Abstract
Estrogen signaling is mediated by ERα and ERβ in hormone dependent, breast cancer (BC). Over the last decade the implication of epigenetic pathways in BC tumorigenesis has emerged: cancer-related epigenetic modifications are implicated in both gene expression regulation, and chromosomal instability. In this review, the epigenetic-mediated estrogen signaling, controlling both ER level and ER-targeted gene expression in BC, are discussed: (1) ER silencing is frequently observed in BC and is often associated with epigenetic regulations while chemical epigenetic modulators restore ER expression and increase response to treatment;(2) ER-targeted gene expression is tightly regulated by co-recruitment of ER and both coactivators/corepressors including HATs, HDACs, HMTs, Dnmts and Polycomb proteins.
Keywords: epigenetic, estrogen, DNA methylation, histones, breast cancer
Introduction
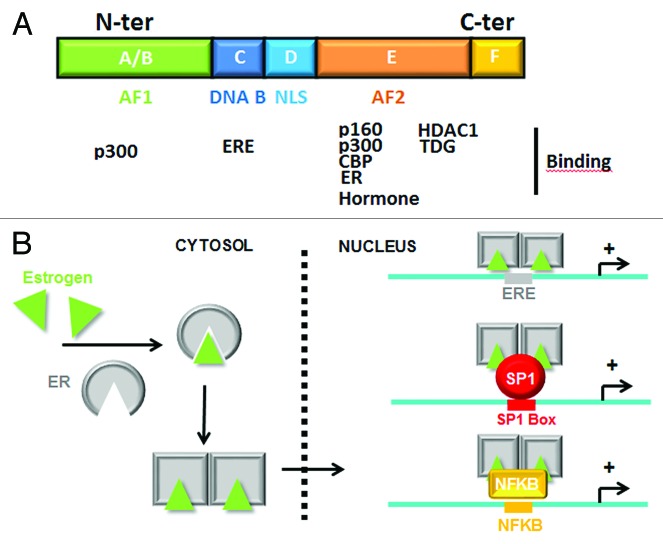
Breast cancer (BC) is the most common malignant tumor among women in the world and is the second cause of death in women between the ages of 35–55 in developed countries. BC can be divided based on molecular criteria into distinct phenotypes: the molecular subtypes are classified by (1) expression of estrogen receptors (ERs) and/or progesterone receptors (PRs), (2) human epidermal receptor 2 (HER2/ERBB2) amplification and (3) a triple negative type (ER-/PR- and normal expression of HER2).1 While estrogen has normal biological roles, such as reproduction, brain development and additional protective effects of sexual steroid hormones, prolonged exposure, combined with high levels of hormone increases the risk of BC by constitutively activating the transcription of genes predominantly implicated in metabolism and cell cycle regulation. ER-mediated mechanisms of gene regulation are well documented. ERs exist as two isoforms (ERα and ERβ) that belong to the family of transcriptional receptors and recognize and bind to a specific DNA consensus sequence to facilitate the transcriptional initiation of hundreds of target genes.2 Following estrogen treatment, the hormone binds to the E-domain of ERs, induces ER dimerization and favors its nuclear translocation, where the dimer finally interacts with DNA on the estrogen response element (ERE) and induces the activation of estrogen regulated genes (Fig. 1). However, following estrogen stimulation, the transcription of additional genes lacking an evident ERE is also activated in response to ERα interaction with particular transcriptional factors (TFs) such as AP-1, SP1 or NFKB. In the latter cases, mechanisms of ER-dependent transcriptional activation are indirect and mediated by the recruitment of ERα on TF boxes3,4.
Figure 1. Structure and activation of Estrogen Receptor. (A) Schematic representation of ERα domains and their potential interaction with co-activators/co-repressors and ERE. (B) Mechanisms of ERα activation by estrogens. Recruitment of liganded ERα on DNA is mediated directly on ERE or not directly via SP1 or NFKB interaction.
Changes in gene expression caused by genetic mutations, which lead to oncogene activation or tumor suppressor gene silencing, have been studied in BC etiology and correlated with BC risk in a recent meta-analysis.5 For example, mutations in BRCA1/2 genes were frequently observed in hereditary BC. Over the last two decades, the idea of an epigenetic control of gene expression in diseases other than genetic disorders has emerged. This includes the deregulation of genes that participate in tumorigenesis initiation and progression. In the latter case, the outcome of both genetic and epigenetic modifications is an aberrant overexpression and/or silencing of genes implicated in cell proliferation and/or in the control of cell death. Epigenetic pathways regulate gene transcription by two different mechanisms that are not mutually exclusive: DNA methylation and post-translational modification of histones. DNA methylation occurs in 2–3% of cytosines in CpG islands and is not randomly distributed throughout the DNA as these sequences are mostly located in the upstream region of promoters. DNA methylation is implemented by a family of enzymes referred to as DNA methyl transferases (Dnmts) 1, 2, 3a, 3b and 3L. Promoters with a high density of CpGs are defined as CG-rich areas and are predominantly subject to DNA methylation. Methylated DNA is generally associated with a decreased TF binding capacity that diminishes/abolishes transcriptional expression of the corresponding gene. Two distinct forms of DNA methylation processes have been described, the first is inherited DNA methylation or maintenance DNA methylation and is predominantly catalyzed by Dnmt1, the second is de novo DNA methylation and is performed mainly by Dnmt3a and Dnmt3b. Maintenance DNA methylation permits the conservation of DNA methylation patterns after DNA replication by copying methylation on the newly synthesized strand using the hemi-methylated DNA as a matrix. Conversely, de novo DNA methylation occurs on both strands of unmethylated DNA (for a review see ref. 6). Global DNA hypomethylation has been observed in many cancers including BC and prostrate tumors.7-9 Artificial disruption of DNA methylation complexes or invalidation of Dnmt1 in normal cells leads to a decrease in global DNA methylation and induces tumor formation in nude mice.10,11 This phenomenon is promoted by reactivation of non-coding repetitive elements leading to chromosomal instability and abnormal gene expression. Besides global DNA hypomethylation, both local hypo and hypermethylation of promoters have also been reported and result in specific gene activation or silencing in cancers.
