Abstract
The objectives of this study were to identify tissue-specific differentially methylated regions (T-DMR’s) in the folate transport genes in placental tissue compared with leukocytes, and from placental tissues obtained from normal infants or with neural tube defects (NTDs). Using pyrosequencing, we developed methylation assays for the CpG islands (CGIs) and the CGI shore regions of the folate receptor α (FOLR1), proton-coupled folate transporter (PCFT) and reduced folate carrier 1 (RFC1) genes. The T-DMRs differed in location for each gene and the difference in methylation ranged between 2 and 54%. A higher T-DMR methylated fraction was associated with a lower mRNA level of the FOLR1 and RFC1 genes. Methylation fractions differed according to RFC1 80G > A genotype in the NTD cases and in leukocytes from subjects with high total plasma homocysteine (tHcy). There were no differences in methylated fraction of folate transporter genes between NTD cases and controls. We suggest that T-DMRs participate in the regulation of expression of the FOLR1 and RFC1 genes, that the RFC1 80G > A polymorphism exerts a gene-nutrition interaction on DNA methylation in the RFC1 gene, and that this interaction appears to be most prominent in NTD-affected births and in subjects with high tHcy concentrations.
Keywords: FOLR1, PCFT, RFC1 80G>A, homocysteine, tissue-specific DNA methylation, GpG island, NTD
Introduction
Cellular intake of folate is crucial for nucleotide synthesis, methylation reactions and production of glycine and serine.1-3 Methylation reactions of DNA, protein and RNA by S-adenosylmethionine (SAM) are normally followed further downstream by the re-methylation of homocysteine (Hcy) to methionine.3 The cell regulates the level of Hcy primarily by excretion via the transsulfuration pathway (homocysteine is converted to cysteine), or by re-methylation.1 An elevated plasma Hcy concentration often results from an insufficient intracellular folate or vitamin B12 supply, impeding the re-methylation of Hcy, and is therefore a convenient biomarker to identify subjects with a functional folate deficiency.4
In human cells, folate transport is facilitated by three types of receptors/transporters: folate receptor α (FOLR1), proton coupled transporter (PCFT) and reduced folate carrier (RFC1).5 The placenta is an organ that mediates uptake of folates to the developing embryo, which are critically important for providing the nutrients needed for cellular proliferation and the development of critical structures, such as the embryonic neural tube.6 In both humans and mice, the placenta expresses all three types of receptors.7 Mice lacking the folate binding protein, ortholog to the human folate receptor (hFRα), develop structural malformations including craniofacial, cardiac and NTDs.8-12 Inhibition of folic acid binding to FRα has been suggested as a risk factor for NTDs.13
DNA methylation is involved in regulating gene expression.14 Most intervention studies of folate intake and DNA methylation are addressing global DNA methylation in blood leukocytes. Findings from the seven previously published folate intervention studies summarized by Terry and colleagues are notable for their inconsistent findings.15 Only 2 of 7 studies found any significant changes in leukocyte global DNA methylation. These studies varied in design; they used different folate intake doses, time frames and ages of the participants. There are still no studies of gene-specific DNA methylation of genes involved in the transport of folate in the placenta or in leukocytes from subjects with differing levels of Hcy. We hypothesized that aberrant DNA methylation of folate transport genes in the placenta could compromise folate supplies to the embryo, which if occurring at a critical developmental window, could result in an increased risk of NTDs. To test this hypothesis, we developed methods to identify tissue-specific differentially methylated DNA regions (T-DMRs)16-18 of folate transport genes using pyrosequencing, which is a quantitative and site-specific method. In addition, we explored whether a common polymorphism, RFC1 80G > A that abolishes a CpG site in the RFC1 gene (CpG → CpA), quantitatively affects DNA methylation in its vicinity. We subsequently applied the methods in a clinical study of DNA samples from newborn infants with or without NTDs.
Results
DNA methylation assay development
The folate transport genes FOLR1, PCFT and RFC1 have differing CpG site content and distribution. The FOLR1 gene has the most limited CpG site distribution, while RFC1 has the broadest. The PCFT and RFC1 genes have more than one CpG island each, and one of them is located at the traditional 5′ end of the gene. In contrast, the FOLR1 gene has one short CpG island located within the coding region of the gene. We designed pyrosequencing methylation assays located in the CpG island and shore regions; the nucleotide positions of analyzed cytosines are defined in Tables 1, 2 and 3. A total number of 24 assays covering 121 CpG sites were developed, 5 in the FOLR1 gene, 8 in the PCFT and 11 in the RFC1 gene. The assay covering CpG sites 50–55 in the RFC1 gene includes a well-known polymorphism RFC1 80G > A (rs 1051266), we therefore also obtained the genotype for this SNP in our DNA samples. The locations of the assays were selected to obtain high quality PCR and sequencing primers. They were tested for potential PCR bias by measuring DNA methylation of standard samples with known percentage methylation and curve analysis of slopes and intercepts. Our assays had curve slope values between 0.93 and 0.98, and intercepts below 12%. Within-assay precision was estimated for CpG sites 3–5 in the FOLR1 gene, CpG site 45–48 in the PCFT gene and 15–17 in the RFC1 gene. The CV was ≤ 4.4% for all CpG sites analyzed in accordance with a previous study.19
Table 1. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the FOLR1 gene Pyrosequencing methylation assays.
| CpG site No. | Nucleotide No.* | Primer sequence, 5′-3′ | Ta (°C) | Size (bp) |
|---|---|---|---|---|
| 1 & 2 |
116, 146 |
F: GGGTTAGGATTGAGTTTTTTAATGTTTG |
59 |
93 |
| 5′CGI shore |
|
R: biotin- AACCCACCTACTCATACAACTT |
|
|
| |
|
S: TGAGTTTTTTAATGTTTGTATGAA |
|
|
| 3–5 |
712, 716, 748 |
F:GTTTTTTTAAAGTGTGGGATTATAGAAATG |
57.5 |
93 |
| 5′CGI shore |
|
R: biotin-AAAAACACAACCCAAAATTTTAC |
|
|
| |
|
S: GTGGGATTATAGAAATGAG |
|
|
|
6−8
|
2508, 2518, 2524
|
F:GGTTAGGATGGTTTTGATTTTTTAGTT |
52.9 |
263 |
|
CGI
|
|
R: biotin-ACCCCAAACTAAATACAATAACTTACTT |
|
|
| |
|
S: AAAATGTTGGGATTATAGG |
|
|
| 9–11 |
3457, 3466, 3477 |
F: GTTGGGATTTTTGAATTTGAGTTT |
61 |
125 |
| 3′CGI shore |
|
R: biotin-TCTTCCCACCATTACTCACAA |
|
|
| |
|
S: TGTATTAAAATTATTTAGGTGGAT |
|
|
| 12–14 |
3843, 3855, 3875 |
F:AGTGGGAGTTGTTTGTTAATTT |
59 |
199 |
| 3′CGI shore |
|
R: biotin-AACCCCACTCATAACTACAACATA |
|
|
| |
|
S: ATTTTTATAAGGTTAGTAATTATAG |
|
|
Nucleotide number is relative to the ATG start site.
Table 2. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the PCFT gene Pyrosequencing methylation assays.
| CpG site No. | Nucleotide No. * | Primer sequence, 5′-3′ | Ta (°C) | Size (bp) |
|---|---|---|---|---|
| 1–5 5′CGI shore |
-4604, -4600,-4598 -4560, -4551 |
F: GGAAAAGAAAATTTGTATTTAGAGTGAGA R: biotin-ATCAAACTACTAACCTCAAATAATCC |
57.5 |
170 |
| |
|
S: ATTTGTAAAAATAATAATATTTGGT |
|
|
| 6–8 |
-2650, -2616, -2594 |
F:TTTGGGTTTTGGATTTTATAGTGTAG |
55 |
177 |
| 5′CGI shore |
|
R: biotin-ACTACCATATCCCCAACATCTAAAATAAT |
|
|
| |
|
S: TTTTTTTTGAGATTGAGAGTT |
|
|
|
9–31
CGI 1 |
-55, -50, -46, -44,
-42, -37, -34, -30 -23, -14, -11, -9 11, 13, 22, 33, 35 39, 46, 53, 63, 70 79 |
F:GGTGGTTTTAGGTTATAGG R: biotin-ATTACTACAACCCCCCCTTTA S1: GTATATGGAGGGGAG S2: GTTGGTTTTAGGTAG |
57.3 |
366 |
|
32–36
CGI 1 |
276, 287, 295, 304
318 |
F: GAGTTTGGTAGGTGGAGGGTT R: biotin-ATCAAACCCCTTTACTCTAATCCC |
60 |
136 |
| |
|
S: AGGGTTTTGGTTTGG |
|
|
|
38–44
CGI 2 |
687, 693, 716, 741
754, 757, 761, 779 |
F: TTTTTTATTGGATTTTTTATATGAA R: biotin-ACACTAAAACCTAAAACAACAAACC |
55 |
147 |
| |
|
S1: TTTATTGGATTTTTTATATG |
|
|
| |
|
S2: TGTTGGGAGTTTGGA |
|
|
| 45–48 3′CGI shore |
138, 1148, 1153 1162 |
F:TATGTAGTTTTTTGTTTTGGTGAGAT R: biotin-CCCAAAATACACAATAATCACCAC |
60 |
177 |
| |
|
S: GGTGAGATTTTAAAGGAGTTA |
|
|
| 49–50 |
2373, 2379 |
F:GTGTTGGGATTATAGGTATGAGT |
60 |
170 |
| 3′CGI shore |
|
R: biotin-AAAATTCCCTTCTACTTAATTATCAAAC |
|
|
| |
|
S: GGGATTATAGGTATGAGTTAT |
|
|
| 51–52 |
3926, 3931 |
F: AGATAGAGTAGGGTGAGTGTTAG |
59 |
100 |
| 3′CGI shore |
|
R: biotin-ACTATTCCCCCAACCATAAC |
|
|
| |
|
S: GGTTTTTTATGTTTAGTGTTGT |
|
|
Nucleotide number is relative to the ATG start site.
