Abstract
The ability to exchange DNA between cells is a molecular process that exists in different species in the domain Archaea. Such horizontal gene transfer events were shown to take place between distant species of archaea and to result in the transfer of large genomic regions. Here we describe recent progress in this field, discuss the potential use of natural gene exchange processes to perform genome shuffling and argue its possible biotechnological applications.
Keywords: Archaea, Cell fusion, Haloferax volcanii, Haloferax mediterranei, Sulfolobus
Introduction
The transfer of genetic information between organisms is a fundamental process, shown to occur in all domains of life. This ability for sexual, or sexual-like, reproduction is vital for the creation of genetic variation. In Eukarya the formation of a diploid cell from two haploid cells is the most obvious case of sexual reproduction. Prokaryotes reproduce asexually, but extensive sequencing of microbial genomes has shown that gene flow (Lateral Gene Transfer, LGT) via parasexual mechanisms between cells of the same, or sometimes even different, species has played a significant role in the evolution of metabolic pathways1-4 and pathogenicity,5-7 for example. In Bacteria LGT involves either phage mediated transduction, assimilation of naked DNA acquired from the environment or unidirectional cell contact-mediated DNA transfer – conjugation.
In Archaea, the ability to transfer DNA between cells was described for two distant genera: Sulfolobus8 (a crenarchaeote) and Haloferax9,10 (a euryarchaeote), and was also observed in two methanogenic archaea.11,12
Mating in Halophilic Archaea
The mating system in the halophilic archaeaon Haloferax volcanii was first characterized nearly three decades ago by Mevarech and Werczberger.13 H. volcanii is a halophilic archaeon (Order Halobacteriales) isolated from the Dead Sea in Israel. Unlike other archaeal species, a broad array of tools for manipulating H. volcanii genetically has been developed.14,15 Mevarech and Werczberger showed that when mixing two different auxotrophic mutants, prototrophic colonies are obtained at a frequency of 10−6. The transfer occurred in a manner that was dependent on establishing physical contact between cells, and increased when the cells were allowed a long incubation time together. Genetic exchange was observed only on solid media, while no transfer was detected when the cells were grown in liquid mediahetero-diploid with continuous shaking. Transformation and transduction were both ruled out, since the transfer was insensitive to DNase treatment and no transfer could be obtained using only the supernatant from one strain. The alternative that remained was conjugation; however, the transfer was bidirectional with no specific donor or recipient. Nevertheless this could still be a form of bi-directional conjugation in the broader sense, assuming that a transient bridge was formed that allowed transfer of single-stranded DNA. However, conjugation was deemed unlikely, because it generally better facilitates the transfer of genes located on plasmids between cells at a significantly higher frequency than genes located on the chromosomes, which was not the case here. Recently, we have compared both plasmid and chromosomal gene exchange in H. volcanii and have shown that the formation of cells containing two episomal plasmids and cells containing two different chromosomes occurs at similar frequencies, indicating that the most probable mechanism is of cell-fusion.10
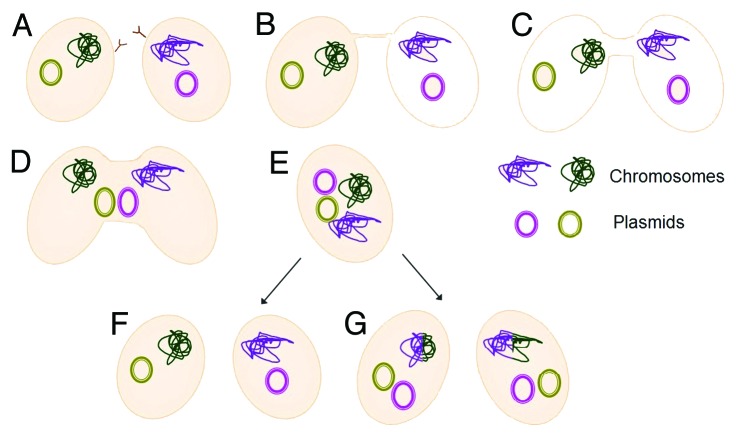
Therefore we suggest a model for a fusion-based mating system in halophilic archaea (Fig. 1). Since the mating system requires physical contact between the cells it would appear that the first step would be adherence of the cells to one another. We further hypothesize that some form of recognition factor is needed for initiation of mating (Fig. 1A).