Nucleosomes are made up of a duplicate of histones H2A, H2B, H3 and H4 enclosed in a DNA loop and regulate chromatin compaction as well as TF accessibility for transcription initiation. Histones are subject to post-translational modifications such as acetylation, methylation and phosphorylation. The “histone code” refers to the sum of these modifications and allows a prediction of a favorable or unfavorable chromatin status for gene transcription. Acetylation of lysines in histones is associated with an uncondensed chromatin status, accessibility of TFs, and is processed by histone acetyl transferase (HATs), while these acetyl groups are removed by histone deacetylases (HDACs). Histone methyl transferase (HMT) or histone demethylases (HDM), respectively, catalyze the methylation or demethylation of lysine or arginine in histones and these modifications favor the compaction or relaxation of chromatin, depending of the methylated residue (for a review see ref. 12).
Epigenetic Silencing of ESR1 and ESR2 Genes in Cancer
Anti-estrogen therapies are used in treatment of BC but are inefficient in ER negative patients. In these therapies, the most used drug over the past 50 years is tamoxifen, a competitive inhibitor of estradiol that binds to ERα. More recent pharmacological molecules include selective ER down-regulators (SEDRs), which inhibit ERα dimerization and nuclear translocation or aromatase inhibitors, which target the enzyme responsible for estrogen synthesis. As described above, estrogen dependent genes are controlled by ERα and ERβ. However, a frequent decrease in ERα expression was observed in BC and may occur during the course of the disease. ER- breast cancers were observed in 20% of low and 50% of high grade BC patients.13 ER expression status is paradoxical in BC. High ERα expression in high grade BC correlates with a better outcome, a lower aggressiveness and a better response to anti-estrogen therapies compared with ER- patients. However, estrogen stimulation in healthy cells increases BC risks. This may be explained by the dual role of ERα in both proliferation and differentiation. Some studies also suggest that DNA methylation-mediated promoter ESR1 (estrogen receptor 1 gene) silencing is found frequently and may participate in tumorigenesis or progression of the disease in other cancers, such as leukemia or colon tumors.14 The etiology of the loss of expression of ERα (about 30% of BC patients are ERα-) is due to DNA hypermethylation in 41% of cases, which correlates with tumor size and histological grade.15-17 Moreover, a recent study on BC patients in India revealed that the proportion of ESR1 hypermethylation was highly increased in triple negative tumors.18 Manipulation of ESR1 hypermethylation can also affect ERα expression. For example, a 5 d Bisphenol A (BpA) exposure in neonatal male rats induces persistent ESR1 promoter hypermethylation in adults, associated with increasing levels of Dnmts.19 Inactivation of Dnmt1 using siRNA or treatment with DNA methylation inhibitors such as 5-azadeoxycytidine, restores ERα expression in ERα negative BC cells.20 As such, in addition to immunodetection of ERα, detection of ESR1 methylation status may aid in predicting a response to anti-estrogen therapies in BC patients.
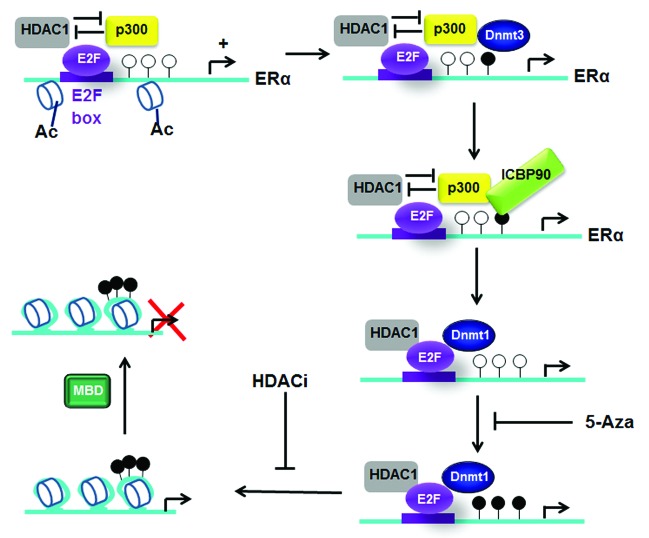
Overexpression of HDAC1 abolishes ESR1 expression in MCF7 cells.21 Macaluso et al. proposed a model of epigenetic inactivation of the ERα promoter (Fig. 2).22,23 In ER+ BC cells such as the MCF7 cell line, an activator complex composed of pRb2/E2F4/5/HDAC1/SUV39H1/p300 binds to a region containing E2F boxes close to the initial transcription site in the ESR1 promoter. The authors proposed that repressor activity of both HDAC1 and the HMT SUV39H1 might be overcome by the HAT activity of p300. Methylation of CpG by Dnmt3a/3b in this promoter may induce the recruitment of ICBP90 (inverted CCAAT box binding protein of 90 kDa) and consequently facilitate the replacement of p300 by Dnmt1 in the repressor complex pRb2/E2F4/5/HDAC1/SUV39H1/Dnmt1 found in ERα- BC, MDA MB231 cells. A further recruitment of MeCP2 to an ERα methylated promoter may also participate in complete ERα repression, as illustrated in Figure 2.24 These epigenetic signals, in particular DNA methylation near the AP-2 binding site, induce a repressive chromatin, blocking the loading of TFAP2C, further RNAP II recruitment and thus transcription of ESR1.25 A recent study in male tissues revealed that among the methylated CpGs close to the ESR1 promoter, the methylation of one particular CpG (located in the +1kb intragenic region of ESR1) correlates with low ESR1 expression. This CpG is included in a TGIF box, and its methylation provokes the recruitment of the repressor TGIF, targeting of HDAC1 and ESR1 silencing. Interestingly, methylation status of ESR1 in these tissues was not sensitive to estrogen exposure.26 Moreover, in MCF7 cells, estrogen treatment induces ESR1 repression in an ERα-mediated mechanism: while coactivators and ERα are found at both distal and proximal ESR1 promoters, Sin3A/ERα complex is specifically recruited on the proximal promoter and represses ESR1 transcription.27
Figure 2. Model of epigenetic inactivation of ESR1. Primary methylation and recruitment of ICBP90 on ERα promoter, provoke histone deacetylation and a large secondary methylation and ERα silencing. Ac, acetylation of histones; white circles symbolize unmethylated CpGs and black circles symbolize methylated CpGs.