Table 3. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the RFC1gene Pyrosequencing methylation assays.
| CpG site No. | Nucleotide No. * | Primer sequence, 5′-3′ | Ta (°C) | Size (bp) |
|---|---|---|---|---|
| 1–3 5′CGI shore |
-8067,-8042,-8025 |
F: GTTAGTGTTGGGAGGTTTGA R: biotin - TACAACTACCCCTCTCTCCA |
59 |
181 |
| |
|
S: GTTTTTGTGATGGGTTAT |
|
|
| 4–7 5′CGI shore |
-7784, -7779, -7687,-7773 |
F: TGGAAATGTGGGAGGAAAAAT R: biotin-ACACCCCATATACAAAAAAAC |
58 |
107 |
| |
|
S: ATTGTATATGTTGGGGTA |
|
|
| 8–14 5′CGI shore |
-6224, -6222,-6218 -6212, -6206,-6202 -6188 |
F: TGTGTGGTTGGGGAATTTT R: biotin-AAACCAATCCCTCACCTATCTC S: AGGGTAGTTTGGGTAGGTTTTT |
57 |
131 |
| 15–17 5′CGI shore |
-5456, -6030, -6010 |
F: GGAAGGGGGTGGGAGTTAT R: CCAAAACAACCTACTCCCTTTAC |
57 |
119 |
| |
|
S: TTTGTGGGAAGGGGTTTA |
|
|
|
18–23
CGI 1 |
-5205, -5199, -5193
-5184, -5182, -5171 |
F: TGGGAGAGTGGTTTAGGT R: biotin-AACCACCAATCCCCATCC |
57 |
107 |
| |
|
S: GTTTTTTTTTTTGAGTGTGAT |
|
|
|
24–30
CGI 1 |
-3713,-3704,-3693
-3688,-3664,-3649 -3629 |
F: GTGTAATTTTTTTTTGGAGTAGTTGTGG R: biotin-AACCCTAAAAAAATAACTTTCCTACTA |
57 |
147 |
| |
|
S1: GTGTAATTTTTTTTTGGAGTAGTTGTGG |
|
|
| |
|
S2: AGAGAGAGTTTGGATAG |
|
|
| 31–33 3′CGI shore |
-2989,-2974,-2953 |
F: TTTGGTTAGTTTTTTAGAGTAGGAGTTG R: biotin-ACCCCACTATCCACAATAAC |
56 |
145 |
| |
|
S: GTAGGAGTTGGTTTAAAT |
|
|
| 34–37 3′CGI shore |
-2065,-2079,-2083 -2092 |
F: biotin-ATAGGTTTTGAGGAGGTATGG R: ACAACCAAAACCCTAAAAATTCTC |
58 |
148 |
| |
|
S: AACCACTACCAAACC |
|
|
| 38–42 5′CGI2 shore |
-1082,-1067,-1049 -1037,-1032 |
F: GTGGTATTTGGAGATATGATTTTGATAAGT R: biotin-ACCCCAAACCACAAAACTTC |
57.3 |
271 |
| |
|
S: ATTTTGATAAGTGGATAGGT |
|
|
| 43–49 5′CGI2 shore |
-971,-982,-988 -950,-939,-930 -918 |
F: biotin-TTGGGTAGAATTAGGATTGAAAG R: ACCCCAAACCACAAAACTT |
57 |
159 |
| |
|
S1: AATAAAACAACAACAACAC |
|
|
| |
|
S2: ACCCAACCTTAAAAACC |
|
|
|
50–55
|
38,59,66,75,78,83
|
F: AGATTATTTTTTAAGGTGTTTTGATTTTAT |
57.3 |
240 |
|
CGI 2
|
|
R: biotin-AAAATAATAAAACTCTCCCCTAACC |
|
|
| S: GGTGGAGAAGTAGGTG |
Nucleotide number is relative to the ATG start site.
Tissue-specific DNA methylation differences and gene expression
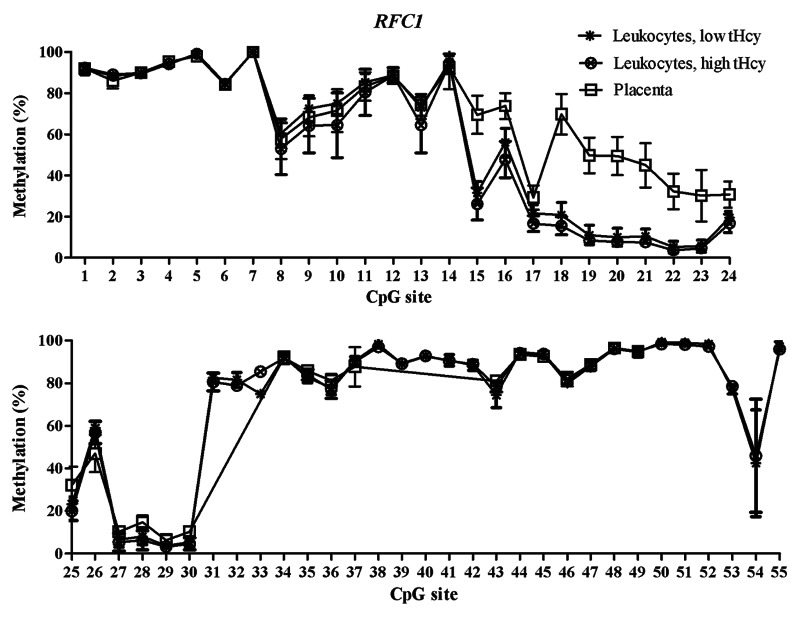
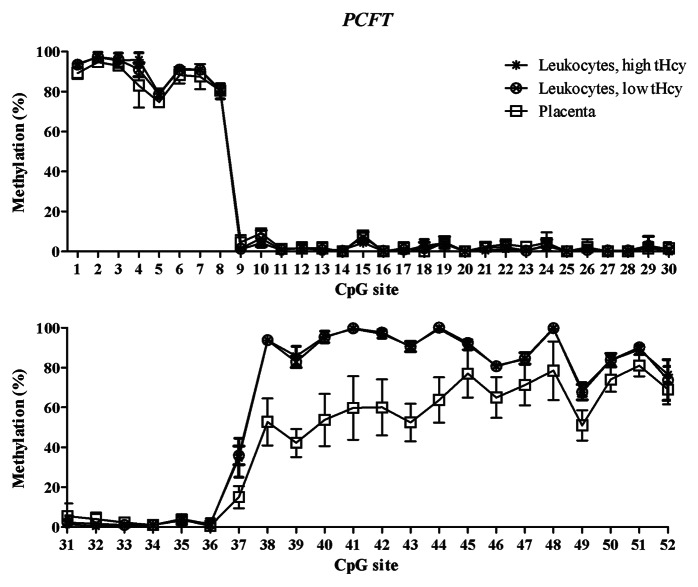
The training sets of normal placental tissues and blood leukocytes differed at specific CpG sites, which tended to cluster in different genomic regions. This allowed us to identify T-DMRs, regions with variable and tissue-specific DNA methylation (Figs. 1–3). In the FOLR1 gene, the mean methylated fractions of CpG sites 1–4 and 12–14 flanking the CpG island in the 5′ shore and 3′ shore regions were statistically significantly (p < 0.05) higher in blood leukocytes. In the PCFT gene, statistically significantly (p < 0.05) higher methylated fractions were found in blood leukocytes in the second CpG island (CGI-2), and in the 3′ shore region of the same island, encompassing CpG sites 38–52. In the RFC1 gene, CpG sites 15–17 in the 5′ shore region and sites 18–30 in the CpG island 1 (CGI-1) showed a statistically significant (p < 0.05) lower methylated fraction in blood leukocytes. Next, we validated these T-DMRs in the 46 normal placental samples from the test cohort and in 25 leukocyte samples with normal tHcy levels from the training set and obtained results, for the FOLR1 and PCFT genes, that were comparable to those in the training set. In the RFC1 gene, we obtained 7 additional CpG sites (data not shown).
