus homeo protein restricted to a narrow band of endoderm. Development 105: 787–794. [DOI] [PubMed] [Google Scholar]
- 47. Horb ME, Slack JM (2002) Expression of amylase and other pancreatic genes in Xenopus. Mech Dev 113: 153–157. [DOI] [PubMed] [Google Scholar]
- 48. Kelly OG, Melton DA (2000) Development of the pancreas in Xenopus laevis. Dev Dyn 218: 615–627. [DOI] [PubMed] [Google Scholar]
- 49. Afelik S, Chen Y, Pieler T (2004) Pancreatic protein disulfide isomerase (XPDIp) is an early marker for the exocrine lineage of the developing pancreas in Xenopus laevis embryos. Gene Expression Patterns 4: 71–76. [DOI] [PubMed] [Google Scholar]
- 50. Chalmers AD, Slack JM, Beck CW (2000) Regional gene expression in the epithelia of the Xenopus tadpole gut. Mech Dev 96: 125–128. [DOI] [PubMed] [Google Scholar]
- 51. Chen Y, Jurgens K, Hollemann T, Claussen M, Ramadori G, et al. (2003) Cell-autonomous and signal-dependent expression of liver and intestine marker genes in pluripotent precursor cells from Xenopus embryos. Mech Dev 120: 277–288. [DOI] [PubMed] [Google Scholar]
- 52. Newman CS, Chia F, Krieg PA (1997) The XHex homeobox gene is expressed during development of the vascular endothelium: overexpression leads to an increase in vascular endothelial cell number. Mech Dev 66: 83–93. [DOI] [PubMed] [Google Scholar]
- 53. Small EM, Vokes SA, Garriock RJ, Li D, Krieg PA (2000) Developmental expression of the Xenopus Nkx2–1 and Nkx2–4 genes. Mech Dev 96: 259–262. [DOI] [PubMed] [Google Scholar]
- 54. Lei Y, Guo X, Liu Y, Cao Y, Deng Y, et al. (2012) Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc Natl Acad Sci U S A 109: 17484–17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schohl A, Fagotto F (2002) Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development 129: 37–52. [DOI] [PubMed] [Google Scholar]
- 56. Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, et al. (1996) XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86: 391–399. [DOI] [PubMed] [Google Scholar]
- 57. Seo YW, Sanyal S, Kim HJ, Won DH, An JY, et al. (2002) FOR, a novel orphan nuclear receptor related to farnesoid X receptor. Journal of Biological Chemistry 277: 17836–17844. [DOI] [PubMed] [Google Scholar]
- 58. Domingos PM, Itasaki N, Jones CM, Mercurio S, Sargent MG, et al. (2001) The Wnt/beta-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signalling. Developmental Biology 239: 148–160. [DOI] [PubMed] [Google Scholar]
- 59. Jang MK, Park HJ, Jung MH (2011) ATF3 represses PDX-1 expression in pancreatic beta-cells. Biochem Biophys Res Commun 412: 385–390. [DOI] [PubMed] [Google Scholar]
- 60. Kim WH, Jang MK, Kim CH, Shin HK, Jung MH (2011) ATF3 inhibits PDX-1-stimulated transactivation. Biochem Biophys Res Commun 414: 681–687. [DOI] [PubMed] [Google Scholar]
- 61. Arima K, Shiotsugu J, Niu R, Khandpur R, Martinez M, et al. (2005) Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Dev Dyn 232: 414–431. [DOI] [PubMed] [Google Scholar]
- 62. Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB (2010) Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Developmental Biology 338: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Spagnoli FM, Brivanlou AH (2006) The RNA-binding protein, Vg1RBP, is required for pancreatic fate specification. Developmental Biology 292: 442–456. [DOI] [PubMed] [Google Scholar]
- 64. Kinkel MD, Eames SC, Alonzo MR, Prince VE (2008) Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development 135: 919–929. [DOI] [PubMed] [Google Scholar]
- 65. Chen F, Cao Y, Qian J, Shao F, Niederreither K, et al. (2010) A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. Journal of Clinical Investigation 120: 2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rankin SA, Gallas AL, Neto A, Gomez-Skarmeta JL, Zorn AM (2012) Suppression of Bmp4 signaling by the zinc-finger repressors Osr1 and Osr2 is required for Wnt/beta-catenin-mediated lung specification in Xenopus. Development 139: 3010–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Neto A, Mercader N, Gomez-Skarmeta JL (2012) The Osr1 and Osr2 genes act in the pronephric anlage downstream of retinoic acid signaling and upstream of Wnt2b to maintain pectoral fin development. Development 139: 301–311. [DOI] [PubMed] [Google Scholar]
- 68.Nieuwkoop PD, Faber J (1967) Normal table of Xenopus laevis (Daudin). Amsterdam, North-Holland.
- 69. Harland RM (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36: 685–695. [DOI] [PubMed] [Google Scholar]
- 70. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, et al. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hausen P, Riebesell M (1991) The early development of Xenopus laevis: an atlas of the histology: Springer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The down-regulated genes upon BMS453 treatment appear dynamic during early development. A Venn diagram indicates that only a few genes showing overlapping at different stages of development. The 9 genes selected for further whole mount in situ analysis are listed in the diagram.
(TIF)
Injection of ndrg1a mRNA into Xenopus embryos generated no phenotype. Xenopus laevis embryos were vegetally injected with 4 ng of ndrg1a mRNA at 4-cell stage and collected at stages 36 (A) and 42 (B) for whole mount staining with probes indicated on the left side. (A5, 6) Dorsal view. The dorsal structures, such as the neural tube, notochord, and somites were removed after whole mount in situ hybridization. All the rest images in A and B are lateral view with head toward the left. The numbers of embryos manipulated are given in the individual images.
(TIF)
Ndrg1a-GFP fusion protein was properly synthesized in Xenopus embryos. 1 ng of ndrg1a-GFP mRNA was injected into the vegetal part of all four blastomeres of 4-cell stage Xenopus laevis embryos. (A) Live GFP signals were observed under an Olympus SZX16 fluorescence microscope when the injected embryos developed to stage 35. (B) Western blot analysis with an anti-GFP antibody confirmed that fusion protein was properly generated in stage 35 embryos. For Western blot analysis, in addition to the control uninjected embryos, we also injected GFP mRNA as a control. CE, control embryos.
(TIF)
atf3 expression is nearly undetectable in developing Xenopus embryos by whole mount in situ hybridization. All images are lateral view. (C–G) Head toward the left.
(TIF)
List of genes down regulated more than 2-fold in BMS453 treated Xenopus laevis embryos collected at stages 12, 23, and 34 based on one Affymetrix Genome Array. The overlapping ones among three stages analyzed are crossed in additional columns. The 9 genes that were chosen for further in situ hybridization analysis are highlighted in bold.
(XLS)