Epigenetic regulation of ESR2 (gene coding for ERβ) has been poorly investigated. However, one study has demonstrated a frequent occurrence of ESR2 promoter methylation in ERβ- BC in Chinese women.28 Indeed, ESR2 methylation was significantly higher in high grade BC (45%) than in starting neoplasia and was strongly correlated with ESR1 methylation, suggesting common epigenetic mechanisms of regulation.28 Overexpression of ERβ in MCF7 cells strongly decreased cell proliferation. Similarly, hypermethylation of ESR2 was also identified in prostate tumors and present on 3 CpG islands during disease progression.29 All of these observations strongly suggest a role of epigenetics in the inactivation of ESR genes in hormone dependent cancers.
HDAC inhibitors (HDACi) such as Entinostat or valproic acid, have been tested in BC cells and efficiently restored both ERα expression and Letrozole sensibility in ER- BC in vitro and in vivo.30,31 The association of HDACi or 5-azadeoxycytidine with a treatment inducing overexpression of TFAP2C might improve ESR1 expression in ER- patients. A combined HDACi and 5-azadeoxycytidine treatment induces the most significant increase in ERα content. Surprisingly however, addition of tamoxifen does not produce a tumorigenic response in ER- BC cells. Hoestetter et al. demonstrated that a better response to tamoxifen in BC cells, correlated with a lower level of the RNA-stabilizing HuR protein. Tamoxifen treatment increased HuR content, and contributed to its own resistance while HDACi/5-azadeoxycytidine decreased HuR. Preliminary treatment with HDACi/5-azadeoxycytidine was given before delivering tamoxifen to attempt to obtain the best tamoxifen sensitivity.32 The precise roles of tamoxifen are complex: although it competes with 17β-estradiol to bind to ERα, ERα bound to tamoxifen is still able to target the TFF1 (also called pS2) promoter without constitutive activation of gene transcription. The loss of transcriptional activity of the tamoxifen-ERα complex is mediated by changes in the balance of co-activators/co-repressors and ERα-interacting partners.
Epigenetic Regulation of Estrogen-Responsive Genes by Estrogen Receptors
Regulation via ER and coactivators
How epigenetic changes affect the transcriptional response of estrogen stimulation in cancer, and particularly in BC, is still poorly understood. However, several groups have shown a connection of both estrogen and ER in epigenetic regulation. Several reports suggest that ERα cooperates with co-activators to epigenetically regulate estrogen responsive genes. Only a small percentage of genes with putative ERE are really activated following estrogen stimulation, suggesting that additional proteins could specifically control ER-responsive gene pathways. Maximal ERα-mediated transcription requires the addition of some epigenetic changes and the removal of others. Estrogen bound ERα orchestrates the recruitment of HATs (p300 and CBP) and HAT coactivators of the p160 family (SRC1/SRC2/SRC3) to modulate chromatin status and allow RNAP II recruitment.33,34 Indeed, overexpression of SRC3 increases BC cell proliferation, while inhibition of SRC1/SRC2 blocks their proliferation. Moreover, in the absence of estrogen stimulation, a direct interaction between HDAC1 and unbound ERα, via its AF2 and DNA binding domains, is constitutive in BC and inhibits its activity.21
Methylation of histones and the enzymes that control this methylation are highly implicated in estrogen signaling. An increase in the epigenetic mark H3K4me3 is generally associated with positive effects on transcription and such an increase on the TFF1 promoter is due to a direct interaction between ERα and the protein linker MEN1. This interaction recruits the coactivators H3K4 methylase MLL1/2 (Mixed Lineage leukemia).35 Based on studies done on JMJD2B/MLL2/ERα interactions, a model was developed in which demethylation of H3K9me by the HDM JMJD2B (Jumonji domain-containing protein 2B) is first required for the further methylation of H3K4 by MML2.36 An increase of H3K4me3 after direct interaction between ERα and MLL 2–4, via its LxxLL domain, was required for activation of cathepsin, liver x-receptor genes.