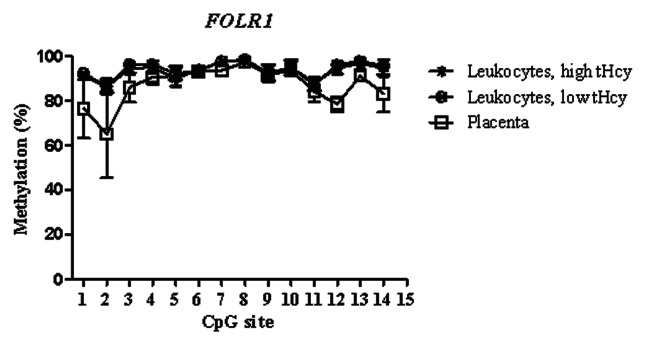
Figure 1. Methylated fraction in the FOLR1 gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 24–25), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 23–25). The error bars show ± 1 SD.
Figure 3. Methylated fraction in the RFC1 gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 22–24), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 21–24). The error bars show ± 1 SD. The upper panel shows CpG sites 1–24 and the lower panel shows CpG sites 25–55.
The mRNA expression of the folate transporter genes PCFT and RFC1 was found to be appreciably lower in the placental tissue than in whole blood leukocytes. In contrast, gene expression of the FOLR1 gene was much higher in the placental tissue than in leukocytes, see Table 4. These expression data are in accordance with annotations in the BioGPS database,20 see http://biogps.org. Moreover, they suggest an inverse relation between DNA methylation in the T-DMRs of these genes and mRNA levels: The higher folate receptor-α mRNA levels in the placenta mirror inversely the significantly lower DNA methylation in the 5′ shore and the 3′ shore of the CGI in the FOLR1 gene (Fig. 1), whereas the lower reduced folate carrier-1 mRNA levels in the placenta inversely mirror a significantly higher DNA methylation of the RFC1 gene in the CGI-1 and its 5′ shore (Fig. 3). In contrast, the lower proton-coupled folate transporter mRNA levels that were found in placental tissue were not accompanied by any higher DNA methylation of the PCFT gene in the 5′ region located in the beginning of the CGI-1, which was fully unmethylated in all studied tissues (Fig. 2). The statistically significant hypomethylation of the PCFT gene that we observed in the placental tissue was located in the CGI-2 and its 3′ shore. The functional significance, if any, of this hypomethylation remains unclear.
Table 4. mRNA expression of FOLR1, PCFT and RFC1 in full-term placental tissue and whole blood leukocytes.
| FOLR1 | PCFT | RFC1 | |
|---|---|---|---|
| Placental tissue |
1.0 × 10−5 ± 1.3 × 10−5 |
1.4 × 10−7 ± 8.5 × 10−8 |
5.0 × 10−6 ± 5.8 × 10−5 |
| WBC |
8.4 × 10−8 ± 1.2 × 10−8 |
7.4 × 10−6 ± 1.5 × 10−5 |
1.3 × 10−4 ± 2.9 × 10−4 |
| Ratio placenta/WBC | 119 | 0.02 | 0.04 |
The relative expression was calculated with the ΔCt method (2^-∆ Ct), using 18S as the reference gene.
Figure 2. Methylated fraction in the PCFT gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 10–25), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 11–25). The error bars show ± 1 SD. The upper panel shows CpG sites 1–30 and the lower panel shows CpG sites 31–52.
Methylated fraction in leukocytes from subjects with high or low tHcy and in placental tissue from normal and NTD fetuses
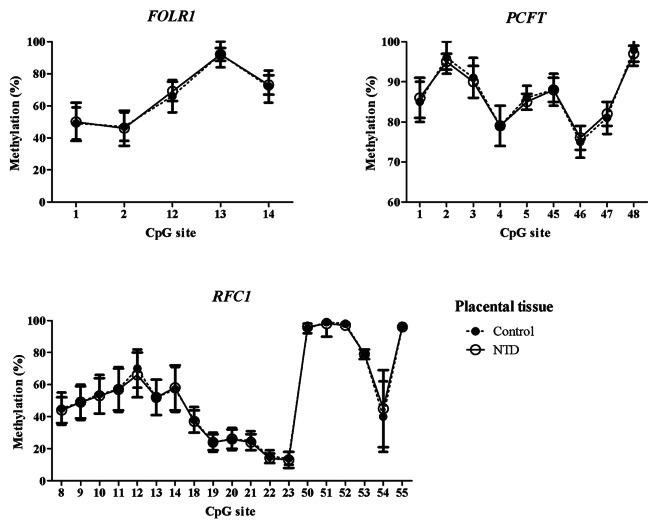
Analysis of methylated fraction of the RFC1 gene in blood leukocytes revealed site-specific differences (p < 0.05) between subjects with low and high tHcy in 17 CpG sites (Table 5). In subjects with high tHcy (used as a proxy for poor nutritional status), the methylated fraction was generally lower (14 CpG sites). Most of these sites were found in the 5′ shore of the CGI-1, where 7 of the CpG sites differed significantly between the groups (p < 0.05). In addition, three CpG sites within the CGI-1 (CpG 18, 21, 25), one CpG site in the 3′ shore CGI-1 (CpG 38), and three CpG sites in the CGI-2 (CpG 50, 52 and 55) were significantly lower. Three CpG sites (CpG site 43, 46 and 53) with significantly higher (p < 0.05) methylated fraction were also observed, located in the 5′ shore and in CGI-2 region, respectively. We did not observe any significant differences between subjects with low or high tHcy in the FOLR1 or PCFT genes. The methylated fraction did not differ significantly between placentas from healthy fetuses and from fetuses with NTDs in either of the FOLR1, PCFT or RFC1 genes (Fig. 4).
Table 5. Mean methylated fraction of CpG sites in the RFC1 gene in subjects with low or high plasma tHcy concentration.
| |
|
Low tHcy |
High tHcy |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG site* | Mean | SD | N | Mean | SD | N | p | ||||
| 5′shore |
8 |
60.5 |
6.7 |
23 |
53.0 |
12.5 |
24 |
0.037 |
|||
| |
9 |
72.5 |
6.3 |
23 |
64.2 |
13.2 |
24 |
0.022 |
|||
| |
10 |
75.0 |
5.4 |
23 |
64.4 |
15.7 |
24 |
0.009 |
|||
| |
13 |
73.0 |
5.5 |
23 |
64.5 |
13.6 |
24 |
0.017 |
|||
| |
15 |
31.5 |
5.6 |
23 |
26.1 |
7.8 |
24 |
0.030 |
|||
| |
16 |
56.0 |
7.0 |
23 |
47.8 |
8.9 |
24 |
0.002 |
|||
| |
17 |
21.7 |
4.2 |
23 |
16.6 |
3.9 |
24 |
0.000 |
|||
| |
|
|
|
|
|
|
|
|
|||
| CGI 1 |
18 |
20.9 |
6.0 |
24 |
15.5 |
4.4 |
24 |
0.005 |
|||
| |
21 |
10.5 |
3.5 |
24 |
7.5 |
1.5 |
24 |
0.024 |
|||
| |
25 |
23.3 |
3.2 |
24 |
20.1 |
4.6 |
24 |
0.038 |
|||
| |
|
|
|
|
|
|
|
|
|||
| 3′ shore |
38 |
98.3 |
1.8 |
24 |
97.1 |
1.7 |
24 |
0.029 |
|||
| |
43 |
74.4 |
5.9 |
23 |
78.5 |
2.6 |
22 |
0.010 |
|||
| |
46 |
80.0 |
1.4 |
23 |
81.5 |
1.8 |
22 |
0.032 |
|||
| |
|
|
|
|
|
|
|
|
|||
| CGI 2 |
50 |
99.3 |
1.2 |
22 |
98.5 |
1.4 |
24 |
0.045 |
|||
| |
52 |
98.5 |
1.4 |
22 |
97.2 |
1.7 |
24 |
0.007 |
|||
| |
53 |
77.1 |
2.2 |
22 |
78.5 |
1.7 |
24 |
0.020 |
|||
| 55 | 97.3 | 2.2 | 22 | 95.9 | 1.9 | 24 | 0.025 | ||||
CpG sites included in the methylation assays, see Table 3. Only sites showing statistically significant differences are show in this table. P, post-hoc significance of difference between mean values in leukocytes from subjects with low or high tHcy, respectively.
Figure 4. Methylated fraction in the FOLR1, PCFT and RFC1 gene in placental tissue from healthy (n = 39–48) and NTD (n = 66–75) subjects. The error bars show 1 ± SD.