Figure 1. Schematic representation of the proposed mechanism for Halophilic archaea mating. Scribbly lines represent the chromosome and the small circles- episomal plasmids (A). Two cells belonging to the same species or related species, are at close proximity. (B) Cells establishing physical contact, through creation of cytoplasmatic bridges. (C and D) Expansion of the brides and subsequent creation of fused cells. (E) Hetero-diploid cells, containing two different chromosome types and all plasmids combined. This state can lead to: (F) segregation of the chromosomes and plasmids that would result in reversion into the original state, or (G). recombination events between the chromosomes that would create a hybrid of the two parental strains. Independently of the chromosomes, the plasmids can be exchanged or divided unequally.
Following cell-cell recognition, the next stage begins with the formation of bridges between the cells. Scanning electron microscopy of H. volcanii cells growing on solid media had shown that the cells were connected by bridges9 (Fig. 1B). We believe that these cytoplasmic bridges are then expanded to allow the bidirectional exchanges of plasmids and whole chromosomes (Fig. 1C+D). At the end of this process, hetero-diploid cells are formed (Fig. 1E), meaning cells that contain two different chromosome types and originating from two different cells (halophilic archaea have been shown to be naturally polyploid16). This temporary state also contains, at least initially, the combined plasmid repertoire of both parent cells. In the absence of selection, hetero-diploid cells can segregate their chromosomes, and/or their plasmids (Fig. 1F). However, in some cases, due to the hetero-diploid content of such cells, the two chromosomes can recombine at different regions resulting in recombinant chromosomes (Fig. 1G). Additionally, plasmids can be shuffled between the two cells, resulting in cells with a novel genetic content without involving recombination.
Interspecies mating and recombination
Owing to this extraordinary ability shown by H. volcanii, to hold two different chromosomes in a single cell, the next step was to attempt to mate two different species. Haloferax mediterranei was isolated from a saltern near Alicante, Spain,17 and its genome has been recently sequenced and annotated.18 H. mediterranei's closest known relative is H. volcanii, however, despite a 98.4% identity in their 16S rRNA sequences, their average nucleotide identity in protein coding genes is merely 86.6%.10 Previous studies have shown that H. volcanii and H. mediterranei are able to exchange plasmids (at least engineered antibiotic resistance-encoding ones).19 We have tested the ability of the two strains to form hetero-diploids and then recombine. For that we have used an H. volcanii strain deleted for the hdrB gene, involved in the synthesis of thymidine, and an H. mediterranei strain deleted for the trpA and pyrE genes, involved in the synthesis of tryptophan and uracil respectively, see Figure 2. The cells were incubated together to allow mating and were then plated on media lacking tryptophan and thymidine, thus selecting for hetero-diploids.
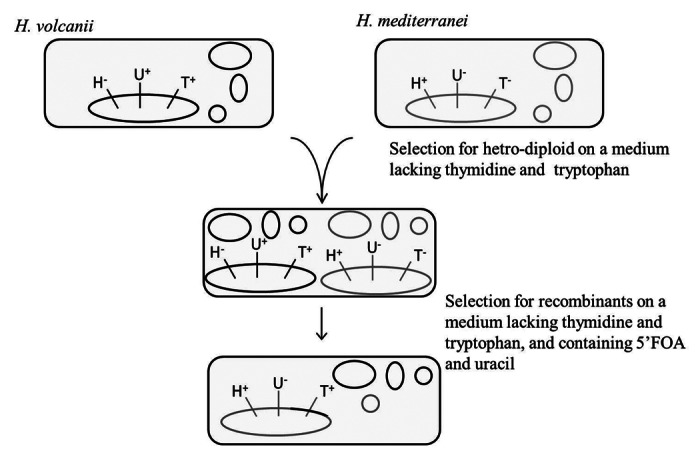
Figure 2. Schematic representation of the creation of specific hybrids between H. volcanii and H. mediterranei. The big circle represents the chromosome and the small circles- the endogenous plasmids. H, hdrB (thymidine); U, pyrE (uracil)T; trpA (tryptophan).