Besides methylated marks, removal of other methylation may also be implicated in estrogen responsive gene regulation. Recruitment of the HMT SMYD3, whose levels increase in BC, was also able to produce the tri-methylation of H3K4me3 and was mediated by both a direct ERα/SMYD3 interaction on the ERE of the TFF1 promoter and/or by the identification of the Ser10 phosphorylation mark on histone H3.37,38 The HDM LSD1 (lysine specific demethylase, also called KDM1) also contributes to H3K9 demethylation on ER targets genes and recruitment of coactivators but this required the presence of activated ERα.39 In some other genes, however, recruitment of LSD1 also follows H3K9 deacetylation and provokes H3K4 demethylation, which is unfavorable to transcription.40 The specificity of H3 methylated substrate on ER target loci such as TFF1 promoter, is orchestrated by the co-recruitment of PELP1/activated ERα/LSD1. PELP1 (proline glutamic acid and leucine rich protein 1) is a reader of methylation marks that recognizes both H3K4me2 and H3K9me2 but its interaction with ERα and LSD1 decreases the LSD1-mediated HDM activity on H3K4me2 in favor of H3K9me2 demethylation and increased ER target gene expression.41 Moreover, the early engagement of some factors on condensed chromatin, in a specific sequence that is dependent of an epigenetic signature, refers to a class called competence or pioneer factors. Pioneer factors are implicated in the opening and activation of transcription. Magnani et al. reported that the association of activated ERα with the pioneer factors PBX1 (pre-B-cell leukemia homeobox 1), and FOXA1 (forkhead box A1) considerably increased estrogen dependent transcriptional response via PBX1-dependent identification of H3K4me2 and chromatin remodeling.42
Expression of CARM1 (coactivator-associated arginine methyltransferase), a coactivator of ERα, correlates with low grade BC and with a decrease in BC cell proliferation. CARM1 is believed to partially govern the proliferation/differentiation balance in BC by controlling 16% of estrogen dependent genes.43 While mechanisms implicating CARM1 are complex and still under investigation, CARM-1-mediated H3R17me and H3R26me seems to be implicated in estrogen response, while methylation of p300 may regulate its activity.44 A direct interaction between free ERα and phosphorylated CARM-1 may be used to recruit other coactivators, while association of CARM-1 with activated ERα may require a p160 coactivator SRC-2.45 Indeed, CARM1-mediated CBP methylation is required for CBP recruitment to some ER target genes and increases its HAT activity.46
Fewer studies have been performed to identify ERβ coactivators. Indeed, as has been observed for ERα/MLL interactions, MLL1-4/ERβ complexes are implicated in HOXC13 gene regulation.47 However, ERβ/eNOS (endothelial nitric oxide synthase) complex was observed in prostate cancer and provoked the activation of hTERT, MSH2, CyclinD1 and TFF1, 4 genes previously identified in prostate cancer grading.48 On the other hand, this complex was also associated with the epigenetic repression of GSTP1 expression, a gene frequently silenced in prostate tumors. Further investigation will be necessary for a better view of the mechanisms controlling epigenetic-mediated ERβ target gene expression.
Regulation via ERs and corepressors
Although the link between ERα and upregulation of gene transcription is well studied, some transcriptome analyses have revealed that about 50% of ERα target genes are downregulated following estrogen treatment.49,50 Indeed, estrogen exposure or ERα loss using both chemical mimetics or siRNA, leads to epigenetic modifications in ER target genes requiring both histone modifying enzymes and Dnmts.
HMT EZH2 (enhancer of zeste homolog 2) is a polycomb protein that catalyzes H3K27me3, a chromatin repressive mark. A high level of EZH2 has been reported in several cancers and is associated with malignancy and the grade in BC. Interestingly, an increase in EZH2 expression both in MCF7 and in vivo, was also reported following estrogen-like exposure.51 Overexpression of EZH2 induces a decrease in the expression of numerous genes, in particular in the ER responsive gene pathway. EZH2-mediated H3K27me3 on ER target genes in BC cells requires EZH2 interaction with REA (repressor of estrogen activity) which preferentially targets ERE and may also recruit HDACs for complete gene silencing.52 On the other hand, Bcl2 is an estrogen responsive gene encoding a major anti-apoptotic protein, upregulation of which is often observed in many cancers including BC. Both genetic and non-genetic ER pathways regulate the expression of Bcl2.53 Bcl2 is normally silenced by EZH2-mediated repressive mark H3K27me3 in its enhancer, promoting the recruitment of other polycomb group proteins (PRC1 and 2). Constitutive S21 phosphorylation-mediated inhibition of EZH2 following PI3K/Akt activation in HER2 positive BC and/or demethylation of H3K27me via ERα/JMJD3 complex recruitment on Bcl2, induce gene expression and contribute to apoptosis resistance in BC. ERα methylation appears essential for non-nuclear functions of ERα such as activation of AKT following ERα/Src/PI3K interaction.52 PRMT1-mediated R260 methylation of cytosolic ERα within its DNA interacting domain occurs rapidly after estrogen treatment and ERα hypermethylation has been reported in 55% of BC. This methylation also required p160 coactivators and is implicated in the non-genomic functions of ERα, leading to a constitutive activation of AKT signaling and a promotion of proliferation and survival signals.54,55
In fact, epigenetic silencing of ER target genes was most frequent in ER- than in ER+ patients and was comparable to the panel of epigenetic modifications observed in MCF7 following ERα inactivationby RNAi.56 According to the literature, DNA methylation and histone modification can cooperate to govern the sequence of epigenetic events leading to the silencing of one gene. DNA methylation and histone modification can be catalyzed within the same complex or successively by independent complexes. Investigations on the kinetics of the addition of epigenetic marks on ER target loci revealed that chromatin remodeling begins 36h after ERα invalidation. First, HDAC1 and the polycomb co-repressors YY1 and EZH2 are recruited while addition of persistent heritable epigenetic marks via Dnmt1 recruitment occurs only at 168h. Local hypermethylation in BC may be the consequence of an increase in Dnmts followed by MeCP2 induction, as was observed in rats treated with high amounts of estrogen.57 Following estrogen exposure, a similar increase in both Dnmt3b expression and activit, in endometrial cancer cells was reported and could be inhibited by an ER antagonist, suggesting a direct implication of ER in Dnmts regulation.58 However, diethylstilbestrol exposure in mice provokes a decrease in Dnmts and SP3 on day 5 while SP1 levels only decreased at day 14, followed by demethylation of several DNA loci.59 Dnmt3a/b expression is under estrogen regulation in normal female tissues.60 To date, the effects of estrogen and the ER pathway on the recruitment of SP1/SP3 to Dnmt promoters have not been investigated. This could be studied with folate treatment, which preferentially permits the recruitment of SP3 in detriment to SP1 and increases Dnmt genes transcription.61 Relative amounts and/or preferential recruitment of SP1/SP3 may explain the tissue-specific response to estrogen exposure. Indeed, hypermethylation of ERCC1, XPC, OGG1 and MLH1 genes, all involved in DNA repair, after estrogen treatment contributed to chromosomal instability and mutations that occur in BC.62 Conversely, expression of HOXA10 was increased following BpA exposure and ERE hypomethylation.63
Moreover, some experiments show that estrogen exposure of breast progenitor cells induces epigenetic modifications and confers a cancer-like methylome in these cells, suggesting a possible role of epigenetic modifications in breast progenitor cells in the initiation of BC.64 Epigenetic modifications (global DNA hypomethylation and histone modifications) occurred as soon as 6 weeks in treated rats, while evident signs of neoplasia could be detected only after 12 weeks. Indeed, estrogen stimulation provoked long-range epigenetic silencing (LRES) in a cluster of 14 genes located at 16p11.2 in normal breast cells.65 The silencing is mediated by ERα translocation into the nucleus and addition of epigenetic marks such as H3K27me3. Prolonged estrogen exposure induces progressive DNA methylation which confers a persistence of epigenetic modifications, similar to those of neoplastic cells.