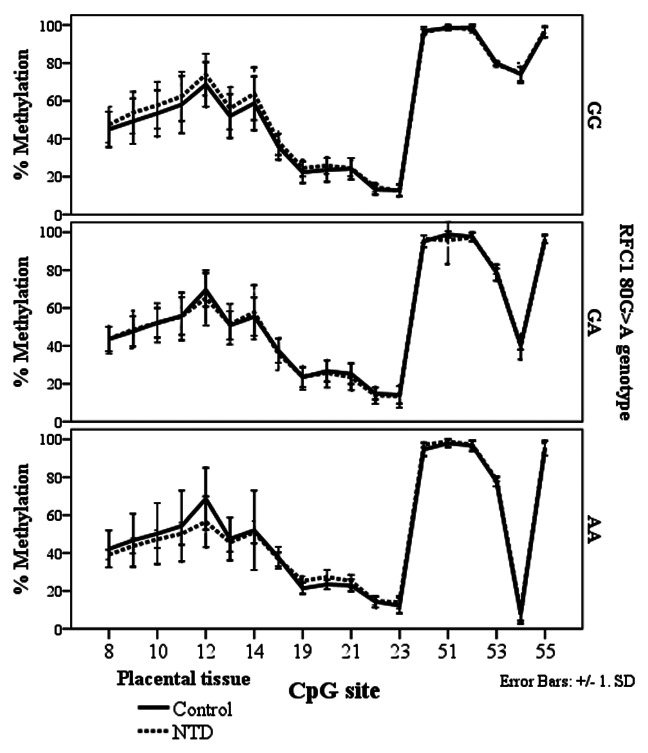
Methylated fraction of CpG sites according to RFC1 80G > A genotypes
The genotyping data in subjects with high or low tHcy and placental tissue from normal and NTD subjects are shown in Table 6. The RFC1 polymorphism was in Hardy-Weinberg equilibrium in all subgroups. The genotype distribution did not differ significantly between subjects with low tHcy and high tHcy (χ2 = 0.199), or between placentas from healthy subjects and those with NTD (χ2 = 1.77).
Table 6.RFC1 80G > A genotype prevalence and allele frequencies in subjects with low and high tHcy and normal and NTD subjects.
| |
Leucocytes |
|
Placenta |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Low tHcy |
|
High tHcy |
|
Normal |
|
NTD |
|||||
| RFC1 80G > A | n | % | n | % | n | % | n | % | ||||
| |
|
|
|
|
|
|
|
|
|
|
|
|
| GG |
6 |
(27) |
|
8 |
(33) |
|
8 |
(21) |
|
22 |
(32) |
|
| GA |
12 |
(54) |
|
12 |
(50) |
|
23 |
(61) |
|
33 |
(49) |
|
| AA |
4 |
(18) |
|
4 |
(17) |
|
7 |
(18) |
|
13 |
(19) |
|
|
Total |
22 |
|
|
24 |
|
|
38 |
|
|
68 |
|
|
| |
|
|
|
|
|
|
|
|
|
|
|
|
| |
χ2 = 0.22 |
|
χ2 = 0.02 |
|
χ2 = 1.7 |
|
χ2 = 0.01 |
|||||
| |
|
|
|
|
|
|
|
|
|
|
|
|
| p(G) |
0.583 |
|
|
0.545 |
|
|
0.513 |
|
0.566 |
|||
| q(A) | 0.417 | 0.454 | 0.487 | 0.434 | ||||||||
The mean methylated fractions of each CpG site in blood leukocytes from subjects with high or low tHcy, grouped according to RFC1 80G > A genotype, are shown in Table 7. When comparing mean methylated fraction between the genotypes by ANOVA, we did not observe any consistent stepwise gene-dose effect due to the A allele in either high- or low-tHcy subjects (see Table 7, p1 columns). In subjects with low tHcy (used as a proxy for nutritional sufficiency), only 3 of the 55 CpG sites were significantly differentially methylated (sites 24, 31 and 34, located in the 3′part of the GCI-1). In subjects with high tHcy, the genotype was significantly related to the mean methylated fraction in 4 CpG sites, located at the 5′ end of CGI-1 and in the 3′ shore of CGI-1 (sites No. 18, 20, 21 and 34); in these subjects, RFC1 80AA genotype was associated with a lower methylated fraction compared with the wild-type in 3 of these CpG sites except No. 34 (Table 7, right-hand p1 column).
Table 7. Methylated fraction of CpG sites in the RFC1 gene in blood leukocytes form subjects with low and high tHcy according to genotype.
| |
|
Low tHcy |
|
High tHcy |
|
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG site | Genotype | Mean | SD | N | p1 | Mean | SD | N | p1 | p2 | |
| |
|
|
|
|
|
|
|
|
|
|
|
|
CpG 3 |
GG |
89.3 |
0.5 |
6 |
|
89.9 |
0.6 |
8 |
|
|
|
| |
GA |
89.3 |
0.7 |
12 |
|
89.8 |
0.8 |
12 |
|
|
|
| |
AA |
89.0 |
0.8 |
4 |
0.656 |
90.0 |
0.8 |
4 |
0.823 |
0.006
|
|
|
CpG 6 |
GG |
85.3 |
0.8 |
6 |
|
83.7 |
1.0 |
7 |
|
|
|
| |
GA |
84.7 |
1.3 |
12 |
|
85.0 |
1.1 |
12 |
|
|
|
| |
AA |
85.3 |
0.5 |
4 |
0.415 |
84.3 |
1.5 |
4 |
0.079 |
0.046
|
|
|
CpG 8 |
GG |
56.8 |
8.6 |
6 |
|
50.3 |
11.4 |
8 |
|
|
|
| |
GA |
61.0 |
6.2 |
12 |
|
56.3 |
8.5 |
12 |
|
|
|
| |
AA |
61.0 |
1.0 |
3 |
0.448 |
48.5 |
23.2 |
4 |
0.434 |
0.029
|
|
|
CpG 9 |
GG |
68.0 |
7.6 |
6 |
|
60.4 |
11.2 |
8 |
|
|
|
| |
GA |
73.2 |
5.2 |
12 |
|
68.1 |
8.8 |
12 |
|
|
|
| |
AA |
73.3 |
0.6 |
3 |
0.195 |
60.3 |
25.3 |
4 |
0.372 |
0.020
|
|
|
CpG 10 |
GG |
72.2 |
7.6 |
6 |
|
58.8 |
16.1 |
8 |
|
|
|
| |
GA |
75.6 |
4.5 |
12 |
|
69.5 |
9.4 |
12 |
|
|
|
| |
AA |
76.0 |
3.6 |
3 |
0.432 |
60.5 |
27.4 |
4 |
0.291 |
0.007
|
|
|
CpG 11 |
GG |
83.5 |
4.9 |
6 |
|
80.3 |
10.5 |
8 |
|
|
|
| |
GA |
85.5 |
4.5 |
12 |
|
82.6 |
6.8 |
12 |
|
|
|
| |
AA |
86.3 |
1.5 |
3 |
0.578 |
74.3 |
21.5 |
4 |
0.454 |
0.049
|
|
|
CpG 13 |
GG |
69.5 |
6.4 |
6 |
|
59.5 |
14.5 |
8 |
|
|
|
| |
GA |
73.5 |
5.3 |
12 |
|
68.8 |
7.9 |
12 |
|
|
|
| |
AA |
74.0 |
0.0 |
3 |
0.307 |
61.3 |
23.1 |
4 |
0.293 |
0.014
|
|
|
CpG 15 |
GG |
26.6 |
2.1 |
5 |
|
26.1 |
7.1 |
8 |
|
|
|
| |
GA |
33.2 |
6.3 |
12 |
|
27.8 |
7.5 |
12 |
|
|
|
| |
AA |
32.5 |
5.4 |
4 |
0.099 |
21.3 |
10.2 |
4 |
0.370 |
0.016
|
|
|
CpG 16 |
GG |
52.2 |
4.4 |
5 |
|
46.9 |
7.8 |
8 |
|
|
|
| |
GA |
56.3 |
7.6 |
12 |
|
50.4 |
7.0 |
12 |
|
|
|
| |
AA |
59.5 |
8.7 |
4 |
0.332 |
41.8 |
14.4 |
4 |
0.233 |
0.001
|
|
|
CpG 17 |
GG |
19.2 |
1.6 |
5 |
|
16.1 |
2.9 |
8 |
|
|
|
| |
GA |
22.3 |
4.9 |
12 |
|
18.1 |
3.7 |
12 |
|
|
|
| |
AA |
22.3 |
4.4 |
4 |
0.407 |
13.3 |
4.3 |
4 |
0.080 |
0.000
|
|
|
CpG 18 |
GG |
16.2 |
2.6 |
6 |
|
15.1 |
4.1 |
8 |
|
|
|
| |
GA |
22.3 |
6.4 |
12 |
|