In the next step, we wanted to select for specific H. mediterranei recombinants that have obtained the region from H. volcanii that contains the trpA gene by recombination. For that purpose we needed to select for the presence of the H. mediterranei chromosome, for the trpA gene from H. volcanii and against the rest of the H. volcanii chromosome. To that end, the hetero-diplod cells were grown on a medium containing uracil and 5 fluoroorotic acid (5′FOA) and lacking thymidine and tryptophan. Deprivation of thymidine allows selection for the H. mediterranei chromosome, since it contains an intact copy of hdrB, and deprivation of tryptophan selects for the trpA gene from H. volcanii. Supplementing the medium with uracil and 5FOA serves as a counter selection for the H. volcanii copy of the intact pyrE gene, and de facto selects for cells that do not contain an intact pyrE gene. The resulting hybrids all contained the mediterranei chromosome with genomic loci from H. volcanii that vary in size from about 300 kbp to over 500 kbp.
Genetic transfer in Sulfolobales
Another archaeal genus that was shown to be capable of DNA transfer belongs to the crenarchaeal order Sulfolobales, and is an aerobic thermoacidophile. In this case several studies recognized the existence of conjugative plasmids, which were shown to spread efficiently in Sulfolobus solfataricus and Sulfolobus islandicus.8,20 These conjugative plasmids were shown to carry the genes required for their transfer,20,21 however the mechanism is still unknown. Furthermore comparative genomic analysis between open reading frames from the archaeal and the bacterial conjugative plasmids showed only remote similarities.22 Moreover, exchange of chromosomal markers was also observed for Sulfolobus acidocaldarius, without any recognized conjugation factor in the cells.23
Interestingly, both DNA transfer mechanisms were shown to occur at higher frequencies following UV exposure.24,25 In a recent study Ajon et al.,24 showed that the UV induced DNA transfer in Sulfolobus species is mediated through type IV pili. Following UV irradiation, these pili mediate the formation of aggregates and the subsequent stimulation of the exchange of chromosomal markers. It is speculated that this is a strategy to assist with cell survival by improving DNA repair following exposure to DNA damage.26
Biotechnological Applications
Using the previously described approached one can, through a series of well-planned selection and enrichment cycles, perform genomic shuffling between closely related species, that would generate a single organism with several desirable properties, similar to plant crop breeding. Such approach would enable the use of whole cell biotechnology using halopilic and thermophilic archaea as has been done before in several microorganisms, and recently reviewed in reference 27.
Since the discovery of microorganisms that can thrive in environments of extreme temperatures, high salinity, extreme pH, and high pressure, there has been much interest in the biotechnological potential of their enzymes.28 Most of the focus has been on thermophiles and hyperthermophiles but a few species, such as Thermococcus aggregans and Pyrococcus furiosus, have received more attention recently because of their biotechnological potential.29
Halophilic archaea have not been extensively developed for whole-cell technological applications, yet their enzymes do possess considerable potential for various industrial processes.30 H. volcanii and H. mediterranei were both shown to produce polyhydroxybutyrate (PHB),31 and H. mediterranei was also shown to produce poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV). These Polymers can be used to synthesize biodegradable plastics,32 and the genetics of the biological processes that produce them have therefore been recently studied in great detail.33 Moreover H. mediterranei is the most promising microbial candidate for industrial scale biodegradable polymer production from surplus feedstock whey.34 Other possible industrial applications also exist, such as ones involving the α-amylase secreted by H. mediterranei when grown on ammonium acetate as nitrogen and carbon source. That particular enzyme was stable and active in temperatures up to 80°C,35 making it a highly versatile catalytic tool.