ERβ has recently been implicated in epigenetic control of estrogen responsive genes. Expression of Glut-4 in MEF cells required the interaction between ERβ and the Glut-4 promoter, which prevented methylation of CpG 11 and therefore allowed the recruitment of SP1 to this region and activation of transcription.65 Epigenetic-mediated neoplastic transition following estrogen exposure has been clearly demonstrated.66,67 Progression of pre-cancerous lesions provoked by estrogen exposure in neoplastic lesions required continuous exposure to estrogen in ACI rats. Indeed, removal of estrogen treatment after 4 weeks followed by 8 weeks of recovery induced a regression of hyperplasia in conjunction with modifications in Dnmts expression.56
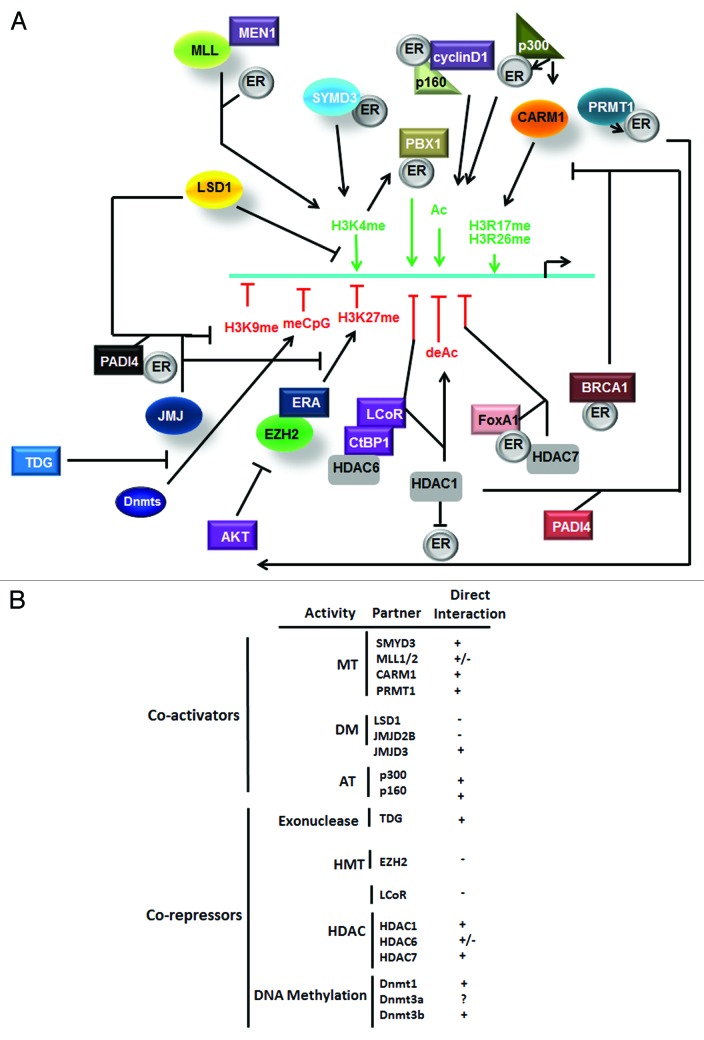
Epigenetic mechanisms implicated in ER target gene silencing seem highly variable and require different co-repressors. Indeed, CTCF (CCCTC-binding factor) recruitment on the CDKN1c promoter following estrogen stimulation is implicated in CDKN1c silencing.68 LCoR, a repressor able to bind to ligand-associated receptors to repress their transcriptional activity also interacts with nuclear HDAC6 and attenuates specific ER target genes, including IGFBP4, but not TFF1 in BC cells. Moreover, the LCoR/CtBP1 repressor complex interacts with HDAC1 and ER on TFF1 and other estrogen responsive promoters.69,70 Malik et al. recently demonstrated a cooperation between HDAC7/FoxA1/ERα in RPRM repression.71 Nevertheless, HDAC7-mediated gene silencing was not related to the weak HDAC activity of HDAC7 but rather to additional properties of this protein. This complex was recruited to both proximal and distal RPRM promoters. HDAC1/PADI4 (peptidylarginine deiminase IV) interaction was also associated with TFF1 silencing. Indeed, this complex provoked H3R deimination, resulting in either a blockade of H3R methylation or in a demethylation of monomethylated H3Rme and therefore inhibited the addition of the positive transcriptional mark H3Rme2, normally processed by CARM1 or PRMT1 (H3R17me and H4R3me). On the other hand, CARM1-mediated H3Rme2 blocks H3R deimination and allowed the recruitment of ERα to the active TFF1 promoter.72 Sin3A is frequently associated with HDAC1/2 in the Sin3 repressor complex and can also be involved in ER-mediated gene silencing (including ESR1) via its multiple interactions with both additional repressors and ERα.27 Similarly, MTA1 (metastasis associated antigen 1), the expression of which correlates with BC progression, can also bind HDAC1/2 and ERα and participate in ER-mediated gene silencing such as BRCA1 silencing.73 BRCA1 was associated with a repression of a subset of estrogen responsive genes in 293T cells while its overexpression induces an almost 90% decrease in ER target gene expression, including TFF1, in MCF7 cells. This inhibition required a direct interaction between active ERα and BRCA1 (aa 338–379 of ERα) and their co-recruitment on ERE which blocked further ERα recognition by coactivators such as p300.74,75 Increasing concentrations of estrogen or overexpression of cyclin D1 which antagonizes and excludes BRCA1, induces estrogen responsive genes. cyclin D1 is frequently overexpressed in BC and its interaction with ERα increases p160 recruitment and promote estrogen signaling. A strong correlation between FoxA1 and ERα recruitment on activated/silenced genes after estrogen exposure was reported following high scale ChIP analysis, suggesting a cooperation between TF and ERα not only in gene activation but also in their silencing. Co-recruitment, however, seemed limited to 7% of ER bound promoters.76,77 Epigenetic mechanisms governing ERα target genes are summarized in Figure 3.