17.4 |
2.8 |
12 |
|
|
|
| |
AA |
23.5 |
7.2 |
4 |
0.089 |
10.8 |
5.7 |
4 |
0.021
|
0.000
|
|
|
CpG 19 |
GG |
8.8 |
1.6 |
6 |
|
8.5 |
2.2 |
8 |
|
|
|
| |
GA |
11.3 |
5.9 |
12 |
|
9.2 |
1.7 |
12 |
|
|
|
| |
AA |
13.5 |
4.8 |
4 |
0.350 |
6.3 |
2.8 |
4 |
0.070 |
0.011
|
|
|
CpG 20 |
GG |
8.0 |
1.4 |
6 |
|
7.6 |
2.0 |
8 |
|
|
|
| |
GA |
10.1 |
5.5 |
12 |
|
8.5 |
1.9 |
12 |
|
|
|
| |
AA |
12.3 |
4.4 |
4 |
0.370 |
5.3 |
2.8 |
4 |
0.042
|
0.012
|
|
|
CpG 21 |
GG |
8.2 |
1.2 |
6 |
|
7.4 |
1.2 |
8 |
|
|
|
| |
GA |
11.2 |
4.1 |
12 |
|
8.3 |
1.1 |
12 |
|
|
|
| |
AA |
11.8 |
3.8 |
4 |
0.190 |
5.8 |
1.9 |
4 |
0.008
|
0.000
|
|
|
CpG 22 |
GG |
3.3 |
0.5 |
6 |
|
3.3 |
1.6 |
8 |
|
|
|
| |
GA |
5.6 |
3.1 |
12 |
|
4.3 |
0.9 |
12 |
|
|
|
| |
AA |
6.5 |
2.4 |
4 |
0.132 |
2.8 |
1.3 |
4 |
0.052 |
0.012
|
|
|
CpG 24 |
GG |
17.2 |
3.0 |
6 |
|
14.8 |
3.5 |
8 |
|
|
|
| |
GA |
21.2 |
2.8 |
12 |
|
18.8 |
4.8 |
12 |
|
|
|
| |
AA |
19.3 |
2.1 |
4 |
0.028
|
14.5 |
3.3 |
4 |
0.081 |
0.009
|
|
|
CpG 25 |
GG |
21.3 |
2.9 |
6 |
|
18.1 |
4.8 |
8 |
|
|
|
| |
GA |
24.3 |
3.4 |
12 |
|
21.6 |
4.1 |
12 |
|
|
|
| |
AA |
22.8 |
1.7 |
4 |
0.186 |
19.8 |
5.1 |
4 |
0.265 |
0.026
|
|
|
CpG 31 |
GG |
80.5 |
3.3 |
6 |
|
78.9 |
4.6 |
8 |
|
|
|
| |
GA |
83.0 |
1.1 |
12 |
|
81.8 |
3.7 |
12 |
|
|
|
| |
AA |
84.3 |
2.1 |
4 |
0.025
|
80.5 |
4.5 |
4 |
0.313 |
0.051 |
|
|
CpG 34 |
GG |
91.0 |
1.2 |
4 |
|
90.0 |
1.3 |
6 |
|
|
|
| |
GA |
92.8 |
0.9 |
10 |
|
92.2 |
1.4 |
11 |
|
|
|
| |
AA |
93.3 |
1.0 |
4 |
0.010
|
93.3 |
1.9 |
4 |
0.006
|
0.232 |
|
|
CpG 38 |
GG |
98.7 |
1.2 |
6 |
|
96.6 |
2.0 |
8 |
|
|
|
| |
GA |
98.2 |
1.7 |
12 |
|
97.4 |
1.7 |
12 |
|
|
|
| |
AA |
98.5 |
2.4 |
4 |
0.833 |
97.0 |
1.6 |
4 |
0.627 |
0.017
|
|
|
CpG 43 |
GG |
73.4 |
8.3 |
5 |
|
77.8 |
2.9 |
6 |
|
|
|
| |
GA |
76.7 |
4.0 |
12 |
|
78.6 |
2.7 |
12 |
|
|
|
| |
AA |
71.3 |
6.4 |
4 |
0.228 |
79.3 |
1.7 |
4 |
0.703 |
0.003
|
|
|
CpG 46 |
GG |
80.4 |
1.8 |
5 |
|
81.0 |
0.6 |
6 |
|
|
|
| |
GA |
79.8 |
1.4 |
12 |
|
81.1 |
1.4 |
12 |
|
|
|
| |
AA |
80.5 |
1.3 |
4 |
0.583 |
83.3 |
3.3 |
4 |
0.093 |
0.007
|
|
|
CpG 50 |
GG |
99.7 |
0.8 |
6 |
|
98.4 |
1.4 |
8 |
|
|
|
| |
GA |
99.2 |
1.3 |
12 |
|
98.7 |
1.6 |
12 |
|
|
|
| |
AA |
99.0 |
1.4 |
4 |
0.646 |
98.0 |
1.4 |
4 |
0.730 |
0.044
|
|
|
CpG 51 |
GG |
99.5 |
0.5 |
6 |
|
98.0 |
1.3 |
8 |
|
|
|
| |
GA |
98.6 |
1.4 |
12 |
|
98.2 |
1.6 |
12 |
|
|
|
| |
AA |
99.5 |
1.0 |
4 |
0.230 |
98.8 |
1.5 |
4 |
0.710 |
0.052 |
|
|
CpG 52 |
GG |
98.0 |
1.7 |
6 |
|
97.0 |
1.5 |
8 |
|
|
|
| |
GA |
98.4 |
1.4 |
12 |
|
97.4 |
1.8 |
12 |
|
|
|
| |
AA |
99.3 |
1.0 |
4 |
0.402 |
96.8 |
1.7 |
4 |
0.756 |
0.006
|
|
|
CpG 53 |
GG |
76.8 |
2.9 |
6 |
|
78.1 |
1.5 |
8 |
|
|
|
| |
GA |
77.3 |
2.1 |
12 |
|
78.6 |
1.8 |
12 |
|
|
|
| |
AA |
77.0 |
1.6 |
4 |
0.902 |
79.0 |
1.4 |
4 |
0.675 |
0.024
|
|
|
CpG 54 |
GG |
76.2 |
2.6 |
6 |
|
77.9 |
1.1 |
8 |
|
|
|
| |
GA |
38.8 |
1.9 |
12 |
|
39.2 |
1.6 |
12 |
|
|
|
| |
AA |
3.3 |
1.0 |
4 |
0.000
|
2.5 |
0.6 |
4 |
0.000
|
0.417 |
|
|
CpG 55 |
GG |
98.2 |
1.9 |
6 |
|
96.3 |
2.4 |
8 |
|
|
|
| |
GA |
97.0 |
2.5 |
12 |
|
96.0 |
1.7 |
12 |
|
|
|
| AA | 97.0 | 1.6 | 4 | 0.568 | 95.0 | 1.4 | 4 | 0.565 | 0.021 | ||
p1 difference between mean methylated fraction in subjects with different RFC1 80G > A genotypes. p2 difference between mean methylated fraction in subjects with high or low tHcy concentration, adjusted for RFC1 80G > A genotype. Please refer to the Statistics section for details of the multivariate model used.
When the mean methylated fraction in subjects with high or low tHcy was analyzed as dependent variable in a multivariate linear regression model with the group variable (high/low tHcy) and RFC1 80G > A genotype as predictors, the group variable was a significant predictor of the mean methylated fraction in a large number of CpG sites (Table 7, column p2), both in the 5′ shore (sites No. 3, 6, 8–11, 13 and 15–17), in CGI-1 (sites No. 18–22 and 24–25), the 3′ shore (sites No. 31, 34, 38, 43 and 46), and in the 5′-end of CGI-2 (sites No. 50–55). Since we used high tHcy as a proxy for poor nutritional status, this suggests an interaction between nutritional factors and DNA methylation in the RFC1 gene.
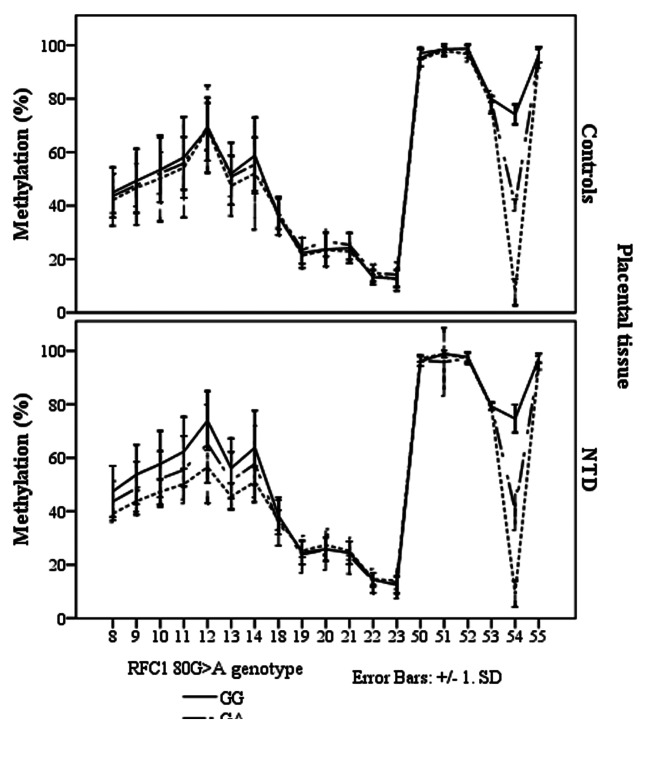
In the NTD placental tissue, we found methylation differences between the RFC1 80 G > A genotypes, at CpG sites 8–14, and CpG site 54 (ANOVA, p < 0.05, Fig. 5). However, there were no statistically significant methylation differences between normal and NTD groups when stratified by genotype (Fig. 6).