The appeal for biotechnological application of thermophiles such as the Sulfolobales, is quite obvious. The ability of proteins to function at elevated temperatures renders them useful for applications that can only be performed by hyper-thermostable enzymes, i.e., reactions where extreme temperatures are necessary. In addition, engineered enzymes may also be capable of maintaining functionality in a broad range of temperatures.36 Although the use of Sulfolobales for such purposes has been limited, a few examples of enzymes of commercial interest already exist, such as the enzymes for methylthioadenosine phosphorylase, for alpha-glucosidase and 2-keto-3-deoxygluconate aldolase, derived from Sulfolobus solfataricus.37-39
Members of the Archaeal kingdom have long been considered an intriguing, yet largely under-studied source of information for the natural solutions for dealing with extreme and harsh environments. As the use of engineered microorganisms expanded, so did the potential of using organisms that naturally possess stable enzymes that operate at extreme temperatures, pH and salt concentrations. The recent advances in understanding the ability of these organisms to naturally exchange genes, or even large genomic regions, will enable researchers to create new gene combinations in their organism of choice. Furthermore, this advance will facilitate screening for functional complexes to improve existing applications or even create new biotechnological uses.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/22649
References
- 1.Boucher Y, Huber H, L’Haridon S, Stetter KO, Doolittle WF. Bacterial origin for the isoprenoid biosynthesis enzyme HMG-CoA reductase of the archaeal orders Thermoplasmatales and Archaeoglobales. Mol Biol Evol. 2001;18:1378–88. doi: 10.1093/oxfordjournals.molbev.a003922. [DOI] [PubMed] [Google Scholar]
- 2.Bolhuis H, Severin I, Confurius-Guns V, Wollenzien UI, Stal LJ. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J. 2010;4:121–30. doi: 10.1038/ismej.2009.99. [DOI] [PubMed] [Google Scholar]
- 3.Gophna U, Bapteste E, Doolittle WF, Biran D, Ron EZ. Evolutionary plasticity of methionine biosynthesis. Gene. 2005;355:48–57. doi: 10.1016/j.gene.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Gophna U, Thompson JR, Boucher Y, Doolittle WF. Complex histories of genes encoding 3-hydroxy-3-methylglutaryl-CoenzymeA reductase. Mol Biol Evol. 2006;23:168–78. doi: 10.1093/molbev/msj019. [DOI] [PubMed] [Google Scholar]
- 5.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–81. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 7.Gophna U, Ron EZ, Graur D. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene. 2003;312:151–63. doi: 10.1016/S0378-1119(03)00612-7. [DOI] [PubMed] [Google Scholar]
- 8.Schleper C, Holz I, Janekovic D, Murphy J, Zillig W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol. 1995;177:4417–26. doi: 10.1128/jb.177.15.4417-4426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenshine I, Tchelet R, Mevarech M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245:1387–9. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 10.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22:1444–8. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 11.Bertani G. Transduction-like gene transfer in the methanogen Methanococcus voltae. J Bacteriol. 1999;181:2992–3002. doi: 10.1128/jb.181.10.2992-3002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meile L, Abendschein P, Leisinger T. Transduction in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1990;172:3507–8. doi: 10.1128/jb.172.6.3507-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mevarech M, Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985;162:461–2. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allers T, Mevarech M. Archaeal genetics - the third way. Nat Rev Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 15.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–53. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breuert S, Allers T, Spohn G, Soppa J. Regulated polyploidy in halophilic archaea. PLoS One. 2006;1:e92. doi: 10.1371/journal.pone.0000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torreblanca M, Rodriguezvalera F, Juez G, Ventosa A, Kamekura M, Kates M. Classification of Non-Alkaliphilic Halobacteria Based on Numerical Taxonomy and Polar Lipid-Composition, and Description of Haloarcula Gen-Nov and Haloferax Gen-Nov. Syst Appl Microbiol. 1986;8:89–99. doi: 10.1016/S0723-2020(86)80155-2. [DOI] [Google Scholar]
- 18.Han J, Zhang F, Hou J, Liu X, Li M, Liu H, et al. Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer. J Bacteriol. 2012;194:4463–4. doi: 10.1128/JB.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tchelet R, Mevarech M. Interspecies Genetic Transfer in Halophilic Archaebacteria. Syst Appl Microbiol. 1994;16:578–81. doi: 10.1016/S0723-2020(11)80328-0. [DOI] [Google Scholar]