Figure 3. Model of ER-mediated epigenetic response in ER target genes. (A) Schematic representation of action of coactivators and corepressors. (B) Direct and indirect interactions of ERα with epigenetic related proteins.
Conclusion
Studies on ERα target gene regulation have introduced a new degree of complexity by reporting cycling of active/repressive states of the TFF1 promoter.78 A combination of interactions between ERα and HAT, HDAC, HMT, MDT, coactivators, corepressors, TFs and RNAP II reveals a complex histone code that regulates competent or transcriptionally engaged TFF1 and CathepsinD promoters with periodic waves of transcription interrupted by clearing of TFs from promoters.79 Recently, a dynamic process of DNA methylation was also reported to be involved in the control of the cyclic expression of ERα target genes. Cycled methylation/demethylation of CpG in the pTFF1 and Wisp-2 promoters following estrogen stimulation revealed the importance of Dnmts control on estrogen dependent gene expression.80 Each cycle, corresponding to an active transcription time, was determined by first, a demethylation of CpGs catalyzed by Dnmt3a/Dnmt3b associated with TDG (thymine DNA glycosylase) complex, and a remethylation and gene silencing assumed by Dnmt1/Dnmt3a/Dnmt3b, in collaboration with NuRD complex. The recruitment of TDG via a direct interaction with the AF2 domain of ERα might occur in some specific targets rather implicated in the co-recruitment of other coactivators than for its glycosylase activity. As the TDG mutant, incapable of DNA repair, still increased ER responsive gene transcription.81 Putative interactions between ER and Dnmts have also been reported.82,83 As methylation and demethylation are cyclic, global methylation status on these promoters is conserved. Interestingly, as discussed above, ERβ seems implicated in the establishment of new and stable methylation.84 All of these results provide strong evidence that estrogen target gene expression is tightly regulated by multiple and highly dynamic machineries implicating ERs, coactivators and corepressors in a classical and epigenetic manner.
Acknowledgments
This article was written while the authors were supported by the University of Franche-Comté, “BQR Jeunes chercheurs of University of Franche-Comté” and the Ministère de l’Enseignement Supérieur et de la Recherche (MESR). The authors thank J. N. Legrand, Dr Ramji R. Rajendran MD PhD (Elk Grove Village, Illinois, USA) and Lisa Oliver PhD (INSERM U892, Nantes, France) for the critical reading of this manuscript and its comments.
Glossary
Abbreviations:
- BC
breast cancer
- BpA
bisphenol A
- Dnmt
DNA methyl transferase
- dnMTase
de novo DNA methylation activity
- ER
estrogen receptor
- ER-
estrogen receptor negative
- ER+
estrogen receptor positive
- ESR1
gene encoding ERα
- ESR2
gene encoding ERβ
- EZH2
enhancer of zeste homolog 2
- HMT
histone methylase
- HDM
histone demethylase
- HAT
histone acetylase
- HDAC
histone deacetylase
- JMJD
Jumonji domain-containing protein
- LSD1
lysine specific demethylase 1
- mMTase
maintaining DNA methylation activity
- PR
progesterone receptor
- TF
transcription factor
- TSG
tumor suppressor gene
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/23790
References
- 1.Lopez-Garcia MA, Geyer FC, Lacroix-Triki M, Marchió C, Reis-Filho JS. Breast cancer precursors revisited: molecular features and progression pathways. Histopathology. 2010;57:171–92. doi: 10.1111/j.1365-2559.2010.03568.x. [DOI] [PubMed] [Google Scholar]
- 2.Kuiper GG, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–70. doi: 10.1210/en.138.3.863. [DOI] [PubMed] [Google Scholar]
- 3.Koos RD, Kazi AA, Roberson MS, Jones JM. New insight into the transcriptional regulation of vascular endothelial growth factor expression in the endometrium by estrogen and relaxin. Ann N Y Acad Sci. 2005;1041:233–47. doi: 10.1196/annals.1282.037. [DOI] [PubMed] [Google Scholar]
- 4.Hockings JK, Degner SC, Morgan SS, Kemp MQ, Romagnolo DF. Involvement of a specificity proteins-binding element in regulation of basal and estrogen-induced transcription activity of the BRCA1 gene. Breast Cancer Res. 2008;10:R29. doi: 10.1186/bcr1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–88. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hervouet E, Hulin P, Vallette FM, Cartron PF. Proximity ligation in situ assay for monitoring the global DNA methylation in cells. BMC Biotechnol. 2011;11:31. doi: 10.1186/1472-6750-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares J, Pinto AE, Cunha CV, André S, Barão I, Sousa JM, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–8. doi: 10.1002/(SICI)1097-0142(19990101)85:1<112::AID-CNCR16>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 9.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–77. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 10.Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, et al. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–92. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 12.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Normanno N, Di Maio M, De Maio E, De Luca A, de Matteis A, Giordano A, et al. NCI-Naple Breast Cancer Group Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr Relat Cancer. 2005;12:721–47. doi: 10.1677/erc.1.00857. [DOI] [PubMed] [Google Scholar]
- 14.Yao J, Zhang XB, Zhang XL, Fu WL. Methylation status of oestrogen receptor alpha-A: a predictor of prognosis in leukaemias. Biosci Rep. 2010;30:217–22. doi: 10.1042/BSR20090044. [DOI] [PubMed] [Google Scholar]
- 15.Wei J, Han B, Mao XY, Wei MJ, Yao F, Jin F. Promoter methylation status and expression of estrogen receptor alpha in familial breast cancer patients. Tumour Biol. 2012;33:413–20. doi: 10.1007/s13277-011-0234-x. [DOI] [PubMed] [Google Scholar]