Figure 5. Methylated fraction in the RFC1 gene according to RFC1 80G > A genotype in placentas, stratified by groups of healthy controls (n = 38) and NTD (n = 68). The error bars show ± SD.
Figure 6. Methylated fraction in the RFC1 gene in placentas of normal control births (n = 38) and NTD births (n = 68) stratified by RFC1 80G > A genotype.
The RFC1 80 G > A genotype status did not affect the methylated fraction of any CpG sites in the FOLR1 or PCFT genes.
Discussion
In this study we used a training set of different tissues to locate tissue-specific differentially methylated regions (T-DMRs) in the folate transporter genes PCFT, RFC1 and FOLR1. In the clinical study, we found genotype-associated differences in the methylated fraction of the RFC1 gene in placentas from cases with NTDs, suggestive of a gene-nutrient interaction affecting DNA methylation of this gene in NTD pregnancies, but not in placentas from healthy deliveries.
DNA methylation differences between cancer cells and normal cells have been shown to co-localize with T-DMRs and to occur with higher frequency outside the CpG islands.17,21,22 We therefore designed our methylation assays to cover both CGIs and CGI shore regions. We found T-DMRs in the CpG island shores of the FOLR1 gene and in the CGI-2 and 3′ shore of the PCFT gene (Figs. 1 and 2). In the RFC1 gene, they were located at the 5′ shore of CGI-1 and in the CGI-1 (Fig. 3). The tissue-specific DNA methylation patterns of these genes indicate that blood leukocytes should not be used routinely as a substitute for other tissues.
The relative gene expression of the folate transporter genes showed that FOLR1 was more abundant in the placental tissue than in leukocytes compared with the PCFT and the RFC1 gene expression, which were less expressed in placenta than in leukocytes. We found an inverse relation between DNA methylation in the T-DMRs of FOLR1 and RFC1 and mRNA levels (Table 4, Figs. 1–3). A larger study is warranted to further analyze the correlation between gene expression and DNA methylation of the folate transport genes. Longitudinal studies throughout pregnancy would be highly informative. A study comparing mRNA expression of folate transporter genes between gestational age found lower mRNA expression of the RFC1 gene in placental tissues from term compared with the 1st trimester, but no DNA methylation data was available.23
We detected DNA methylation differences in the RFC1 gene between subjects with high and low tHcy concentration, but not in the FOLR1 or PCFT genes. This suggests that DNA methylation in the RFC1 gene may be more sensitive to a deficiency of methyl donors, such as low folate concentrations, than the other genes. In ethanol-induced folate deficient rats, the DNA methylated fraction in CGI of the RFC1 gene was analyzed, and suggested to be lower compared with the control rats.24 The mechanism of this ethanol effect is unclear, but it suggests that nutrition may have an impact on the DNA methylation of the RFC1 gene. Our results showed the greatest difference in methylation in CpG sites in the 5′-shore of the RFC1 gene: subjects with high tHcy had a lower methylated fraction compared with low tHcy subjects.
The human RFC1 gene has several promoter regions located ≈4 kB upstream of the transcriptional start site.25-28 Promoter regions A1/A2, A and B are GC rich, lack TATA boxes, bind Sp transcription factors, and include E-box and Ikaros elements.25,26 Our assays covering CpG sites 38–42 are located in the promoter region A1/A2, assays including CpG sites 8–14, 18–23 and 24–30 are close to the promoter regions A, B and C. We observed the main difference between subjects with high or low tHcy in the CpG sites 8–17, located upstream of the promoter B region which is also the 5′ - shore region related to the CGI-1.
Correlation between DNA methylation in the promoter region of the RFC1 gene and mRNA expression has been analyzed in various cell lines.29 In diffuse large B-cell lymphoma cell lines, DNA methylation was suggested to alter mRNA expression, but not in acute lymphoblastic cell lines.30-32 In contrast to these studies that used qualitative or semi-quantitative assays, we applied quantitative assays to measure DNA methylation in the human folate transport genes.
A crucial question that arises is whether the expression of the three folate transport genes is regulated by methylation in response to folate availability. In order to systematically answer this question, a cohort receiving a controlled folate diet should be analyzed. Regulation of folate transport genes can occur at both the pre- and post-transcriptional levels. Homocysteine itself has been suggested to upregulate the translation of the FOLR1 receptor.33 Colon adenoma cancer cells and MCF7 breast cancer cells grown in folate depleted medium showed a reduction of mRNA expression of the RFC1 gene.34,35 The authors suggested that it could be a response to prevent efflux of folate through the RFC1. Our finding of a higher methylated fraction of the RFC1 gene in subjects with low tHcy fits with this proposed mechanism of preventing folate-efflux.
The mechanisms by which folate reduces the prevalence of NTDs are still unclear, but it has been confirmed that folic acid consumption by mothers reduces the risk of NTD.3 Placental tissue express all three folate transport genes, and the suggested metabolic pathway of utilizing Hcy in the placenta is thru the methionine cycle, with folate serving as the methyl donor.23 We were interested in whether DNA methylation is a potential modulator of folate transport genes in placentas from deliveries of fetuses with NTD. We did not observe any group differences in DNA methylation in PCFT, RFC1 or FOLR1 genes between placentas from healthy deliveries and those with NTD. If the transport of folate into the cells is defective in NTDs, then the regulation could still be compromised at the mRNA or protein levels, but aberrant DNA methylation of these genes is unlikely to be causative. It would be ideal to explore the effect of DNA methylation in the relevant cell type and at the critical developmental time, in this case neuroepithelial cells and yolk sac cells during the period of neural tube closure, but such collection is ethically and practically undoable and we maintain that the placenta remains a reasonably valid surrogate for the yolk sac in this type of study.
We did observe lower DNA methylated fractions of the RFC1 gene in placental tissue from infants with NTDs carrying the RFC1 80AA genotype when compared with 80GG. Similarly, subjects with high tHcy were found to have lower mean methylated DNA fraction in leukocytes, if they were homozygous for the 80AA allele, than the low tHcy subjects. The importance of the RFC1 80G > A polymorphism for plasma tHcy levels has not been clear-cut to date, as some studies showed that AA homozygosity is associated with higher levels of serum folate in women only, while others saw no effect.36-38 Its association with NTD has also not been consistently reproduced.39 In a rat model, folate status in females was found to influence the global DNA methylation in placentas by altering the available methyl pool.40 It is possible that the alteration in methylation status of the RFC1 gene epigenetically impacts its pattern of expression in such a way as to impair normal neural tube closure. If the folate transport genes simply moved molecules of one carbon donors into and out of cells, that would be one way in which to envision a mechanism underlying NTD formation. However, there is every likelihood that these transport proteins have a cell signaling function which is every bit as important in regulating critical signal transduction events during neurulation. Altering the expression of RFC1 is perhaps only an initiating step in impacting a network of genes involved in cell morphogenetic movements that are essential to completing neural tube closure. We do not rule out that DNA methylation changes in other one-carbon metabolism genes or planar cell polarity genes may be contributing, perhaps in interaction with DNA methylation changes in folate transport genes, to an increased NTD risk, but we are filling a significant data gap on T-DMRs in folate transport genes.
In conclusion, we have identified tissue-specific differentially methylated DNA regions (T-DMRs) in three folate transport genes, and shown an inverse relation between methylation and mRNA abundance for the FOLR1 and RFC1 genes. RFC1 mean methylated fraction was found to depend on the RFC1 80G > A genotype in subjects with high tHcy, used as a proxy for poor nutritional status, and in placentas from NTD subjects. We therefore postulate a gene-nutrition interaction between folate intake, RFC1 genotype, RFC1 DNA methylation, and RFC1 mRNA transcription, which could account for part of the spectrum of NTD births. Recognition of this interaction in future research may lead to a fuller appreciation of the role of the folate receptors in the etiology of NTDs.
Material and Methods
Study population
A training set consisting of 56 discarded blood samples and four placental tissues collected from our routine clinical activities (Dept. of Laboratory Medicine, Örebro University Hospital) was used for method development purposes and to identify regions of differential DNA methylation. We selected and de-identified 25 blood samples with tHcy in the range of 5–10 μmol/L (serving as a proxy to define presumably well-nourished subjects), 25 blood samples in the range of 20–113 μmol/L (serving as a proxy to define presumably poorly nourished subjects), in the following designated “low tHcy” and “high tHcy” respectively, and finally, four de-identified placental tissues collected immediately after uncomplicated vaginal delivery. The testing cohort consisted of placental tissues from deliveries of healthy fetuses (n = 48) or fetuses with neural tube defects (n = 75), collected with appropriate informed consent in Shanxi Province, China. All clinical samples were obtained with appropriate IRB approval. The Regional Ethics Review Board, Uppsala, approved the Swedish part of the project.