- 20.Stedman KM, She Q, Phan H, Holz I, Singh H, Prangishvili D, et al. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J Bacteriol. 2000;182:7014–20. doi: 10.1128/JB.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prangishvili D, Albers SV, Holz I, Arnold HP, Stedman K, Klein T, et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid. 1998;40:190–202. doi: 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- 22.She Q, Phan H, Garrett RA, Albers SV, Stedman KM, Zillig W. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles. 1998;2:417–25. doi: 10.1007/s007920050087. [DOI] [PubMed] [Google Scholar]
- 23.Grogan DW. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J Bacteriol. 1996;178:3207–11. doi: 10.1128/jb.178.11.3207-3211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ajon M, Fröls S, van Wolferen M, Stoecker K, Teichmann D, Driessen AJ, et al. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol Microbiol. 2011;82:807–17. doi: 10.1111/j.1365-2958.2011.07861.x. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt KJ, Beck KE, Grogan DW. UV stimulation of chromosomal marker exchange in Sulfolobus acidocaldarius: implications for DNA repair, conjugation and homologous recombination at extremely high temperatures. Genetics. 1999;152:1407–15. doi: 10.1093/genetics/152.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allers T. Swapping genes to survive - a new role for archaeal type IV pili. Mol Microbiol. 2011;82:789–91. doi: 10.1111/j.1365-2958.2011.07860.x. [DOI] [PubMed] [Google Scholar]
- 27.de Carvalho CC. Enzymatic and whole cell catalysis: finding new strategies for old processes. Biotechnol Adv. 2011;29:75–83. doi: 10.1016/j.biotechadv.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Sellek GA, Chaudhuri JB. Biocatalysis in organic media using enzymes from extremophiles. Enzyme Microb Technol. 1999;25:471–82. doi: 10.1016/S0141-0229(99)00075-7. [DOI] [Google Scholar]
- 29.Demirjian DC, Morís-Varas F, Cassidy CS. Enzymes from extremophiles. Curr Opin Chem Biol. 2001;5:144–51. doi: 10.1016/S1367-5931(00)00183-6. [DOI] [PubMed] [Google Scholar]
- 30.Litchfield CD. Potential for industrial products from the halophilic Archaea. J Ind Microbiol Biotechnol. 2011;38:1635–47. doi: 10.1007/s10295-011-1021-9. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Castillo R, Rodriguez-Valera F, Gonzalez-Ramos J, Ruiz-Berraquero F. Accumulation of Poly (beta-Hydroxybutyrate) by Halobacteria. Appl Environ Microbiol. 1986;51:214–6. doi: 10.1128/aem.51.1.214-216.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hezayen FF, Rehm BH, Eberhardt R, Steinbüchel A. Polymer production by two newly isolated extremely halophilic archaea: application of a novel corrosion-resistant bioreactor. Appl Microbiol Biotechnol. 2000;54:319–25. doi: 10.1007/s002530000394. [DOI] [PubMed] [Google Scholar]
- 33.Zhao D, Cai L, Wu J, Li M, Liu H, Han J, et al. Improving polyhydroxyalkanoate production by knocking out the genes involved in exopolysaccharide biosynthesis in Haloferax mediterranei. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4415-3. [DOI] [PubMed] [Google Scholar]
- 34.Koller M, Hesse P, Bona R, Kutschera C, Atlić A, Braunegg G. Potential of various archae- and eubacterial strains as industrial polyhydroxyalkanoate producers from whey. Macromol Biosci. 2007;7:218–26. doi: 10.1002/mabi.200600211. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Pomares F, Bautista V, Ferrer J, Pire C, Marhuenda-Egea FC, Bonete MJ. Alpha-amylase activity from the halophilic archaeon Haloferax mediterranei. Extremophiles. 2003;7:299–306. doi: 10.1007/s00792-003-0327-6. [DOI] [PubMed] [Google Scholar]
- 36.Unsworth LD, van der Oost J, Koutsopoulos S. Hyperthermophilic enzymes--stability, activity and implementation strategies for high temperature applications. FEBS J. 2007;274:4044–56. doi: 10.1111/j.1742-4658.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 37.Cacciapuoti G, Porcelli M, Bertoldo C, De Rosa M, Zappia V. Purification and characterization of extremely thermophilic and thermostable 5′-methylthioadenosine phosphorylase from the archaeon Sulfolobus solfataricus. Purine nucleoside phosphorylase activity and evidence for intersubunit disulfide bonds. J Biol Chem. 1994;269:24762–9. [PubMed] [Google Scholar]
- 38.Buchanan CL, Connaris H, Danson MJ, Reeve CD, Hough DW. An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated substrates. Biochem J. 1999;343:563–70. doi: 10.1042/0264-6021:3430563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer MW, Driskill LE, Kelly RM. Glycosyl hydrolases from hyperthermophilic microorganisms. Curr Opin Biotechnol. 1998;9:141–5. doi: 10.1016/S0958-1669(98)80106-7. [DOI] [PubMed] [Google Scholar]