- 16.Ramos EA, Camargo AA, Braun K, Slowik R, Cavalli IJ, Ribeiro EM, et al. Simultaneous CXCL12 and ESR1 CpG island hypermethylation correlates with poor prognosis in sporadic breast cancer. BMC Cancer. 2010;10:23. doi: 10.1186/1471-2407-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen JC, Garrett-Mayer E, Pettit C, Mack KM, Manning J, Herman JG, et al. Epigenetic regulation of protein phosphatase 2A (PP2A), lymphotactin (XCL1) and estrogen receptor alpha (ER) expression in human breast cancer cells. Cancer Biol Ther. 2004;3:1304–12. doi: 10.4161/cbt.3.12.1458. [DOI] [PubMed] [Google Scholar]
- 18.Prabhu JS, Wahi K, Korlimarla A, Correa M, Manjunath S, Raman N, et al. The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women. Tumour Biol. 2012;33:315–23. doi: 10.1007/s13277-012-0343-1. [DOI] [PubMed] [Google Scholar]
- 19.Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011;289:74–82. doi: 10.1016/j.tox.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 2001;61:7025–9. [PubMed] [Google Scholar]
- 21.Kawai H, Li H, Avraham S, Jiang S, Avraham HK. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer. 2003;107:353–8. doi: 10.1002/ijc.11403. [DOI] [PubMed] [Google Scholar]
- 22.Macaluso M, Cinti C, Russo G, Russo A, Giordano A. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene. 2003;22:3511–7. doi: 10.1038/sj.onc.1206578. [DOI] [PubMed] [Google Scholar]
- 23.Macaluso M, Montanari M, Noto PB, Gregorio V, Bronner C, Giordano A. Epigenetic modulation of estrogen receptor-alpha by pRb family proteins: a novel mechanism in breast cancer. Cancer Res. 2007;67:7731–7. doi: 10.1158/0008-5472.CAN-07-1476. [DOI] [PubMed] [Google Scholar]
- 24.Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor alpha gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;151:731–40. doi: 10.1210/en.2009-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodfield GW, Hitchler MJ, Chen Y, Domann FE, Weigel RJ. Interaction of TFAP2C with the estrogen receptor-alpha promoter is controlled by chromatin structure. Clin Cancer Res. 2009;15:3672–9. doi: 10.1158/1078-0432.CCR-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fürst RW, Kliem H, Meyer HH, Ulbrich SE. A differentially methylated single CpG-site is correlated with estrogen receptor alpha transcription. J Steroid Biochem Mol Biol. 2012;130:96–104. doi: 10.1016/j.jsbmb.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Ellison-Zelski SJ, Solodin NM, Alarid ET. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol Cell Biol. 2009;29:4949–58. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Yu Z, Li Y, Wen X, Ma W, Wang L, et al. Clinical implications of ERβ methylation on sporadic breast cancers in Chinese women. Med Oncol. 2012;29:1569–75. doi: 10.1007/s12032-011-0107-6. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, et al. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–12. doi: 10.1016/S0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabnis GJ, Goloubeva O, Chumsri S, Nguyen N, Sukumar S, Brodie AM. Functional activation of the estrogen receptor-α and aromatase by the HDAC inhibitor entinostat sensitizes ER-negative tumors to letrozole. Cancer Res. 2011;71:1893–903. doi: 10.1158/0008-5472.CAN-10-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortunati N, Bertino S, Costantino L, De Bortoli M, Compagnone A, Bandino A, et al. Valproic acid restores ER alpha and antiestrogen sensitivity to ER alpha-negative breast cancer cells. Mol Cell Endocrinol. 2010;314:17–22. doi: 10.1016/j.mce.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Hostetter CL, Licata LA, Keen JC. Timing is everything: order of administration of 5-aza 2′ deoxycytidine, trichostatin A and tamoxifen changes estrogen receptor mRNA expression and cell sensitivity. Cancer Lett. 2009;275:178–84. doi: 10.1016/j.canlet.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen MP, et al. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–18. doi: 10.1210/me.12.10.1605. [DOI] [PubMed] [Google Scholar]
- 34.Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011;30:4764–76. doi: 10.1038/emboj.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreijerink KM, Mulder KW, Winkler GS, Höppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–35. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 36.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:7541–6. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, Heo K, Kim JH, Kim K, Choi J, An W. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J Biol Chem. 2009;284:19867–77. doi: 10.1074/jbc.M109.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, et al. The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem. 2011;286:13925–36. doi: 10.1074/jbc.M111.223198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238–49. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schüle R, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–44. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Dhaheri M, Wu J, Skliris GP, Li J, Higashimato K, Wang Y, et al. CARM1 is an important determinant of ERα-dependent breast cancer cell differentiation and proliferation in breast cancer cells. Cancer Res. 2011;71:2118–28. doi: 10.1158/0008-5472.CAN-10-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102:3611–6. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carascossa S, Dudek P, Cenni B, Briand PA, Picard D. CARM1 mediates the ligand-independent and tamoxifen-resistant activation of the estrogen receptor alpha by cAMP. Genes Dev. 2010;24:708–19. doi: 10.1101/gad.568410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ceschin DG, Walia M, Wenk SS, Duboé C, Gaudon C, Xiao Y, et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25:1132–46. doi: 10.1101/gad.619211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansari KI, Kasiri S, Hussain I, Mandal SS. Mixed lineage leukemia histone methylases play critical roles in estrogen-mediated regulation of HOXC13. FEBS J. 2009;276:7400–11. doi: 10.1111/j.1742-4658.2009.07453.x. [DOI] [PubMed] [Google Scholar]