DNA extraction, Bisulphite treatment
Genomic DNA was extracted from 200 µL whole EDTA blood using QIAamp EZ1 DNA blood 200 μl Kit according to the manufacturer’s instruction (Qiagen Inc.) utilizing a BioRobot EZ1 (Qiagen Inc.). Genomic DNA from placental tissue was manually extracted with Gentra PureGene or QIAmp DNA mini kit (Qiagen Inc.). Approximately 1,000 ng extracted DNA was used for the bisulphite treatment performed with EZ DNA Methylation Gold kit according to the instructions by the manufacturer and eluted in 25 µL elution buffer (Zymo Research, Orion Diagnostica).
mRNA extraction and Quantitative Real-time PCR (qPCR)
Expression analysis of the folate transporters was performed from the training set tissues of four placental tissues and six healthy normal blood donors. RNA was extracted from 4 placental tissues with a standard TRIzol RNA extraction protocol (TRIzol®Reagent, Invitrogen, Sweden). Extractions from whole blood samples were made with the QIAmp RNA Blood Mini Kit (QIAGEN Inc.).
qPCR was performed with an ABI 7500 Fast Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. All samples and negative controls were run as triplicates and the qPCR protocol was as follows. The baseline was set between 3–15 cycles and the threshold to 0.2, a total of 50 cycles was run.
Gene expression assays for the folate receptor α (FOLR1, hs01124177_m1) and 18S (Hs99999901_s1) were purchased from Applied Biosystems. Assays for: SLC46A1, proton coupled folate transporter (PCFT), and SLC19A1, reduced folate carrier (RFC1) were designed in-house, with the Primer Express 3.0 software (Applied Biosystems) see Table 8 for details. All in-house assays are labeled with Fam-Tamra dyes and were purchased from www.biomers.net (Ulm/Donau). Normalization was performed with 18S using the ΔCt-method.41
Table 8. In-house design of mRNA expression assays for the PCFT and RFC1 genes.
| Gene | Sequence 5′-3′ | Size (bp) | ||
|---|---|---|---|---|
|
RFC1 |
Fw |
CGTCAAGACCATCATCACTTTCA |
100 |
|
| |
Rev |
CAGGATCAGGAAGTACACGGAGTA |
|
|
| |
Pr |
CCGGTCCGCAAGCAGTTCCAGTT |
|
|
|
PCFT |
Fw |
TTCACAGGATATGGGTTGCTTTT |
120 |
|
| |
Rev |
CACACAGGCCACAGCAGAAA |
|
|
| Pr | CTGTCATCCGGGCTAAACTCTCCAAGCT |
Fw, forward primer; Rw, reverse primer; Pr, probe
Methylation assay development and PCR amplification
Prior to primer design, we used the online web tool CpG island searcher (http://cpgislands.usc.edu/) to locate the CpG sites and putative CpG islands (CGI). We used the nucleotide sequence GenBank U20391.1 for the FOLR1 gene and Ensemble sequence ENSG00000076351 for the PCFT gene, including an additional 5,000 bp upstream of the 5′ end of the gene. The RFC1 genomic sequence and 3861 bp up-stream of the 5′ end was obtained from Ensemble (ENSG00000173638), and was used for the development of the methylation assays. Regions of interest were located and primers were designed using Pyromark assay design software 2.0 (Qiagen). Touch-down PCR has been recommended for amplifying bisulphite treated DNA and was used here.42 The PCR program was as follows: initial denaturation step of 5 min at 95°C, followed by 12 cycles of 30 sec denaturation at 94°C, 45 sec of varying annealing temperature (Ta + 10°C, decreasing 1°C each cycle) and extension of 45 sec at 72°C. Further 40 cycles were as follows: denaturation of 30 sec at 94°C, specific annealing Ta for 45 sec and extension for 45 sec at 72°C and one cycle for 7 min at 72°C. 40 µL PCR reaction was performed with the HotStarTaq DNA Polymerase Kit (QIAGEN Inc.), containing 0.15 µmol/L of each primer, 1.25 unit of Taq polymerase, 1.5–3.0 mM MgCl2, and 0.1 mM each of dGTP, dATP, dTTP, dCTP and 30 ng of bisulphite treated DNA was added as template. The PCR primers, annealing temperatures, and amplicon sizes are shown in Table 1–3. All primers were purchased from www.biomers.net (Ulm/Donau).
To detect biased amplification of the un-methylated allele, we performed a titration assay on samples with known methylation levels of 0%, 10%, 25%, 50%, 85% and 100% methylation.43 Synthetically methylated and non-methylated DNA standards were purchased from Zymo Research (Orion Diagnostica) and Qiagen. The standards were mixed to obtain the percentages of methylation for titration and used in subsequent bisulfite- PCR- and pyrosequencing reactions.
Pyrosequencing
To obtain a quantitative measure of site-specific methylation, the CpG sites were analyzed using Pyrosequencing® technology. Following PCR (see above, and Table 1–3), the samples were prepared using the Vacuum Prep Workstation (Qiagen): 25–30 µL of the amplicon, 3 µL Streptavidin Sepharose HP Beads (Amersham Biosciences), 37 µL binding buffer (10 mmol/L TRIS-HCl, 2 mol/L NaCl, 1 mmol/L EDTA, 0.1% Tween 20, Milli-Q (18.2 MΩ × cm) water, pH 7.6) and 10–15 µL Milli-Q water were mixed and used in the Vacuum Prep workstation. The biotinylated amplicons were immobilized onto the Streptavidin sepharose beads and then passed through one denaturation and two washing steps using the Vacuum Prep Workstation according to a standard protocol. The amplicons were subsequently transferred to a plate containing sequencing primer (0.4 µmol/L) in 40 µL annealing buffer (20 mmol/L Tris-Acetate, 2 mmol/L Magnesium acetate, pH 7.6). Sequencing was performed using a Pyromark Gold Q96 Reagent Kit and a PSQ 96ID system (Qiagen). The nucleotide addition order was optimized by the Pyro Q-CpG software version 1.0.9 (Qiagen). Results were automatically analyzed using the same software.
Assay precision
To determine the precision of the method we studied CpG sites 3–5 in the FOLR1 gene, CpG sites 45–48 in the PCFT gene and sites 15–17 in the RFC1 gene. One leukocyte DNA sample was bisulphite treated 8 times. Each aliquot was then PCR amplified three times and analyzed by pyrosequencing. The CV of the assays was calculated as the standard deviation divided by the mean value for each separate CpG site.
Statistical analysis
Means were compared using ANOVA. The Hardy-Weinberg equilibrium of genotypes was tested with χ2 test and performed for subjects within a specific group. The statistical significance of 80G > A genotype and of study group (subjects with high or low tHcy, respectively) as determinants of the mean methylated fraction of CpG sites was tested in a general linear model, with RFC1 genotype and study group as fixed factors and methylated fraction as dependent variable. All calculations were performed by the SPSS software version 15.0 (SPSS Inc.).
Acknowledgments
Financial support by Lions cancerfond, Nyckelfonden, and Örebro läns landsting is gratefully acknowledged. RHF was supported in part by NIH grants HD067244 and ES021390. AR was supported in part by a grant from the State Key Development Program for Basic Research (2007CB5119001), People’s Republic of China.
Glossary
Abbreviations:
- FOLR1
folate receptor alpha
- PCFT
proton-coupled folate transporter
- RFC1
reduced folate carrier 1
- tHcy
total plasma homocysteine
- CGI
CpG island
- T-DMR
tissue-specific differentially methylated region
- NTD
neural tube defect
- SAM
S-adenosylmethionine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Financial Disclosures
S.A.F., A.K.B., H.S.I. and T.K.N. were supported by Lions cancerfond, Nyckelfonden and Örebro läns landsting. RHF was supported in part by NIH grants HD067244 and ES021390. A.R. was supported in part by a grant from the State Key Development Program for Basic Research (2007CB5119001), People’s Republic of China.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/23988
References
- 1.Blom HJ, Smulders Y. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J Inherit Metab Dis. 2011;34:75–81. doi: 10.1007/s10545-010-9177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 3.Blom HJ. Folic acid, methylation and neural tube closure in humans. Birth Defects Res A Clin Mol Teratol. 2009;85:295–302. doi: 10.1002/bdra.20581. [DOI] [PubMed] [Google Scholar]
- 4.Jacob RA, Wu MM, Henning SM, Swendseid ME. Homocysteine increases as folate decreases in plasma of healthy men during short-term dietary folate and methyl group restriction. J Nutr. 1994;124:1072–80. doi: 10.1093/jn/124.7.1072. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Diop-Bove N, Visentin M, Goldman ID. Mechanisms of Membrane Transport of Folates into Cells and Across Epithelia. Annual Review of Nutrition, Vol 31 2011; 31:177-201. [DOI] [PMC free article] [PubMed]
- 6.Saitsu H, Ishibashi M, Nakano H, Shiota K. Spatial and temporal expression of folate-binding protein 1 (Fbp1) is closely associated with anterior neural tube closure in mice. Dev Dyn. 2003;226:112–7. doi: 10.1002/dvdy.10203. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda S, Hasui S, Yamamoto C, Yoshioka C, Kobayashi M, Itagaki S, et al. Placental folate transport during pregnancy. Biosci Biotechnol Biochem. 2008;72:2277–84. doi: 10.1271/bbb.80112. [DOI] [PubMed] [Google Scholar]
- 8.Spiegelstein O, Mitchell LE, Merriweather MY, Wicker NJ, Zhang Q, Lammer EJ, et al. Embryonic development of folate binding protein-1 (Folbp1) knockout mice: Effects of the chemical form, dose, and timing of maternal folate supplementation. Dev Dyn. 2004;231:221–31. doi: 10.1002/dvdy.20107. [DOI] [PubMed] [Google Scholar]
- 9.Piedrahita JA, Oetama B, Bennett GD, van Waes J, Kamen BA, Richardson J, et al. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23:228–32. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- 10.Tang LS, Finnell RH. Neural and orofacial defects in Folbp1 knockout mice. Birth Defects Research Part a-Clinical and Molecular Teratology 2003; 67. [DOI] [PubMed] [Google Scholar]
- 11.Tang LS, Wlodarczyk BJ, Santillano DR, Miranda RC, Finnell RH. Developmental consequences of abnormal folate transport during murine heart morphogenesis. Birth Defects Research Part a-Clinical and Molecular Teratology 2004; 70. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Wlodarczyk BJ, Scott M, Yu W, Merriweather M, Gelineau-van Waes J, et al. Cardiovascular abnormalities in Folr1 knockout mice and folate rescue. Birth Defects Research Part a-Clinical and Molecular Teratology 2007; 79. [DOI] [PubMed] [Google Scholar]
- 13.Boyles AL, Ballard JL, Gorman EB, McConnaughey DR, Cabrera RM, Wilcox AJ, et al. Association between inhibited binding of folic acid to folate receptor alpha in maternal serum and folate-related birth defects in Norway. Hum Reprod. 2011;26:2232–8. doi: 10.1093/humrep/der144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M. Human Cancer Epigenetics. European Journal of Cancer. 2011;47:S25–S. doi: 10.1016/S0959-8049(11)70311-2. [DOI] [Google Scholar]
- 15.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saied MH, Marzec J, Khalid S, Smith P, Down TA, Rakyan VK, et al. Genome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeats. PLoS One. 2012;7:e33213. doi: 10.1371/journal.pone.0033213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löf-Ohlin ZM, Nilsson TK. Pyrosequencing assays to study promoter CpG site methylation of the O6-MGMT, hMLH1, p14ARF, p16INK4a, RASSF1A, and APC1A genes. Oncol Rep. 2009;21:721–9. [PubMed] [Google Scholar]
- 20.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, et al. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–9. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- 23.Solanky N, Requena Jimenez A, D’Souza SW, Sibley CP, Glazier JD. Expression of folate transporters in human placenta and implications for homocysteine metabolism. Placenta. 2010;31:134–43. doi: 10.1016/j.placenta.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Wani NA, Nada R, Kaur J. Biochemical and molecular mechanisms of folate transport in rat pancreas; interference with ethanol ingestion. PLoS One. 2011;6:e28599. doi: 10.1371/journal.pone.0028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whetstine JR, Matherly LH. The basal promoters for the human reduced folate carrier gene are regulated by a GC-box and a cAMP-response element/AP-1-like element. Basis for tissue-specific gene expression. J Biol Chem. 2001;276:6350–8. doi: 10.1074/jbc.M008074200. [DOI] [PubMed] [Google Scholar]
- 26.Liu MJ, Whetstine JR, Payton SG, Ge Y, Flatley RM, Matherly LH. Roles of USF, Ikaros and Sp proteins in the transcriptional regulation of the human reduced folate carrier B promoter. Biochem J. 2004;383:249–57. doi: 10.1042/BJ20040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whetstine JR, Flatley RM, Matherly LH. The human reduced folate carrier gene is ubiquitously and differentially expressed in normal human tissues: identification of seven non-coding exons and characterization of a novel promoter. Biochem J. 2002;367:629–40. doi: 10.1042/BJ20020512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Payton SG, Liu MJ, Ge YB, Matherly LH. Transcriptional regulation of the human reduced folate carrier A1/A2 promoter: Identification of critical roles for the USF and GATA families of transcription factors. Biochim Biophys Acta. 2005;1731:115–24. doi: 10.1016/j.bbaexp.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Yang R, Li W-W, Hoang BH, Kim H, Banerjee D, Kheradpour A, et al. Quantitative correlation between promoter methylation and messenger RNA levels of the reduced folate carrier. BMC Cancer. 2008;8:8. doi: 10.1186/1471-2407-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kastrup IB, Worm J, Ralfkiaer E, Hokland P, Guldberg P, Grønbaek K. Genetic and epigenetic alterations of the reduced folate carrier in untreated diffuse large B-cell lymphoma. Eur J Haematol. 2008;80:61–6. doi: 10.1111/j.1600-0609.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu MJ, Ge YB, Payton SG, Aboukameel A, Buck S, Flatley RM, et al. Transcriptional regulation of the human reduced folate carrier in childhood acute lymphoblastic leukemia cells. Clin Cancer Res. 2006;12:608–16. doi: 10.1158/1078-0432.CCR-05-1954. [DOI] [PubMed] [Google Scholar]
- 32.Rothem L, Stark M, Kaufman Y, Mayo L, Assaraf YG. Reduced folate carrier gene silencing in multiple antifolate-resistant tumor cell lines is due to a simultaneous loss of function of multiple transcription factors but not promoter methylation. J Biol Chem. 2004;279:374–84. doi: 10.1074/jbc.M309092200. [DOI] [PubMed] [Google Scholar]
- 33.Antony AC, Tang YS, Khan RA, Biju MP, Xiao XL, Li QJ, et al. Translational upregulation of folate receptors is mediated by homocysteine via RNA-heterogeneous nuclear ribonucleoprotein E1 interactions. J Clin Invest. 2004;113:285–301. doi: 10.1172/JCI11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi I, Sohn K-J, Stempak JM, Croxford R, Kim Y-I. Folate deficiency induces cell-specific changes in the steady-state transcript levels of genes involved in folate metabolism and 1-carbon transfer reactions in human colonic epithelial cells. J Nutr. 2007;137:607–13. doi: 10.1093/jn/137.3.607. [DOI] [PubMed] [Google Scholar]
- 35.Ifergan I, Jansen G, Assaraf YG. The reduced folate carrier (RFC) is cytotoxic to cells under conditions of severe folate deprivation. RFC as a double edged sword in folate homeostasis. J Biol Chem. 2008;283:20687–95. doi: 10.1074/jbc.M802812200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devlin AM, Clarke R, Birks J, Evans JG, Halsted CH. Interactions among polymorphisms in folate-metabolizing genes and serum total homocysteine concentrations in a healthy elderly population. Am J Clin Nutr. 2006;83:708–13. doi: 10.1093/ajcn.83.3.708. [DOI] [PubMed] [Google Scholar]
- 37.Stanisławska-Sachadyn A, Mitchell LE, Woodside JV, Buckley PT, Kealey C, Young IS, et al. The reduced folate carrier (SLC19A1) c.80G>A polymorphism is associated with red cell folate concentrations among women. Ann Hum Genet. 2009;73:484–91. doi: 10.1111/j.1469-1809.2009.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chango A, Emery-Fillon N, de Courcy GP, Lambert D, Pfister M, Rosenblatt DS, et al. A polymorphism (80G->A) in the reduced folate carrier gene and its associations with folate status and homocysteinemia. Mol Genet Metab. 2000;70:310–5. doi: 10.1006/mgme.2000.3034. [DOI] [PubMed] [Google Scholar]
- 39.Greene NDE, Stanier P, Copp AJ. Genetics of human neural tube defects. Hum Mol Genet. 2009;18(R2):R113–29. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J-M, Hong K, Lee JH, Lee S, Chang N. Effect of folate deficiency on placental DNA methylation in hyperhomocysteinemic rats. J Nutr Biochem. 2009;20:172–6. doi: 10.1016/j.jnutbio.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Shen LL, Guo Y, Chen XL, Ahmed S, Issa JPJ. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques. 2007;42:48–, 50, 52 passim. doi: 10.2144/000112312. [DOI] [PubMed] [Google Scholar]
- 43.Reed K, Poulin ML, Yan L, Parissenti AM. Comparison of bisulfite sequencing PCR with pyrosequencing for measuring differences in DNA methylation. Anal Biochem. 2010;397:96–106. doi: 10.1016/j.ab.2009.10.021. [DOI] [PubMed] [Google Scholar]