- 48.Re A, Aiello A, Nanni S, Grasselli A, Benvenuti V, Pantisano V, et al. Silencing of GSTP1, a prostate cancer prognostic gene, by the estrogen receptor-β and endothelial nitric oxide synthase complex. Mol Endocrinol. 2011;25:2003–16. doi: 10.1210/me.2011-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–74. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 50.Lin CY, Ström A, Vega VB, Kong SL, Yeo AL, Thomsen JS, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doherty LF, Bromer JG, Zhou Y, Aldad TS, Taylor HS. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: an epigenetic mechanism linking endocrine disruptors to breast cancer. Horm Cancer. 2010;1:146–55. doi: 10.1007/s12672-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang C, Giri VN, Wilkinson JC, Wright CW, Wilkinson AS, Cooney KA, et al. EZH2 regulates the transcription of estrogen-responsive genes through association with REA, an estrogen receptor corepressor. Breast Cancer Res Treat. 2008;107:235–42. doi: 10.1007/s10549-007-9542-7. [DOI] [PubMed] [Google Scholar]
- 53.Svotelis A, Bianco S, Madore J, Huppé G, Nordell-Markovits A, Mes-Masson AM, et al. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERα ligand dependency. EMBO J. 2011;30:3947–61. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol Endocrinol. 2010;24:993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, et al. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–21. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 56.Leu YW, Yan PS, Fan M, Jin VX, Liu JC, Curran EM, et al. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–92. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- 57.Kutanzi KR, Koturbash I, Kovalchuk O. Reversibility of pre-malignant estrogen-induced epigenetic changes. Cell Cycle. 2010;9:3078–84. doi: 10.4161/cc.9.15.12516. [DOI] [PubMed] [Google Scholar]
- 58.Cui M, Wen Z, Yang Z, Chen J, Wang F. Estrogen regulates DNA methyltransferase 3B expression in Ishikawa endometrial adenocarcinoma cells. Mol Biol Rep. 2009;36:2201–7. doi: 10.1007/s11033-008-9435-9. [DOI] [PubMed] [Google Scholar]
- 59.Sato K, Fukata H, Kogo Y, Ohgane J, Shiota K, Mori C. Neonatal exposure to diethylstilbestrol alters expression of DNA methyltransferases and methylation of genomic DNA in the mouse uterus. Endocr J. 2009;56:131–9. doi: 10.1507/endocrj.K08E-239. [DOI] [PubMed] [Google Scholar]
- 60.Yamagata Y, Asada H, Tamura I, Lee L, Maekawa R, Taniguchi K, et al. DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum Reprod. 2009;24:1126–32. doi: 10.1093/humrep/dep015. [DOI] [PubMed] [Google Scholar]
- 61.Hervouet E, Debien E, Campion L, Charbord J, Menanteau J, Vallette FM, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res. 2009;15:3519–29. doi: 10.1158/1078-0432.CCR-08-2062. [DOI] [PubMed] [Google Scholar]
- 62.Singh KP, Treas J, Tyagi T, Gao W. DNA demethylation by 5-aza-2-deoxycytidine treatment abrogates 17 beta-estradiol-induced cell growth and restores expression of DNA repair genes in human breast cancer cells. Cancer Lett. 2012;316:62–9. doi: 10.1016/j.canlet.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 63.Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–80. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–96. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rüegg J, Cai W, Karimi M, Kiss NB, Swedenborg E, Larsson C, et al. Epigenetic regulation of glucose transporter 4 by estrogen receptor β. Mol Endocrinol. 2011;25:2017–28. doi: 10.1210/me.2011-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovalchuk O, Tryndyak VP, Montgomery B, Boyko A, Kutanzi K, Zemp F, et al. Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications and aberrant microRNA expression. Cell Cycle. 2007;6:2010–8. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 67.Starlard-Davenport A, Tryndyak VP, James SR, Karpf AR, Latendresse JR, Beland FA, et al. Mechanisms of epigenetic silencing of the Rassf1a gene during estrogen-induced breast carcinogenesis in ACI rats. Carcinogenesis. 2010;31:376–81. doi: 10.1093/carcin/bgp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez BA, Weng YI, Liu TM, Zuo T, Hsu PY, Lin CH, et al. Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis. 2011;32:812–21. doi: 10.1093/carcin/bgr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palijan A, Fernandes I, Verway M, Kourelis M, Bastien Y, Tavera-Mendoza LE, et al. Ligand-dependent corepressor LCoR is an attenuator of progesterone-regulated gene expression. J Biol Chem. 2009;284:30275–87. doi: 10.1074/jbc.M109.051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palijan A, Fernandes I, Bastien Y, Tang L, Verway M, Kourelis M, et al. Function of histone deacetylase 6 as a cofactor of nuclear receptor coregulator LCoR. J Biol Chem. 2009;284:30264–74. doi: 10.1074/jbc.M109.045526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malik S, Jiang S, Garee JP, Verdin E, Lee AV, O’Malley BW, et al. Histone deacetylase 7 and FoxA1 in estrogen-mediated repression of RPRM. Mol Cell Biol. 2010;30:399–412. doi: 10.1128/MCB.00907-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 73.Molli PR, Singh RR, Lee SW, Kumar R. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–80. doi: 10.1038/sj.onc.1210839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65:6557–67. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 75.Ma Y, Fan S, Hu C, Meng Q, Fuqua SA, Pestell RG, et al. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol Endocrinol. 2010;24:76–90. doi: 10.1210/me.2009-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 77.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Denis H, Deplus R, Putmans P, Yamada M, Métivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Mol Cell Biol. 2009;29:4982–93. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/S0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 80.Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 81.Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem. 2003;278:38586–92. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 82.Hervouet E, Vallette FM, Cartron PF. Dnmt1/Transcription factor interactions: an alternative mechanism of DNA methylation inheritance. Genes Cancer. 2010;1:434–43. doi: 10.1177/1947601910373794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–99. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 84.Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–44. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]